A novel Bcl6 reporter mouse is used to dissect the developmental requirements, plasticity, and genetic profile of Tfh cells.
Abstract
T follicular helper cells (Tfh cells) play a pivotal role in germinal center reactions, which require B cell lymphoma 6 (Bcl6) transcription factor. To analyze their relationships with other effector T cell lineages and their stability in vivo, we developed and analyzed a new Bcl6 reporter mouse alone or together with other lineage reporter systems. Assisted with genome-wide transcriptome analysis, we show substantial plasticity of T cell differentiation in the early phase of immune response. At this stage, CXCR5 appears to be expressed in a Bcl6-independent manner. Once Bcl6 is highly expressed, Tfh cells can persist in vivo and some of them develop into memory cells. Together, our results indicate Bcl6 as a bona fide marker for Tfh polarized program.
After activation, naive CD4+ T cells differentiate into effector T cells with distinct immunoregulatory function. T follicular helper cells (Tfh cells), a newly defined T cell subset, provide essential help to B cells, especially in the germinal center (GC) reactions (Breitfeld et al., 2000; Schaerli et al., 2000; Yu and Vinuesa, 2010; Crotty, 2011). Tfh cells, originally characterized by their chemokine (C-X-C motif) receptor 5 (CXCR5) expression, exhibit a unique gene expression profile distinct from Th1, Th2, and Th17 cells (Chtanova et al., 2004; Nurieva et al., 2008). Recently, we and other groups have identified B cell lymphoma 6 (Bcl6) as a transcriptional factor necessary for Tfh cell differentiation and GC reactions; bcl6−/− CD4+ T cells are impaired in the generation of CXCR5+ Tfh cells in vivo (Johnston et al., 2009; Nurieva et al., 2009a; Yu et al., 2009).
The relationship of Tfh cells with other T cell lineages has been a topic of debates. Although our group and others have shown that Tfh cells expressed greatly reduced levels of Th1-, Th2-, or Th17-related cytokines and transcription factors (Breitfeld et al., 2000; Nurieva et al., 2008), several studies reported that Tfh cells, using the CXCR5 marker, still expressed cytokines including IFN-γ (Smith et al., 2004; Johnston et al., 2009; Reinhardt et al., 2009), IL-4 (Smith et al., 2000; King et al., 2008; Reinhardt et al., 2009), and IL-17 (Bauquet et al., 2009). Whether these cytokine-producing cells are coexpressing Bcl6 is unclear. With the discovery of Bcl6 as an obligatory factor for Tfh cell generation, the relationship between Tfh cells and other CD4+ T helper cells needs reevaluation at the single-cell level.
After the primary immune response, effector CD8+ T cells undergo rapid contraction, leaving behind a long-lived antigen-specific memory pool. In this process, Bcl6 was shown to play an important role in maintaining memory CD8+ T cells (Ichii et al., 2002). However, whether the same is true for Tfh cells is largely unknown. In recent studies, several groups showed that Tfh cells in local lymphoid tissues rapidly enhance the recall response (Fazilleau et al., 2007; MacLeod et al., 2011; Marshall et al., 2011). Although these studies suggest the possible presence of Tfh-like memory cells, their nature and origin are unclear.
In this present study, we developed a Bcl6-RFP reporter mouse and analyzed Tfh development and the relationship of Tfh cells with other effector T cell lineages in vivo by using dual reporter mice and genome-wide transcriptome analysis. We show considerable plasticity in the early phase of T cell differentiation in vivo. However, Bcl6hi Tfh cells are sustained in their phenotypes, and some of them develop as memory-like cells. Thus, Bcl6 expression in T cells specifies the Tfh program.
RESULTS
Generation and characterization of a novel Bcl6-RFP reporter mouse
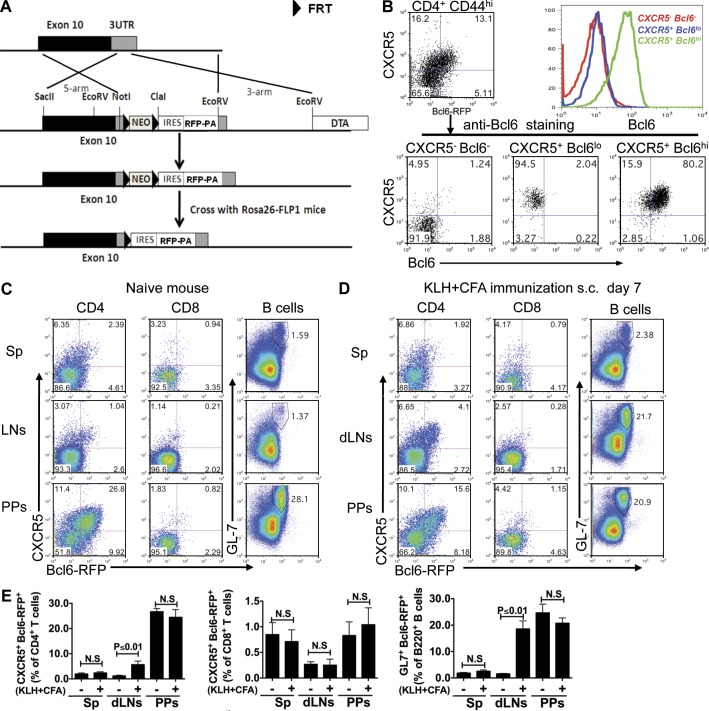
We have developed a Bcl6-RFP (bcl6rfp/+ or bcl6rfp/rfp) reporter mouse strain to allow analysis of Tfh cells in vivo. A cassette containing internal ribosome entry site (IRES)–driven monomeric RFP and a bovine growth hormone poly (A) tail was inserted after the stop codon in the 3′ untranslated region (UTR) of Bcl6 gene through homologous recombination in mouse embryonic stem cells (Fig. 1 A). This strategy was used to create a transcriptional reporter mouse for Bcl6 without interfering with the function of Bcl6 protein. After germline transmission, animals carrying the targeted allele were crossed with mice expressing the flippase recombination enzyme (FLP) to remove the neomycin (Neo) gene. To validate Bcl6-RFP reporter mice, we immunized them with antigen KLH emulsified in CFA s.c. 7 d after immunization, and three populations (CXCR5−RFP−, CXCR5+RFPlo, and CXCR5+RFPhi) of cells were observed in activated (CD4+CD44hi) T cell population (Fig. 1 B). After purification by FACS and restaining with anti-Bcl6 antibody, we found that the level of RFP expression correlated well with Bcl6 protein expression (Fig. 1 B). To assess whether bcl6 gene expression is affected by rfp gene insertion at the 3′ UTR, naive T (CD4+CD62hiCD44loCD25lo) cells were sorted from OT-II mice with chicken OVA-specific TCR and Bcl6-RFP×OT-II mice and transferred in equal numbers into CD45.1 congenic mice. 7 d after immunization, we compared Bcl6 expression in donor cells at both protein and messenger RNA (mRNA) levels and found that Bcl6 expression was not affected by rfp insertion (not depicted).
Figure 1.
Generation and validation of Bcl6-RFP reporter mice. (A) Schematic map for the mouse Bcl6 gene locus, targeting DNA construct, and targeted Bcl6 locus. (B) Bcl6 intracellular staining on sorted cells from Bcl6-RFP reporter mice. Histogram overlay of three populations of T cells: CXCR5−Bcl6−, CXCR5+Bcl6lo, and CXCR5+Bcl6hi. Data are a representative of two independent experiments. (C and D) In naive (C) and KLH/CFA s.c. immunized (D) reporter mice, CXCR5 and Bcl6-RFP expression was assessed on gated CD4+ T cells and CD8+ T cells from spleens (Sps), dLNs, and Peyer’s patches (PPs) by flow cytometry. Also, analysis of GL7+Bcl6-RFP+ cells is shown in gated B220+ B cells. Data are representative of two independent experiments. (E) Summary of data from C and D. Bar graphs display the number of donor cells as mean ± SD. n = 3 per group. N.S., no statistically significant difference.
We then further analyzed Bcl6-RFP expression in peripheral lymphoid tissues before and after immunization. 7 d after immunization with KLH in CFA, Bcl6-RFP expression was significantly up-regulated in both CD4+ T cells and B cells (GL-7, a marker for GC B cells) from draining LNs (dLNs) but not in spleens (Fig. 1, C–E). High percentages of CXCR5+Bcl6-RFP+ CD4+ T cells and GL7+Bcl6-RFP+ B cells were observed in Peyer’s patches, regardless of immunization (Fig. 1, C–E), which may reflect highly active GCs for constitutive IgA production (McGhee, 2005). In contrast to CD4+ T cells and B cells, CD8+ T cells expressed very low Bcl6-RFP (Fig. 1, C–E). These data collectively showed that RFP served as a reliable marker for Bcl6 expression.
CXCR5 expression is initiated in a Bcl6-independent manner
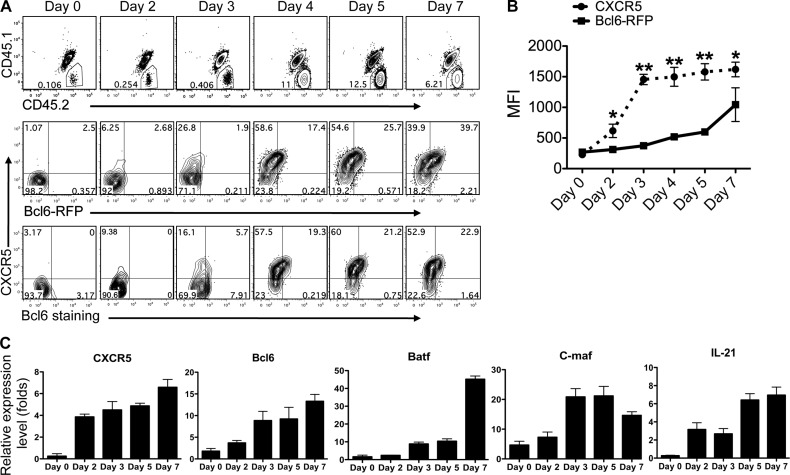
To probe antigen-specific Tfh cell differentiation in vivo, we purified naive CD4+ T cells from Bcl6-RFP OT-II mice (CD45.2+CD45.1−) and transferred into recipient mice (CD45.1+CD45.2+). After s.c. immunization with OVA emulsified in CFA, recipient mice were sacrificed at the indicated time points, followed by flow cytometry analysis. In the dLNs, the expansion of donor cells peaked at days 4–5 (Fig. 2 A), and CXCR5 up-regulation on T cells was detected to occur at day 2 after immunization and to reach a plateau at day 3 (Fig. 2, A and B). In contrast, Bcl6 expression appeared to gradually increase from day 2 to 7 (Fig. 2, A and B), as shown by both RFP measurement and anti-Bcl6 staining (Fig. 2 A). To further characterize the transcriptional changes during effector T cell development, we sorted donor OT-II cells from OVA/CFA-immunized mice at different time points and examined the mRNA expression of Tfh-related genes. Consistent with the staining data, expression of CXCR5 mRNA was greatly increased at day 2 after immunization, whereas Bcl6 mRNA was slightly up-regulated at this time point but more substantially on day 3 (Fig. 2 C). Batf, a transcriptional factor indispensible for Tfh development (Betz et al., 2010; Ise et al., 2011) was slowly up-regulated from day 0 to day 5, whereas substantial enhancement was detected at day 7 (Fig. 2 C). Another important transcriptional factor, c-maf, reported to be regulated directly by Batf and required for IL-21 and CD40L production (Ellyard and Vinuesa, 2011), reached its peak expression on day 3 (Fig. 2 C).
Figure 2.
Ontogeny analysis of Bcl6 expression and Tfh cell development. (A) Naive Bcl6-RFP OT-II CD4+ T cells (CD45.1−CD45.2+) were transferred into CD45.1+CD45.2+ congenic mice, which were subsequently immunized s.c. with OVA in CFA. Donor cell expansion (top) and CXCR5 and Bcl6-RFP expression in donor cells (middle) were determined by flow cytometry. Intracellular staining of Bcl6 was also performed (bottom). Data are representative of two independent experiments. (B) Expression of CXCR5 and Bcl6-RFP in donor cells was determined at various time points after immunization and shown in line graph as median fluorescence intensity (MFI) ± SD. *, P < 0.05; **, P < 0.01. (C) Quantitative RT-PCR measurement of Tfh-specific genes in sorted donor cells. The graphs display mean ± SD. (A–C) n = 3 per group.
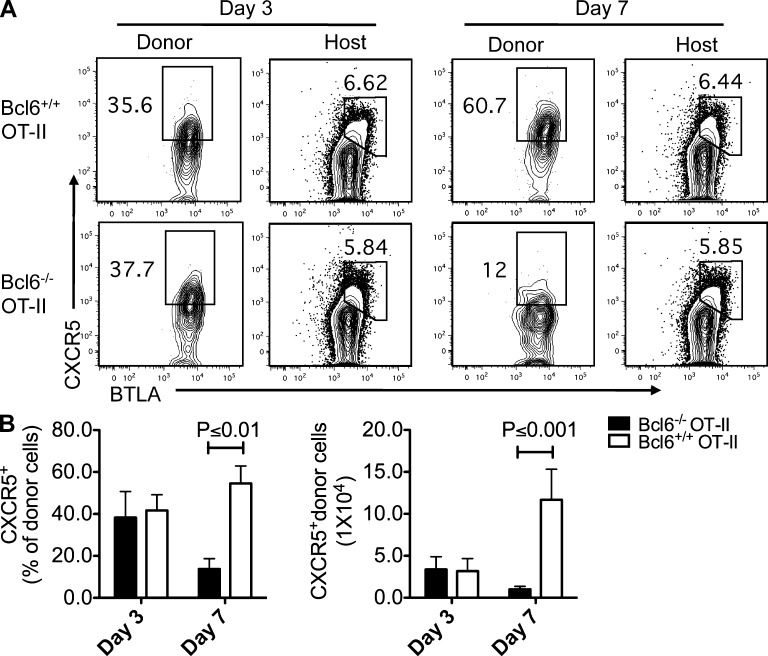
The apparent difference in the kinetics of CXCR5 and Bcl6 expression in the early phase of T cell activation raised a possibility that CXCR5 expression in Tfh cell was not initiated by Bcl6. To test this hypothesis, we transferred naive CD4+ T cells from WT or bcl6−/− mice into CD45.1 recipient mice, followed by s.c. immunization with KLH + CFA. On day 3 after the immunization, CXCR5 expression was equally enhanced in both WT and Bcl6−/− donor cells (not depicted). To further confirm this result, we transferred naive WT and bcl6−/− OT-II T cells into recipient mice, followed by s.c. immunization with OVA/CFA. As shown in Fig. 3 (A and B), the expression of CXCR5 expression at day 3 after the immunization was comparable between WT and Bcl6−/− OT-II donor cells but severely decreased in Bcl6−/− OT-II cells at day 7 (Fig. 3, A and B), indicating that Bcl6 expression in T cells is required for maintaining but not initiating CXCR5 expression.
Figure 3.
Bcl6 is not required for CXCR5 initiation. (A) Naive bcl6−/− and bcl6+/+ OT-II T cells were transferred into CD45.1+ recipient mice, respectively, and then followed with OVA/CFA immunization s.c., and 3 and 7 d after immunization, donor cells and endogenous host CD4+ T cells from dLNs were characterized by anti-CXCR5 and BTLA staining. Data are representative of two independent experiments. n = 3 per group. (B) Summary of data from A. Bar graphs display the frequency and number of CXCR5+ donor cell as mean ± SD.
Polarized transcriptional program associated with Bcl6 expression
From our aforementioned analysis, antigen-specific CXCR5+ T cells seem to exhibit Bcl6lo/− and Bcl6hi phenotypes. The former appeared earlier in immune responses and the latter had further increases of both Bcl6 and CXCR5 expression. To analyze the differences of these two subsets or states of T cells, we sorted three populations of T cells (CXCR5−Bcl6−, CXCR5+Bcl6lo, and CXCR5+Bcl6hi) at day 7 after immunization and conducted surface staining and gene-profiling analysis. Bcl6 expression correlated positively with several known Tfh surface markers, including B and T lymphocyte attenuator (BTLA), inducible T cell co-stimulator (ICOS), and PD-1, and negatively with P-selectin glycoprotein ligand 1 (PSGL1; not depicted). However, 25% of non-Tfh (CXCR5−Bcl6−) cells also expressed high amounts of ICOS (not depicted), which was consistent with a previous report that ICOS deficiency affected the differentiation of multiple CD4+ T cell lineages (Dong and Nurieva, 2003).
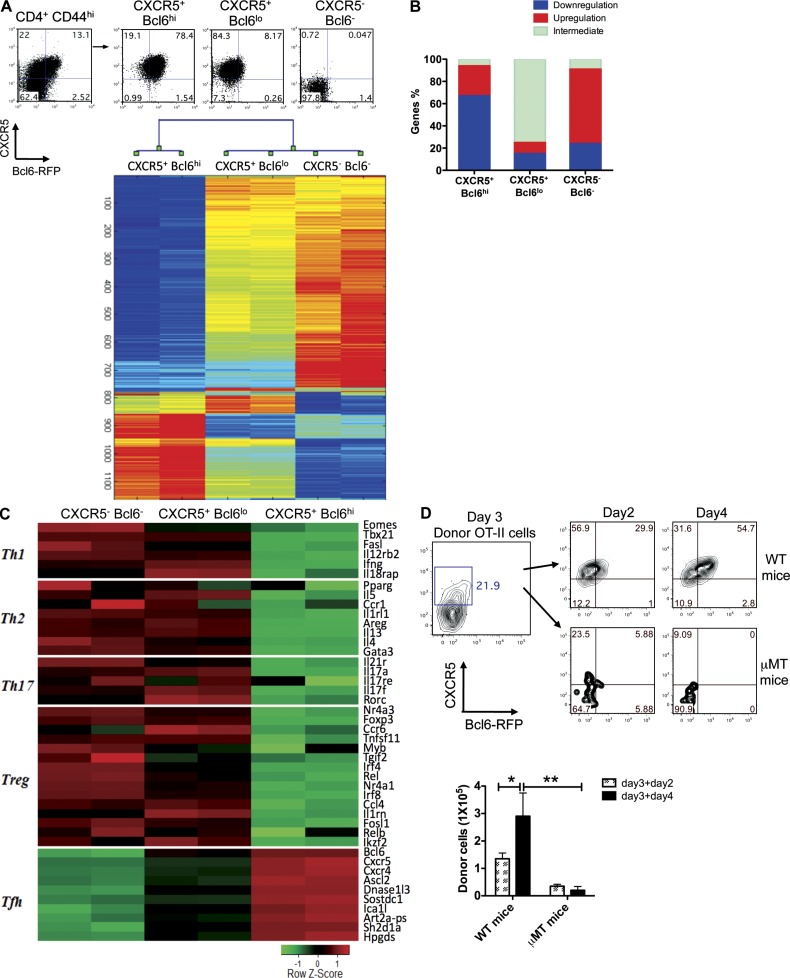
We next analyzed the gene profiling on three different subpopulations of CD4+CD44+ T cells via Affymetrix gene chips. After normalization of microarray data, 1,130 genes whose expression changed over twofold across samples were selected and used for hierarchical clustering (Fig. 4 A and Table S1). Based on this analysis, CXCR5+Bcl6hi cells appeared to have a distinct gene expression profile from both CXCR5−Bcl6− and CXCR5+Bcl6lo cells (Fig. 4 A). Among the analyzed genes, ∼70% were down-regulated in CXCR5+Bcl6hi cells compared with CXCR5−Bcl6− and CXCR5+Bcl6lo cells, whereas ∼75% of them were maintained at intermediate expression levels in CXCR5+Bcl6lo cells (Fig. 4 B). To validate the microarray data, we first analyzed the signature genes representing different T cell lineages and found that Th1 cell–, Th2 cell–, and Th17 cell–associated genes were substantially suppressed in CXCR5+Bcl6hi Tfh cells (Fig. 4 C), consistent with our previous data and other groups’ studies (Breitfeld et al., 2000; Nurieva et al., 2008; Crotty, 2011). In contrast, Tfh-related genes including cxcr5, bcl6, sh2d1a (SAP), and cxcr4 were highly expressed in CXCR5+Bcl6hi cells (Fig. 4 C). Some of the genes were further confirmed by real-time RT-PCR (not depicted).
Figure 4.
Global transcriptional changes are associated with Tfh cell development. Bcl6-RFP reporter mice were s.c. immunized with KLH in CFA. 7 d later, mice were sacrificed, and activated CD44+CD4+ T cell from dLNs were sorted into three populations: CXCR5+Bcl6hi, CXCR5+Bcl6lo, and CXCR5−Bcl6− cells. (A) Microarray analysis of sorted cells (duplicated samples). Hierarchical clusters of 1,130 genes for three groups of cells. The color coding applies to the expression level of 1,130 genes with 0 as a median. Each of the samples was duplicated. (B) The percentages of genes associated with relative down-regulation, up-regulation, and intermediate for each population of cells. (C) Heat map of signature genes for Th1, Th2, Th17, Treg, and Tfh cells. (D) CXCR5+Bcl6lo T cells were sorted on day 3 after immunization and transferred into WT or µMT recipient mice. 2 and 4 d after immunization with OVA + IFA s.c., flow cytometry analysis of CXCR5 and Bcl6 expression was conducted. Data are representative of two independent experiments. Bar graph displays the number of donor cells as mean ± SD. n = 3 per group. *, P < 0.05; **, P < 0.01.
Upon closer examination, notably, a group of differentially up-regulated genes in Tfh cells encoding transferases (16 of 313 genes), hydrolases (29 of 313 genes), DNA-binding proteins (20 of 313 genes), and cell surface receptors (21 of 313 genes) were detected (Table S1). More interestingly, hierarchical clustering analysis showed that CXCR5+Bcl6lo cells had an intermediate pattern of gene expression: they expressed certain levels of Th1 cell–, Th2 cell–, Th17 cell–, and regulatory T cell (Treg cell)–related genes and also had up-regulated Tfh marker genes (Fig. 4 C). These results suggested that CXCR5+Bcl6lo cells represented a transitional stage during Tfh development with mixed gene expression patterns.
Thus, CXCR5+Bcl6lo cells developed before CXCR5+Bcl6hi cells and exhibited a nonpolarized gene expression pattern. To further test their relationship, we sorted CXCR5+Bcl6lo OT-II T cells on day 3 after immunization and transferred them into WT or B cell–deficient (µMT) mice, followed by immunization with OVA emulsified in IFA. In WT mice, Bcl6 expression in donor cells was significantly increased, and the numbers of CXCR5+Bcl6hi donor cells were expanded at days 2 and 4 after immunization (Fig. 4 D). Strikingly, the donor cells did not expand or further mature in µMT mice (Fig. 4 D). These data suggest that B cells were required for intermediate CXCR5+Bcl6lo T cells to develop into mature CXCR5+Bcl6hi Tfh cells. However, this result is different from previous studies using a viral infection model (Choi et al., 2011; Fahey et al., 2011). This discrepancy may be caused by different in vivo models. Also we do not exclude the possibility that B cells are not required for CXCR5 up-regulation, but we suggest that mature Tfh program, associated with Bcl6 expression, may be dependent on B cells for their maintenance. Indeed, another study using a protein/adjuvant immunization mouse model showed that B cells were necessary for Tfh maturation (Goenka et al., 2011).
Plasticity of early T cell differentiation in vivo
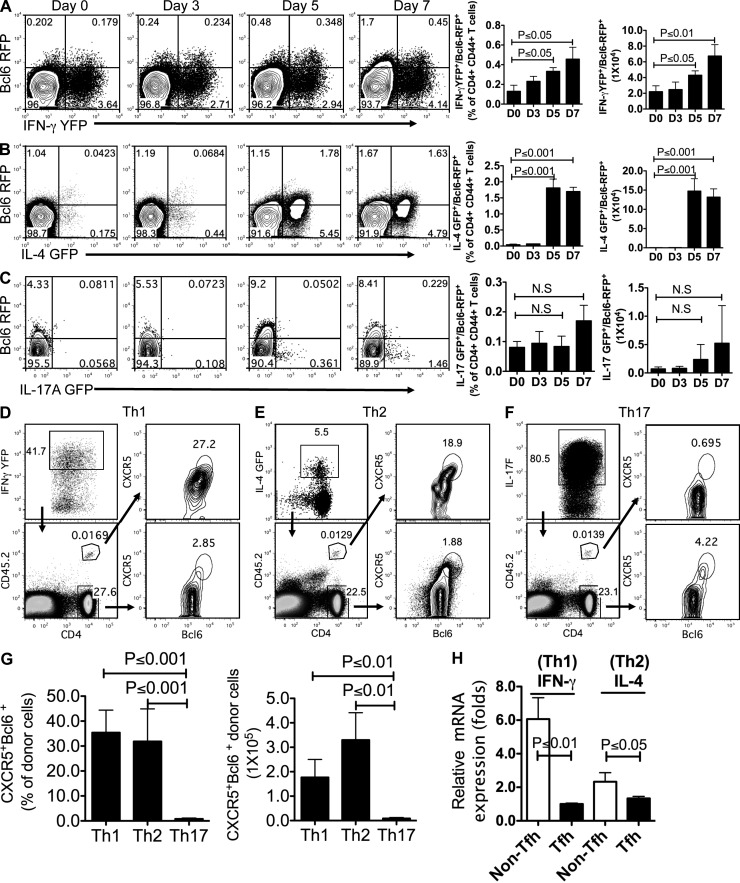
These aforementioned observations suggest that Tfh development was associated with gradual transcriptional changes: the signature genes representing Th1, Th2, and Th17 were reduced, and Tfh signature genes were increased. To better analyze the developmental relationships between Tfh and other T cell lineages at the single-cell level, dual reporter mice were generated by crossing Bcl6-RFP mice with three other known reporter mice, i.e., IL-4–GFP (il4gfp; 4get), IFN-γ–YFP (ifngyfp; Yeti), and IL-17A–GFP (il17agfp; Mohrs et al., 2001; Stetson et al., 2003; Esplugues et al., 2011). These double reporter mice were s.c. immunized with KLH in CFA for 7 d, followed by FACS analysis with CD4+CD44hi gating.
In Bcl6-RFP/IFN-γ–YFP dual reporter mice, we observed a gradual increase of both Bcl6-RFP+YFP+ and Bcl6-RFP+YFP− T cells in a time-dependent manner (Fig. 5 A). Interestingly, Bcl6-RFP+YFP− T cells appeared to dominate over Bcl6-RFP+YFP+ T cells on day 7 (Fig. 5 A). When three populations of activated CD4+ T cells (RFP+YFP−, RFP+YFP+, and RFP−YFP+) from dLNs were sorted and subjected to measurement of Th1-related genes by quantitative RT-PCR, we found the intermediate expression of T-bet and IFN-γ in RFP+YFP+ Tfh cells, compared with that in RFP+YFP− Tfh cells and RFP−YFP+ T (Th1) cells (not depicted). Likewise, in Bcl6-RFP/IL-4–GFP mice, RFP+GFP+ CD4+ T cells (1.63–1.78%) were observed at days 5 and 7 after immunization, consisting of >50% of total Bcl6-RFP+ T cells (Fig. 5 B), which is consistent with previous studies from a Th2-biased animal model (King and Mohrs, 2009; Reinhardt et al., 2009). Furthermore, we compared the expression of Th2 signature genes among RFP+GFP−, RFP+GFP+, and RFP−GFP+ T cells and found that Gata3 and IL-13 expression was negatively correlated with Bcl6-RFP expression (not depicted). In contrast, IL-4 expression was not decreased in RFP+GFP+ Tfh cells in comparison with RFP−GFP+ T (Th2) cells (not depicted), suggesting that IL-4 expression in Tfh cells is not dependent on Gata3 (Liang et al., 2012).
Figure 5.
Th1 and Th2 but few of Th17 cells are able to become Tfh cells. (A–C) Dual reporter mice were immunized s.c. with KLH in CFA for 0, 3, 5, and 7 d. Flow cytometry analysis of Bcl6-RFP and eGFP/YFP on gated CD4+CD44+ T cells from dLNs. Data are representative of two independent experiments. Bar graphs display the percentage and number of double-positive cells as mean ± SD. (A) Flow cytometry analysis of Bcl6-RFP and IFN-γ–YFP from CD4+CD44+ T cells in Bcl6-RFP/Yeti double reporter mice. (B) Flow cytometry analysis of Bcl6-RFP and IL-4–GFP from CD4+CD44+ T cells in Bcl6-RFP/4get double reporter mice. (C) Flow cytometry analysis of Bcl6-RFP and IL-17A–eGFP from CD4+CD44+ T cells in Bcl6-RFP/IL-17A–GFP double reporter mice. (D) The in vitro committed YFPhi OT-II T (Th1) cells were collected from 4-d culture and intravenously transferred into congenic recipient mice (CD45.1+). 7 d after immunization with OVA in CFA s.c., donor and host CD4+ T cells were subject to flow cytometry analysis of CXCR5 and Bcl6. (E) Flow cytometry analysis of the CXCR5 and Bcl6 expression in donor IL-4–GFPhi OT-II T (Th2) cells and host CD4+ T cells. (F) Flow cytometry analysis of the CXCR5 and Bcl6 expression in donor IL-17F–RFPhi OT-II T (Th17) cells and host CD4+ T cells after transfer and immunization. (D–F) Data are representative of two independent experiments. (G) Summary of data from D–F. Bar graphs display the percentage and number of donor cells as mean ± SD. (H) In the assay of conversion from Th1 to Tfh cell (D) and from Th2 to Tfh cell (E), 7 d after immunization, donor cell–derived CXCR5+Bcl6hi (Tfh) and CXCR5−Bcl6− (Non-Tfh) were sorted and subjected to the measurement of cytokine expression by quantitative RT-PCR. Data are representative of two independent experiments displayed as mean ± SD. n = 3 per group. N.S., no statistically significant difference.
Distinctly, the immunized IL-17A–GFP/Bcl6-RFP dual reporter mice displayed few signal overlays between IL-17A–GFP+ and Bcl6-RFP+ T cells (Fig. 5 C). To rule out the possibility that the lack of GFP+RFP+ T cells was caused by weak IL-17A–GFP, we immunized dual reporter mice with a higher concentration of adjuvant CFA (5 mg/ml) + KLH. 7 d later, although the percentage of IL-17A–GFP+ T cells was increased, the population of GFP+RFP+ T cells was still negligible (not depicted). Therefore, the aforementioned data indicated that although Tfh cell commitment required the down-regulation of Th1 and Th2 signature genes, Bcl6hi Tfh cells might share intrinsic developmental regulation with Th1 and Th2 cells but not with Th17 cells.
To further test the plasticity of T cells, we generated OVA-specific Th1, Th2, and Th17 cells in vitro with naive OT-II reporter T cells under polarizing conditions and then transferred IFN-γ–YFPhi (Th1), IL-4–GFPhi (Th2), or IL-17F–RFPhi (Th17) into recipient mice, respectively (Fig. 5, D–F). Consistent with the observation obtained from dual reporter mice, on day 7 after OVA/CFA immunization, 27.2% of IFN-γ–YFPhi Th1 cells and 19% of IL-4–GFPhi Th2 cells gained Tfh cell phenotypes (CXCR5+Bcl6+) in the dLNs (Fig. 5, D, E, and G), whereas no significant Bcl6 and CXCR5 expression was detected in donor IL-17F–RFPhi Th17 cells (Fig. 5, F and G), which is in agreement with our previous study on the stable phenotype of Th17 cells in vivo (Nurieva et al., 2009b). Furthermore, we examined the changes in cytokine expression between Th1/Th2 and Th1/Th2-derived Tfh cells. As shown in Fig. 5 H, IFN-γ and IL-4 expression was substantially decreased in T cells once they gained the Tfh phenotype, suggesting a Tfh-specific transcriptional suppression. Collectively, our results demonstrated that both Th1 and Th2 but few of Th17 cells were able to become Tfh cells.
Tfh cells can be sustained and develop into memory cells
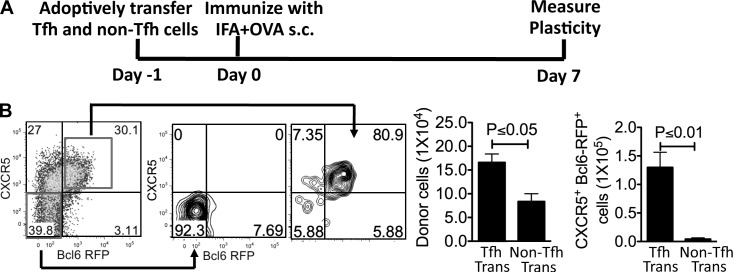
Our aforementioned results have unveiled considerable plasticity in early T cell differentiation in vivo. To address whether Tfh cells can be reprogrammed into other Th lineages, we sorted Tfh and non-Tfh cells from Bcl6-RFP reporter mice immunized with KLH in CFA. When they were cultured under different polarizing conditions in vitro, they were able to express genes associated with other T cell lineages (not depicted), similar to a recent study using CXCR5+PD-1+ Tfh cells (Lu et al., 2011). We thus tested their plasticity in vivo by transferring equal numbers of Tfh and non-Tfh cells into naive recipient mice, followed by KLH/IFA immunization s.c. (Fig. 6 A). Surprisingly, 7 d after immunization, we found that >80% of donor Tfh and non-Tfh cell in dLNs maintained their phenotype based on the expression of CXCR5 and Bcl6 (Fig. 6 B), suggesting that Tfh and non-Tfh cells were stable in vivo.
Figure 6.
The stability and plasticity of Tfh cells in vivo. (A) Experimental design for testing Tfh stability in vivo. (B) The sorted effector Tfh and non-Tfh (CD45.2+) cells were adoptively transferred into congenic recipient mice (CD45.1+). 7 d after s.c. immunization with KLH in IFA, the expression of CXCR5 and Bcl6 in donor Tfh and non-Tfh cells was measured by flow cytometry. Data are representative of two independent experiments. Bar graphs display the number of donor cells as mean ± SD. n = 3 per group.
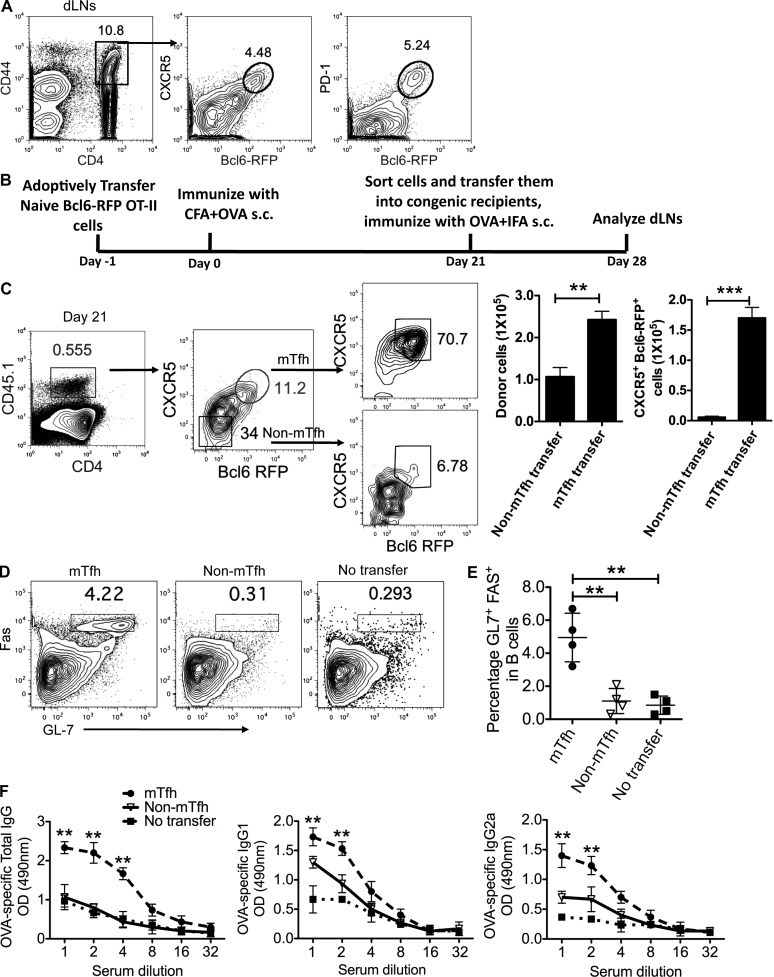
Because effector Tfh cells remained stable in vivo, we then assessed whether CXCR5+Bcl6hi Tfh cells can form memory cells. First, we analyzed Bcl6-RFP reporter mice at a later stage after immunization. 30 d after immunization with KLH in CFA s.c., a small population of CXCR5+Bcl6hi (∼4.5%) cells was still detectable in dLNs, and Bcl6-RFP expression was highly correlated with PD-1 (Fig. 7 A). Consistently, a similar CXCR5+Bcl6hi population was observed in mice 30 d after receiving i.p. injection of KLH emulsified in CFA (not depicted), and these cell lacked surface IL-2Ra (Choi et al., 2011) and Ly6C (a surface marker down-regulated in memory CD4+ T cells; not depicted; Marshall et al., 2011). Thus, these results indicated the existence of memory Tfh cells. To further confirm this, we transferred naive Bcl6-RFP OT-II T cells into congenic recipient mice, followed with OVA/CFA immunization. 21 d after immunization, 11% of donor cells expressed both CXCR5 and Bcl6 (Fig. 7, B and C). Also, those CXCR5+ donor cells lacked Ly6C expression (not depicted), exhibiting a feature of memory CD4+ T cells. When quantifying the gene expression of persistent CXCR5+Bcl6hi cells, we found that these cells maintained effector Tfh signature gene expression (not depicted).
Figure 7.
Memory Tfh cells promote humoral recall responses. (A) 30 d after immunization with KLH in CFA s.c., flow cytometry analysis of CXCR5 and Bcl6-RFP expression in activated T cells from Bcl6-RFP reporter mice was performed. Data are representative of two independent experiments. (B) Experimental design for functional analysis of mTfh and non-mTfh cells. (C) CXCR5+Bcl6hi or CXCR5−Bcl6− OT-II CD4+ T cells (CD45.1+) were purified 21 d after immunization and adoptively transferred at equal number (1.5 × 105) into naive congenic recipients (CD45.2+). 7 d later, the phenotype of donor cells in the dLNs and their population were determined by flow cytometry. (D) Flow cytometry analysis of GL7 and Fas on B220+ B cells from mTfh and non-mTfh transferred recipients; the nontransfer mice were set as controls. (E) The percentage of GL7+Fas+ B cells in dLNs was quantified. (F) OVA-specific antibody levels (total IgG, IgG1, and IgG2a) were determined by ELISA. (C–F) Cumulative data are representative of two independent experiments displayed as mean ± SD. n = 4 per group. **, P < 0.01; ***, P < 0.001.
To investigate whether long-lived memory Tfh cells can contribute to recall responses for subsequent antigen challenge, we sorted and transferred day 21 CXCR5−Bcl6− and CXCR5+Bcl6hi memory T cells into naive recipients, respectively, and assessed their capacity to help B cells in vivo (Fig. 7 B). 7 d after challenge with OVA in IFA, we observed a larger number of CXCR5+Bcl6hi donor-derived OT-II cells in the dLNs compared with the CXCR5−Bcl6− counterpart (Fig. 7, B and C). Of note, in both groups, the majority of donor-derived cells maintained their initial phenotype: ∼70% of CXCR5+Bcl6hi donor-derived cells maintained high levels of CXCR5 and Bcl6 expression; >90% of CXCR5−Bcl6− donor-derived cells lacked CXCR5 or Bcl6 expression (Fig. 7 C). As expected, transfer of CXCR5+Bcl6hi OT-II cells increased the frequencies of GC B cells compared with the CXCR5−Bcl6− OT-II counterpart (Fig. 7, D and E). As an additional measure of B cell help, we examined the anti-OVA IgG titers in the sera and found that anti-OVA IgGs including total IgG, IgG1, and IgG2a were significantly enhanced in mice receiving memory Tfh cells (Fig. 7 F). Thus, long-lived CXCR5+Bcl6hi T cells as memory cells contribute to robust GC reaction in humoral recall responses.
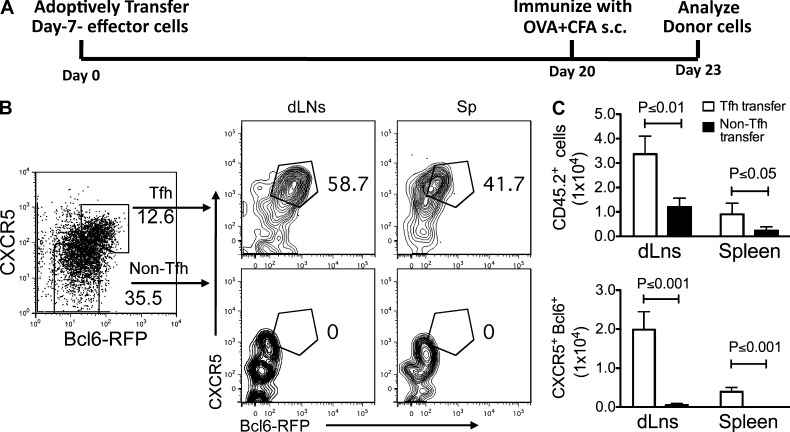
To test whether long-lived CXCR5+Bcl6hi cells are directly derived from effector Tfh cells, naive Bcl6-RFP OT-II cells were transferred into recipient mice. 7 d after immunization, CXCR5+Bcl6hi Tfh cells were purified from dLNs and transferred into naive recipient mice, followed by OVA + IFA immunization. 30 d after immunization, ∼50% donor cells were still present as CXCR5+Bcl6hi cells (not depicted). To test whether effector Tfh cells can survive in the absence of antigens, we transferred effector Tfh and non-Tfh OT-II cells into recipient mice. 20 d later, the recipient mice were immunized to analyze memory T cells at present (Fig. 8 A). Flow cytometry analysis showed that transferring effector Tfh cells resulted in a substantially larger amount of CXCR5+Bcl6hi cells in both dLNs and spleens of recipient mice compared with that in non-Tfh transferred mice (Fig. 8, B and C). Consistent with a recent study using IL-21 reporter mice (Lüthje et al., 2012), our findings showed that Tfh cells were able to survive in an antigen-free environment as memory Tfh cells.
Figure 8.
Effector Tfh cells can develop into memory Tfh cells. Naive Bcl6-RFP OT-II cells (CD45.2+) were transferred into CD45.1+ recipient mice, followed by s.c. immunization with OVA in CFA for 7 d. CXCR5+Bcl6hi OT-II T cells were then purified and transferred into naive mice (1.5 × 105 cells/mouse). 20 d later, recipients were immunized with OVA + CFA for another 3 d, and then the phenotypes of donor cells in the dLNs and spleens were determined by flow cytometry. Data are representative of two independent experiments. n = 3 per group. (A) Experimental strategy of examining the transition from effector Tfh to memory Tfh cell. (B) Flow cytometry analysis of CXCR5 and Bcl6 in donor Tfh and non-Tfh cells is shown. (C) After adoptive transfer and immunization, the cell numbers of CXCR5+Bcl6-RFP+ and CXCR5−Bcl6-RFP− donor-derived cells were summarized and compared. Cumulative data are representative of two independent experiments displayed as mean ± SD.
DISCUSSION
The cellular and molecular regulation of Tfh cell development has recently been extensively studied. The transcription factor Bcl6 not only is selectively expressed in Tfh cells but also controls Tfh cell generation. Here, by using Bcl6-RFP reporter mice, we performed phenotypic, functional and genome-wide transcriptome analysis of Tfh cells generated in vivo and found that the initial up-regulation of CXCR5 was not dependent on Bcl6. Moreover, once T cells up-regulated Bcl6, they could be sustained in vivo with or without antigen and form functional memory cells.
Because Bcl6 was shown as the master regulator of Tfh cells, it is important to understand the regulation of Bcl6 expression. In a recent work, a Bcl6-YFP reporter mouse was generated via fusing Bcl6 with YFP (Kitano et al., 2011). It was acknowledged by the authors that the Bcl6-YFP fusion protein has compromised function. In their study, Bcl6-YFP was shown to be initially up-regulated together with CXCR5 on all T cells as early as day 2 after immunization, at its peak at day 3, and then subsequently down-regulated. This phenotype was also observed by another group using the same strategy of cotransferring both antigen-specific T cells and B cells, which may lead to accelerated Tfh cell differentiation (Kerfoot et al., 2011; King, 2011). In this study, we developed a new Bcl6-RFP reporter mouse in which an ires-rfp sequence was inserted into the 3′ UTR of the Bcl6 gene to allow for reporting its transcription without interfering with the function of the native protein. We found that RFP faithfully correlates with endogenous Bcl6 expression. We also found that, from day 2 to 7, Bcl6 expression was gradually up-regulated along with Tfh development and was maintained for a certain period of time.
Different from the sequential increase in Bcl6 expression during Tfh development, we found that CXCR5 expression was rapidly enhanced and maintained at high level in activated T cells. This finding suggested that initial CXCR5 expression may be Bcl6 independent. Indeed, CXCR5 expression was found intact in bcl6−/− OT-II cells on day 3 after immunization. Thus, the initiation of mouse CXCR5 expression in T cells is independent of Bcl6, whereas its maintenance requires Bcl6. The mechanisms by which CXCR5 and Bcl6 are differentially regulated remain unclear and require further investigations.
At an early stage of Tfh development, some of the activated T cells up-regulated the expression of Tfh genes while maintaining expression of Th1, Th2, and Th17 genes. These intermediate Tfh cells can be further matured into CXCR5+Bcl6hi Tfh cells with the help of cognate B cells. The resulting Bcl6hi expression appeared to correlate with the formation of GC structures (unpublished data). Earlier studies from other groups and ours demonstrated that Tfh cells expressed low levels of Th1, Th2, and Th17 cytokines (Chtanova et al., 2004; Vinuesa et al., 2005; Nurieva et al., 2008). However, studies from pathogen-infected mouse models observed IFN-γ– and IL-4–secreting Tfh cells in the GCs (King and Mohrs, 2009; Reinhardt et al., 2009). Using dual reporter mice (Bcl6-RFP/Yeti and Bcl6-RFP/4get) immunized with KLH in CFA, we found that a certain amount of Bcl6hi Tfh cells were simultaneously expressing the reporter genes inserted into the Ifng or Il4 gene locus. Also, both Th1 and Th2 cells were able to become Tfh cells. After being converted, the expressions of Th1/Th2 signature genes in Bcl6+ cells were significantly reduced, suggesting a mechanism of Bcl6-dependent Tfh programming. Nonetheless, Th17 cells hardly became Tfh cells, which is consistent with an earlier study of human Th17 cells (Acosta-Rodriguez et al., 2007). However, our data do not exclude the possibility that Th17 cells function in supporting B cell responses at extrafollicular sites, as suggested by identification of IL-21+ T cells located outside of follicles (King et al., 2008).
After maturation, the fate of Tfh cells remains an important question. Several recent studies have shown that the persistent deports of peptide–MHC class II after the primary immune response allow CXCR5+ T cells to survive the contraction phase and eventually become the long-lived memory compartment, albeit with diverse characteristics (Fazilleau et al., 2007; MacLeod et al., 2011; Marshall et al., 2011). In this study, we found that long-lived memory Tfh cells (either polyclonal or monoclonal) could be derived from effector Tfh cells and were able to accelerate GC response. Our study also provided direct evidence that the transition from effector Tfh to memory Tfh cells occurred in the absence of antigen, which was in agreement with a very recent observation from IL-21 reporter mice (Lüthje et al., 2012).
Overall, we have used a novel Bcl6 reporter system to analyze Tfh cell development and memory Tfh cell generation. Our results have dissected the process of Tfh development by showing the Bcl6-independent up-regulation of CXCR5 expression and revealed the transition from effector Tfh to memory Tfh cell. Our data together specified Tfh cells in vivo with Bcl6 expression.
MATERIALS AND METHODS
Generation of Bcl6-RFP reporter mice.
The Bcl6-RFP mice were generated by the insertion of IRES-mRFP-polyA construct after the stop codon of Bcl6 by homologous recombination using the 129/TC1 embryonic stem cell line. An Frt-flanked NeoR and diphtheria toxin served as positive and negative selection markers in the targeting vector, respectively. Targeted embryonic stem clones were injected into C57BL/6 blastocysts to generate chimeras. Bcl6-RFP reporter mice were obtained after deletion of NeoR cassette by crossing with Flper strain. Bcl6-RFP reporter mice were backcrossed to C57BL/6 background for six to eight generations. Bcl6-RFP reporter mice on C57BL/6 background were used along with age- and sex-matched B6, B6sjl, and B6×B6sjl mice.
The genotyping primers for Bcl6-RFP reporter mice were as follows: forward, 5′-GTCTCTTCAGTGTGAGAAGTG-3′; and reverse, 5′-CTTCGGCCAGTAACGTTAG-3′, amplifying an ∼350-bp band. Primers for WT control mice were as follows: forward, 5′-AGTCCGAGGGACCCGTGAGC-3′; and reverse, 5′-TATGATTTGCACTAGTGGATG-3′, amplifying an ∼500-bp band.
The other mice.
OT-II, 4get, µMT, Rosa26-FLP1, and B6sjl (CD45.1) mice were purchased from the Jackson Laboratory. Yeti mice were provided by R.M. Locksley (University of California, San Francisco, San Francisco, CA; Stetson et al., 2003). IL-17A–GFP and IL-17F–RFP reporter mice were previously reported (Yang et al., 2008; Esplugues et al., 2011). bcl6−/− mice were provided by R. Dalla-Favera (Columbia University, New York, NY; Ye et al., 1997). Mice were housed in a specific pathogen–free animal facility at the University of Texas MD Anderson Cancer Center, and animals were used according to the protocols approved by the Institutional Animal Care and Use Committee.
Th cell differentiation and cell transfer.
Naive CD4+CD25−CD44loCD62Lhi T cells from Yeti/OT-II, 4get/OT-II, and IL-17F–RFP/OT-II mice were sorted by flow cytometry and activated with irradiated APCs plus 10 µg/ml OT-II peptide under the conditions for Th1, Th2, and Th17 cell differentiation. 4 d after activation, GFPhi or RFPhi cells were purified before transfer. Naive Bcl6-RFP/OT-II T cells, in vivo generated OT-II Tfh (CD4+ Bcl6-RFP+CXCR5+) cells, or in vitro differentiated Th1, Th2, and Th17 cells were intravenously transferred into congenic recipient mice. Cell transfer numbers for different cells were as follows: 106 for naive T cells, 106 for Th helper cells, and 105 for Tfh cells.
Microarray.
Total cellular RNA was extracted from sorted Tfh (CD4+CD44+Bcl6-RFP+CXCR5+), intermediate Tfh (CD4+CD44+Bcl6-RFP−CXCR5+), and non-Tfh (CD4+CD44+Bcl6-RFP−CXCR5−) cells with TRIZOL reagent (Invitrogen). DNA microarray labeling and analysis were performed by the microarray core at the Institute for Systems Biology. Approximately 10 µg RNA was labeled and hybridized to GeneChip Mouse Genome 430.2 arrays (Affymetrix) according to the manufacturer’s protocols. Expression values were defined with GeneChip Operating Software (GCOS; Affymetrix). The microarray data have been deposited into the NCBI GEO database under accession no. GSE40068.
Real-time RT-PCR analysis.
Total RNA was extracted with TRIZOL reagent. Oligo (dT) and MMLV reverse transcription (Invitrogen) were used to generate cDNA. The expression levels of genes were normalized to a reference gene β-actin. The primer pairs for real-time RT-PCR analysis of Bcl6, CXCR5, IL-21, IL-4, Gata3, IFN-γ, T-bet, Il-17A, and RORγ were previously described (Nurieva et al., 2008). The primer pair for detection of Batf is forward, 5′-GGCAAACAGGACTCATCTGATGATG-3′; and reverse, 5′-GGCAGCCCGGCCTCAGTTTACATG-3′. Primers for C-maf are forward, 5′-GCAGAGACACGTCCTGGAGTCG-3′; and reverse, 5′-CGAGCTTGGCCCTGCAACTAGC-3′.
Online supplemental material.
Table S1, included as a separate PDF file, shows gene expression profiles of Tfh (CXCR5+Bcl6-RFPhi), intermediate Tfh (CXCR5+Bcl6-RFPlo), and non-Tfh (CXCR5−Bcl6-RFP−) cells. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20120219/DC1.
Supplementary Material
Acknowledgments
We thank the MD Anderson Genetic Engineering Mouse Facility for their assistance in the generation of Bcl6-RFP reporter mice, Dr. Richard M. Locksley for Yeti mice, Dr. Ricchardo Dalla-Favera for Bcl6 knockout mice, Dr. Lai Wei and Danielle Yi for microarray analysis, and the Dong laboratory members for their help.
The work is supported by research grants from the National Institutes of Health (to C. Dong) and an Odyssey fellowship from the University of Texas MD Anderson Cancer Center (to X. Liu and B. Zhong). C. Dong is an Olga and Harry Weiss Distinguished University Chair in Cancer Research of the University of Texas MD Anderson Cancer Center and a Leukemia and Lymphoma Society Scholar.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- dLN
- draining LN
- GC
- germinal center
- IRES
- internal ribosome entry site
- mRNA
- messenger RNA
- Tfh cell
- T follicular helper cell
- UTR
- untranslated region
References
- Acosta-Rodriguez E.V., Rivino L., Geginat J., Jarrossay D., Gattorno M., Lanzavecchia A., Sallusto F., Napolitani G. 2007. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat. Immunol. 8:639–646 10.1038/ni1467 [DOI] [PubMed] [Google Scholar]
- Bauquet A.T., Jin H., Paterson A.M., Mitsdoerffer M., Ho I.C., Sharpe A.H., Kuchroo V.K. 2009. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat. Immunol. 10:167–175 10.1038/ni.1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz B.C., Jordan-Williams K.L., Wang C., Kang S.G., Liao J., Logan M.R., Kim C.H., Taparowsky E.J. 2010. Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. J. Exp. Med. 207:933–942 10.1084/jem.20091548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitfeld D., Ohl L., Kremmer E., Ellwart J., Sallusto F., Lipp M., Förster R. 2000. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J. Exp. Med. 192:1545–1552 10.1084/jem.192.11.1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.S., Kageyama R., Eto D., Escobar T.C., Johnston R.J., Monticelli L., Lao C., Crotty S. 2011. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity. 34:932–946 10.1016/j.immuni.2011.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chtanova T., Tangye S.G., Newton R., Frank N., Hodge M.R., Rolph M.S., Mackay C.R. 2004. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. J. Immunol. 173:68–78 [DOI] [PubMed] [Google Scholar]
- Crotty S. 2011. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29:621–663 10.1146/annurev-immunol-031210-101400 [DOI] [PubMed] [Google Scholar]
- Dong C., Nurieva R.I. 2003. Regulation of immune and autoimmune responses by ICOS. J. Autoimmun. 21:255–260 10.1016/S0896-8411(03)00119-7 [DOI] [PubMed] [Google Scholar]
- Ellyard J.I., Vinuesa C.G. 2011. A BATF-ling connection between B cells and follicular helper T cells. Nat. Immunol. 12:519–520 10.1038/ni.2042 [DOI] [PubMed] [Google Scholar]
- Esplugues E., Huber S., Gagliani N., Hauser A.E., Town T., Wan Y.Y., O’Connor W., Jr, Rongvaux A., Van Rooijen N., Haberman A.M., et al. 2011. Control of TH17 cells occurs in the small intestine. Nature. 475:514–518 10.1038/nature10228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey L.M., Wilson E.B., Elsaesser H., Fistonich C.D., McGavern D.B., Brooks D.G. 2011. Viral persistence redirects CD4 T cell differentiation toward T follicular helper cells. J. Exp. Med. 208:987–999 10.1084/jem.20101773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazilleau N., Eisenbraun M.D., Malherbe L., Ebright J.N., Pogue-Caley R.R., McHeyzer-Williams L.J., McHeyzer-Williams M.G. 2007. Lymphoid reservoirs of antigen-specific memory T helper cells. Nat. Immunol. 8:753–761 10.1038/ni1472 [DOI] [PubMed] [Google Scholar]
- Goenka R., Barnett L.G., Silver J.S., O’Neill P.J., Hunter C.A., Cancro M.P., Laufer T.M. 2011. Cutting edge: dendritic cell-restricted antigen presentation initiates the follicular helper T cell program but cannot complete ultimate effector differentiation. J. Immunol. 187:1091–1095 10.4049/jimmunol.1100853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichii H., Sakamoto A., Hatano M., Okada S., Toyama H., Taki S., Arima M., Kuroda Y., Tokuhisa T. 2002. Role for Bcl-6 in the generation and maintenance of memory CD8+ T cells. Nat. Immunol. 3:558–563 10.1038/ni802 [DOI] [PubMed] [Google Scholar]
- Ise W., Kohyama M., Schraml B.U., Zhang T., Schwer B., Basu U., Alt F.W., Tang J., Oltz E.M., Murphy T.L., Murphy K.M. 2011. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat. Immunol. 12:536–543 10.1038/ni.2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R.J., Poholek A.C., DiToro D., Yusuf I., Eto D., Barnett B., Dent A.L., Craft J., Crotty S. 2009. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 325:1006–1010 10.1126/science.1175870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerfoot S.M., Yaari G., Patel J.R., Johnson K.L., Gonzalez D.G., Kleinstein S.H., Haberman A.M. 2011. Germinal center B cell and T follicular helper cell development initiates in the interfollicular zone. Immunity. 34:947–960 10.1016/j.immuni.2011.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C. 2011. A fine romance: T follicular helper cells and B cells. Immunity. 34:827–829 10.1016/j.immuni.2011.06.007 [DOI] [PubMed] [Google Scholar]
- King C., Tangye S.G., Mackay C.R. 2008. T follicular helper (TFH) cells in normal and dysregulated immune responses. Annu. Rev. Immunol. 26:741–766 10.1146/annurev.immunol.26.021607.090344 [DOI] [PubMed] [Google Scholar]
- King I.L., Mohrs M. 2009. IL-4–producing CD4+ T cells in reactive lymph nodes during helminth infection are T follicular helper cells. J. Exp. Med. 206:1001–1007 10.1084/jem.20090313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano M., Moriyama S., Ando Y., Hikida M., Mori Y., Kurosaki T., Okada T. 2011. Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. Immunity. 34:961–972 10.1016/j.immuni.2011.03.025 [DOI] [PubMed] [Google Scholar]
- Liang H.E., Reinhardt R.L., Bando J.K., Sullivan B.M., Ho I.C., Locksley R.M. 2012. Divergent expression patterns of IL-4 and IL-13 define unique functions in allergic immunity. Nat. Immunol. 13:58–66 10.1038/ni.2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K.T., Kanno Y., Cannons J.L., Handon R., Bible P., Elkahloun A.G., Anderson S.M., Wei L., Sun H., O’Shea J.J., Schwartzberg P.L. 2011. Functional and epigenetic studies reveal multistep differentiation and plasticity of in vitro-generated and in vivo-derived follicular T helper cells. Immunity. 35:622–632 10.1016/j.immuni.2011.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüthje K., Kallies A., Shimohakamada Y., TBelz G.T., Light A., Tarlinton D.M., Nutt S.L. 2012. The development and fate of follicular helper T cells defined by an IL-21 reporter mouse. Nat. Immunol. 13:491–498 10.1038/ni.2261 [DOI] [PubMed] [Google Scholar]
- MacLeod M.K., David A., McKee A.S., Crawford F., Kappler J.W., Marrack P. 2011. Memory CD4 T cells that express CXCR5 provide accelerated help to B cells. J. Immunol. 186:2889–2896 10.4049/jimmunol.1002955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall H.D., Chandele A., Jung Y.W., Meng H., Poholek A.C., Parish I.A., Rutishauser R., Cui W., Kleinstein S.H., Craft J., Kaech S.M. 2011. Differential expression of Ly6C and T-bet distinguish effector and memory Th1 CD4(+) cell properties during viral infection. Immunity. 35:633–646 10.1016/j.immuni.2011.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J.R. 2005. Peyer’s patch germinal centers: the elusive switch site for IgA. J. Immunol. 175:1361–1362 [DOI] [PubMed] [Google Scholar]
- Mohrs M., Shinkai K., Mohrs K., Locksley R.M. 2001. Analysis of type 2 immunity in vivo with a bicistronic IL-4 reporter. Immunity. 15:303–311 10.1016/S1074-7613(01)00186-8 [DOI] [PubMed] [Google Scholar]
- Nurieva R.I., Chung Y., Hwang D., Yang X.O., Kang H.S., Ma L., Wang Y.H., Watowich S.S., Jetten A.M., Tian Q., Dong C. 2008. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 29:138–149 10.1016/j.immuni.2008.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva R.I., Chung Y., Martinez G.J., Yang X.O., Tanaka S., Matskevitch T.D., Wang Y.-H., Dong C. 2009a. Bcl6 mediates the development of T follicular helper cells. Science. 325:1001–1005 10.1126/science.1176676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva R., Yang X.O., Chung Y., Dong C. 2009b. Cutting edge: in vitro generated Th17 cells maintain their cytokine expression program in normal but not lymphopenic hosts. J. Immunol. 182:2565–2568 10.4049/jimmunol.0803931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt R.L., Liang H.E., Locksley R.M. 2009. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat. Immunol. 10:385–393 10.1038/ni.1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaerli P., Willimann K., Lang A.B., Lipp M., Loetscher P., Moser B. 2000. CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. J. Exp. Med. 192:1553–1562 10.1084/jem.192.11.1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.M., Pottage L., Thomas E.R., Leishman A.J., Doig T.N., Xu D., Liew F.Y., Garside P. 2000. Th1 and Th2 CD4+ T cells provide help for B cell clonal expansion and antibody synthesis in a similar manner in vivo. J. Immunol. 165:3136–3144 [DOI] [PubMed] [Google Scholar]
- Smith K.M., Brewer J.M., Rush C.M., Riley J., Garside P. 2004. In vivo generated Th1 cells can migrate to B cell follicles to support B cell responses. J. Immunol. 173:1640–1646 [DOI] [PubMed] [Google Scholar]
- Stetson D.B., Mohrs M., Reinhardt R.L., Baron J.L., Wang Z.E., Gapin L., Kronenberg M., Locksley R.M. 2003. Constitutive cytokine mRNAs mark natural killer (NK) and NK T cells poised for rapid effector function. J. Exp. Med. 198:1069–1076 10.1084/jem.20030630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuesa C.G., Cook M.C., Angelucci C., Athanasopoulos V., Rui L., Hill K.M., Yu D., Domaschenz H., Whittle B., Lambe T., et al. 2005. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature. 435:452–458 10.1038/nature03555 [DOI] [PubMed] [Google Scholar]
- Yang X.O., Chang S.H., Park H., Nurieva R., Shah B., Acero L., Wang Y.H., Schluns K.S., Broaddus R.R., Zhu Z., Dong C. 2008. Regulation of inflammatory responses by IL-17F. J. Exp. Med. 205:1063–1075 10.1084/jem.20071978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B.H., Cattoretti G., Shen Q., Zhang J., Hawe N., de Waard R., Leung C., Nouri-Shirazi M., Orazi A., Chaganti R.S., et al. 1997. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat. Genet. 16:161–170 10.1038/ng0697-161 [DOI] [PubMed] [Google Scholar]
- Yu D., Vinuesa C.G. 2010. The elusive identity of T follicular helper cells. Trends Immunol. 31:377–383 10.1016/j.it.2010.07.001 [DOI] [PubMed] [Google Scholar]
- Yu D., Rao S., Tsai L.M., Lee S.K., He Y., Sutcliffe E.L., Srivastava M., Linterman M., Zheng L., Simpson N., et al. 2009. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 31:457–468 10.1016/j.immuni.2009.07.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.