Abstract
Tom40 is the major subunit of the translocase of the outer mitochondrial membrane (the TOM complex). To study the assembly pathway of Tom40, we have followed the integration of the protein into the TOM complex in vitro and in vivo using wild-type and altered versions of the Neurospora crassa Tom40 protein. Upon import into isolated mitochondria, Tom40 precursor proteins lacking the first 20 or the first 40 amino acid residues were assembled as the wild-type protein. In contrast, a Tom40 precursor lacking residues 41 to 60, which contains a highly conserved region of the protein, was arrested at an intermediate stage of assembly. We constructed mutant versions of Tom40 affecting this region and transformed the genes into a sheltered heterokaryon containing a tom40 null nucleus. Homokaryotic strains expressing the mutant Tom40 proteins had growth rate defects and were deficient in their ability to form conidia. Analysis of the TOM complex in these strains by blue native gel electrophoresis revealed alterations in electrophoretic mobility and a tendency to lose Tom40 subunits from the complex. Thus, both in vitro and in vivo studies implicate residues 41 to 60 as containing a sequence required for proper assembly/stability of Tom40 into the TOM complex. Finally, we found that TOM complexes in the mitochondrial outer membrane were capable of exchanging subunits in vitro. A model is proposed for the integration of Tom40 subunits into the TOM complex.
INTRODUCTION
Transport of proteins into and across the two mitochondrial membranes is achieved through the concerted action of translocation machineries: the TOM complex in the outer membrane and either of the two TIM complexes in the inner membrane (Glick and Schatz, 1991; Lill et al., 1996; Schatz, 1996; Neupert, 1997; Pfanner et al., 1997; Koehler et al., 1999; Bauer et al., 2000). Targeting and initial translocation of most preproteins that are destined to the mitochondrial matrix are dependent on amino-terminal, cleavable presequences (Haucke and Schatz, 1997; Neupert, 1997). In contrast, proteins of the outer membrane and a number of proteins of the inner membrane and the intermembrane space contain noncleavable targeting signals (Shore et al., 1995; Stuart and Neupert, 1996; Neupert, 1997). Currently, the nature of most of these latter signals is obscure, though a few such as Tom70 (McBride et al., 1992), Tom22 (Rodriguez-Cousino et al., 1998), BCS1 (Fölsch et al., 1996), and cytochrome c heme lyase (Diekert et al., 1999) have been analyzed in detail.
The TOM complex contains import receptors for the initial recognition of preproteins (Tom20, Tom22, and Tom70) and membrane-embedded components that form the general import pore, which facilitates the translocation of preproteins across the outer membrane (Tom40, Tom5, Tom6, and Tom7). Tom40, a protein essential for viability of yeast and Neurospora crassa cells, was found to be the most abundant component of the TOM complex (Dekker et al., 1998; Künkele et al., 1998) and the core element of the preprotein-conducting pore (Hill et al., 1998; Künkele et al., 1998). The protein forms oligomers, with dimers as the basic structure, and interacts with polypeptide chains in transit (Vestweber et al., 1989; Kiebler et al., 1990; Rapaport et al., 1997; Rapaport et al., 1998). During preprotein translocation, the Tom40 oligomer undergoes conformational changes that affect both the structure of the Tom40 dimer and its interaction with other constituents of the TOM complex (Rapaport et al., 1998). Tom40 has been predicted to traverse the outer membrane as a series of 14 antiparallel β-strands that form a β-barrel (Court et al., 1995; Mannella et al., 1996). In contrast, all other TOM components are postulated to be anchored to the outer membrane by helical transmembrane segments. The import signals of these latter components were suggested to be located in the membrane anchor itself or in the sequences that flank the anchor (McBride et al., 1992; Cao and Douglas, 1995; Rodriguez-Cousino et al., 1998).
Tom20 and Tom70 act as receptors in the recognition of Tom40 precursors, whereas the translocation pore of the TOM complex is utilized for insertion (Keil et al., 1993; Rapaport and Neupert, 1999). Previous studies have suggested the presence of a targeting signal in a yet undefined internal part of the Tom40 precursor and a signal required for assembly at the N-terminal region of the protein (Rapaport and Neupert, 1999). Understanding the biogenesis of the TOM complex requires more information on the mechanisms by which Tom40 is inserted into the membrane, how it achieves its final structure, and how it interacts with the other components in the assembled complex. In the present study we have addressed the question of Tom40 assembly and have identified a region at the N-terminus of the protein that is involved in the process. The region is highly conserved in Tom40 sequences from various species. We also give evidence that TOM complexes in the outer membrane can dynamically exchange subunits, and we discuss possible models for the insertion of Tom40 subunits into preexisting TOM complexes.
MATERIALS AND METHODS
Strains, Media, and Growth
Growth and handling of N. crassa strains was as described (Davis and De Serres, 1970). Race tubes were constructed in sterile 25-ml pipettes or 80-cm glass tubes as described (Davis and De Serres, 1970; White and Woodward, 1995). The extent of mycelial elongation was recorded every 24 h.
Construction of N. crassa tom40 Mutant Strains
Mutant alleles of tom40 were created by site directed mutagenesis of single-stranded DNA derived from a plasmid containing the genomic version of N. crassa tom40 and a bleomycin resistance gene in a Bluescript plasmid. After confirmation of the desired mutation, plasmids were transformed (Schweizer et al., 1981; Akins and Lambowitz, 1985) into spheroplasts of a tom40RIP sheltered heterokaryon (to be described elsewhere) that was generated by the standard N. crassa genetic procedure (Metzenberg and Grotelueschen, 1992; Harkness et al., 1994) of sheltered RIP (repeat induced point mutation). Rescue of the RIPed nucleus was confirmed by testing for biochemical requirements (see RESULTS), and integration of the mutant alleles was confirmed by DNA sequence analysis of PCR products from genomic DNA of the transformants.
Isolation of the Suppressor Mutant of Yeast tom40ts
The suppressor mutant of tom40ts was isolated by performing random mutagenesis. Yeast strain KKY-Isp42-6 (Kassenbrock et al., 1993), a haploid strain containing a complete deletion of the genomic tom40 coding sequence and a mutated tom40 gene on the centromere plasmid, pRS314, was mutagenized (Lawrence, 1991) with methanesulfonic acid ethylester and incubated on plates at the nonpermissive temperature (37°C). After several days, 77 suppressor-containing colonies were tested for plasmid-linked mutations in the tom40ts gene. For this purpose, the ability of plasmids isolated from these colonies to confer the suppressor phenotype was tested. In 66 cases the suppressor phenotype was found to be plasmid linked. The strongest suppressor mutant, tom40tsSup, had a back mutation at position 66 from a proline residue in the temperature-sensitive strain to the wild-type leucine residue. This back mutation restored the steady-state level and stability of the Tom40 protein.
Import of Preproteins into Isolated Mitochondria
For import of Tom40 precursors in vitro, mitochondria from N. crassa were isolated as described (Mayer et al., 1993). Radiolabeled preproteins were synthesized in rabbit reticulocyte lysate in the presence of [35S]methionine (ICN Biomedicals, Costa Mesa, CA) after in vitro transcription by SP6 polymerase from pGEM4 vectors containing the gene of interest. Import reactions were performed by incubation of radiolabeled preproteins with 30–50 μg mitochondria in import buffer (0.5% BSA [wt/vol], 250 mM sucrose, 80 mM KCl, 5 mM MgCl2, 2 mM ATP, 10 mM MOPS-KOH, pH 7.2) at the indicated temperature. Proteinase K (PK) or trypsin treatment of samples was performed by incubation with the protease for 15 min on ice, followed by addition of 1 mM phenylmethylsulfonyl fluoride (PMSF) for 5 min. Import was analyzed by SDS-PAGE, and the gels were viewed by autoradiography or quantified using a phosphorimaging system (BAS 1500; Fuji Medical Systems, Stamford, CT). Immunodecoration was according to standard procedures and was visualized by the ECL method (Amersham).
Carbonate extraction was performed to determine if imported precursor proteins were inserted into membranes. We used sucrose flotation gradients to avoid the possibility of having nonintegrated protein aggregates pelleting with membranes. After import reactions, mitochondria were pelleted, resuspended in 100 μl 0.1 M Na2CO3, and incubated for 30 min on ice. Then, a solution of 2.4 M sucrose, 0.1 M Na2CO3 was added to a final concentration of 1.5 M sucrose (final volume, 266 μl). This was overlayed first with 250 μl of buffer containing 1.4 M sucrose, 0.1 M Na2CO3 and then with 200 μl of buffer containing 0.25 M sucrose, 0.1 M Na2CO3. The gradient was centrifuged for 2 h at 337,000 × g in a Beckman SW60 rotor (Fullerton, CA) at 2°C, which causes the membranes to float to the upper layer of the gradient. Gradients were analyzed by removing 250 μl from the top zone, 150 μl from the middle zone, and 200 μl from the bottom zone of the gradient. Proteins in these fractions were precipitated with trichloroacetic acid and analyzed by SDS-PAGE and autoradiography or Western blotting.
Construction of tom40 Mutants for In Vitro Import
pGEM4-Tom40(Δ2–20) DNA and pGEM4-Tom40(Δ2–40) DNA were constructed by PCR amplification of the relevant DNA from pGEM4-Tom40, which contains the N. crassa wild-type tom40 gene. For pGEM4-Tom40(Δ2–20) and pGEM4-Tom40(Δ2–40) the upstream primers 5′-AGAAAAGAATTCACCATGAGCCTTTCCGATGCCTTC-3′ and 5′-AGAAAAGAATTCACCATGCCCGGCACGATCGAGACC-3′, respectively, were used. In both cases the downstream primer 5′-CTCTAAGCTTTTAAAAGGGGATGTTGAGG-3′ was used. Both PCR products were digested with EcoRI and HindIII and subcloned into pGEM4. pGEM4-Tom40(Δ41–60) DNA was constructed by a method involving the simultaneous ligation of two inserts. The first insert, representing amino acid residues 1–40, and the second insert, representing amino acid residues 61–349, were constructed by PCR amplification of the relevant DNA from pGEM4-Tom40. The upstream primer for the first insert represents the sequence containing the NheI from the pGEM4 vector, whereas the downstream primer 5′-AAAAAATCATATGGTTGGAAAGACCGAACTGTTT-3′ contained an NdeI site. This PCR product was digested with EcoRI (site derived from the multiple cloning site of pGEM4) and NdeI. For the second insert, the upstream primer was 5′-AAA GAA TTC CAT ATG TTC TCT GGC CTC CGC GCC GAC-3′, whereas the downstream primer was the same as used for the cloning of pGEM4-Tom40(Δ2–20) and pGEM4-Tom40(Δ2–40). This PCR product was digested with NdeI and HindIII. The digested products were ligated into pGEM4 that had been digested with EcoRI and HindIII.
Tom40 variants for in vitro import that contained smaller deletions and amino acid substitutions were generated by site-directed mutagenesis of a Tom40 cDNA cloned in the pGEM-7Zf(+) vector.
Cross Linking and Coimmunoprecipitation
For cross-linking experiments, radiolabeled precursors were incubated with isolated mitochondria under various conditions. After the import reaction mitochondria were isolated and resuspended in import buffer followed by addition of 440 μM of the cross-linking reagent disuccinimidyl glutarate (DSG; Pierce Chemical Co., Rockford, IL) for 40 min at 0°C. Excess cross-linker was quenched by the addition of 80 mM glycine, pH 8.0, and incubation for 15 min at 0°C. Aliquots were removed before and after addition of the cross-linking reagents. For coimmunoprecipitation, samples were dissolved in lysis buffer (0.5% digitonin, 150 mM NaCl, 10 mM Tris-HCl, pH 7.2). After a clarifying spin (15 min at 20,000 × g), the supernatant was incubated with antibodies that were coupled to protein A-Sepharose beads.
Blue Native Gel Electrophoresis (BNGE)
Mitochondria (50–100 μg) were lysed in 50 μl detergent-containing buffer (1% digitonin, 0.3% dodecylmaltoside, or 1% dodecylmaltoside in 20 mM Tris-HCl, 0.1 mM EDTA, 50 mM NaCl, 10% glycerol, 1 mM PMSF, pH 7.4). After incubation on ice for 10 min and a clarifying spin (20 min, 22,000 × g), 5 μl sample buffer (5% [wt/vol] Coomassie brilliant blue G-250, 100 mM Bis-Tris, 500 mM 6-aminocaproic acid, pH 7.0) were added, and the mixture was analyzed on a 6 to 13% gradient blue native gel (Schägger et al., 1994; Schägger and von Jagow, 1991).
RESULTS
A Tom40 Precursor Lacking Amino Acid Residues 41 to 60 Does Not Integrate into the TOM Complex
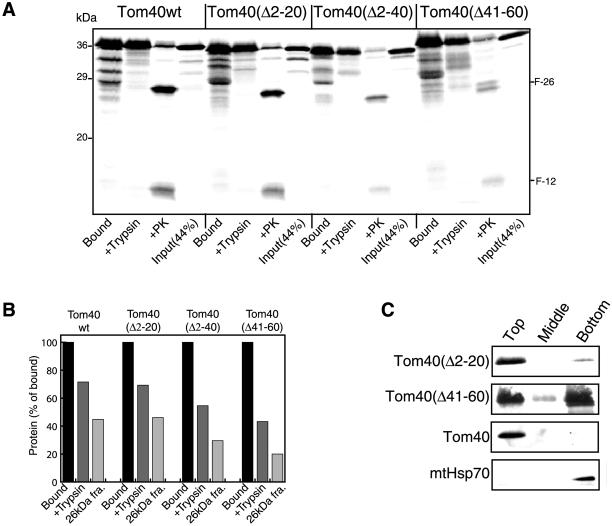
The assembly pathway of Tom40 can be divided into three stages: binding to the mitochondrial surface, insertion into the membrane, and assembly into the TOM complex (Rapaport and Neupert, 1999). Here we have investigated Tom40 with respect to the structural features required for integration into the complex. Precursor proteins lacking residues 2 to 20, 2 to 40, or 41 to 60 were analyzed for their ability to be integrated into the TOM complex, because previous studies had indicated a role for the N-terminal region in the process (Rapaport and Neupert, 1999). All three mutant forms were targeted to mitochondria with efficiencies similar to wild-type (Figure 1, Bound). The ability of the variants to insert correctly into the mitochondrial outer membrane was determined by assessing the acquisition of protection against added trypsin and the formation of proteinase K cleavage fragments characteristic of the inserted wild-type protein. Deletion of residues 2 to 20 did not affect proper insertion, whereas removing residues 2 to 40 had a moderate effect (Figure 1). The variant lacking residues 41 to 60 was ca. 50% less efficient in its ability to properly insert than the wild-type precursor. Treatment of mitochondria with proteinase K after import of the Δ41–60 variant resulted in the formation of an additional cleavage fragment visible just above the usual F-26 fragment that is observed for wild-type Tom40. The additional fragment was likely a digestion product from the fraction of the Tom40Δ41–60 molecules that did not insert properly.
Figure 1.
Insertion of Tom40 variants into the mitochondrial outer membrane. (A) Radiolabeled precursors of Tom40 wild-type (Tom40wt) and the Tom40 deletion mutants were incubated with isolated mitochondria for 15 min at 25°C. After import, each sample was divided into three parts, and mitochondria in all parts were reisolated by centrifugation and resuspended in import buffer. One part was left untreated (Bound), and the other two parts were treated with 100 μg/ml either trypsin (+Trypsin) or Proteinase K (+PK). Samples were analyzed by SDS-PAGE and autoradiography. F-26 and F-12: characteristic N-terminal and C-terminal fragments of Tom40 formed after proteinase K treatment, respectively. (B) The results from A were quantified and presented as a percentage of total bound material. Bound, total bound precursor; +Trypsin, precursor resistant to trypsin treatment; 26-kDa fra., quantification of the N-terminal 26-kDa fragment (F-26) after correction for loss of [35S]methionine residues. (C) Radiolabeled precursors of Tom40 deletion mutants were imported into isolated mitochondria as for A. After import, the mitochondria were pelleted and then suspended in 0.1 M Na2CO3. After 30 min on ice, the samples were analyzed on sucrose flotation gradients where membranes float to the top of the gradient and soluble proteins remain at the bottom (see MATERIALS AND METHODS). The top two boxes show the fractionation pattern of the radiolabeled precursors. The bottom two boxes show immunodecoration of the endogenous Tom40 and mtHsp70 proteins.
Insertion of the precursor proteins into the mitochondrial outer membrane was further assessed by carbonate extraction after in vitro import (Figure 1C). The extraction products were analyzed on sucrose gradients, which results in flotation of membranes to the top of the gradient (see MATERIALS AND METHODS). Thus, integral membrane proteins (e.g., Tom40) will be found in the upper zone of the gradient, whereas soluble proteins (e.g., Hsp70) will be found in the bottom zone. Tom40Δ2–20 was found in the upper zone of the gradient, demonstrating its insertion into mitochondrial membranes. About half of the Tom40Δ41–60 molecules were integrated into the membrane, whereas the remaining half fractionated with the soluble proteins. These data are in agreement with the results of the protease treatment experiments (Figure 1, A and B), where about half of the protein molecules acquired the correct conformation. Taken together, these results demonstrate that the deletions did not impair membrane insertion or cause dramatic changes in the conformation of the inserted protein.
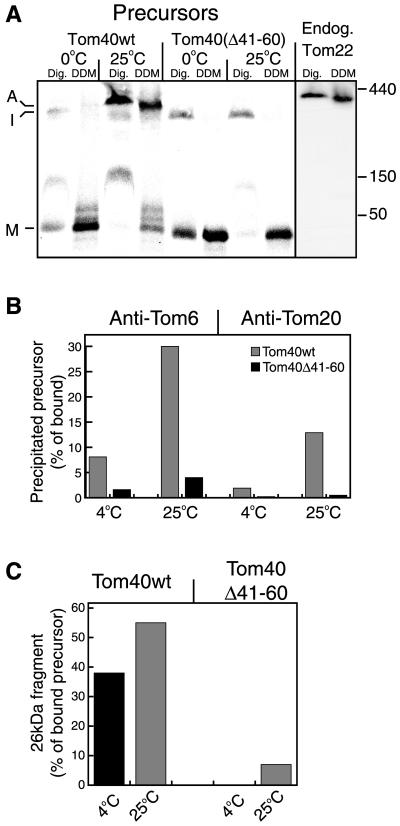
Integration of the variants into the endogenous TOM complex was studied by BNGE. The variant lacking the first 20 amino acid residues was found to integrate into the fully assembled complex with an efficiency similar to the wild-type form (see Figure 4D; shown as a control), whereas the variant lacking the first 40 residues assembled at slightly reduced efficiency (our unpublished results). However, the precursor lacking residues 41 to 60 accumulated at a stage shown previously to be an intermediate (Figure 2A, I) on the assembly pathway (Rapaport and Neupert, 1999) and was not assembled into an authentic TOM complex (Figure 2A). In the intermediate, both the wild-type and mutant forms were only loosely associated with the TOM complex and they dissociated from the complex upon solubilization of mitochondria with dodecylmaltoside (Figure 2A). The band between the monomer and intermediate bands (Figure 2A, lanes 1, 3, and 7) is unproductively bound material (Rapaport and Neupert, 1999).
Figure 4.
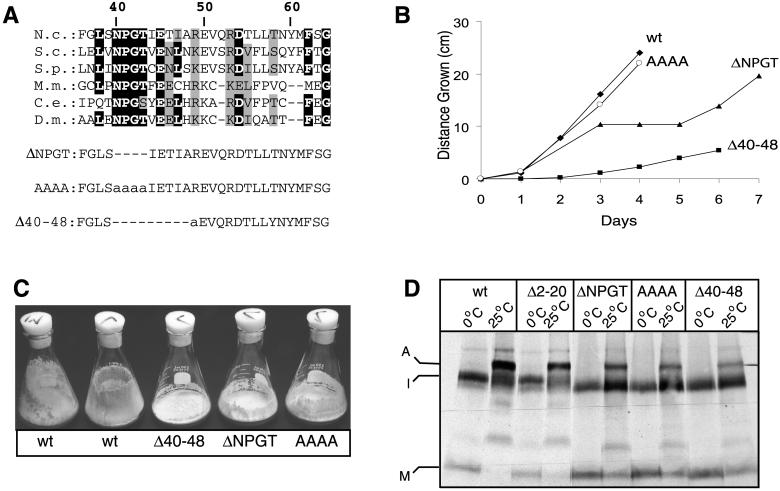
Tom40 mutants and their growth phenotypes. (A) Protein sequence alignment of N. crassa Tom40 (residues 36–64) with homologues from other organisms. Regions modified in the mutant variants are indicated below the alignment. Deleted residues are indicated by dashes, and altered residues are shown in lower case. N.c., Neurospora crassa; S.c. Saccharomyces cerevisiae; S.p., Schizosaccharomyces pombe; C.e., Caenorhabditis elegans; M.m., Mus musculus; D.m, Drosophila melanogaster. Identical residues occurring in four or more sequences are indicated in black highlight. Similar residues are indicated by gray highlights. (B) Mycelial elongation rate of wild-type (wt) and mutant strains as measured in race tubes. Representative strains of the AAAA and ΔNPGT strains are shown, but it should be noted that some strains containing the AAAA mutation grow in the manner shown for ΔNPGT, whereas some ΔNPGT strains grow in the manner depicted for AAAA (see text). (C) Strains containing mutations in the region encompassing residues 40 to 50 are inefficient at climbing the walls of flasks and in the formation of conidia compared with the wild-type strains (wt) that were the parents in the sheltered RIP cross that gave rise to the sheltered heterokaryon shown in Figure 3. (D) Radiolabeled Tom40 variants have reduced ability to assemble into authentic TOM complex. Radiolabeled precursors of wild-type Tom40 (wt) and mutant variants were incubated at 0°C or 25°C with 50 μg mitochondria for 20 min. All samples were reisolated and solubilized in buffer containing 1% digitonin. Further treatment was as described in the legend to Figure 2A. Molecular weights are as indicated in Figure 2A.
Figure 2.
Tom40 variant lacking amino acid residues 41–60 is not integrated into the TOM complex. (A) Radiolabeled precursors were incubated at 0°C or 25°C with 50 μg mitochondria for 20 min. All samples were reisolated and solubilized in buffer containing either 1% digitonin (Dig.) or 0.3% dodecylmaltoside (DDM). After a clarifying spin, the solubilized mitochondria were loaded on a blue native gel. For detection of the endogenous TOM complex, antibodies against Tom22 were used. The three main stages of import are indicated: M, bound monomer; I, high-molecular-weight intermediate; and A, assembled TOM core complex. The positions of molecular weight markers (kDa) are indicated on the right side of A. (B) Tom40Δ41–60 is not stably associated with Tom20 and Tom6. Tom40 (Tom40wt) and Tom40Δ41–60 radiolabeled precursors were incubated with mitochondria for 20 min at 4°C or 25°C and mitochondria were reisolated. One aliquot from each import reaction was directly analyzed by SDS-PAGE and was taken as total bound precursor. A second aliquot was solubilized with 0.5% digitonin and split into three aliquots that were subjected to immunoprecipitation with antibodies against either Tom6, Tom20, or with antibodies from preimmune serum. The latter precipitated only background levels of the precursors (our unpublished results). The lower amounts of assembled precursor observed with Tom20 antibodies are likely due to the fact that Tom20 is only loosely bound to the TOM complex (Ahting et al., 1999). (C) Tom40 can acquire its correctly folded state despite lack of integration into the TOM complex. A third aliquot of the import reactions described in B was treated with proteinase K, and the formation of the characteristic 26-kDa fragment was quantified.
The efficiency of integration into the TOM complex was tested further by immunoprecipitation with antibodies against either Tom6 or Tom20. When full-length Tom40 was imported into mitochondria at 4°C, only low levels of the protein were assembled into the TOM complex. However, the efficiency was high when import was performed at 25°C (Figure 2B). In contrast, very low levels of assembly of Tom40Δ41–60 were detected upon import at either temperature (Figure 2B). These levels might reflect association with the TOM complex as an insertion intermediate rather than actual assembly. The levels of integration of precursors lacking the first 20 or 40 residues were similar to those of wild-type precursor (our unpublished results). Thus, the immunoprecipitation experiments confirm the results of the BNGE analysis and demonstrate the inability of Tom40Δ41–60 to progress into fully assembled TOM complexes. Interestingly, significant levels of the characteristic 26-kDa proteinase K fragment from the wild-type precursor were observed after incubation at 4°C (Figure 2C). This implies that the Tom40 precursor readily acquires its native (or near native) folding even before it is stably integrated into the authentic TOM complex. In previous experiments we observed that folding did not occur in experiments performed at 0°C with short incubation times (Rapaport and Neupert, 1999).
Mutations in Amino Acid Residues 40 to 50 of Tom40 Result in Growth Defects
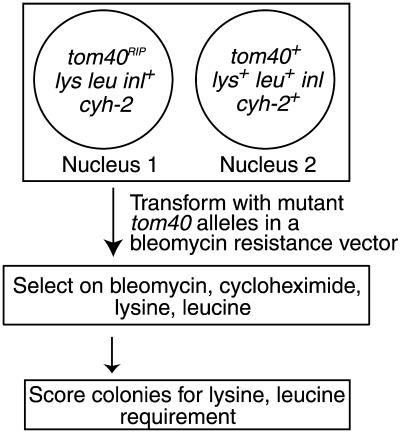
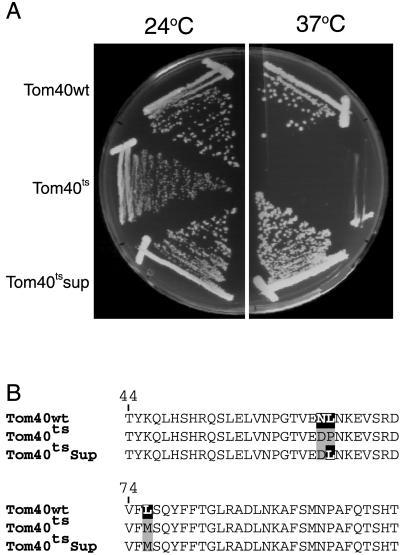
To investigate further the role of the N-terminal portion of the protein in assembly and function of Tom40 in vivo, we examined the ability of mutant derivatives of tom40 to complement a nonfunctional RIP allele of the gene (Figure 3). Because Tom40 is an essential protein, we used the procedure of sheltered RIP (Metzenberg and Grotelueschen, 1992; Harkness et al., 1994) to create a strain of N. crassa (to be described elsewhere), in which a nucleus lacking a functional tom40 gene is maintained in a heterokaryon with a nucleus containing a wild-type version of the gene (Figure 3). A tom40 gene encoding a protein lacking amino acid residues 2 to 60 was not able to restore viability to the nucleus harboring the RIPed version of tom40, supporting the in vitro findings (Rapaport and Neupert, 1999) that the first 60 residues of the N-terminal domain contain crucial information for Tom40 assembly. To identify regions in the N-terminal part of the protein that might play a role in the assembly process, we compared Tom40 sequences from various organisms. We observed no conservation of sequence between organisms prior to amino acid 38 of the N. crassa protein, which is in agreement with the results from in vitro experiments. However, we found a number of highly conserved residues in the region of residues 40 to 60 with the greatest level of similarity within residues 40 to 50 (Figure 4A). Three mutant derivatives of tom40 affecting the conserved region were created (Figure 4A). One mutant (ΔNPGT) had a deletion of the four highly conserved residues NPGT at positions 40 to 43 of the N. crassa sequence, whereas a second mutant (AAAA) had four alanine residues at those sites. The third mutant (Δ40–48) had a deletion of residues 40 to 48, with an additional single amino acid change, R49A. Each of these mutant alleles was able to rescue the tom40RIP nucleus and give rise to homokaryons requiring lysine and leucine (Figure 3).
Figure 3.
Creation of N. crassa strains expressing Tom40 variants. A sheltered heterokaryon containing a nucleus with a tom40RIP allele (Nucleus 1) and a nucleus with a wild-type tom40 allele (Nucleus 2) is depicted in the large box at the top of the figure. The heterokaryon was created by the procedure of sheltered RIP using strains described previously for linkage group V of N. crassa (Metzenberg and Grotelueschen, 1992). Additional markers in nucleus 1 impart cycloheximide resistance and requirements for lysine and leucine. Transformation of the strain with mutant alleles of tom40, carried on a bleomycin resistance plasmid, followed by selection on media containing bleomycin (3.7 μg/ml), cycloheximide (50 μg/ml), lysine, and leucine resulted in homokaryons expressing only the predicted mutant alleles of tom40.
The ΔNPGT and AAAA mutants displayed a complex growth phenotype. We analyzed 18 different lysine- and leucine-requiring transformants of the AAAA type and 13 of the ΔNPGT type for growth characteristics on race tubes. Nine of the AAAA strains and five of the ΔNPGT strains displayed a “stop-start” growth phenotype. These strains grew at near normal rates for a few days and then grew very slowly or not at all for 1 or 2 days before they resumed their initial growth rate. The strains exhibiting this behavior are remarkably consistent, because impaired growth was observed on the same day in up to six different race tubes of an individual strain. The rest of the strains analyzed from both groups grew slightly slower than control strains for 12 days without evidence of stopping. An example of each type of growth is shown in Figure 4B. Tom40 levels in all strains were similar, regardless of their growth phenotype (our unpublished results). Still, it is conceivable that subtle differences in expression, not identified in the analysis of Western blots, may explain the different growth characteristics. This could be due to locus-specific effects at different integration points of the transformed mutant alleles in the individual strains. Regardless, we have found no differences between the strains except for this behavior. For further analysis, one stopper type of the ΔNPGT strains and one normal growth type of the AAAA strains were chosen. The Δ40–48 strain had a slower growth rate that was easily distinguished from the other mutants (Figure 4B). The ability of all three mutant strains to climb the walls of flasks and to form conidia in these flasks was significantly reduced (Figure 4C). Thus, the data from the mutant strains suggest a crucial role for residues 40–49 in the function of Tom40.
The fact that viable strains were obtained by transforming the tom40RIP nucleus with the AAAA, ΔNPGT, and Δ40–48 variants implies that these forms are at least partially capable of assembling into mature, functional TOM complex. This was confirmed by using BNGE to assess assembly after in vitro import of the mutant precursors. All three mutants show partial assembly into the TOM complex, whereas a significant proportion remains in the high molecular weight intermediate (Figure 4D). Interestingly, the Δ40–48 mutant appears to have the least amount of precursor in the fully assembled form, which correlates well with the more severe growth phenotype observed for strains bearing this mutation (Figure 4B).
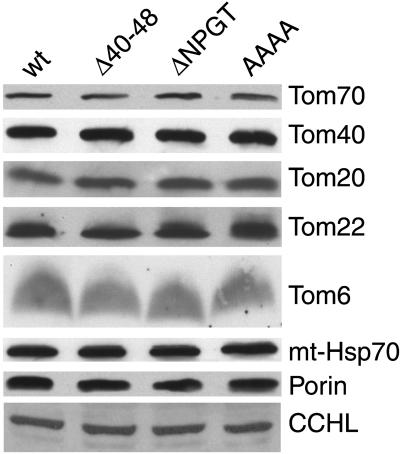
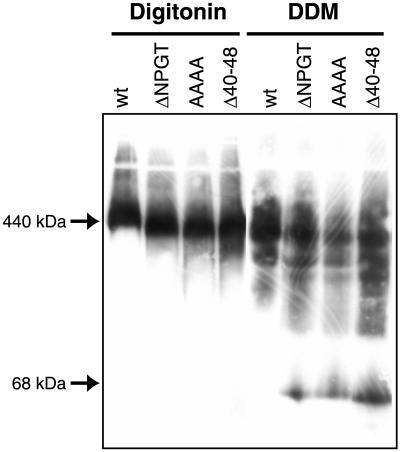
Mutations in Residues 40 to 50 of Tom40 Result in a More Fragile TOM complex
The level of TOM complex components and other mitochondrial proteins from the ΔNPGT, AAAA, and Δ40–48 strains was found to be similar to those in wild-type controls (Figure 5). The TOM complex in the mutants was further examined by BNGE and immunoblotting with antibodies directed against individual TOM complex components. When mitochondria were dissolved in 1% digitonin and subjected to BNGE and the blots decorated with antibody to Tom40, all three mutants were found to contain a TOM complex with slightly increased electrophoretic mobility (Figure 6). When the experiment was repeated with mitochondria dissolved in 1% dodecylmaltoside, the TOM complex in the three mutants was found to be more fragile than the wild-type strain, and at least some fraction of the Tom40 molecules in the mutants migrated as monomers (Figure 6). These results demonstrate that residues 40 to 49 play an important role in the stability of the TOM complex. Blots of blue native gels were also examined using antibodies directed against two other TOM core complex components (Ahting et al., 1999), Tom6 and Tom22. In both cases, the samples solubilized in digitonin showed the same electrophoretic mobility alteration seen in the blots examined with Tom40 antibodies (our unpublished results). For samples dissolved in 1% dodecylmaltoside, the patterns were similar to those seen with Tom40 antibodies, except that only trace amounts of Tom6 monomers were released from the mutant complexes.
Figure 5.
Mitochondrial proteins in wild-type (wt) and mutant strains. Mitochondria isolated from each strain were subjected to SDS-PAGE, blotted to nitrocellulose membrane, and decorated with antisera to TOM complex proteins as well as the mitochondrial outer membrane protein porin, the intermembrane space protein cytochrome c heme lyase (CCHL), and the matrix protein mitochondrial Hsp70 (mt-Hsp70).
Figure 6.
Stability of TOM complexes containing mutant Tom40 molecules. Mitochondria were dissolved in 1% digitonin or 1% dodecylmaltoside (DDM) and subjected to BNGE. The gels were blotted and stained with antiserum to Tom40. In the dodecylmaltoside experiments, the low-molecular-weight form of Tom40 in the lanes containing mutant mitochondria comigrated with Tom40 monomer released when mitochondria were dissolved in SDS (our unpublished results). The positions of molecular weight markers are indicated on the left.
Reversion of a Yeast Tom40 Temperature-sensitive Mutant Restores an Amino Acid in the Conserved N-terminal Region
A study of temperature-sensitive strains of the yeast Saccharomyces cerevisiae further supports the notion that amino acid residues in the 40-to-50 region are important for the biogenesis of Tom40. Kassenbrock et al. (1993) isolated several temperature-sensitive strains carrying mutations in the tom40 gene. In one of the strains (KKY-Isp42-6), DNA sequencing identified 10 mutations in the tom40 gene. We isolated revertants of this temperature-sensitive strain and found that a single reversion in the temperature-sensitive strain, Pro66, back to the wild-type Leu66, was sufficient to allow the revertant strain to grow at the wild-type rate at the restrictive temperature (Figure 7). This result suggests a crucial role for Leu66 in the function of yeast Tom40. The yeast Tom40 Leu66 corresponds to the Ile residue at position 47 in the N. crassa protein (Figure 4A).
Figure 7.
A revertant suppresses the growth phenotype of the tom40 temperature-sensitive mutant KKY-Isp42–6. (A) Growth on agar plates of wild-type cells (Tom40wt), the Tom40 temperature-sensitive strain (Tom40ts) and a revertant (Tom40tssup). (B) The tom40 gene in the revertant was sequenced and one back mutation from Pro66 to the original wild-type Leu66 was identified. Two additional amino acid substitutions in the temperature-sensitive protein that are located in the vicinity of residue 66 are indicated.
Integration of Tom Components into the TOM Complex
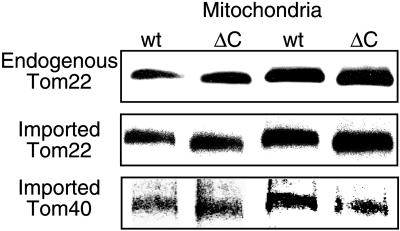
To gain further insight into the structure and assembly of the TOM complex, we wished to determine if newly incorporated precursors can integrate into preexisting complexes. In a mutant strain of N. crassa that expresses only a truncated form of Tom40 lacking the C-terminal 20 amino acid residues, the TOM complex is expected to be ∼13 to 17 kDa smaller than the wild-type complex, based on estimates of six to eight molecules of Tom40 per complex (Ahting et al., 1999). We found that this size difference could be detected by BNGE (Figure 8). The difference was exploited to determine if imported TOM complex subunits can be inserted into preexisting complexes. When full-length precursors of Tom22 and Tom40 were imported into mitochondria isolated from the C-terminal deletion strain, they were rapidly integrated into complexes with the molecular weight characteristic of this strain (Figure 8). These observations support previously suggested models in which precursors are either taken up into a small pool of nearly completed complexes or integrated directly into existing functional complexes (Rapaport and Neupert, 1999).
Figure 8.
Precursors of Tom40 and Tom22 integrate into preexisting complexes. Radiolabeled precursors of either Tom22 or Tom40 were incubated for 20 min at 25°C with 50 μg mitochondria isolated from either a wild-type strain (wt) or a strain lacking the C-terminal 20 amino acid residues of Tom40 (ΔC). The mitochondria were then reisolated and solubilized in a buffer containing 1% digitonin. After a clarifying spin, the solubilized mitochondria were loaded onto a blue native gel. The endogenous TOM core complex was detected by antibodies against Tom22. The location of the imported precursors was detected by exposure of the blot to x-ray film. For easier observation of the small size differences, the same samples were loaded twice in alternating lanes.
Exchange of TOM Complex Subunits in Isolated Outer Membrane Vesicles (OMVs)
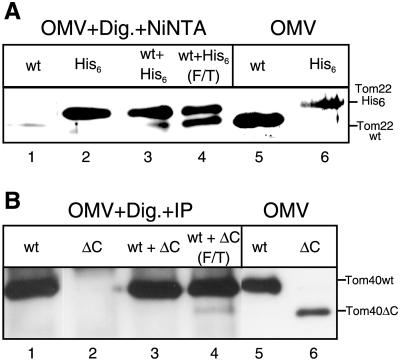
Because the stoichiometry of subunits in a functional TOM complex is likely to be constant, direct integration into functional complex would imply displacement of preexisting subunits. To determine if exchange of subunits can take place between different complexes in vitro, we mixed OMVs isolated from a wild-type N. crassa strain with OMVs from a strain whose only form of Tom22 contained a hexahistidinyl tag. The two OMV samples were induced to undergo fusion by three cycles of freeze/thaw, which is known to induce fusion of lipid vesicles (Hincha et al., 1998). The samples were then solubilized with digitonin and incubated with Ni-NTA sepharose beads to isolate TOM complex containing his-tagged Tom22. Complexes containing his-tagged Tom22 were also found to contain the wild-type form of the protein, indicating that mixing of the two original complexes had occurred (Figure 9A, lane 4). Optimal formation of mixed complexes required fusion of the vesicles, whereas simple mixing, without freeze/thaw cycles, resulted in low levels of mixed TOM complex (Figure 9A, lane 3) only slightly above background (Figure 9A, lane 1).
Figure 9.
TOM complexes can exchange subunits. (A) OMV from a wild-type strain or from a strain expressing a His-tagged version of Tom22 instead of native Tom22 (40 μg each per lane) were incubated separately or together for 2 h at 25°C in a buffer composed of Tris-HCl (10 mM, pH 7.2). In one case, three cycles of rapid freezing in liquid N2 followed by a 5-min thaw at 25°C were performed (F/T). The OMV were then centrifuged (20 min, 125,000 × g) and resuspended in buffer containing 1% digitonin and 10 mM imidazole. Ni-NTA sepharose beads were added for 20 min. The beads were washed once, and bound proteins were eluted with sample buffer. The proteins were analyzed by SDS-PAGE and immunodecoration with antibody against Tom22. For comparison, OMV from both strains were directly loaded on the gel (lanes 5 and 6). (B) OMV (80 μg per lane) from a wild-type strain (wt) or from a strain expressing a C-terminal deletion of Tom40 (ΔC) were incubated separately or together for 2 h at 25°C in a buffer composed of Tris-HCl 10 mM, pH 7.2. In one case three cycles of freeze/thaw (as above) were included in the incubation period (F/T). The OMV were centrifuged (20 min, 125,000 × g), resuspended in buffer containing 1% digitonin, and immunoprecipitated with antibodies against a C-terminal peptide of Tom40. The precipitated proteins were analyzed by SDS-PAGE and immunodecoration with antibody against an N-terminal peptide of Tom40. For comparison, OMV from both strains were directly loaded on the gel (lanes 5 and 6).
In a related set of experiments, antibodies against a C-terminal peptide of Tom40 were used as a tool to analyze exchange of Tom40 subunits. OMVs from a wild-type strain and OMVs from the strain harboring Tom40 with the C-terminal deletion were fused, solubilized with digitonin, and subjected to immunoprecipitation with the C-terminal antibodies. Both forms of Tom40 were present in immunoprecipitates when OMVs underwent fusion but not in immunoprecipitates from controls where OMVs were mixed but not subjected to the fusion treatment (Figure 9B, lanes 3 and 4). Thus, both Tom22 and Tom40 subunits can be exchanged between complexes. The data suggest that Tom22 subunits can be exchanged more easily than Tom40 subunits. Tom40 molecules may be more tightly associated in the complex than Tom22 so that exchange of these subunits may be less frequent.
DISCUSSION
We have examined the mechanisms by which the polytopic mitochondrial outer membrane protein Tom40 is inserted into the mitochondrial outer membrane and assembled into the TOM complex of N. crassa. The highly conserved region containing amino acid residues 41 to 60 of Tom40 was found to be expendable for binding receptors and membrane insertion but crucial for the assembly of the protein into the complex. Deletion of residues 41–60 completely abolished assembly in vitro, whereas smaller mutations resulted in the formation of TOM complexes with altered electrophoretic mobility and stablility in vivo. The changes in mobility likely reflect changes in conformation of the TOM complex because all mutants analyzed displayed similar changes in mobility, whereas the molecular weight of Tom40 in the AAAA mutant is not significantly different from that of the wild-type form. Tom40 monomers were lost from the mutant strains after solubilization in dodecylmaltoside. Because the three mutants exhibited similar behavior with respect to loss of Tom40, this suggests that the NPGT sequence, which is affected in all three strains, may play a role in mediating proper assembly and stability of the TOM complex. The N. crassa strains expressing the Tom40 variants were altered in their ability to form conidia and exhibited growth defects. Furthermore, a suppressing back mutation of a yeast tom40 temperature-sensitive allele occurred within the same amino acid residues, emphasizing the importance of the conserved region in a separate organism. Taken together, these observations show that the sequence containing residues 41–60 plays an important role in the assembly of Tom40. The sequence seems to function as an assembly signal only in its native context, because fusion proteins between this region of Tom40 and a cytosolic protein DHFR (Tom40(41–60)-DHFR) or Tom20 (Tom40(41–60)-Tom20) did not integrate into the Tom core complex (our unpublished results). Thus, these residues are necessary but not sufficient to achieve assembly.
Several observations support the view that the impaired assembly of Tom40 mutant variants reflects a specific function of the affected amino acid residues rather than simple misfolding of the mutant forms. Tom40 in the TOM complex yields characteristic fragments upon treatment of mitochondria with proteinase K (Künkele et al., 1998). The same fragments can be generated after integration of full-length Tom40 precursor into the membrane of isolated mitochondria at a stage when Tom40 is not yet assembled into the complex. Similarly, the Δ41–60 variant, which is not assembled into the complex, still gave rise to the characteristic proteolytic cleavage fragments after integration into the membrane. Thus, Tom40 appears to reach its final, or near final, conformation rather early in its assembly pathway, and the actual integration of Tom40 precursor into the core of the TOM complex may not induce major conformational changes.
Wild-type Tom40 precursors do not persist in a pool of monomers in the outer membrane but are quickly assembled into full-size complexes via a short-lived, high molecular weight intermediate. The relatively small number of radiolabeled molecules of either Tom40 or Tom22 taken up by mitochondria during in vitro import are rapidly integrated into complexes with preexisting subunits. Newly imported subunits could be directly assembled into the preexisting functional complexes by which they are imported. Alternatively, the newly imported subunits could be moved into the lipid phase of the outer membrane and quickly associate with a small pool of partially assembled complexes. In the first model, assuming a fixed stoichiometry of the complex, incorporation of new subunits must be coupled to the release of preexisting subunits. This suggests the existence of a pool of partially assembled TOM complexes made up of the released subunits. Thus, the two models are related in that both require a pool of partially assembled subunits that can interact with other subunits. The inability to detect such a pool implies that such pools would be small and formation of the functional complex rapid. Our observation of subunits being exchanged between existing TOM complexes is consistent with models suggesting that incorporation of new subunits into the complex could result in the displacement of preexisting subunits. Regardless of the mechanism of assembly of new subunits into the TOM complex, a dynamic equilibrium of completely assembled and partially assembled TOM complex may exist in the outer membrane.
ACKNOWLEDGMENTS
We are grateful to Petra Heckmeyer, Thomas Waizenegger, Bonnie Crowther, and Allison Kennedy for excellent technical assistance. This work was supported by a grant from the Medical Research Council of Canada and the Sonderforschungsbereich 184 of the Deutsche Forschungsgemeinschaft. RDT was the recipient of financial support from the Natural Sciences and Engineering Research Council of Canada and the Alberta Heritage Foundation for Medical Research.
Abbreviations used:
- BNGE
blue native gel electrophoresis
- OMV
outer membrane vesicles
- RIP
repeat induced point mutation
- TOM
translocase of the outer mitochondrial membrane
REFERENCES
- Ahting U, Thun C, Hegerl R, Typke D, Nargang F, Neupert W, Nussberger S. The TOM core complex: the general protein import pore of the outer membrane of mitochondria. J Cell Biol. 1999;147:959–968. doi: 10.1083/jcb.147.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akins RA, Lambowitz AM. General method for cloning Neurospora crassa nuclear genes by complementation of mutants. Mol Cell Biol. 1985;5:2272–2278. doi: 10.1128/mcb.5.9.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer MF, Hofmann S, Neupert W, Brunner M. Protein translocation into mitochondria: the role of the TIM complexes. Trends Cell Biol. 2000;10:25–31. doi: 10.1016/s0962-8924(99)01684-0. [DOI] [PubMed] [Google Scholar]
- Cao W, Douglas MG. Biogenesis of ISP6, a small carboxyl-terminal anchored protein of the receptor complex of the mitochondrial outer membrane. J Biol Chem. 1995;270:5674–5679. doi: 10.1074/jbc.270.10.5674. [DOI] [PubMed] [Google Scholar]
- Court DA, Lill R, Neupert W. The protein import apparatus of the mitochondrial outer membrane. Can J Bot. 1995;73(suppl 1):S193–S197. [Google Scholar]
- Davis RH, De Serres FJ. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 1970;17:79–143. [Google Scholar]
- Dekker PJT, Ryan MT, Brix J, Müller H, Hönlinger A, Pfanner N. Preprotein translocase of the outer mitochondrial membrane: molecular dissection and assembly of the general import pore complex. Mol Cell Biol. 1998;18:6515–6524. doi: 10.1128/mcb.18.11.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekert K, Kispal G, Guiard B, Lill R. An internal targeting signal directing proteins into the mitochondrial intermembrane space. Proc Natl Acad Sci USA. 1999;96:11752–11757. doi: 10.1073/pnas.96.21.11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fölsch H, Guiard B, Neupert W, Stuart RA. Internal targeting signal of the BCS1 protein: a novel mechanism of import into mitochondria. EMBO J. 1996;15:479–487. [PMC free article] [PubMed] [Google Scholar]
- Glick BS, Schatz G. Import of proteins into mitochondria. Annu Rev Genet. 1991;25:21–44. doi: 10.1146/annurev.ge.25.120191.000321. [DOI] [PubMed] [Google Scholar]
- Harkness TAA, Metzenberg RL, Schneider H, Lill R, Neupert W, Nargang FE. Inactivation of the Neurospora crassa gene encoding the mitochondrial protein import receptor MOM19 by the technique of “sheltered RIP.”. Genetics. 1994;136:107–118. doi: 10.1093/genetics/136.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haucke V, Schatz G. Import of proteins into mitochondria and chloroplasts. Trends Cell Biol. 1997;7:103–106. doi: 10.1016/S0962-8924(96)10052-0. [DOI] [PubMed] [Google Scholar]
- Hill K, Modell K, Ryan M, Dietmeier K, Martin F, Wagner R, Pfanner N. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature. 1998;395:516–521. doi: 10.1038/26780. [DOI] [PubMed] [Google Scholar]
- Hincha DK, Oliver AE, Crowe JH. The effects of chloroplast lipids on the stability of liposomes during freezing and drying. Biochim Biophys Acta. 1998;1368:150–160. doi: 10.1016/s0005-2736(97)00204-6. [DOI] [PubMed] [Google Scholar]
- Kassenbrock CK, Cao W, Douglas MG. Genetic and biochemical characterization of ISP6, a small mitochondrial outer membrane protein associated with the protein translocation complex. EMBO J. 1993;12:3023–3034. doi: 10.1002/j.1460-2075.1993.tb05971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil P, Weinzierl A, Kiebler M, Dietmeier K, Söllner T, Pfanner N. Biogenesis of the mitochondrial receptor complex. Two receptors are required for binding of MOM38 to the outer membrane surface. J Biol Chem. 1993;268:19177–19180. [PubMed] [Google Scholar]
- Kiebler M, Pfaller R, Söllner T, Griffiths G, Horstmann H, Pfanner N, Neupert W. Identification of a mitochondrial receptor complex required for recognition and membrane insertion of precursor proteins. Nature. 1990;348:610–616. doi: 10.1038/348610a0. [DOI] [PubMed] [Google Scholar]
- Koehler CM, Merchant S, Schatz G. How membrane proteins travel across the mitochondrial intermembrane space. Trends Biochem Sci. 1999;24:428–432. doi: 10.1016/s0968-0004(99)01462-0. [DOI] [PubMed] [Google Scholar]
- Künkele K-P, Heins S, Dembowski M, Nargang FE, Benz R, Thieffry M, Walz J, Lill R, Nussberger S, Neupert W. The preprotein translocation channel of the outer membrane of mitochondria. Cell. 1998;93:1009–1019. doi: 10.1016/s0092-8674(00)81206-4. [DOI] [PubMed] [Google Scholar]
- Künkele K-P, Juin P, Pompa C, Nargang FE, Henry J-P, Neupert W, Lill R, Thieffry M. The isolated TOM complex of the mitochondrial outer membrane: characterization of the cation-selective and voltage-gated preprotein-conducting pore. J Biol Chem. 1998;273:31032–31039. doi: 10.1074/jbc.273.47.31032. [DOI] [PubMed] [Google Scholar]
- Lawrence CW. Classical mutagenesis techniques. In: Guthrie C, Frank GR, editors. Methods in Enzymology. San Diego: Academic Press; 1991. pp. 273–281. [DOI] [PubMed] [Google Scholar]
- Lill R, Nargang FE, Neupert W. Biogenesis of mitochondrial proteins. Curr Opin Cell Biol. 1996;8:505–512. doi: 10.1016/s0955-0674(96)80028-7. [DOI] [PubMed] [Google Scholar]
- Mannella CA, Neuwald AF, Lawrence CE. Detection of likely transmembrane beta strand regions in sequences of mitochondrial pore proteins using the Gibbs sampler. J Bioenerg Biomembr. 1996;28:163–169. doi: 10.1007/BF02110647. [DOI] [PubMed] [Google Scholar]
- Mayer A, Lill R, Neupert W. Translocation and insertion of precursor proteins into isolated outer membranes of mitochondria. J Cell Biol. 1993;121:1233–1243. doi: 10.1083/jcb.121.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride HM, Millar DG, Li JM, Shore GC. A signal-anchor sequence selective for the mitochondrial outer membrane. J Cell Biol. 1992;119:1451–1457. doi: 10.1083/jcb.119.6.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzenberg RL, Grotelueschen JS. Disruption of essential genes in Neurospora by RIP. Fungal Genet Newsl. 1992;39:37–49. [Google Scholar]
- Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- Pfanner N, Craig EA, Hönlinger A. Mitochondrial preprotein translocase. Annu Rev Cell Dev Biol. 1997;13:25–51. doi: 10.1146/annurev.cellbio.13.1.25. [DOI] [PubMed] [Google Scholar]
- Rapaport D, Künkele K-P, Dembowski M, Ahting U, Nargang FE, Neupert W, Lill R. Dynamics of the TOM complex of mitochondria during binding and translocation of preproteins. Mol Cell Biol. 1998;18:5256–5262. doi: 10.1128/mcb.18.9.5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D, Neupert W. Biogenesis of Tom40, core component of the TOM complex of mitochondria. J Cell Biol. 1999;146:321–331. doi: 10.1083/jcb.146.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D, Neupert W, Lill R. Mitochondrial protein import: Tom40 plays a major role in targeting and translocation of preproteins by forming a specific binding site for the presequence. J Biol Chem. 1997;272:18725–18731. doi: 10.1074/jbc.272.30.18725. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Cousino N, Nargang FE, Baardman R, Neupert W, Lill R, Court DA. An import signal in the cytosolic domain of the Neurospora mitochondrial outer membrane protein TOM22. J Biol Chem. 1998;273:11527–11532. doi: 10.1074/jbc.273.19.11527. [DOI] [PubMed] [Google Scholar]
- Schägger H, Cramer WA, von Jagow G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. Blue native electrophoresis for isolation of membrane complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- Schatz G. The protein import system of mitochondria. J Biol Chem. 1996;271:31763–31766. doi: 10.1074/jbc.271.50.31763. [DOI] [PubMed] [Google Scholar]
- Schweizer M, Case ME, Dykstra CC, Giles NH, Kushner SR. Identification and characterization of recombinant plasmids carrying the complete qa gene cluster from Neurospora crassa including the qa-1+ regulatory gene. Proc Natl Acad Sci USA. 1981;78:5086–5090. doi: 10.1073/pnas.78.8.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore GC, McBride HM, Millar DG, Steenaart NAM, Nguyen M. Import and insertion of proteins into the mitochondrial outer membrane. Eur J Biochem. 1995;227:9–18. doi: 10.1111/j.1432-1033.1995.tb20354.x. [DOI] [PubMed] [Google Scholar]
- Stuart RA, Neupert W. Topogenesis of inner membrane proteins of mitochondria. Trends Biochem Sci. 1996;21:261–267. [PubMed] [Google Scholar]
- Vestweber D, Brunner KP, Baker A, Schatz G. A 42K outer membrane protein is a component of the yeast mitochondrial import site. Nature. 1989;341:205–209. doi: 10.1038/341205a0. [DOI] [PubMed] [Google Scholar]
- White B, Woodward D. A simple method for making disposable race tubes. Fungal Genet Newsl. 1995;42:79. [Google Scholar]