Inhibition of macrophage SIRPα–CD47 interactions mediates phagocytosis and clearance of acute myeloid leukemia stem cells.
Abstract
Although tumor surveillance by T and B lymphocytes is well studied, the role of innate immune cells, in particular macrophages, is less clear. Moreover, the existence of subclonal genetic and functional diversity in some human cancers such as leukemia underscores the importance of defining tumor surveillance mechanisms that effectively target the disease-sustaining cancer stem cells in addition to bulk cells. In this study, we report that leukemia stem cell function in xenotransplant models of acute myeloid leukemia (AML) depends on SIRPα-mediated inhibition of macrophages through engagement with its ligand CD47. We generated mice expressing SIRPα variants with differential ability to bind human CD47 and demonstrated that macrophage-mediated phagocytosis and clearance of AML stem cells depend on absent SIRPα signaling. We obtained independent confirmation of the genetic restriction observed in our mouse models by using SIRPα-Fc fusion protein to disrupt SIRPα–CD47 engagement. Treatment with SIRPα-Fc enhanced phagocytosis of AML cells by both mouse and human macrophages and impaired leukemic engraftment in mice. Importantly, SIRPα-Fc treatment did not significantly enhance phagocytosis of normal hematopoietic targets. These findings support the development of therapeutics that antagonize SIRPα signaling to enhance macrophage-mediated elimination of AML.
Innate immune receptors can discriminate cell surface ligands expressed on aged, virally infected, or malignant cells, triggering effector mechanisms that aid in their clearance. NK cells lyse diseased cells and spare normal cells through the combined function of their activating and inhibitory receptors whose broadly expressed ligands, such as MHC class 1, can be altered qualitatively or quantitatively by infection or malignant transformation (Raulet, 2004). Similarly, macrophages and dendritic cell receptors discriminate altered-self molecules on aged or dying cells from normal-self markers on healthy cells, resulting in activation or inhibition of the phagocytosis response (Taylor et al., 2005). However, the role of these altered self discrimination mechanisms in the maintenance of normal blood cell homeostasis and in surveillance of transformed cells in hematologic malignancies is not fully understood.
Our understanding of surveillance mechanisms in hematologic malignancies such as human acute myeloid leukemia (AML) is further complicated by the extensive functional heterogeneity that exists among the individual cells that make up the leukemic clone. AML is organized as a cellular hierarchy sustained by a subpopulation of leukemia stem cells (LSCs; Lapidot et al., 1994; Bonnet and Dick, 1997; Hope et al., 2004). LSCs are the only AML cells that possess the canonical stem cell properties of self-renewal and the capacity to generate vast numbers of leukemic progenitors and blasts. Experimentally, LSCs are assayed by their ability to initiate engraftment in immunodeficient mouse recipients after i.v. or direct intrafemoral (i.f.) injection (Bonnet and Dick, 1997; Jin et al., 2006). There is accumulating evidence that LSCs are inherently resistant to conventional antiproliferative chemotherapy and lie at the heart of posttreatment relapse (Ishikawa et al., 2007; Yeung et al., 2010). These observations underscore the importance of defining surveillance mechanisms that target LSCs in addition to the blasts that make up the bulk of the leukemic clone.
Xenotransplantation into nonobese diabetic (NOD)–SCID (NOD/ShiLtJ-Prkdcscid; henceforth abbreviated NS) and related mouse strains has become the gold standard assay for human hematopoietic stem cells (HSCs) as well as LSCs. During development of these xenotransplantation assays, we observed that strain-specific factors governed the success of HSC engraftment in NS mice versus equally immunodeficient strains not on the NOD background. The parental NOD strain has been extensively studied by our group and others as a model of type 1 diabetes (Fox et al., 2000). Type 1 diabetes is genetically complex, requiring multiple NOD-derived Idd (insulin-dependent diabetes) genomic regions. The genetically related nonobese diabetes-resistant (NOR) strain differs from NOD only at four Idd loci. In our prior work, we identified the NOD-derived allele of Sirpa (signal regulatory protein α), located in the Idd13 locus on chromosome 2 (Fox et al., 2000), as the gene that contributed to the superior engraftment of HSCs in NS mice (Takenaka et al., 2007).
Sirpa encodes an Ig superfamily receptor expressed on macrophages, dendritic cells, and neurons. SIRPα and its ubiquitously expressed ligand CD47 interact through their respective Ig variable region (IgV)–like domains (Hatherley et al., 2007). Upon binding CD47, SIRPα immunoreceptor tyrosine-based inhibition motifs mediate inhibitory signals via recruitment of the src homology-2 domain containing protein tyrosine phosphatases SHP-1 and SHP-2 (Fujioka et al., 1996; Kharitonenkov et al., 1997; Veillette et al., 1998), leading to decreased phagocytosis by macrophages, inhibition of neutrophil migration, and attenuated production of the inflammatory cytokine TNF (Lindberg et al., 1996; Neznanov et al., 2003). We showed that the Sirpa IgV-like domain is polymorphic in mice and only NOD-derived SIRPα is able to bind to human CD47. Furthermore, we found that engraftment of normal HSCs in NS mice depends on the interaction between SIRPα on mouse macrophages and CD47 on human HSCs (Takenaka et al., 2007). In xenotransplantation assays, NS mice congenic for NOR alleles at the Idd13 locus (NOD.NOR-Idd13.Prkdcscid mice; henceforth abbreviated NS-Idd13) express a SIRPα variant that does not bind to human CD47, resulting in macrophage-mediated elimination and abrogation of human HSC engraftment (Takenaka et al., 2007).
Given that normal HSCs and LSCs share many properties including self-renewal and dependence on a niche (Lapidot et al., 1994; Hope et al., 2004; Clarke et al., 2006; Jin et al., 2006), we reasoned that SIRPα inhibitory signaling in macrophages might also play an important role in LSC engraftment, niche interactions, and/or maintenance in AML. This hypothesis is supported by evidence that high CD47 expression in AML is associated with poor clinical outcome and that treatment of NS mice bearing human AML grafts with an anti-CD47 antibody (Ab) reduces leukemic engraftment (Majeti et al., 2009). However, the mechanism of action of anti-CD47 Ab has not been fully clarified (Zhao et al., 2012). CD47 (first called integrin-associated protein; Lindberg et al., 1993, 1994) binds to multiple ligands other than SIRPα, including SIRPγ, thrombospondin, heterodimeric G proteins, and integrins (Gao et al., 1996a,b; Chung et al., 1997; Wang et al., 1999; Barazi et al., 2002; Brooke et al., 2004). These CD47 interactions govern processes in both normal tissues and cancer, including cell adhesion and migration, cell cycling, angiogenesis, apoptosis, and immune regulation (Lindberg et al., 1996; Isenberg et al., 2006; Olsson et al., 2007; Wang et al., 2007; Sarfati et al., 2008). Given the complexity of CD47 interactions, the observed antileukemia effects of anti-CD47 Ab treatment do not provide direct evidence that SIRPα inhibitory signaling in macrophages plays a role in regulating LSC function in AML.
In this study, we used a combined genetic and functional approach to investigate directly the role of SIRPα in AML. Xenotransplantation assays using NS congenic strains expressing different Sirpa alleles established that interaction between SIRPα on macrophages and CD47 on AML cells is critical for leukemic engraftment and migration by allowing evasion of immune surveillance by host macrophages. The genetic requirement for SIRPα signaling was validated by hSIRPα-Fc fusion protein–mediated disruption of SIRPα–CD47 interactions, which led to increased macrophage phagocytosis in vitro and impaired leukemic engraftment in treated NS mice. Importantly, we demonstrate that hSIRPα-Fc treatment significantly and preferentially enhances phagocytosis of AML targets over normal hematopoietic cells by human macrophages, supporting the development of antileukemia therapies that antagonize SIRPα signaling.
RESULTS
SIRPα–CD47 interaction is critical for LSC function in vivo
Our previous work demonstrating that normal HSCs depend on SIRPα–CD47 interaction (Takenaka et al., 2007) raised the question of whether this axis may also be important for LSC function in AML. Conclusions regarding the role of SIRPα in AML based on prior studies from other groups using an anti-CD47 Ab (Jaiswal et al., 2009; Majeti et al., 2009) are confounded by the fact that this Ab (B6H12) not only blocks CD47 interaction with SIRPα, but also with other binding partners including thrombospondin (Voit et al., 2003) and SIRPγ (Stefanidakis et al., 2008). Moreover, our data show that B6H12 induces mouse macrophage-mediated phagocytosis of AML cells even when the macrophages express a SIRPα variant that fails to bind human CD47 (Fig. 1). We therefore took a genetic approach to study the role of SIRPα in LSC function directly. Similar to the assay for normal human HSCs, the ability to initiate leukemic growth after transplantation into NS mice and related immunodeficient strains such as NOD/ShiLtJ-Prkdcscid.Il2rgtm1.1Flv (NSG) mice (Shultz et al., 2005) represents the only way to assay LSCs. To determine the importance of SIRPα–CD47 interaction for engraftment of LSCs, we injected primary AML cells (n = 4; see Table 1 for characteristics of patient samples used in this study) i.v. into sublethally irradiated NS or NS-Idd13 mice. 8 wk after transplantation, engrafted human AML cells were detected in the BM and spleen (SP) of NS but not NS-Idd13 mice (Fig. 2, a and b). Prior studies from our group have established that i.f. injection provides a more sensitive stem cell assay than i.v. injection and enables assessment of migration and graft establishment at distant sites, both key properties of normal HSCs and LSCs (Mazurier et al., 2003, 2004). Transplantation of bulk primary AML samples (n = 7) i.f. into NS mice resulted in engraftment in the injected femur as well as noninjected BM sites (Fig. 2, c and d). As is typical with this xenotransplant model, the level of engraftment was sample dependent (Bonnet and Dick, 1997; Hope et al., 2004; Jin et al., 2006, 2009). In contrast, three of seven AML samples were not able to repopulate either the injected or noninjected bones of NS-Idd13 mice after i.f. transplantation. Four samples were able to generate a local graft in the injected femur of NS-Idd13 mice; however, none of these was able to migrate and engraft noninjected BM sites or the SP, in contrast to their behavior in NS hosts. We performed a more detailed analysis of two AML samples, transplanting cells i.f. over a wide dose range, and observed high level engraftment at all BM sites in NS mice at all cell doses. In contrast, transplantation of NS-Idd13 mice at low cell doses resulted in lower levels of engraftment in the injected bone and reduced or absent engraftment at noninjected BM sites (Fig. 2 e). Both AML samples tested were able to engraft at higher transplanted doses in NS-Idd13 mice, suggesting that a high leukemic burden can overcome the SIRPα-dependent restriction of engraftment.
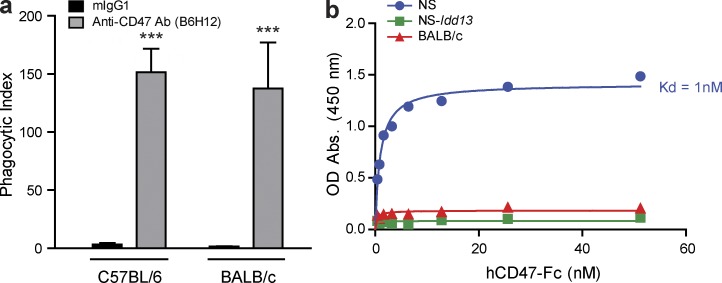
Figure 1.
Anti-CD47 Ab induces phagocytosis of primary human AML cells by macrophages expressing a SIRPα variant that does not bind human CD47. (a) Phagocytosis of human AML cells (AML19) by untreated C57BL/6 or BALB/c macrophages with addition of 7 µg mouse IgG1 or anti-CD47 Ab (B6H12), as assessed by spinning disk confocal microscopy. The phagocytic index was determined as the number of engulfed AML cells per 100 macrophages, performed in triplicate. Bars indicate mean ± SD. ***, P < 0.001 (Student’s t test). (b) Analysis of binding of NS, NS-Idd13, or BALB/c macrophages to human CD47-Fc. NS-Idd13 and C57BL/6 macrophages express an identical SIRPα variant.
Table 1.
Characteristics of AML patient samples
| Sample | Age at diagnosis | Sex | FAB | WHO | Cytogenetics | Blasts | CD34 | Flt3-ITD | NPM1 | CD47-RFI |
| yr | % | % | ||||||||
| 1 | 35 | M | M4 | AML not otherwise specified | inv(3)(q21q26.2),-7 | 70 | 91 | NA | NA | 4.6 |
| 2 | 64 | M | M1 | AML not otherwise specified | normal | 90 | 83 | + | WT | 3.1 |
| 3 | 83 | F | M5a | AML not otherwise specified | +8,+20 | 70 | 0 | NA | NA | 12.4 |
| 4 | 70 | M | M0 | AML not otherwise specified | +13 | 90 | 92 | NA | NA | 17.8 |
| 5 | 72 | F | M1 | AML not otherwise specified | normal | 90 | 27 | NA | NA | 6.7 |
| 6 | 39 | F | M5a | AML not otherwise specified | +8 | 90 | 16 | NA | NA | 4.4 |
| 7 | 45 | F | M1 | AML not otherwise specified | normal | 80 | 70 | NAa | NA | 12.5 |
| 8 | 93 | M | NA | AML not otherwise specified | normal | NA | 22 | + | mut | 11.7 |
| 9 | 51 | M | NA | AML with multilineage dysplasia following a MDS | +8 | 30 | 3 | − | WT | 5.7 |
| 10 | 51 | M | M5 | AML not otherwise specified | normal | 29 | 1 | + | mut | 20.2 |
| 11 | 23 | M | M0 | AML not otherwise specified | +i(8)(q10)/-8,+r1/-8,+r1,+r2 | 28 | 96 | + | WT | 7.4 |
| 12 | 52 | F | M2 | AML not otherwise specified | +8 | 95 | 70 | + | WT | 9.4 |
| 13 | 47 | F | M6 | AML not otherwise specified | normal | 40 | 6 | − | NA | 6.2 |
| 14 | 70 | M | M0 | AML not otherwise specified | +8 | 90 | 84 | NA | NA | 5.5 |
| 15 | 20 | F | M4 | AML not otherwise specified | normal | 50 | 14 | NA | NA | 4.1 |
| 16 | 63 | F | M4 | AML not otherwise specified | normal | 40 | 6 | − | mut | 7.9 |
| 17 | 61 | M | M4 | AML not otherwise specified | normal | 90 | 15 | + | NA | 3.9 |
| 18 | NA | NA | NA | NA | NA | NA | 76 | NA | NA | 16.3 |
| 19 | 56 | F | M5a | AML not otherwise specified | NA | 95 | 9 | NA | NA | 15 |
| 20 | 80 | F | NA | AML with multilineage dysplasia following a MDS | t(X;3)(q22;p26),add(7)(q22)/idem,del(16)(q22) | 95 | 3 | + | WT | 12.7 |
| 21 | 83 | M | M2 | AML with multilineage dysplasia following a MDS | +3,+9,+12 | 90 | NA | NA | NA | 12 |
| 22 | 69 | M | M0 | AML with multilineage dysplasia following a MDS | -Y | 97 | 93 | NA | NA | 14.2 |
| 23 | 67 | M | M1 | AML not otherwise specified | NA | 80 | 83 | + | WT | 13.1 |
| 24 | 47 | F | M1 | AML not otherwise specified | normal | 85 | 1 | + | NA | NA |
| 25 | 60 | F | M2 | AML not otherwise specified | normal | 23 | 96 | − | WT | NA |
| 26 | 40 | M | M4 | AML not otherwise specified | 46,XY,add(8)(q24.3),t(16;16)(p13.1;q22)[20].ish t(16;16)(3′ CBFB+;5′ CBFB+)[2] and INV (16) | 66 | 99 | NA | NA | 10.6 |
| 27 | 46 | M | M5b | AML not otherwise specified | normal | 90 | 1 | + | mut | 3.6 |
| 28 | 33 | M | M1 | AML not otherwise specified | 46,XY,ider(7)(q10)del(7)(q21) | >90 | 99 | NA | NA | 6.3 |
| 29 | 74 | M | M0 | AML not otherwise specified | NA | 83 | >95 | NA | NA | NA |
F, female; FAB, French-American-British; M, male; mut, mutated; NA, not available; RFI, relative fluorescence intensity compared with unstained (no Ab added).
Inconclusive results.
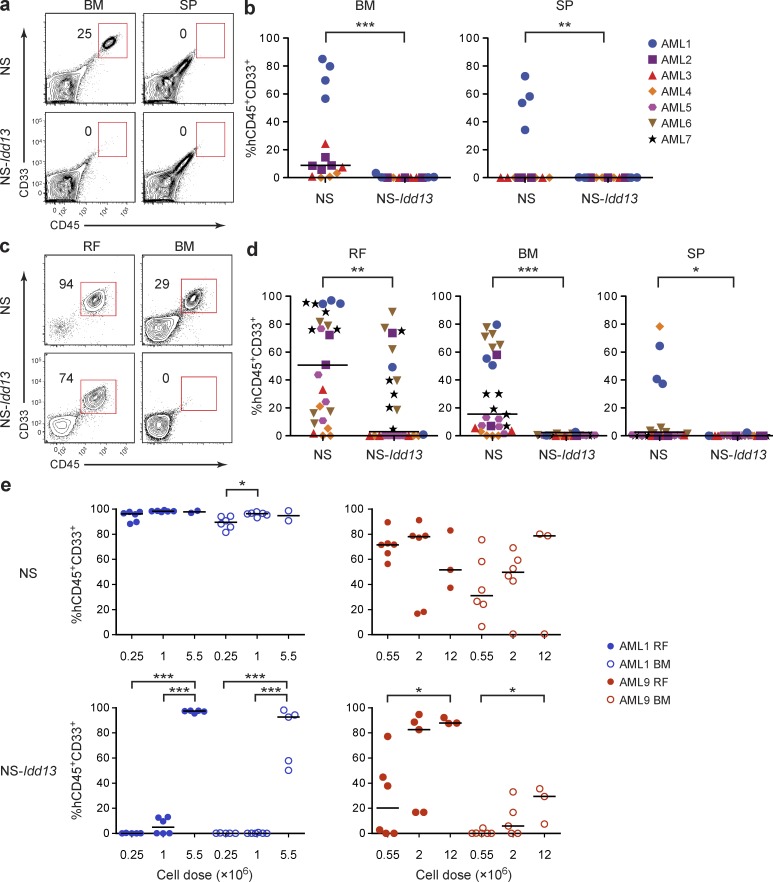
Figure 2.
LSC engraftment in the NOD.SCID xenograft model depends on NOD-derived SIRPα. (a) Flow cytometric analysis of live cells in the BM (two femurs plus two tibias) and SP of NS and NS-Idd13 mice 8 wk after i.v. transplantation of human AML cells. Representative plots are from sample AML3. (b) Summary of human leukemic engraftment (CD45+CD33+) in murine BM and SP 8 wk after i.v. transplantation of AML cells from four patient samples (AML1–4) into NS (n = 14) and NS-Idd13 (n = 11) mice. (c) Flow cytometric analysis of live cells in the right femur (RF) and noninjected BM (left femur plus two tibias) of NS and NS-Idd13 mice 8 wk after transplantation of human AML cells into the RF. Representative plots are from sample AML7. (d) Summary of human leukemic engraftment in the RF, noninjected BM, and SP 8 wk after transplantation of AML cells from seven patient samples (AML1–7) into the RF of NS (n = 23) and NS-Idd13 (n = 21) mice. (e) Human leukemic engraftment in the RF and noninjected BM 8 wk after transplantation of AML cells from samples AML1 and AML9 at three different cell doses into the RF of NS and NS-Idd13 mice, as determined by flow cytometry. (a and c) The percentage of human CD45+CD33+ cells in the live cell gate is indicated. (b, d, and e) Each symbol represents one mouse. Bars indicate median values. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 (b and d, Wald test; e, Tukey’s HSD).
We next transplanted CD34+CD38− cells from two patients i.f. into NS or NS-Idd13 mice. Although LSCs may have variable expression of CD34 and CD38, the CD34+CD38− cell fraction is enriched for LSCs in most AML samples (Lapidot et al., 1994; Bonnet and Dick, 1997; Hope et al., 2004; Eppert et al., 2011). We observed leukemic engraftment at both injected and noninjected BM sites in NS mice but no engraftment in NS-Idd13 mice (Fig. 3, a and b). Finally, we performed serial transplantation experiments in which cells from injected and noninjected BM sites of primary NS and NS-Idd13 mouse recipients were harvested and transplanted i.f. into secondary NS mice (Fig. 3 c). After 10 wk, AML engraftment in noninjected BM and SP was significantly lower in secondary NS mice transplanted with cells from NS-Idd13 primary recipients compared with cells from NS primary recipients (Fig. 3, d and e), indicating that in primary transplanted NS-Idd13 mice, LSCs are reduced in number or have impaired function. Collectively, these results establish that initiation of engraftment by LSCs and subsequent migration of AML cells are regulated by Sirpa polymorphism in the xenotransplantation model and provide the first direct evidence that SIRPα plays a role in LSC function.
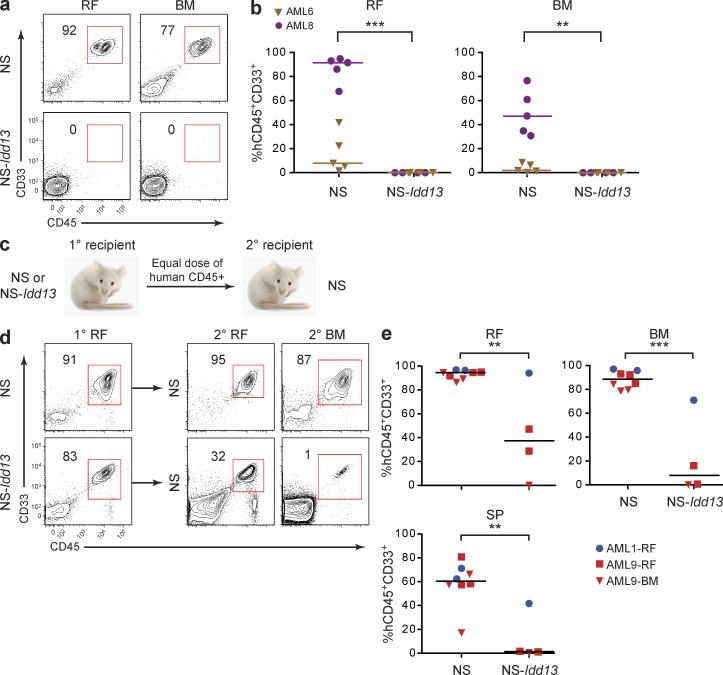
Figure 3.
LSC engraftment and serial transplantation ability depend on NOD-derived SIRPα. (a) Flow cytometric analysis of live cells in the right femur (RF) and noninjected BM of NS and NS-Idd13 mice 8 wk after transplantation of CD34+CD38− cells from sample AML8 into the RF. (b) Summary of human leukemic engraftment in the RF and noninjected BM 8 wk after transplantation of AML cells from two patient samples (AML6 and AML8) into the RF of NS (n = 10) and NS-Idd13 (n = 8) mice. (c) Protocol for serial transplantation of human AML cells harvested from primary NS and NS-Idd13 mice into secondary NS mice. Equal numbers of human CD45+ cells harvested from the injected RF or noninjected BM of primary engrafted NS or NS-Idd13 were injected into the RF of secondary NS mice. (d) Flow cytometric analysis of live cells harvested from the injected RF of primary NS and NS-Idd13 mice and the injected RF and noninjected BM of secondary NS recipient mice. Representative plots are from sample AML9-RF. (e) Summary of human leukemic engraftment in the injected RF, noninjected BM, and SP of secondary NS mice 10 wk after transplantation of AML cells from two patient samples harvested from primary NS (n = 8) and NS-Idd13 (n = 4) mice. (a and d) The percentage of human CD45+CD33+ cells in the live cell gate is indicated. (b and e) Each symbol represents one mouse. Bars indicate median values. **, P < 0.01; and ***, P < 0.001 (Wald test).
Macrophages limit AML engraftment in NS-Idd13 mice
To gain insight into the cell types contributing to restriction of AML engraftment in NS-Idd13 mice, we investigated macrophages and NK cells, both of which are critical effectors of the innate immune system. NK cells can inhibit engraftment of AML in NS mice (Jin et al., 2009). To study the role of NK cells in modulating leukemic engraftment via SIRPα–CD47 interaction, we pretreated NS and NS-Idd13 mice with anti-CD122 (IL-2 receptor β-chain) Abs before AML cell transplantation. Anti-CD122 Ab treatment targets NK cells and a small proportion of macrophages, resulting in marked reduction of F4/80−CD49b+ NK cells in the BM and SP, but no reduction of F4/80+CD49b− macrophages (Table 2). We transplanted primary cells from 10 AML patients i.f. into NS or NS-Idd13 mice pretreated with anti-CD122 Ab (Fig. 4 a). All AML samples engrafted both injected and noninjected BM sites of NS mice. 8 of 10 samples generated a graft in the injected femur of NS-Idd13 mice; however, migration and repopulation of noninjected sites were observed in only 3 of 45 NS-Idd13 mice (transplanted with 2 of 10 AML samples). Three samples were able to generate a graft in the injected femur of NS-Idd13 mice that was comparable with the graft seen in NS mice, but were not able to engraft noninjected BM or SP. Overall, these results are similar to what we observed in NS-Idd13 mice that were not pretreated with CD122 Ab (Fig. 2 d) and indicate that for most samples, NK cell depletion in vivo does not overcome the barrier to AML engraftment and migration imposed by a SIRPα variant that does not bind human CD47.
Table 2.
Depletion of NK cells and macrophages by anti-CD122 Ab or CLO treatment
| Treatment | NK | MAC | ||
| BM | SP | BM | SP | |
| % | % | % | % | |
| NS mice | ||||
| PBS | 2.7 ± 0.1 (3) | 19.7 ± 1.6 (3) | 11.0 ± 2.9 (3) | 46.3 ± 1.0 (3) |
| CD122 | 0.9 ± 0.1 (3)c | 10.3 ± 1.0 (3)c | 11.0 ± 1.4 (3) | 54.5 ± 1.4 (3)c |
| CLO | 3.0 ± 0.2 (5) | 34.0 ± 2.1 (5)c | 1.7 ± 0.2 (4)b | 2.1 ± 0.2 (4)c |
| NS-Idd13 mice | ||||
| PBS | 2.7 ± 0.7 (3) | 22.9 ± 1.3 (3) | 4.8 ± 1.2 (3) | 37.3 ± 1.5 (3) |
| CD122 | 0.8 ± 0.2 (3)a | 7.8 ± 1.1 (3)c | 6.6 ± 1.5 (3) | 48.1 ± 1.4 (3)c |
| CLO | 3.0 ± 0.4 (5) | 25.7 ± 3.2 (5) | 1.6 ± 0.2 (5)a | 2.6 ± 0.3 (5)c |
Percentage of NK cells (F4/80−CD49b+) and macrophages (MAC; F4/80+CD49b−) harvested from the BM and SP of NS and NS-Idd13 mice after treatment with control liposomes containing PBS, anti-CD122 Ab (CD122), or CLO liposomes. Data are expressed as mean ± SEM (n mice). CD122 and CLO treatment resulted in depletion of NK cells and macrophages, respectively, with concomitant enrichment of the opposite cell population in some cases.
*, P < 0.05 versus PBS (ANOVA).
**, P < 0.01 versus PBS (ANOVA).
***, P < 0.001 versus PBS (ANOVA).
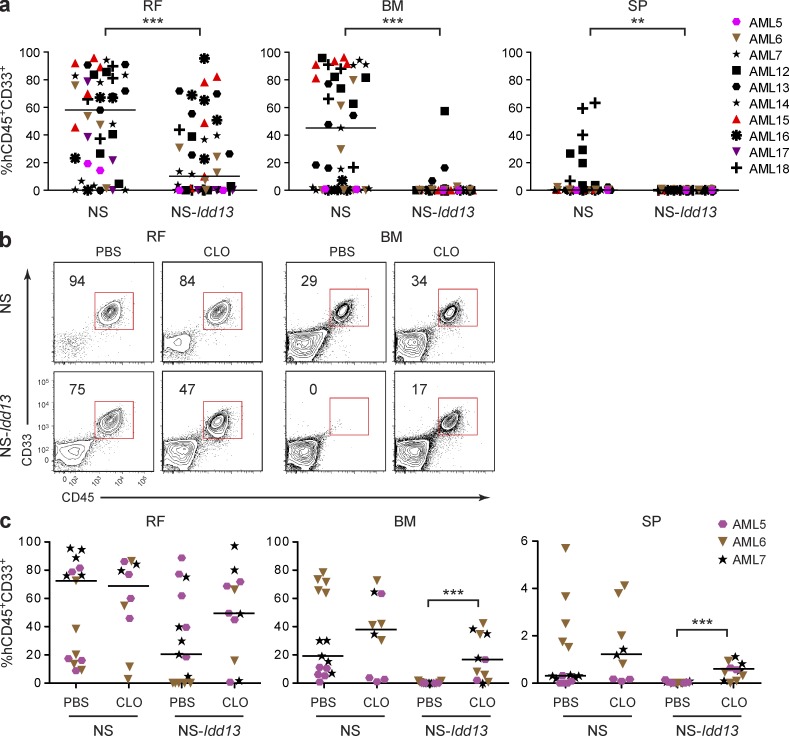
Figure 4.
Macrophages but not NK cells limit AML engraftment in NS-Idd13 mice. (a) Summary of human leukemic engraftment (CD45+CD33+) in the right femur (RF), noninjected BM (left femur plus two tibias), and SP 8 wk after transplantation of 10 human AML samples into the RF of NS (n = 43) and NS-Idd13 (n = 45) mice treated with anti-CD122 Ab. (b) Flow cytometric analysis of live cells in the RF and noninjected BM of NS and NS-Idd13 mice 8 wk after transplantation of human AML cells into the RF. Mice were treated with PBS liposomes or CLO liposomes 48 h before transplantation and then once per week until sacrificed. The percentage of human CD45+CD33+ cells in the live gate is indicated. Representative plots are from sample AML7. (c) Summary of human leukemic engraftment in RF, noninjected BM, and SP 8 wk after transplantation of AML cells from three patient samples (AML5–7) into the RF of NS (n = 25) and NS-Idd13 (n = 26) mice. (a and c) Each symbol represents one mouse. Bars indicate median values. **, P < 0.01; and ***, P < 0.001 (Wald test).
We previously identified macrophages as the effector cells that modulate normal human HSC engraftment via SIRPα–CD47 interactions (Takenaka et al., 2007). Macrophages, but not NK cells, express high levels of SIRPα on their surface (not depicted). To investigate the specific contribution of macrophages in limiting AML in NS-Idd13 mice, we pretreated NS and NS-Idd13 mice with clodronate (CLO)-containing liposomes. CLO is a bisphosphonate that induces apoptosis of phagocytic cells when delivered via liposomes (van Rooijen et al., 1996). CLO treatment selectively depleted F4/80+CD49b− macrophages without depleting NK cells in mouse BM and SP (Table 2). We performed xenotransplantation experiments using three AML samples selected for their ability to generate a disseminated leukemic graft in NK cell–depleted NS but not NS-Idd13 mice. AML cells were transplanted i.f. into mice of both strains treated with either PBS or CLO before transplantation and then weekly for 8 wk. Macrophage depletion in CLO-treated mice was confirmed by flow cytometry (percent reduction in CD45+F4/80+ cells compared with PBS-treated mice: 65 ± 6%, 73 ± 12%, and 74 ± 6% in injected femur, noninjected BM, and SP, respectively, of NS mice; and 82 ± 2%, 90 ± 3%, and 77 ± 10% in injected femur, noninjected BM, and SP, respectively, of NS-Idd13 mice). In NS mice, CLO treatment did not significantly alter engraftment at local or noninjected BM sites compared with controls (Fig. 4, b and c). In contrast, CLO-mediated macrophage depletion in NS-Idd13 mice enabled significantly higher levels of leukemic engraftment by all AML samples not only in the injected femur but also in noninjected BM and SP. These findings demonstrate that the SIRPα-dependent genetic restriction of AML engraftment in NS-Idd13 mice is alleviated by macrophage depletion, establishing a role for macrophages in governing the function of LSCs in xenograft models.
Enhanced phagocytosis of AML cells by NS-Idd13 macrophages
SIRPα signaling mediates inhibition of several cellular processes including macrophage phagocytosis, neutrophil migration, and TNF secretion (Neznanov et al., 2003). CLO depletion experiments (Fig. 4, b and c) indicated that macrophages are critical effector cells restricting AML engraftment in NS-Idd13 mice. We therefore investigated whether phagocytosis was a mechanism of elimination of AML cells. We adapted established protocols (Leidi et al., 2009) to develop an in vitro phagocytosis assay using BM-derived macrophages from NS and NS-Idd13 mice prestimulated with LPS and IFN-γ. Prestimulation did not alter macrophage cell surface SIRPα expression (not depicted). Macrophages were incubated with CFSE-labeled primary human AML cells, and their engulfment was assessed by both flow cytometry and spinning disc confocal microscopy (Fig. 5). Macrophages from NS-Idd13 mice showed consistently higher phagocytic activity against all AML samples tested compared with NS macrophages (Fig. 5 c). These results indicate that effective interaction between NOD-derived SIRPα and human CD47 reduces phagocytosis of AML cells by activated macrophages.
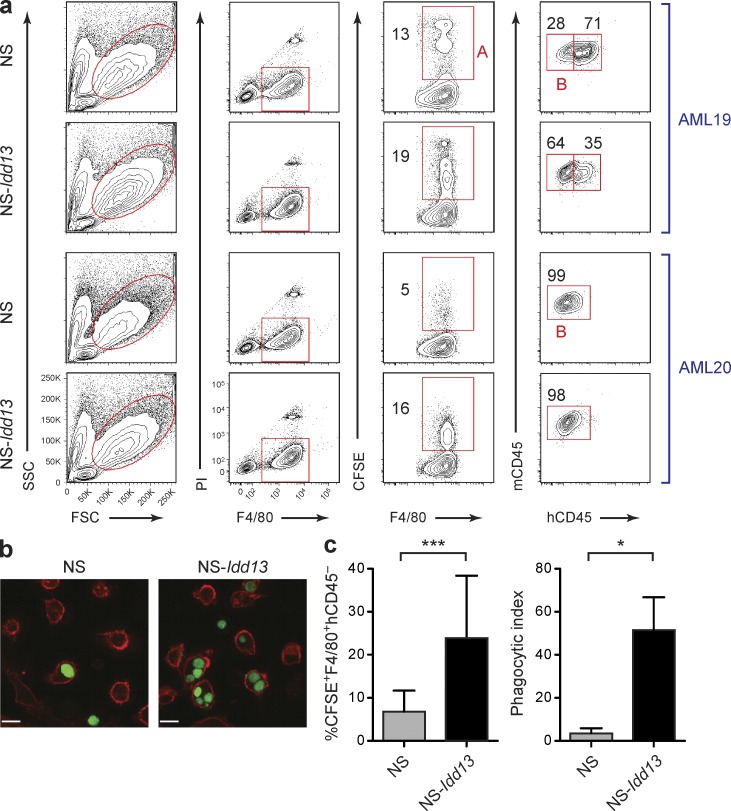
Figure 5.
Enhanced phagocytosis of AML cells by NS-Idd13 macrophages. (a) Quantification of phagocytosis of human AML cells by LPS/IFN-γ–treated NS and NS-Idd13 macrophages using flow cytometry. Phagocytosis of CFSE-labeled human primary AML targets was defined as the percentage of mouse macrophages (F4/80+) positive for CFSE (gate A) and negative for human CD45 (gate B). The percentage of mouse macrophages in each gate is indicated. Representative plots are from samples AML19 and AML20. (b) Visualization of engulfment of CFSE-labeled human AML cells by NS and NS-Idd13 LPS/IFN-γ–stimulated macrophages by spinning disk confocal microscopy. Bars, 25 µm. (c) Summary of human AML cell phagocytosis by NS and NS-Idd13 macrophages assessed by flow cytometry (left; n = 6 AML samples) and by spinning disk confocal microscopy (right; n = 3 AML samples). The phagocytic index was determined as the number of engulfed AML cells per 100 macrophages. Bars indicate mean ± SD. *, P < 0.05; and ***, P < 0.001 (Wald test).
hSIRPα-Fc induces phagocytosis of human AML by activated NS macrophages
To provide an independent line of evidence that deficiency in interaction between SIRPα on macrophages and CD47 on AML cells restricts LSC function through macrophage-mediated elimination, we developed a blocking agent suitable for use in vitro and in vivo. We generated a fusion protein from the IgV domain of human SIRPα fused to a human IgG4-Fc moiety (hSIRPα-Fc) designed to bind CD47 on AML cells and disrupt interaction with SIRPα on macrophages. The hSIRPα-Fc fusion protein efficiently blocked SIRPα–CD47 binding and did not induce apoptosis of primary AML cells (Fig. 6 a and not depicted). We then tested whether hSIRPα-Fc was functional and could promote phagocytosis of human AML cells by NS macrophages in vitro. The macrophages were either untreated or preactivated with LPS and IFN-γ before addition of CFSE-labeled primary AML targets. We observed a significant increase in phagocytic activity by activated NS macrophages against AML cells with addition of hSIRPα-Fc compared with addition of IgG4 control Fc protein (Fig. 6, b–d). These results confirm our genetic evidence that impairment of SIRPα–CD47 interaction leads to enhanced phagocytosis of AML cells by macrophages. Interestingly, hSIRPα-Fc–specific induction of phagocytosis was significantly lower in untreated compared with preactivated macrophages. The inability of hSIRPα-Fc to enhance phagocytosis by nonstimulated macrophages implies that disruption of SIRPα–CD47 interaction alone is insufficient to activate macrophage phagocytosis of AML cells.
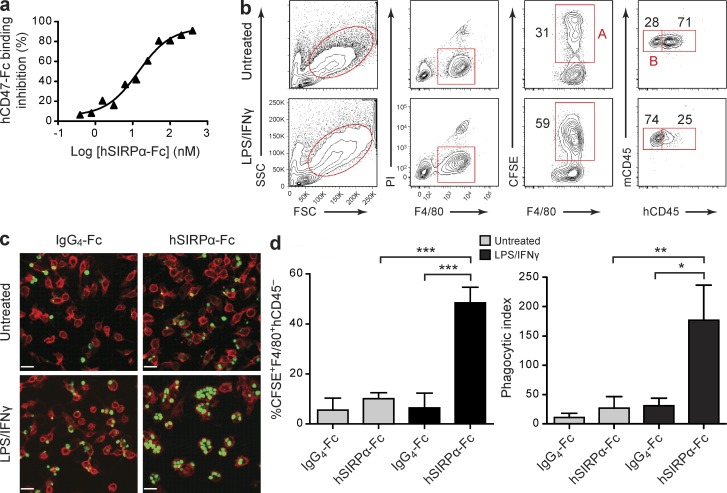
Figure 6.
hSIRPα-Fc fusion protein promotes phagocytosis of human AML cells by activated NS macrophages. (a) hSIRPα-Fc–mediated inhibition of human CD47 binding to SIRPα expressed on NOD macrophages. Data are expressed as percent reduction of hCD47 binding compared with NOD macrophages incubated with hCD47-Fc alone. Representative results from one of two experiments are shown. (b) Flow cytometric analysis of untreated or LPS/IFN-γ–stimulated NS macrophages incubated with CFSE-labeled AML cells from patient AML20 with addition of hSIRPα-Fc. Phagocytosis of AML cells was defined as the percentage of mouse macrophages (F4/80+) positive for CFSE (gate A) and negative for human CD45 (gate B). The percentage of mouse macrophages in each gate is indicated. (c) Visualization of engulfment of CFSE-labeled human AML cells by NS macrophages by spinning disk confocal microscopy. AML cells were coincubated with untreated or LPS/IFN-γ–stimulated macrophages with addition of IgG4-Fc or hSIRPα-Fc. Bars, 25 µm. (d) Summary of human AML cell phagocytosis by untreated or LPS/IFN-γ–stimulated NS macrophages assessed by flow cytometry (left; AML19, AML20, and AML23) and by spinning disk confocal microscopy (right; AML22–26). The phagocytic index was determined as the number of engulfed AML cells per 100 macrophages. Bars indicate mean ± SD. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 (Wald test).
Disruption of SIRPα–CD47 interaction by hSIRPα-Fc impairs AML engraftment
To investigate whether hSIRPα-Fc–mediated blocking of interactions between CD47 on AML cells and SIRPα on macrophages impacts AML engraftment, we treated NS mice bearing established human AML grafts with hSIRPα-Fc or an IgG4-Fc control protein at a dose of 8 mg/kg three times per week for 4 wk (Fig. 7 a). This dosing protocol was determined by pharmacokinetic experiments (Fig. 7 b). When treatment was started at day 10 after transplantation (Fig. 7 a, Approach 1), we observed robust leukemic engraftment in IgG4-Fc control–treated mice at all BM sites but did not detect engraftment in the injected femur, noninjected BM, or SP of hSIRPα-Fc–treated mice (Fig. 7 c). When treatment was delayed until 28 d after AML transplantation (Fig. 7 a, Approach 2), we observed significant reduction of leukemic engraftment in mice treated with hSIRPα-Fc compared with IgG4-Fc control–treated mice in both injected and noninjected BM sites (Fig. 7, d–f). These results demonstrate that disruption of SIRPα–CD47 interactions by hSIRPα-Fc in vivo impairs human AML engraftment and dissemination, providing independent confirmation of the findings in our genetic models linking LSC function with SIRPα signaling in macrophages.
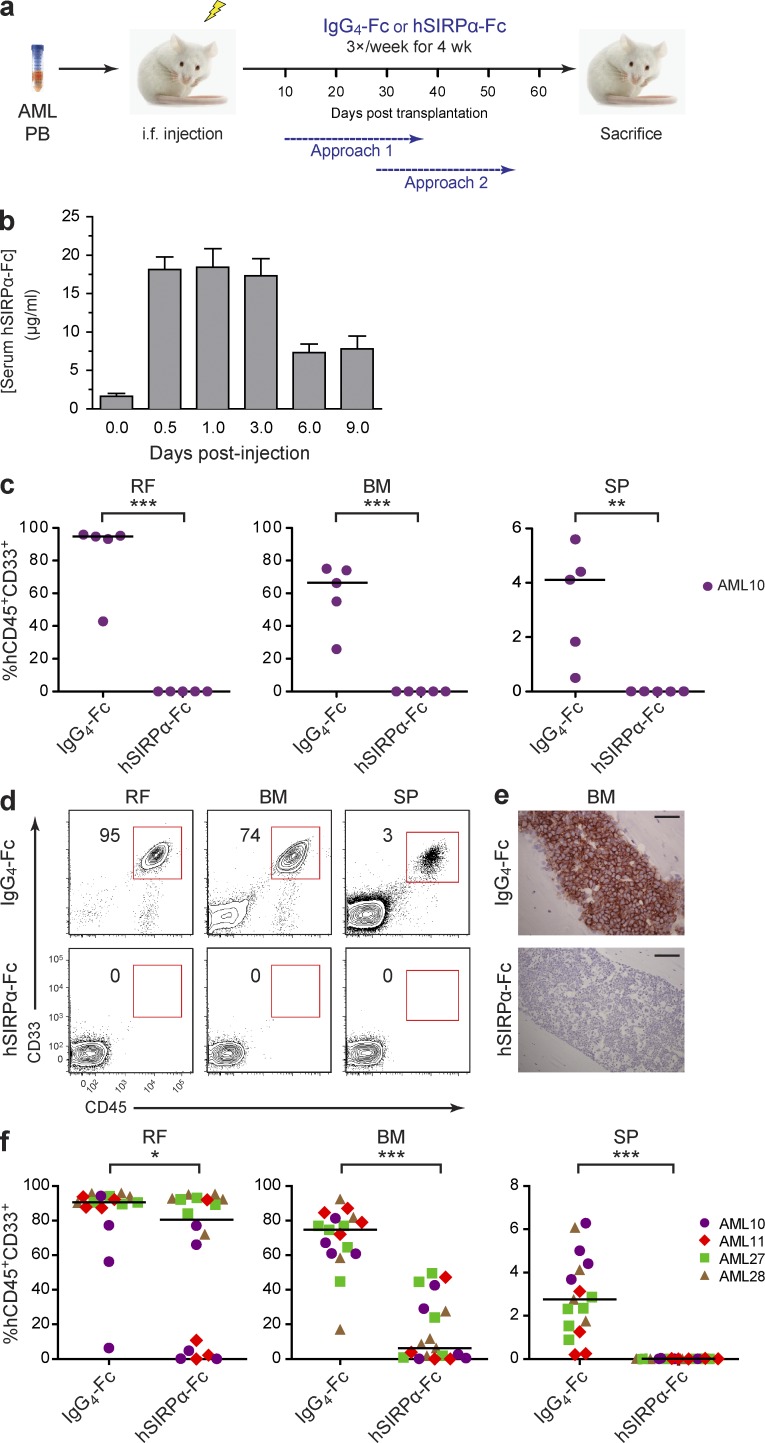
Figure 7.
Disruption of SIRPα–CD47 interaction by hSIRPα-Fc impairs AML engraftment. (a) Overview of the experimental protocol. Mice were sacrificed 1–3 d after completion of hSIRPα-Fc treatment. (b) Pharmacokinetics of hSIRPα-Fc in NOD mice. Six groups of NOD mice (n = 4 mice/group) were injected intraperitoneally with a single dose of 50 µg hSIRPα-Fc. The mice were sacrificed at the indicated time points after injection, and serum hSIRPα-Fc protein concentration was determined by ELISA. Half-life for hSIRPα-Fc was calculated as 120 h. Bars indicate mean values ± SD. (c) Summary of human leukemic engraftment in the injected right femur (RF), noninjected BM (left femur plus two tibias), and SP of NS mice treated with IgG4-Fc control or hSIRPα-Fc for 4 wk starting 10 d after transplantation of cells from sample AML10 (Approach 1; n = 5 mice per treatment group). (d) Flow cytometric analysis of live cells in the injected RF, noninjected BM, and SP of NS mice treated with IgG4-Fc control or hSIRPα-Fc for 4 wk starting 28 d after transplantation of cells from patient AML11 (Approach 2). The percentage of human CD45+CD33+ cells in the live gate is indicated. (e) Murine BM sections stained for human CD45 from IgG4-Fc– and hSIRPα-Fc–treated mice. Bars, 50 µm. (f) Summary of human leukemic engraftment in the injected RF, noninjected BM, and SP of mice treated with IgG4-Fc control or hSIRPα-Fc for 4 wk starting 28 d after transplantation of cells from four AML samples (AML10, AML11, AML27, and AML28; Approach 2; n = 4 or 5 mice per treatment group). (c and f) Each symbol represents one mouse. Bars indicate median values. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 (Wald test).
Disruption of SIRPα–CD47 interaction enhances phagocytosis of AML but not normal hematopoietic targets by activated human macrophages
Currently, no assay system allows the in vivo study of SIRPα–CD47 interactions between LSCs and human macrophages; therefore we examined them in vitro. SIRPα is strongly expressed on human macrophages and binds with low nanomolar affinity to dimeric human CD47-Fc fusion protein (Fig. 8, a and b). CD47 expression was higher on CD34+CD38− AML cells, the fraction which includes the LSCs in the majority of functionally tested patient samples (Eppert et al., 2011), compared with HSC-enriched CD34+CD38− cells from normal adult human BM (Fig. 8 c). There was no statistically significant difference in CD47 expression between CD34+CD38− cells from umbilical cord blood (CB) and AML. Bulk and CD34+CD38− AML cells had similar CD47 expression levels (not depicted). To determine whether AML cells depend on SIRPα–CD47 interactions to evade macrophage-mediated elimination, we tested whether addition of hSIRPα-Fc fusion protein promoted phagocytosis in vitro. Similar to what we observed with mouse macrophages, unstimulated human macrophages did not exhibit phagocytic activity against either AML or normal hematopoietic targets with addition of hSIRPα-Fc (not depicted). In contrast, human macrophages prestimulated with LPS and IFN-γ exhibited increased phagocytic activity against AML targets with the addition of hSIRPα-Fc fusion protein compared with control IgG4-Fc (P < 0.001; Fig. 8 d). Importantly, phagocytosis of AML targets was significantly greater than that observed against normal hematopoietic targets after hSIRPα-Fc treatment. Indeed, hSIRPα-Fc did not significantly enhance phagocytosis of normal hematopoietic cells by stimulated macrophages compared with control IgG4-Fc (Fig. 8 d). To confirm the observed preferential targeting of AML cells after hSIRPα-Fc treatment, AML and normal CB targets were incubated together with prestimulated human macrophages. Macrophages preferentially phagocytosed AML cells, even when they were outnumbered two to one by CB cells (Fig. 8 e). The enhanced phagocytosis of AML compared with normal targets cannot be explained solely by differences in CD47 expression, as there was no significant difference between CB and AML and the difference between AML and BM was less than twofold (Fig. 8 c). Collectively, these data indicate that AML cells rely on SIRPα–CD47 interactions to avoid macrophage-mediated killing and predict that LSCs would be more susceptible than normal HSCs to therapies that disrupt SIRPα–CD47 interactions.
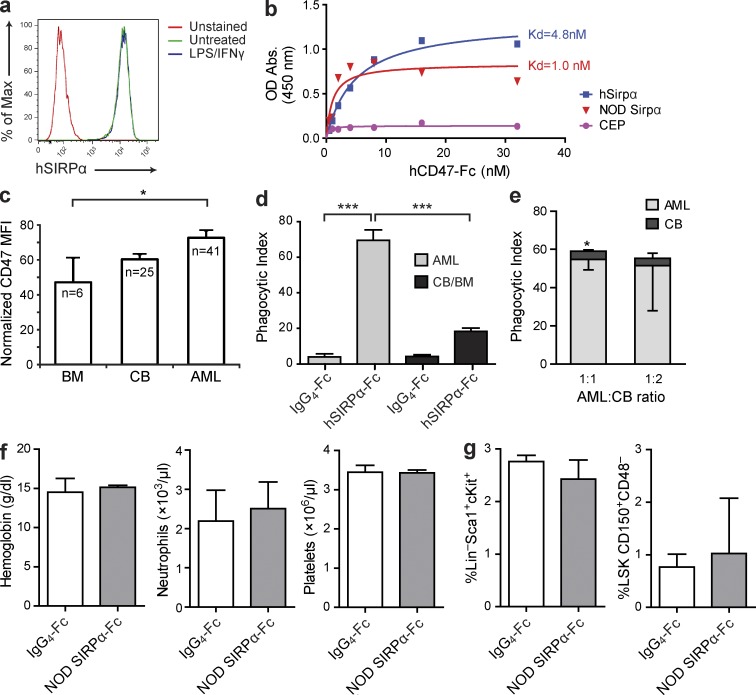
Figure 8.
Disruption of SIRPα–CD47 interaction by hSIRPα-Fc enhances phagocytosis of AML but not normal hematopoietic targets. (a) SIRPα expression on human macrophages, untreated or stimulated with IFN-γ for 24 h and LPS for 1 h before flow cytometric analysis using anti–human SIRPα Ab. (b) Analysis of binding of NOR macrophages infected with lentiviral vectors expressing human SIRPα or NOD mouse SIRPα or empty vector (CEP), as indicated, to human CD47-Fc. (c) Flow cytometric analysis of CD47 expression on CD34+CD38− cells from normal human adult BM, CB, and primary AML patient samples. The number of samples analyzed is indicated. CD47 mean fluorescence intensity (MFI) for each sample was normalized for cell size and CD47 expression level on lymphocytes from the same sample. Error bars indicate SE. *, P < 0.05 (ANOVA). (d) Summary of phagocytosis of human AML cells (AML9, AML19, AML22, AML23, and AML26) or normal mononuclear cells from human CB (n = 2) or BM (n = 4) by LPS/IFN-γ–stimulated human CB-derived macrophages with addition of IgG4-Fc or hSIRPα-Fc as indicated. ***, P < 0.001 (ANOVA). (e) Phagocytosis of human AML (AML29) and CB mononuclear cells coincubated with LPS/IFN-γ–stimulated human CB-derived macrophages with addition of hSIRPα-Fc. *, P < 0.05 (Student’s t test). (d and e) The phagocytic index was determined by spinning disk confocal microscopy as the number of engulfed cells per 100 macrophages, with two to five replicates/sample. Bars indicate mean ± SD. (f) Peripheral blood hemoglobin concentration and neutrophil and platelet counts of NOD mice treated with NOD SIRPα-Fc (n = 4) or IgG4-Fc control (n = 3) for 4 wk. (g) Percentage of Sca1+cKit+ cells in the Lin− population (LSK) and CD150+CD48− cells in the LSK fraction in the BM of NOD mice after 4 wk of treatment with NOD SIRPα-Fc or IgG4-Fc control (n = 3 mice/group). (f and g) Bars indicate mean ± SD. No statistically significant differences were observed (Student’s t test).
Disruption of SIRPα–CD47 interactions in vivo does not adversely affect normal hematopoiesis
To study whether healthy hematopoietic cells are targeted for elimination after disruption of SIRPα–CD47 interactions in vivo, we treated immunocompetent NOD mice with a fusion protein composed of the NOD mouse SIRPα IgV domain fused to a human IgG4 Fc moiety (mSIRPα-Fc) at a dose of 8 mg/kg three times per week for 4 wk. After 2 and 4 wk of treatment, peripheral blood neutrophil and platelet counts as well as hemoglobin concentration were not different in mSIRPα-Fc– compared with control Fc–treated mice (Fig. 8 f and not depicted). Furthermore, there was no difference in the percentage of Lin−Sca1+cKit+ or LSK CD150+CD48− HSC populations in the BM after 4 wk of treatment (Fig. 8 g). These findings are consistent with our in vitro human macrophage data showing preferential targeting of AML cells over normal hematopoietic cells and indicate that under homeostatic conditions in nontransplanted hosts, healthy hematopoietic cells are not appreciably targeted for elimination upon disruption of SIRPα–CD47 interactions.
DISCUSSION
Our findings provide direct evidence that SIRPα is the key CD47 binding partner modulating LSC survival in xenotransplant models and demonstrate that the interaction between SIRPα on macrophages and CD47 on AML cells is critical for engraftment of LSCs and the migration of AML cells. Impaired SIRPα–CD47 interaction, as a result of either Sirpa polymorphism in the immunodeficient mouse recipients or disruption with a hSIRPα-Fc blocking agent, increased phagocytosis of AML cells in vitro and reduced AML engraftment in vivo. Importantly, these data establish that macrophage immune surveillance of human AML in the xenotransplant setting eliminates not only the bulk blast cell population but also the disease-sustaining LSCs. The relevance of our findings to human disease is strongly supported by the enhanced phagocytosis of AML targets by human macrophages in vitro after disruption of SIRPα–CD47 interactions by hSIRPα-Fc. Interestingly, hSIRPα-Fc promoted phagocytosis of AML cells only when macrophages had previously received stimulation through the toll-like receptor 4 LPS and IFN-γ receptors, indicating that disruption of SIRPα–CD47 interactions alone does not activate macrophages, which must receive independent activating signals to trigger phagocytosis of target cells. This observation is concordant with previous reports that in the absence of SIRPα-dependent inhibition, phagocytosis by macrophages requires additional signals through Fcγ or complement receptors (Oldenborg et al., 2000, 2001; Okazawa et al., 2005). The fact that hSIRPα-Fc was effective in reducing leukemic burden in mice engrafted with AML suggests that in the xenotransplantation setting, macrophages have been activated and are competent to eliminate leukemic cells but are inhibited because of engagement of SIRPα with CD47 expressed on AML targets. If macrophages play a similar role in AML immune surveillance in patients, these findings provide a strong rationale for the development of agents that disrupt SIRPα–CD47 interactions, such as the hSIRPα-Fc fusion protein used here, to target LSCs for macrophage-mediated elimination. Given the significant polymorphism in human SIRPA (Takenaka et al., 2007), it will be interesting to examine in future studies whether this genetic variability results in functional diversity in SIRPα–CD47 interactions that could affect disease outcome or response to SIRPα-targeted therapies in AML.
Evidence of genetic and functional diversity within the leukemia-initiating cell compartment in human acute lymphoblastic leukemia (Anderson et al., 2011; Notta et al., 2011) and in AML (Hope et al., 2004) predict that antileukemia therapies targeting intrinsic LSC survival pathways may fail if not all subclones depend on the same drug-targeted pathway. One approach to circumvent this mechanism of therapeutic resistance is to target systemic microenvironmental interactions that LSCs require for survival. In this study, we observed dependence of LSC function on SIRPα–CD47 interactions in all AML samples tested, which represented multiple subtypes with diverse karyotypic and molecular abnormalities. Thus, SIRPα–CD47 interaction may represent a broadly relevant therapeutic target in AML. The dependence of LSCs on SIRPα–CD47 interaction appears to be mitigated when the local leukemic burden is high, as indicated by the ability of some primary AML samples to engraft the injected BM of NS-Idd13 mice, particularly at high transplanted cell doses. However, despite generation in some cases of a local graft in NS-Idd13 mice or in NS mice treated with hSIRPα-Fc, migration and engraftment at distant sites were significantly impaired, suggesting that LSCs were targeted for elimination when they exited the BM microenvironment. The impact of leukemic burden on the requirement for SIRPα–CD47 interaction is consistent with our prior observation that macrophage-mediated suppression of normal human hematopoietic progenitors depends on macrophage/hematopoietic cell ratios (Takenaka et al., 2007) and suggests that the ability of macrophages to eliminate AML cells can be saturated. Thus, treatments to disrupt SIRPα–CD47 interactions will be most effective at eliminating LSCs if administered to patients after debulking of disease by standard induction chemotherapy.
Ideally, antileukemia therapies should also have a sufficient therapeutic window to allow elimination of LSCs while preserving normal HSCs. The lower levels of CD47 expression on HSCs from normal adult BM compared with LSCs in AML suggest that HSCs may be less affected by therapies that disrupt SIRPα–CD47 interactions. However, CD47 expression level is likely not the only, or even most important, factor modulating the consequences of SIRPα–CD47 disruption, as we observed significantly more phagocytosis of AML targets by human macrophages compared with normal CB cells after treatment with hSIRPα-Fc, despite similar CD47 expression levels. This observation suggests that AML cells provide stronger and/or distinct prophagocytic signals to macrophages than do healthy hematopoietic cells, rendering them more reliant on SIRPα–CD47 interactions to evade elimination by this innate immune surveillance mechanism. Indeed, addition of hSIRPα-Fc did not significantly promote phagocytosis of normal hematopoietic targets in vitro compared with control IgG4-Fc. Collectively, these findings support the existence of a therapeutic window for agents that disrupt SIRPα–CD47 interactions in vivo.
Our findings provide direct evidence that macrophages are the effector cells restricting LSC engraftment in xenotransplant recipients when SIRPα–CD47 interactions are attenuated or absent, demonstrating a role for these innate immune cells in leukemia immune surveillance. Recent studies have linked anti-CD47 Ab therapy with reduced survival of xenotransplanted leukemia and lymphoma cells through increased phagocytosis of cancerous cells by macrophages (Jaiswal et al., 2009; Majeti et al., 2009; Chao et al., 2010, 2011). However, this Ab promotes phagocytosis of human leukemic cells by BALB/c mouse macrophages which, like other inbred strains except NOD (129/Sv, C57BL/6, NOR/Lt), express a SIRPα variant that does not bind to human CD47 (Subramanian et al., 2006, 2007; Takenaka et al., 2007; Tsai et al., 2010). Thus, additional SIRPα-independent mechanisms, such as induction of Ab-dependent cell-mediated cytotoxicity (Zhao et al., 2011), may contribute to the observed anti-CD47 Ab therapeutic effect. A role for macrophages in leukemia immune surveillance accords with recent evidence that macrophages participate in immune surveillance of pancreatic cancer (Beatty et al., 2011). After treatment with an agonist Ab directed against CD40, a TNF receptor superfamily member, tumor-infiltrating macrophages up-regulated activation markers, secreted elevated levels of cytokines such as IL-12, TNF, and IFN-γ, and mediated tumor regression. It will be of interest to determine whether enhanced activation of macrophages through agonists such as anti-CD40 Ab can augment the antileukemic effects of hSIRPα-Fc treatment in vivo.
Macrophages play complex roles in tumor biology. In contrast to the function of classically activated macrophages in tumor immune surveillance, tumor-associated macrophages have been implicated in the progression of both solid tumors and hematologic malignancies through promotion of tumor vascularization, invasion, metastasis, and resistance to chemotherapy (Ruhrberg and De Palma, 2010; Steidl et al., 2010; DeNardo et al., 2011). Recent evidence that macrophages promote retention of normal HSCs within their BM niche (Winkler et al., 2010; Chow et al., 2011) raises the intriguing possibility that a subset of macrophages may also have supportive interactions with LSCs. Despite their aberrant developmental programming, LSCs in AML do require interaction with a microenvironmental niche (Jin et al., 2006). This interaction can be targeted therapeutically, for example with anti-CD44 Ab (Jin et al., 2006), but little is known about the cellular components of the LSC niche. Mesenchymal progenitors, which have been identified as components of the perivascular HSC niche (Sacchetti et al., 2007), also express SIRPα (Vogel et al., 2003), although the effects of SIRPα signaling in these cells are unknown. A better understanding of the complex interactions between normal HSCs and LSCs with macrophages and other components of the normal and leukemic stem cell niche will aid in the development of novel antileukemia agents that can target or modulate these interactions for therapeutic benefit.
MATERIALS AND METHODS
Collection of normal hematopoietic and AML patient samples.
CB samples were obtained according to procedures approved by the Research Ethics Board (REB) of the University Health Network (UHN) and the Research Review Team of Trillium Health Centre. Normal adult BM mononuclear cell preparations were purchased from Lonza or obtained from healthy volunteers with informed consent according to procedures approved by the UHN REB. Peripheral blood cells were collected from patients with newly diagnosed AML after obtaining informed consent according to procedures approved by the UHN REB. Low-density mononuclear cells were frozen viably in FCS plus 10% DMSO.
Transplantation of human leukemia cells into NS and NS-Idd13 mice.
NS and NS-Idd13 mice were bred and housed at the UHN/Princess Margaret Hospital (PMH). Animal experiments were performed in accordance with institutional guidelines approved by the UHN/PMH Animal Care Committee. The mouse repopulation assay was performed as previously described (Bonnet and Dick, 1997; Hope et al., 2004; Jin et al., 2006, 2009). In brief, 8–12-wk-old NS or NS-Idd13 mice were sublethally irradiated with 275 cGy from a 137Cs γ-irradiator 24 h before i.f. or i.v. transplantation of human AML cells. NS and NS-Idd13 mice in the same experiment received equal numbers of AML cells (range 1.5–15 × 106 cells depending on sample). Mice were sacrificed 8 wk after transplantation, and human engraftment was evaluated by flow cytometry using human-specific Abs. For dose range, CD34+CD38−, and secondary transplantation experiments, mice were treated with anti-CD122 Ab. AML samples used for in vivo experiments were selected based on availability of adequate cell numbers and ability to engraft NS mice as determined by prescreening.
Cell staining, sorting, flow cytometry, immunohistochemistry, and peripheral blood analysis.
Human AML engraftment in transplanted mice was assessed with the following human-specific mouse Abs: anti–CD45-APC, anti–CD33-PE, anti–CD34-APC-Cy7, anti–CD47-FITC, anti–CD38-PE-Cy7 (BD), and anti–CD19-PE-Cy5 (Beckman Coulter). CD47 expression on human hematopoietic cells was assessed using anti–CD47-FITC (BD). SIRPα expression on human macrophages was assessed using anti–human CD172a (SIRPα)–PE-Cy7 Abs (BioLegend). Analysis of mouse tissues was performed using the following anti–mouse rat or hamster Abs: anti–CD172a (SIRPα)-PE, anti–CD49b-FITC, anti–CD45-APC-Cy7 (BD) and anti–NK1.1-APC, and anti–F4/80-PE-Cy7 (eBioscience). To obtain CD34+38− leukemic cells, AML blasts were stained with anti–CD34-APC and anti–CD38-PE (BD) and then sorted using a Mo-Flo (BD). For immunohistochemistry, one mouse tibia was fixed in formalin, and tissue sections were performed. Sections were stained with hematoxylin and eosin and hCD45 (Dako). For analysis of mouse peripheral blood counts and hemoglobin concentration, 50 µl of whole blood was collected from the jugular vein and analyzed using a HEMAVET 950 analyzer (Drew Scientific). For flow cytometric analysis of mouse BM, we used the following mouse-specific Abs: Lineage (anti-CD3, CD4, CD8, CD19, CD127, B220, TER-119, Gr-1)-PE-Cy5, anti–c-Kit–APC, anti–Sca-1–PE-Cy7, anti–CD150-PE, and anti–CD48-FITC (all BioLegend except anti-CD127 from eBioscience).
SIRPα-Fc fusion protein generation and application.
hSIRPα-Fc fusion protein was generated by cloning cDNA of the IgV of human SIRPα into pINFUSE-hIgG4-Fc1 (InvivoGen). 293F cells were transiently transfected with the cloned plasmid and hSIRPα-Fc fusion protein was harvested from the supernatant and purified by protein G column (Thermo Fisher Scientific). NOD mouse SIRPα-Fc was generated by cloning cDNA of the IgV of NOD mouse SIRPα into pINFUSE-hIgG4-Fc1, and protein was produced as described for hSIRPα-Fc. For in vitro phagocytosis experiments, hSIRPα-Fc fusion protein or IgG4-Fc was added at a concentration of 50 µg/ml. For in vivo AML experiments, hSIRPα-Fc fusion protein or IgG4-Fc was injected intraperitoneally at a dose of 200 µg per injection three times per week for 4 wk starting 10 or 28 d after transplantation. For NOD mouse experiments without human cell transplants, NOD SIRPα-Fc or IgG4-Fc was injected intraperitoneally at a dose of 8 mg/kg per injection three times per week for 4 wk.
In vitro phagocytosis assay.
NS and NS-Idd13 mouse macrophages were generated by culturing BM progenitors in DME (Invitrogen) containing 10% FCS supplemented with murine M-CSF (R&D Systems) for 7 d. Macrophages were harvested by incubating in Accumax (Innovative Cell Technologies) at 37°C for 20 min, detached by gentle scraping, and replated into 24-well plates at a concentration of 2.5 × 105/well. For macrophage activation, macrophages were incubated with 100 ng/ml murine IFN-γ (R&D Systems) for 24 h and/or 0.3 µg/ml LPS (Sigma-Aldrich) for 1 h and then washed three times with PBS. Human AML cells were labeled with CFSE (2.5 µM for flow cytometry or 10 µM for microscopy; Invitrogen) in PBS containing 0.1% FCS at room temperature for 8 min. An equal volume of FCS (prewarmed to 37°C) was then added, and the AML cells were incubated at 37°C for 10 min, washed three times with PBS containing 2% FCS, and then added to the wells at a target to effector cell ratio of 5:1. Macrophages and AML cells were coincubated for 2 h in serum-free DME without cytokines, washed three times with PBS, and then harvested for analysis. For flow cytometric analysis, phagocytosis of AML cells was defined as the percentage of F4/80+CFSE+hCD45− cells. For visualization of phagocytic activity, macrophages were washed and stained with Cholera toxin subunit B conjugated to Alexa Fluor 555 (Invitrogen). Images were acquired using a 20×/0.75 objective on a Quorum spinning disk confocal, based on an IX81 microscope (Olympus), equipped with a CSU-X1 scan head (Yokogawa), laser module (405/491/561/640 nm; Spectral Applied Research), and C9100-13 electron-magnifying charge-coupled device (Hamamatsu Photonics). Images were acquired and processed using Volocity 6.0.1 software (PerkinElmer). Engulfed AML cells were counted, and the phagocytic index was determined as the number of engulfed cells per 100 macrophages. 200 macrophages were counted for each condition. As a negative control for active engulfment of AML cells, 15 µM Cytochalasin D (Invitrogen) was used to inhibit the phagocytic function of murine macrophages. In some experiments, fusion proteins were added at a concentration of 50 µg/ml at the same time as the AML cells.
For experiments using human macrophages, macrophages were obtained from CB by culturing adherent mononuclear cells in RPMI (Gibco) supplemented with 10 ng/ml human M-CSF (R&D Systems) for 7–10 d before use in phagocytosis assays as described above, except that 100 ng/ml human rather than mouse IFN-γ (PeproTech) was used for activation, cells were cultured in RPMI instead of DME, and in some experiments, mononuclear cells from normal CB or BM were used as targets at the same macrophage/target cell ratio as for AML. For experiments in which AML and CB targets were coincubated in the same well, CB cells were labeled with 5 µM Cell Tracker Blue CMAC (Invitrogen) in RPMI prewarmed to 37°C, incubated at 37°C for 30 min, washed once with prewarmed RPMI, incubated at 37°C for another 30 min, and then washed once in RPMI before adding to wells.
Macrophage depletion with CLO and NK cell depletion with anti-CD122 Ab.
CLO was a gift of Roche. For macrophage depletion experiments, 200 µl per mouse of CLO or PBS liposome suspension was injected intraperitoneally 48 h before transplantation of AML cells, followed by 100 µl once weekly until sacrifice. The degree of depletion of mouse macrophages was assessed in BM and SP by flow cytometry. Murine NK cell depletion was performed by injection of 200 µg purified anti-CD122 Ab immediately after irradiation. The anti-CD122 monoclonal Ab generated from the hybridoma cell line TM-β1 (gift of T. Tanaka, Osaka University Medical Center, Osaka, Japan; Tanaka et al., 1991) was purified using the High Trap Protein G Column (GE Healthcare).
Pharmacokinetic experiments.
Six groups of four NOD mice were given single intraperitoneal injections of 50 µg hSIRPα-Fc. Each group of mice was sacrificed at different time points (0, 0.5, 1, 3, 6, and 9 d after injection), and blood was collected by cardiac puncture. Serum hSIRPα-Fc protein concentration was measured using the Easy-titer hIgG (γ-chain) assay kit (Thermo Fisher Scientific). In brief, sensitized beads and serum were mixed in a 96-well microplate, and absorbance was measured at 340 nm using an ELISA plate reader. Standard curves were generated, and hSIRPα-Fc protein concentration was determined for each time point. Each sample was measured in three technical replicates.
Apoptosis assay.
2 × 105 AML cells were incubated in duplicate for 3 h with IgG4-Fc, hSIRPα-Fc, or 25 nM staurosporine (Sigma-Aldrich) in serum-free IMDM. Cells were then harvested, washed, and stained with 7-AAD and Annexin V–FITC (BD). The percentage of 7-AAD–negative, Annexin V–positive cells was determined by flow cytometry.
SIRPα–CD47 binding assay.
Confluent mouse BM-derived macrophages, either untreated or infected with lentiviral vectors expressing NOD mouse or human SIRPα or GFP alone (control) as previously described (Takenaka et al., 2007), were incubated in a 96-well plate in the presence of increasing concentrations of human CD47-Fc for 30 min at 37°C, washed three times with cold PBS, and then incubated with HRP-conjugated goat polyclonal Ab specific for the human IgG Fcγ fragment (1:2,000; Jackson ImmunoResearch Laboratories, Inc.) for 30 min at 4°C. Cells were washed three times with PBS, and hCD47-Fc binding was determined by peroxidase activity assay using 3,3′,5,5′-tetramethylbenzidine (KPL) as a substrate and measuring absorbance at 450 nm on a microplate reader. Data analysis was performed using PRISM version 4.0 software (GraphPad Software).
CD47 binding inhibition assay.
Binding assays were performed as described above using confluent NOD mouse BM-derived macrophages incubated with 1 nM hCD47-Fc, alone or in the presence of increasing concentrations of hSIRPα-Fc (0.4–400 nM). hCD47-Fc binding was determined as described above.
Statistical analysis.
Comparison of AML engraftment in NS and NS-Idd13 mice and in IgG4-Fc– and hSIRPα-Fc–treated mice was performed using Wald test in the general linear models framework or Tukey’s HSD, where appropriate. Statistical tests for in vitro assays are indicated in the text.
Acknowledgments
We thank Dr. Shaheena Bashir (Ontario Institute for Cancer Research, Toronto, Ontario, Canada) for providing statistical analyses.
This work was supported by funds from the Leukemia and Lymphoma Society (to J.S. Danska and J.C.Y. Wang), Ontario Institute for Cancer Research with funds from the province of Ontario (to J.S. Danska, J.E. Dick, and J.C.Y. Wang), Trillium Therapeutics Inc. (TTI; to J.S. Danska and J.C.Y. Wang), Canadian Institutes for Health Research (to J.E. Dick), the Canadian Cancer Society Research Institute (to J.E. Dick), the Terry Fox Foundation (to J.E. Dick), Genome Canada through the Ontario Genomics Institute (to J.S. Danska, J.E. Dick, and J.C.Y. Wang), the Swiss Cancer League and the Swiss National Science Foundation (to A.P.A. Theocharides), the University of Ottawa (to J.M. Ho), and a Canada Research Chair (to J.E. Dick). This research was funded in part by the Ontario Ministry of Health and Long Term Care (OMOHLTC; to J.E. Dick and J.C.Y. Wang). The views expressed do not necessarily reflect those of the OMOHLTC.
There is an existing license agreement between TTI and University Health Network/SickKids Hospital, and A.P.A. Theocharides, L. Jin, T.K. Prasolava, J.S. Danska, J.E. Dick, and J.C.Y. Wang may be entitled to receive financial benefits further to this license and in accordance with their respective institutions’ intellectual property policies. The authors have no additional conflicting financial interests.
Footnotes
Abbreviations used:
- Ab
- antibody
- AML
- acute myeloid leukemia
- CB
- cord blood
- CLO
- clodronate
- HSC
- hematopoietic stem cell
- i.f.
- intrafemoral(ly)
- IgV
- Ig variable region
- LSC
- leukemia stem cell
- NOD
- nonobese diabetic
- NOR
- nonobese diabetes resistant
- SP
- spleen
References
- Anderson K., Lutz C., van Delft F.W., Bateman C.M., Guo Y., Colman S.M., Kempski H., Moorman A.V., Titley I., Swansbury J., et al. 2011. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 469:356–361 10.1038/nature09650 [DOI] [PubMed] [Google Scholar]
- Barazi H.O., Li Z., Cashel J.A., Krutzsch H.C., Annis D.S., Mosher D.F., Roberts D.D. 2002. Regulation of integrin function by CD47 ligands. Differential effects on alpha vbeta 3 and alpha 4beta1 integrin-mediated adhesion. J. Biol. Chem. 277:42859–42866 10.1074/jbc.M206849200 [DOI] [PubMed] [Google Scholar]
- Beatty G.L., Chiorean E.G., Fishman M.P., Saboury B., Teitelbaum U.R., Sun W., Huhn R.D., Song W., Li D., Sharp L.L., et al. 2011. CD40 agonists alter tumor stroma and show efficacy against pancreatic carcinoma in mice and humans. Science. 331:1612–1616 10.1126/science.1198443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D., Dick J.E. 1997. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 3:730–737 10.1038/nm0797-730 [DOI] [PubMed] [Google Scholar]
- Brooke G., Holbrook J.D., Brown M.H., Barclay A.N. 2004. Human lymphocytes interact directly with CD47 through a novel member of the signal regulatory protein (SIRP) family. J. Immunol. 173:2562–2570 [DOI] [PubMed] [Google Scholar]
- Chao M.P., Alizadeh A.A., Tang C., Myklebust J.H., Varghese B., Gill S., Jan M., Cha A.C., Chan C.K., Tan B.T., et al. 2010. Anti-CD47 antibody synergizes with rituximab to promote phagocytosis and eradicate non-Hodgkin lymphoma. Cell. 142:699–713 10.1016/j.cell.2010.07.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao M.P., Alizadeh A.A., Tang C., Jan M., Weissman-Tsukamoto R., Zhao F., Park C.Y., Weissman I.L., Majeti R. 2011. Therapeutic antibody targeting of CD47 eliminates human acute lymphoblastic leukemia. Cancer Res. 71:1374–1384 10.1158/0008-5472.CAN-10-2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow A., Lucas D., Hidalgo A., Méndez-Ferrer S., Hashimoto D., Scheiermann C., Battista M., Leboeuf M., Prophete C., van Rooijen N., et al. 2011. Bone marrow CD169+ macrophages promote the retention of hematopoietic stem and progenitor cells in the mesenchymal stem cell niche. J. Exp. Med. 208:261–271 10.1084/jem.20101688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J., Gao A.G., Frazier W.A. 1997. Thrombspondin acts via integrin-associated protein to activate the platelet integrin alphaIIbbeta3. J. Biol. Chem. 272:14740–14746 10.1074/jbc.272.23.14740 [DOI] [PubMed] [Google Scholar]
- Clarke M.F., Dick J.E., Dirks P.B., Eaves C.J., Jamieson C.H., Jones D.L., Visvader J., Weissman I.L., Wahl G.M. 2006. Cancer stem cells—perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 66:9339–9344 10.1158/0008-5472.CAN-06-3126 [DOI] [PubMed] [Google Scholar]
- DeNardo D.G., Brennan D.J., Rexhepaj E., Ruffell B., Shiao S.L., Madden S.F., Gallagher W.M., Wadhwani N., Keil S.D., Junaid S.A., et al. 2011. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 1:54–67 10.1158/2159-8274.CD-10-0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppert K., Takenaka K., Lechman E.R., Waldron L., Nilsson B., van Galen P., Metzeler K.H., Poeppl A., Ling V., Beyene J., et al. 2011. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat. Med. 17:1086–1093 10.1038/nm.2415 [DOI] [PubMed] [Google Scholar]
- Fox C.J., Paterson A.D., Mortin-Toth S.M., Danska J.S. 2000. Two genetic loci regulate T cell-dependent islet inflammation and drive autoimmune diabetes pathogenesis. Am. J. Hum. Genet. 67:67–81 10.1086/302995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka Y., Matozaki T., Noguchi T., Iwamatsu A., Yamao T., Takahashi N., Tsuda M., Takada T., Kasuga M. 1996. A novel membrane glycoprotein, SHPS-1, that binds the SH2-domain-containing protein tyrosine phosphatase SHP-2 in response to mitogens and cell adhesion. Mol. Cell. Biol. 16:6887–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao A.G., Lindberg F.P., Dimitry J.M., Brown E.J., Frazier W.A. 1996a. Thrombospondin modulates alpha v beta 3 function through integrin-associated protein. J. Cell Biol. 135:533–544 10.1083/jcb.135.2.533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao A.G., Lindberg F.P., Finn M.B., Blystone S.D., Brown E.J., Frazier W.A. 1996b. Integrin-associated protein is a receptor for the C-terminal domain of thrombospondin. J. Biol. Chem. 271:21–24 10.1074/jbc.271.1.21 [DOI] [PubMed] [Google Scholar]
- Hatherley D., Harlos K., Dunlop D.C., Stuart D.I., Barclay A.N. 2007. The structure of the macrophage signal regulatory protein alpha (SIRPalpha) inhibitory receptor reveals a binding face reminiscent of that used by T cell receptors. J. Biol. Chem. 282:14567–14575 10.1074/jbc.M611511200 [DOI] [PubMed] [Google Scholar]
- Hope K.J., Jin L., Dick J.E. 2004. Acute myeloid leukemia originates from a hierarchy of leukemic stem cell classes that differ in self-renewal capacity. Nat. Immunol. 5:738–743 10.1038/ni1080 [DOI] [PubMed] [Google Scholar]
- Isenberg J.S., Ridnour L.A., Dimitry J., Frazier W.A., Wink D.A., Roberts D.D. 2006. CD47 is necessary for inhibition of nitric oxide-stimulated vascular cell responses by thrombospondin-1. J. Biol. Chem. 281:26069–26080 10.1074/jbc.M605040200 [DOI] [PubMed] [Google Scholar]
- Ishikawa F., Yoshida S., Saito Y., Hijikata A., Kitamura H., Tanaka S., Nakamura R., Tanaka T., Tomiyama H., Saito N., et al. 2007. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat. Biotechnol. 25:1315–1321 10.1038/nbt1350 [DOI] [PubMed] [Google Scholar]
- Jaiswal S., Jamieson C.H., Pang W.W., Park C.Y., Chao M.P., Majeti R., Traver D., van Rooijen N., Weissman I.L. 2009. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell. 138:271–285 10.1016/j.cell.2009.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L., Hope K.J., Zhai Q., Smadja-Joffe F., Dick J.E. 2006. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat. Med. 12:1167–1174 10.1038/nm1483 [DOI] [PubMed] [Google Scholar]
- Jin L., Lee E.M., Ramshaw H.S., Busfield S.J., Peoppl A.G., Wilkinson L., Guthridge M.A., Thomas D., Barry E.F., Boyd A., et al. 2009. Monoclonal antibody-mediated targeting of CD123, IL-3 receptor alpha chain, eliminates human acute myeloid leukemic stem cells. Cell Stem Cell. 5:31–42 10.1016/j.stem.2009.04.018 [DOI] [PubMed] [Google Scholar]
- Kharitonenkov A., Chen Z., Sures I., Wang H., Schilling J., Ullrich A. 1997. A family of proteins that inhibit signalling through tyrosine kinase receptors. Nature. 386:181–186 10.1038/386181a0 [DOI] [PubMed] [Google Scholar]
- Lapidot T., Sirard C., Vormoor J., Murdoch B., Hoang T., Caceres-Cortes J., Minden M., Paterson B., Caligiuri M.A., Dick J.E. 1994. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 367:645–648 10.1038/367645a0 [DOI] [PubMed] [Google Scholar]
- Leidi M., Gotti E., Bologna L., Miranda E., Rimoldi M., Sica A., Roncalli M., Palumbo G.A., Introna M., Golay J. 2009. M2 macrophages phagocytose rituximab-opsonized leukemic targets more efficiently than m1 cells in vitro. J. Immunol. 182:4415–4422 10.4049/jimmunol.0713732 [DOI] [PubMed] [Google Scholar]
- Lindberg F.P., Gresham H.D., Schwarz E., Brown E.J. 1993. Molecular cloning of integrin-associated protein: an immunoglobulin family member with multiple membrane-spanning domains implicated in alpha v beta 3-dependent ligand binding. J. Cell Biol. 123:485–496 10.1083/jcb.123.2.485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg F.P., Lublin D.M., Telen M.J., Veile R.A., Miller Y.E., Donis-Keller H., Brown E.J. 1994. Rh-related antigen CD47 is the signal-transducer integrin-associated protein. J. Biol. Chem. 269:1567–1570 [PubMed] [Google Scholar]
- Lindberg F.P., Bullard D.C., Caver T.E., Gresham H.D., Beaudet A.L., Brown E.J. 1996. Decreased resistance to bacterial infection and granulocyte defects in IAP-deficient mice. Science. 274:795–798 10.1126/science.274.5288.795 [DOI] [PubMed] [Google Scholar]
- Majeti R., Chao M.P., Alizadeh A.A., Pang W.W., Jaiswal S., Gibbs K.D., Jr, van Rooijen N., Weissman I.L. 2009. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 138:286–299 10.1016/j.cell.2009.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurier F., Doedens M., Gan O.I., Dick J.E. 2003. Rapid myeloerythroid repopulation after intrafemoral transplantation of NOD-SCID mice reveals a new class of human stem cells. Nat. Med. 9:959–963 10.1038/nm886 [DOI] [PubMed] [Google Scholar]
- Mazurier F., Gan O.I., McKenzie J.L., Doedens M., Dick J.E. 2004. Lentivector-mediated clonal tracking reveals intrinsic heterogeneity in the human hematopoietic stem cell compartment and culture-induced stem cell impairment. Blood. 103:545–552 10.1182/blood-2003-05-1558 [DOI] [PubMed] [Google Scholar]
- Neznanov N., Neznanova L., Kondratov R.V., Burdelya L., Kandel E.S., O’Rourke D.M., Ullrich A., Gudkov A.V. 2003. Dominant negative form of signal-regulatory protein-alpha (SIRPalpha /SHPS-1) inhibits tumor necrosis factor-mediated apoptosis by activation of NF-kappa B. J. Biol. Chem. 278:3809–3815 10.1074/jbc.M210698200 [DOI] [PubMed] [Google Scholar]
- Notta F., Mullighan C.G., Wang J.C., Poeppl A., Doulatov S., Phillips L.A., Ma J., Minden M.D., Downing J.R., Dick J.E. 2011. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature. 469:362–367 10.1038/nature09733 [DOI] [PubMed] [Google Scholar]
- Okazawa H., Motegi S.I., Ohyama N., Ohnishi H., Tomizawa T., Kaneko Y., Oldenborg P.A., Ishikawa O., Matozaki T. 2005. Negative regulation of phagocytosis in macrophages by the CD47-SHPS-1 system. J. Immunol. 174:2004–2011 [DOI] [PubMed] [Google Scholar]
- Oldenborg P.A., Zheleznyak A., Fang Y.F., Lagenaur C.F., Gresham H.D., Lindberg F.P. 2000. Role of CD47 as a marker of self on red blood cells. Science. 288:2051–2054 10.1126/science.288.5473.2051 [DOI] [PubMed] [Google Scholar]
- Oldenborg P.A., Gresham H.D., Lindberg F.P. 2001. CD47-signal regulatory protein α (SIRPalpha) regulates Fcgamma and complement receptor-mediated phagocytosis. J. Exp. Med. 193:855–862 10.1084/jem.193.7.855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson M., Nilsson A., Oldenborg P.A. 2007. Dose-dependent inhibitory effect of CD47 in macrophage uptake of IgG-opsonized murine erythrocytes. Biochem. Biophys. Res. Commun. 352:193–197 10.1016/j.bbrc.2006.11.002 [DOI] [PubMed] [Google Scholar]
- Raulet D.H. 2004. Interplay of natural killer cells and their receptors with the adaptive immune response. Nat. Immunol. 5:996–1002 10.1038/ni1114 [DOI] [PubMed] [Google Scholar]
- Ruhrberg C., De Palma M. 2010. A double agent in cancer: deciphering macrophage roles in human tumors. Nat. Med. 16:861–862 10.1038/nm0810-861 [DOI] [PubMed] [Google Scholar]
- Sacchetti B., Funari A., Michienzi S., Di Cesare S., Piersanti S., Saggio I., Tagliafico E., Ferrari S., Robey P.G., Riminucci M., Bianco P. 2007. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 131:324–336 10.1016/j.cell.2007.08.025 [DOI] [PubMed] [Google Scholar]
- Sarfati M., Fortin G., Raymond M., Susin S. 2008. CD47 in the immune response: role of thrombospondin and SIRP-alpha reverse signaling. Curr. Drug Targets. 9:842–850 10.2174/138945008785909310 [DOI] [PubMed] [Google Scholar]
- Shultz L.D., Lyons B.L., Burzenski L.M., Gott B., Chen X., Chaleff S., Kotb M., Gillies S.D., King M., Mangada J., et al. 2005. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J. Immunol. 174:6477–6489 [DOI] [PubMed] [Google Scholar]
- Stefanidakis M., Newton G., Lee W.Y., Parkos C.A., Luscinskas F.W. 2008. Endothelial CD47 interaction with SIRPgamma is required for human T-cell transendothelial migration under shear flow conditions in vitro. Blood. 112:1280–1289 10.1182/blood-2008-01-134429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steidl C., Lee T., Shah S.P., Farinha P., Han G., Nayar T., Delaney A., Jones S.J., Iqbal J., Weisenburger D.D., et al. 2010. Tumor-associated macrophages and survival in classic Hodgkin’s lymphoma. N. Engl. J. Med. 362:875–885 10.1056/NEJMoa0905680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S., Tsai R., Sen S., Dahl K.N., Discher D.E. 2006. Membrane mobility and clustering of Integrin Associated Protein (IAP, CD47)—major differences between mouse and man and implications for signaling. Blood Cells Mol. Dis. 36:364–372 10.1016/j.bcmd.2006.01.012 [DOI] [PubMed] [Google Scholar]
- Subramanian S., Boder E.T., Discher D.E. 2007. Phylogenetic divergence of CD47 interactions with human signal regulatory protein alpha reveals locus of species specificity. Implications for the binding site. J. Biol. Chem. 282:1805–1818 10.1074/jbc.M603923200 [DOI] [PubMed] [Google Scholar]
- Takenaka K., Prasolava T.K., Wang J.C., Mortin-Toth S.M., Khalouei S., Gan O.I., Dick J.E., Danska J.S. 2007. Polymorphism in Sirpa modulates engraftment of human hematopoietic stem cells. Nat. Immunol. 8:1313–1323 10.1038/ni1527 [DOI] [PubMed] [Google Scholar]
- Tanaka T., Tsudo M., Karasuyama H., Kitamura F., Kono T., Hatakeyama M., Taniguchi T., Miyasaka M. 1991. A novel monoclonal antibody against murine IL-2 receptor beta-chain. Characterization of receptor expression in normal lymphoid cells and EL-4 cells. J. Immunol. 147:2222–2228 [PubMed] [Google Scholar]
- Taylor P.R., Martinez-Pomares L., Stacey M., Lin H.H., Brown G.D., Gordon S. 2005. Macrophage receptors and immune recognition. Annu. Rev. Immunol. 23:901–944 10.1146/annurev.immunol.23.021704.115816 [DOI] [PubMed] [Google Scholar]
- Tsai R.K., Rodriguez P.L., Discher D.E. 2010. Self inhibition of phagocytosis: the affinity of ‘marker of self’ CD47 for SIRPalpha dictates potency of inhibition but only at low expression levels. Blood Cells Mol. Dis. 45:67–74 10.1016/j.bcmd.2010.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooijen N., Sanders A., van den Berg T.K. 1996. Apoptosis of macrophages induced by liposome-mediated intracellular delivery of clodronate and propamidine. J. Immunol. Methods. 193:93–99 10.1016/0022-1759(96)00056-7 [DOI] [PubMed] [Google Scholar]
- Veillette A., Thibaudeau E., Latour S. 1998. High expression of inhibitory receptor SHPS-1 and its association with protein-tyrosine phosphatase SHP-1 in macrophages. J. Biol. Chem. 273:22719–22728 10.1074/jbc.273.35.22719 [DOI] [PubMed] [Google Scholar]
- Vogel W., Grünebach F., Messam C.A., Kanz L., Brugger W., Bühring H.J. 2003. Heterogeneity among human bone marrow-derived mesenchymal stem cells and neural progenitor cells. Haematologica. 88:126–133 [PubMed] [Google Scholar]
- Voit S., Udelhoven M., Lill G., Aktas B., Nieswandt B., Schrör K., Weber A.A. 2003. The C-terminal peptide of thrombospondin-1 stimulates distinct signaling pathways but induces an activation-independent agglutination of platelets and other cells. FEBS Lett. 544:240–245 10.1016/S0014-5793(03)00472-1 [DOI] [PubMed] [Google Scholar]
- Wang H., VerHalen J., Madariaga M.L., Xiang S., Wang S., Lan P., Oldenborg P.A., Sykes M., Yang Y.G. 2007. Attenuation of phagocytosis of xenogeneic cells by manipulating CD47. Blood. 109:836–842 10.1182/blood-2006-04-019794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.Q., Lindberg F.P., Frazier W.A. 1999. Integrin-associated protein stimulates alpha2beta1-dependent chemotaxis via Gi-mediated inhibition of adenylate cyclase and extracellular-regulated kinases. J. Cell Biol. 147:389–400 10.1083/jcb.147.2.389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler I.G., Sims N.A., Pettit A.R., Barbier V., Nowlan B., Helwani F., Poulton I.J., van Rooijen N., Alexander K.A., Raggatt L.J., Lévesque J.P. 2010. Bone marrow macrophages maintain hematopoietic stem cell (HSC) niches and their depletion mobilizes HSCs. Blood. 116:4815–4828 10.1182/blood-2009-11-253534 [DOI] [PubMed] [Google Scholar]
- Yeung J., Esposito M.T., Gandillet A., Zeisig B.B., Griessinger E., Bonnet D., So C.W. 2010. β-Catenin mediates the establishment and drug resistance of MLL leukemic stem cells. Cancer Cell. 18:606–618 10.1016/j.ccr.2010.10.032 [DOI] [PubMed] [Google Scholar]
- Zhao X.W., van Beek E.M., Schornagel K., Van der Maaden H., Van Houdt M., Otten M.A., Finetti P., Van Egmond M., Matozaki T., Kraal G., et al. 2011. CD47-signal regulatory protein-α (SIRPα) interactions form a barrier for antibody-mediated tumor cell destruction. Proc. Natl. Acad. Sci. USA. 108:18342–18347 10.1073/pnas.1106550108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X.W., Kuijpers T.W., van den Berg T.K. 2012. Is targeting of CD47-SIRPα enough for treating hematopoietic malignancy? Blood. 119:4333–4334, author reply :4334–4335 10.1182/blood-2011-11-391367 [DOI] [PubMed] [Google Scholar]