Neuropilin-1 surface expression discriminates between nT reg cells with stable expression and Nrp1 low iT reg cells showing inducible expression under inflammatory conditions.
Abstract
Foxp3 activity is essential for the normal function of the immune system. Two types of regulatory T (T reg) cells express Foxp3, thymus-generated natural T reg (nT reg) cells, and peripherally generated adaptive T reg (iT reg) cells. These cell types have complementary functions. Until now, it has not been possible to distinguish iT reg from nT reg cells in vivo based solely on surface markers. We report here that Neuropilin 1 (Nrp1) is expressed at high levels by most nT reg cells; in contrast, mucosa-generated iT reg and other noninflammatory iT reg cells express low levels of Nrp1. We found that Nrp1 expression is under the control of TGF-β. By tracing nT reg and iT reg cells, we could establish that some tumors have a very large proportion of infiltrating iT reg cells. iT reg cells obtained from highly inflammatory environments, such as the spinal cords of mice with spontaneous autoimmune encephalomyelitis (EAE) and the lungs of mice with chronic asthma, express Nrp1. In the same animals, iT reg cells in secondary lymphoid organs remain Nrp1low. We also determined that, in spontaneous EAE, iT reg cells help to establish a chronic phase of the disease.
The powerful effects of Foxp3+ regulatory T cells are illustrated by the devastating inflammatory diseases caused by Foxp3 mutations in mice and humans (Bennett et al., 2001; Brunkow et al., 2001; Wildin et al., 2001). As a consequence, experimental or clinical manipulation of the entire Foxp3+ T reg compartment could have catastrophic consequences (Kim et al., 2007).
It has been proposed that, because of their different developmental origin and TCR repertoires, Foxp3+ nT reg and iT reg cells could have some nonoverlapping regulatory functions in vivo (Bluestone and Abbas, 2003; Curotto de Lafaille and Lafaille, 2009; Haribhai et al., 2009). It was recently shown that to completely prevent mortality and inflammation in Foxp3-deficient mice, both nT reg and iT reg cells were necessary (Bilate and Lafaille, 2011; Haribhai et al., 2011).
The nonoverlapping functions of nT reg and iT reg cells raise the possibility of selective intervention strategies that would not affect all T reg cells—only nT reg or iT reg cells, or subsets of them. A major barrier to such an approach is the lack of suitable surface markers that distinguish nT reg and iT reg cell populations. The aforementioned studies addressing the issue of division of labor required very specialized strains of mice. However, these experimental systems cannot be used to identify nT reg and iT reg cells in unmanipulated WT mice. In this study, we show that surface expression of Neuropilin 1 (Nrp1) is preferentially up-regulated by nT reg cells in WT mice, and that, in contrast, iT reg cells generated under several in vivo conditions, including the physiologically relevant mucosal route, express low levels of surface Nrp1.
RESULTS
Lack of Nrp1 surface expression characterizes iT reg cells generated in vivo by mucosal or intravenous routes
T cell receptor transgenic mice crossed to RAG-deficient mice lack nT reg cells (Lafaille et al., 1994; Olivares-Villagómez et al., 1998; Curotto de Lafaille et al., 2001), but Foxp3+ iT reg cells can be induced. Oral antigen administration results in the induction of Foxp3+ iT reg cells (Mucida et al., 2005), which are essential for the establishment of oral tolerance (Curotto de Lafaille et al., 2008; Hadis et al., 2011). Foxp3+ iT reg cells can be generated in vivo by other routes (Apostolou and von Boehmer, 2004; Cobbold et al., 2004; Curotto de Lafaille et al., 2004; Kretschmer et al., 2005), but the gut environment appears to be particularly suited for the physiological generation of iT reg cells (Faria and Weiner, 2005; Coombes et al., 2007; Mucida et al., 2007; Sun et al., 2007; Atarashi et al., 2011).
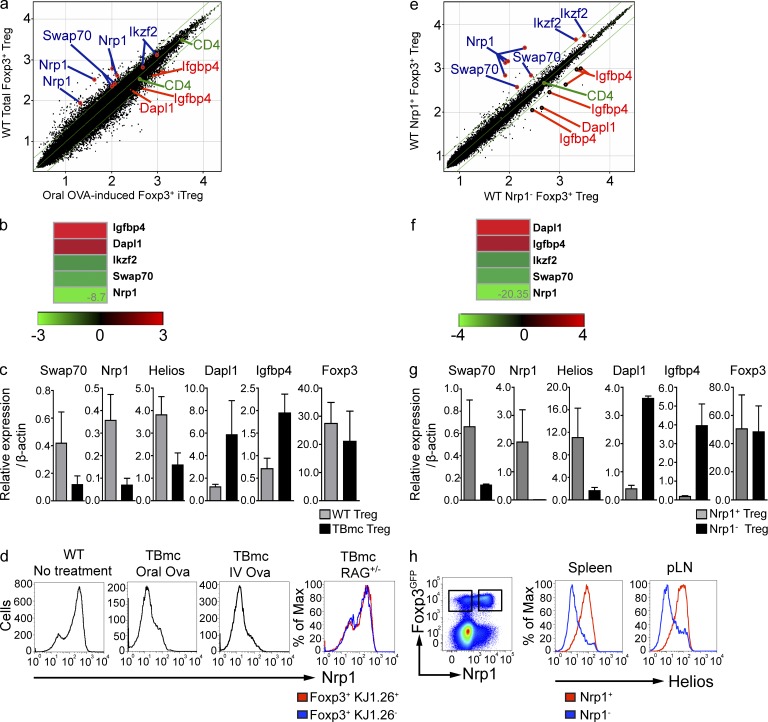
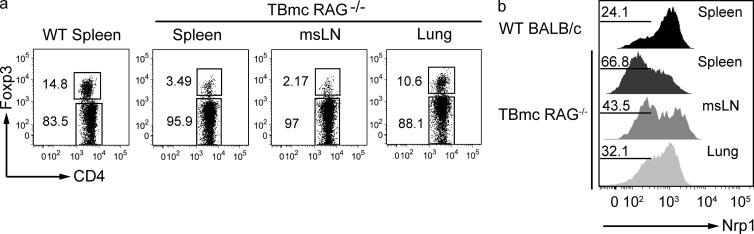
We took advantage of the generation of pure Foxp3+ iT reg cells via mucosal route to determine the gene expression pattern of iT reg cells by microarray. Compared with total Foxp3+ T cells in WT mice, iT reg cells expressed lower levels of Nrp1, Swap70, and Ikzf2 (Helios) mRNA. In contrast, iT reg cells expressed higher levels of Igfbp4 and Dapl1 (Fig. 1, a and b). The microarray data were confirmed by qPCR (Fig. 1 c). As previously established, Foxp3 expression levels were similar between iT reg and nT reg cells. The Nrp1 data were particularly appealing, as Nrp1 is a surface protein and, to date, no surface marker capable of distinguishing iT reg from nT reg cells has been identified. Nrp1 surface staining indicated that the mucosa-generated iT reg cells found in the mesenteric LN (mLN) and spleen were largely Nrp1-negative/low, whereas total Foxp3+ cells from WT mice were largely positive, with a distinct minor peak of Nrp1-negative/low Foxp3+ T cells (Fig. 1 d). Hereafter, we will refer to Nrp1-negative/low cells as Nrp1−. iT reg cells generated by a different route, intravenous injection of antigen without adjuvant, were also Nrp1− (Fig. 1 d) indicating that Nrp1− iT reg cells can also be induced through nonmucosal routes. In our studies of iT reg cell induction, we used T cell/B cell monoclonal (TBmc) mice, whose T cells are from D011.10 mice crossed to RAG1-deficient mice. We sought to ensure that the presence of the D011.10 transgenic TCR did not determine the lack of Nrp1 expression in OVA-specific T reg cells. We therefore analyzed the Nrp1 surface expression in D011.10 RAG1+ mice. D011.10 RAG+ mice have two populations of Foxp3+T reg cells, one OVA-specific (KJ1-26+), which is selected in the thymus on the basis of the expression of endogenous TCR chains, and the other population that express no OVA-specific TCR (KJ1-26−). As anticipated, in D011.10 RAG+ mice the proportions of Foxp3+Nrp1+ and Foxp3+Nrp1− cells were similar to that observed in WT BALB/c mice (Fig. 1 d).
Figure 1.
Lack of Nrp1 surface expression characterizes iT reg cells generated in vivo by mucosal or intravenous route. (a) Log10 raw expression plots showing differences in gene expression between CD4+ Foxp3GFP+ cells sorted from mLN of BALB/c or OVA-fed TBmc mice. Green diagonal lines indicate twofold expression difference between datasets. Selected genes are indicated in blue (up in WT T reg) or in red (up in TBmc T reg). Data are the average of 3 experiments, each corresponding to a pool of mLN of 15–20 mice per condition. (b) Heat maps showing fold difference of selected genes in panel a. (c) Real-time PCR analysis comparing mRNA level expression of selected genes between total T reg from WT mice and iT reg cells from OVA-fed TBmc mice. Dapl1, P = 0.0022; Nrp1, P = 0.0022; Helios, P = 0.0043; Swap70, P = 0.0043; Igfbp4, P = 0.0022; Foxp3, P = 1.0; nonparametric Mann-Whitney U test. Graph contains results from three biological replicates (two independent experiments). (d) Flow cytometric analysis of Nrp1 expression by CD4+ Foxp3+ T reg cells from WT BALB/c and TBmc mice after OVA treatment by the indicated routes. Histograms are representative of at least five different spleens (WT and TBmc mice injected i.v. with OVA) or mLNs of TBmc mice treated orally with OVA and TBmc RAG+ mice gated on KJ1.26-positive and -negative T reg cells. (e) Log10 raw expression plots showing differences in gene expression between CD4+ Foxp3GFP+ Nrp1+ or Nrp1− cells sorted from spleens of BALB/c mice. Green diagonal lines indicate twofold expression difference between datasets. Selected genes are indicated in blue (up in Nrp1+ T reg) or in red (up in Nrp1− T reg). Data are the average of 2 experiments, each corresponding to a pool of 6–13 spleens per condition. (f) Heat maps showing fold difference of selected genes in e. (g) Real-time PCR analysis of mRNA level expression of genes selected in e. The graph shows the results from five biological replicates (run in duplicate). Dapl1, P = 0.0021; Nrp1, P = 0.0079; Helios, P = 0.0159; Swap70, P = 0.0079; Igfbp4, P = 0.0079; Foxp3, P = 0.6857; nonparametric Mann-Whitney U test. (h) Flow cytometric analysis of Nrp1 and Helios expression by WT spleen and peripheral LNs (pLN). Data are representative of at least five different mice.
We hypothesized that the Foxp3+Nrp1− cells in WT spleens were iT reg, and Foxp3+Nrp1+ cells were nT reg cells. To determine whether this was the case, another microarray analysis was performed, this time with purified splenic Foxp3+Nrp1− and Foxp3+Nrp1+ cells from WT BALB/c Foxp3GFP mice. As we had seen with the pure iT reg cells, Nrp1− cells expressed low levels of Nrp1, Swap70, and Helios mRNA, and high levels of Igfbp4 and Dapl1 (Fig. 1, e and f). We also confirmed these microarray data by qPCR (Fig. 1 g). Of the latter genes, we focused more on Dapl1 (death-associated protein-like 1) as its mRNA expression correlates well with the iT reg origin of Foxp3+ cells under a variety of conditions, as shown below. Thus, there is good agreement in the gene expression between splenic bona fide iT reg cells generated through oral antigen administration and splenic Nrp1− cells purified from WT mice.
It has been recently reported that Helios, an Ikaros family transcription factor, was differentially expressed in nT reg cells (Thornton et al., 2010). We sought to determine whether there was correspondence between Helios and Nrp1 protein expression. While there was an excellent correlation between Nrp1 and Helios expression on Foxp3+ cells, there were some Helios+Nrp1− cells and Helios−Nrp1+ cells (Fig. 1 h). Because high quality RNA cannot be extracted from cells that underwent permeabilization for Helios staining, it is not currently possible to determine whether expression of other nT reg and iT reg–specific genes better correspond to Helios or Nrp1 expression. In any event, the level of discordance between the expression of the two proteins is small.
The intestinal lamina propria harbors Nrp1− iT reg cells in high proportion, which is partly dependent on the microbiota
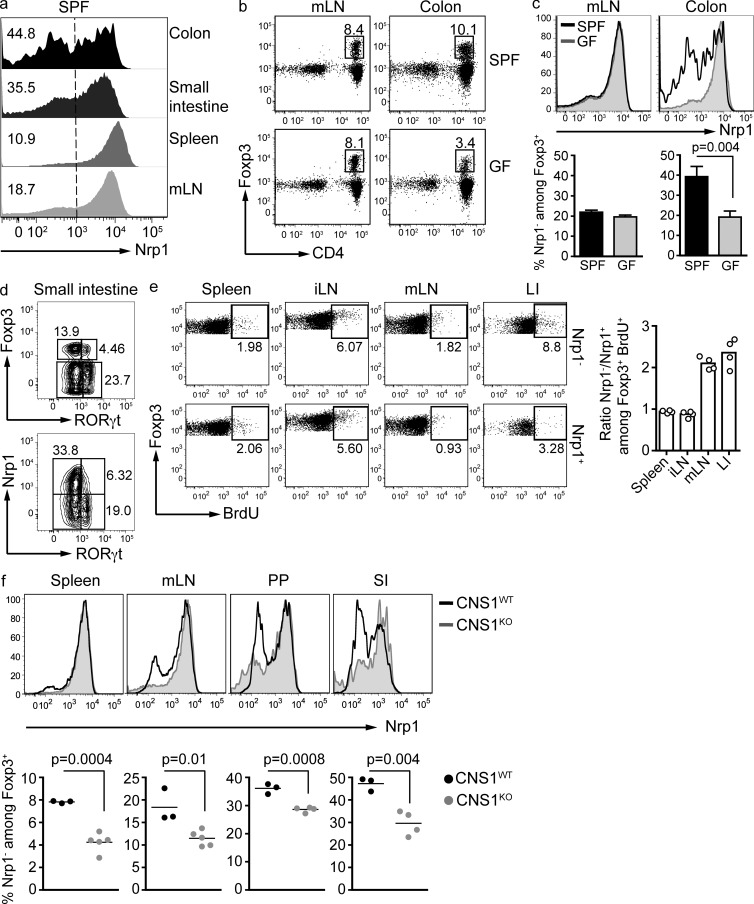
The mucosal immune system is considered a primary site for iT reg cell generation (Curotto de Lafaille and Lafaille, 2009). In fact, it has recently been shown that components of the gut commensal microbiota can direct conversion of naive T cells to iT reg cells, and that some iT reg cells recognize microbiota antigens (Round and Mazmanian, 2010; Atarashi et al., 2011; Lathrop et al., 2011). We therefore investigated the expression of Nrp1 in Foxp3+ cells in secondary lymphoid organs, as well as in the small and large intestine of WT mice. Consistent with the idea that Nrp1− cells are iT reg cells, the percentage of Nrp1− cells among Foxp3+ T reg cells in both small and large intestines was higher than in secondary lymphoid organs (Fig. 2 a).
Figure 2.
The intestinal lamina propria harbors Nrp1− iT reg cells in high proportions, which is partly dependent on the microbiota. (a) Flow cytometric analysis of Nrp1 expression by CD4+ Foxp3+ T reg present in the indicated organs of SPF mice. Histograms are representative of at least five different mice. Numbers on the left indicate the percentage of Nrp1− cells among Foxp3+ CD4+ cells. Vertical dashed line indicates the negative and positive populations. LI, large intestine; SI, small intestine. (b) Frequency of Foxp3+ cells within the CD4+ cell population isolated from mLNs or large intestines lamina propria (LP) of SPF and germ-free mice. FACS plots are gated on CD45high TCRβ+. Dot plots are representative of three SPF and three germ-free mice (two independent experiments). (c) Analysis of Nrp1 expression by CD4+ Foxp3+ T reg cells from mLNs and LI of SPF and germ-free Swiss-Webster mice (same mice as in b). (d) Analysis of RORγt and Nrp1 expression by Foxp3+ T reg cells present in the small intestine LP of SPF mice. FACS plots are gated on CD4+ TCRβ+ (top FACS plot) and CD4+ TCRβ+ Foxp3+ (bottom FACS plot). Data are representative of two independent experiments (n = 3 mice/group). (e) BrdU incorporation by Nrp1+ and Nrp1− T reg cells 4 h after BrdU injection. FACS plots are gated on CD4+ CD45+ Foxp3+, and each plot shows 50,000 events. Plots are representative of two independent experiments (n = 2 mice/group). Graph shows the data of the two experiments. (f) Nrp1 expression by CD4+ Foxp3+ T reg cells extracted from the indicated organs of CNS1-deficient mice compared with WT littermates. PP, Peyer’s patches. Data are representative of two independent experiments (n = 3–5 mice/group). Significance was determined by unpaired Student’s t test.
Furthermore, when we analyzed the colonic lamina propria, we found that germ-free mice displayed a lower proportion of Foxp3+ cells than specific pathogen–free (SPF) mice, a decrease that was not found in secondary lymphoid organs (Fig. 2 b). When we assessed the surface expression of Nrp1 in intestinal Foxp3+ T reg cells, we observed that the percentage of Nrp−Foxp3+ iT reg cells was considerably decreased in germ-free mice (Fig. 2 c), accounting for nearly the total decrease in colonic Foxp3+ cells. These results suggest that Nrp1− T reg cells are, at least in part, induced by the microbiota.
A population of RORγt+Foxp3+ double-positive T cells was described in the small intestine of WT mice (Zhou et al., 2008). It has been proposed that this population could give rise to either T reg cells or Th17 cells. As a percentage of total CD4+ cells in the small intestine, this population was increased in germ-free mice (Ivanov et al., 2009). We studied Nrp1 expression on this microbiota-independent population. Interestingly, we found that RORγt+Foxp3+ cells were Nrp1− (Fig. 2 d), suggesting a local but microbiota-independent induction of Foxp3. Alternatively, RORγt could interfere with the expression of Nrp1.
Because Nrp1− iT reg cells are abundant in the gut mucosa, and iT reg cells can recognize microbiota-derived or -induced antigens (Atarashi et al., 2011; Lathrop et al., 2011) we assessed the proliferation status of Nrp1− and Nrp1+ T reg cells by BrdU incorporation. WT mice were injected with BrdU and, 4 h later, spleen, LN, and large intestine were harvested. In spleen and peripheral LN, the fraction of BrdU+ cells was similar between Nrp1+ and Nrp1− T reg cells. Interestingly, Nrp1− cells from large intestine and mLN incorporated nearly twice as much BrdU as Nrp1+ T reg cells (Fig. 2 e). This result is consistent with the fact that the gut and the GALT are active sites of iT reg generation/expansion.
To confirm the usefulness of Nrp1 as a marker to distinguish iT reg and nT reg cells, we analyzed mice lacking one of the conserved noncoding DNA elements at the Foxp3 locus (CNS1). CNS1-null mice display normal development of thymic nT reg cells, but have a severe impairment in the development of TGF-β–dependent iT reg cells at mucosal surfaces (Zheng et al., 2010; Josefowicz et al., 2012). CNS1-null mice harbored significantly less Nrp1−Foxp3+ T reg cells, which is consistent with the known defect of iT reg cell generation in these mice. This difference was striking in mucosa-associated tissues such as lamina propria and Peyer’s patches (Fig. 2 f).
Thus, low Nrp1 expression appears to be a characteristic of iT reg cells generated by different tolerogenic routes, in normal and mutated mouse strains.
A population of Nrp1− Foxp3high cells is absent in WT thymus
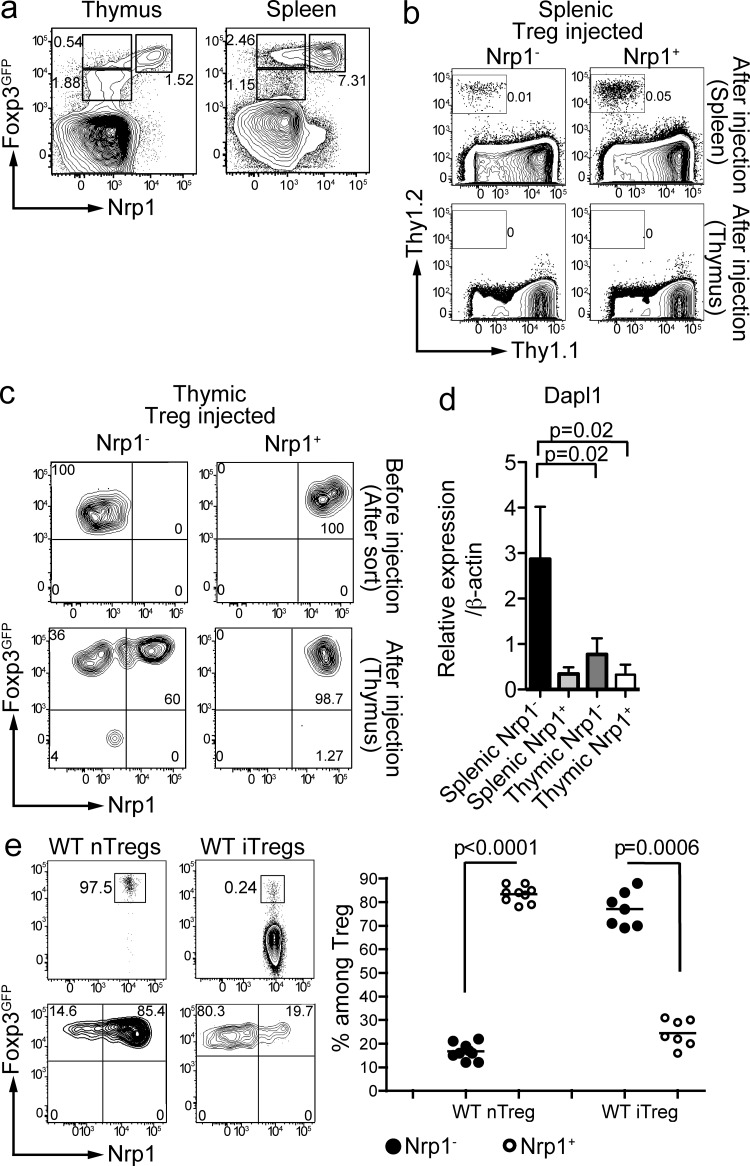
One of the definitions of iT reg cells is that they can be generated in the absence of a thymus (Kretschmer et al., 2005; Curotto de Lafaille et al., 2008). If lack of Nrp1 expression were a marker for iT reg cells, all thymic Foxp3+ cells would be Nrp1+. Indeed, a population of Foxp3+Nrp1− cells was absent in the thymus. However, there was a population of Foxp3lowNrp1− cells (Fig. 3 a). This Foxp3low population was more clearly distinguished in the Foxp3GFP reporter mice that we used (Bettelli et al., 2006) than in other Foxp3GFP mice.
Figure 3.
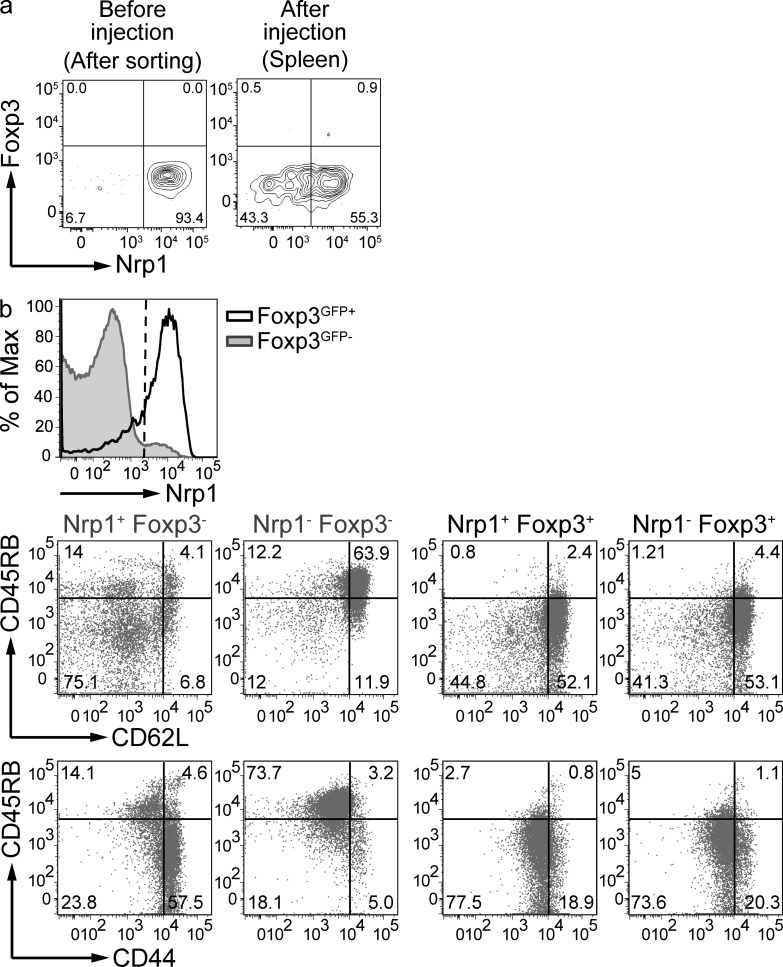
A population of Nrp1− Foxp3high cells is absent in WT thymus. (a) Phenotype of CD4 single-positive T cells in the thymus and the spleen of Foxp3GFP mice. Gates show the percentage of Nrp1− (Foxp3low in the thymus; left), Foxp3high in the spleen (right) and Nrp1+ T reg cells. Dot plots are representative of at least five mice. (b) 1–2 × 105 Thy1.2+ Nrp1− (left) or Nrp1+ (right) spleen T reg cells were injected i.v. into Thy1.1+ recipient mice, and their recovery was analyzed in the spleen (top) or thymus (bottom) 1 or 2 wk later by flow cytometry (FACS plots show CD4+ cells). Dot plots are representative of 4 different experiments (n = 2 mice/group); a 2-wk time-point experiment is shown. (c) Sorted thymic Thy1.2+ Nrp1− (left) or Nrp1+ (right) T reg cells were intrathymically injected and thymi were analyzed 4 d later. FACS plots are gated on CD4+ Thy1.2+ Thy1.1− cells. Data are representative of 2 independent experiments (n = 2 mice/group). (d) qPCR analysis of Dapl1 expression by sorted thymic and splenic T reg populations (n = 3 biological samples run in duplicate). P-values were calculated with nonparametric Mann-Whitney U test. (e; top) Thy1.2+ total thymic CD4 SP Foxp3+ T cells (referred to as WT nT reg) were intrathymically injected, whereas splenic Foxp3− T cells were injected i.v. (referred to as WT iT reg) into congenic hosts. 2 wk later, pooled spleen and LNs were analyzed for Foxp3 and Nrp1 expression on transferred cells. FACS plots are gated on CD4+ Thy1.2+ Thy1.1− cells (top dot plots); bottom plots show the aforementioned Foxp3+ gate. Data are the result of 3 independent experiments for the nT reg group (n = 2–4 mice/experiment) and 3 experiments for the iT reg group (n = 2–3 mice/experiment). Unpaired Student’s t test was used to determine significance.
Thymic Foxp3lowNrp1− cells could have potentially been iT reg cells that migrated into the thymus and down-regulated Foxp3 expression in the process; however, this possibility was deemed highly unlikely, as neither sorted Foxp3+Nrp1− nor Foxp3+Nrp1+ splenic T reg cells were found in significant numbers in the thymus of Thy1 congenic mice upon i.v. injection (Fig. 3 b).
To determine the properties of thymic Foxp3lowNrp1− cells, we performed a series of ultrasound-guided intrathymic injections. Thymic Foxp3lowNrp1− and Foxp3+Nrp1+ cells were injected into Thy1 congenic WT mice, and the injected thymi were dissected 4 d later. Virtually all of the injected Foxp3+Nrp1+ T cells retained their Nrp1 and Foxp3 expression levels. In contrast, 60% of the Foxp3lowNrp1− cells had up-regulated Nrp1 and Foxp3 expression, and 36% (±9.5%) had started to up-regulate Foxp3 expression without up-regulating Nrp1 (Fig. 3 c). Thus, thymic Foxp3lowNrp1− cells are precursors of Foxp3+Nrp1+ cells, but not the opposite.
We also determined the expression of Dapl1 mRNA in thymic T reg cells (by definition, nT reg cells) and compared it with the expression in secondary lymphoid organs. Neither Foxp3lowNrp1− nT reg progenitors nor Foxp3+Nrp1+ mature nT reg cells expressed high levels of Dapl1 mRNA, which is consistent with the selective expression of Dapl1 in iT reg cells (Fig. 3 d).
To further assess the contribution of the thymus to the Nrp1+ population in secondary lymphoid organs, we injected total thymic Foxp3+ cells into Thy1 congenic thymi. Most of the cells found in secondary lymphoid organs 2 wk after injection were Nrp1+ (83% ± 3.5%; Fig. 3 d). From all these studies, we conclude that the thymus lacks a population of Foxp3+Nrp− iT reg cells, providing further support to the use of Nrp1 as a marker of nT reg cells.
To probe the usefulness of Nrp1 as a marker capable of distinguishing nT reg and iT reg cells in a WT experimental system, sorted splenic CD4+Foxp3− T cells were injected i.v. into Thy1 congenic mice. 2 wk later, we analyzed the Nrp1 expression on the newly generated Foxp3+ iT reg cells. In contrast with thymic-derived T reg cells, the vast majority of WT iT reg cells were Nrp1− (78 ± 5%; Fig. 3 e). Therefore, in both WT and transgenic systems, iT reg cells generated in the absence of inflammation are Nrp1−.
Both Foxp3+Nrp1+ nT reg cells and Foxp3+Nrp1− iT reg cells are stable cell populations in the steady state in vivo
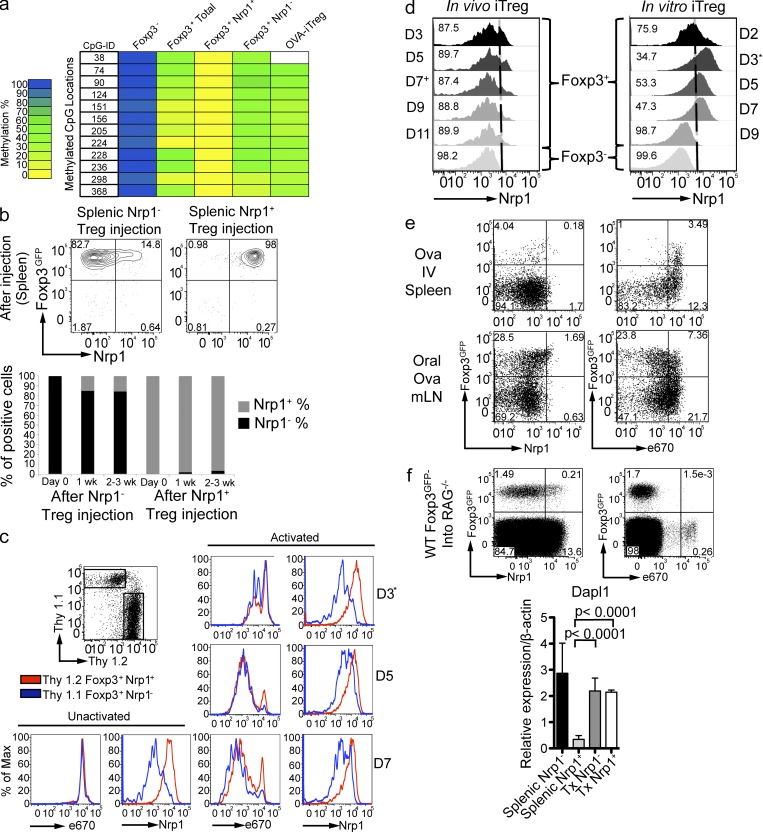
Changes in the level of DNA methylation at T reg–specific demethylated region (TSDR) of the Foxp3 locus reflect the stability of Foxp3 expression (Huehn et al., 2009). We sought to determine the stability of Foxp3+Nrp1+ and Foxp3+Nrp1− cells by analyzing the TSDR methylation state. Naive T cells were fully methylated at the TSDR, whereas total Foxp3+ T reg cells from WT mice displayed low methylation, consistent with previous studies (Floess et al., 2007; Fig. 4 a and Table S1). When total Foxp3+ cells from WT mice were sorted into Nrp1+ and Nrp1− cells, Nrp1− iT reg cells were demethylated to a similar degree as the oral OVA-induced iT reg cells (Fig. 4 a and Table S2), which were similar to other in vivo–generated iT reg cells (Polansky et al., 2008). Interestingly, Nrp1+ nT reg cells were fully demethylated (methylation score of 0), suggesting a high degree of stability regarding Foxp3 expression.
Figure 4.
Both Foxp3+Nrp1+ nT reg cells and Foxp3+Nrp1− iT reg cells are stable cell populations in the steady state in vivo. (a) The indicated cell populations were sorted from spleens of BALB/c Foxp3GFP or TBmc Foxp3GFP mice. Each lane represents a pool of 3–5 mice (2 independent experiments). OVA-iT reg cells were generated by oral OVA treatment for 1 wk, and T reg cells were sorted 2 wk later. Methylation analysis of the T reg–specific demethylation region (TSDR) was performed by bisulfite sequencing. Color indicates methylation percentage of each CpG island in the TSDR. Actual percentages are shown in Table S1. (b) Splenic Thy1.2+ Nrp1− (left) or Nrp1+ (right) T reg cells were injected i.v., and spleens were analyzed at the indicated time points. FACS plots are gated on CD4+ Thy1.2+ Thy1.1− cells. Shown in graph are the frequencies of Nrp1− or Nrp1+ cells among transferred Foxp3+ cells. Dot plots are representative of 2 experiments (n = 2 mice/group). (c) Stability of Nrp1 expression by T reg cells. Thy 1.2+ Nrp1+ and Thy 1.1+ Nrp1− T reg cells were FACS sorted, labeled with 2.5 µM of eFluor 670, and co-cultured with plate-bound anti-CD3, anti-CD28, and IL-2. On day 3, cells were transferred to new wells with no plate-bound antibody, but containing IL-2 and analyzed at the indicated time points. All plots are gated on CD4+ cells. Histograms are representative of two independent experiments run in duplicate with similar results. (d) Kinetics of Nrp1 expression by in vivo– and in vitro–generated iT reg cells. (right) Sorted naive T cells were cultured with plate-bound anti-CD3 and anti-CD28, IL-2, and active TGF-β. Nrp1 and Foxp3 expression were analyzed at the indicated time points. *, On day 3, cells were transferred to new wells, as in c. Histograms are gated on CD4+ and either Foxp3GFP+ or Foxp3GFP−, as indicated. The experiment was performed twice, in duplicate. (left) In vivo generation of iT reg cells. Thy1.1 recipient mice were transferred with 106 CD4+ Foxp3GFP− cells sorted from TBmc RAG−/−. Left histograms show Nrp1 expression on CD4+ Thy 1.1−, Thy 1.2+ Foxp3GFP+ T reg, and Foxp3GFP− cells extracted from mLN at the indicated time points after oral OVA treatment. +On day 7, oral OVA treatment was stopped. Histograms are representative of two independent experiments with n = 2 mice per experiment. (e) Correlation of route of iT reg generation and proliferation. WT mice were transferred with 106 naive D011.10 CD4+ T cells from TBmc Foxp3GFP mice labeled with 5 µM of eFluor 670 and treated with OVA by the indicated routes. Nrp1 expression and proliferation were analyzed on day 4 after OVA treatment. FACS plots are gated on CD4+ cells. Data are representative of 3 experiments (2 mice per experiment). (f) BALB/c RAG−/− or TCR α−/− β−/− mice were transferred with 5 × 106 splenic CD4+ Foxp3− T cells from WT Foxp3GFP mice (3 experiments, 2 mice per group), and cells transferred into RAG−/− were labeled with 5 µM eFluor 670. Nrp1 expression and proliferation were analyzed 1 wk after injection (pooled spleen and LN). FACS plots are gated on CD4+ Thy1.1+ Thy1.2− cells (transferred cells). Additionally BALB/c TCR α−/− β−/− mice were treated for 1 wk with broad spectrum antibiotics and transferred with 5 × 106 splenic CD4+ Foxp3− T cells from WT Foxp3GFP mice. After 1 wk of continued antibiotic treatment, T reg populations were sorted from pooled spleen and LNs. The graph in the bottom panel shows qPCR of Dapl1 expression by the in vivo–generated iT reg cells. (n = 2 biological replicates run in duplicate pooled from 2 groups of 3 mice). Significance was determined by nonparametric Mann-Whitney U test.
Next, we determined the stability of Foxp3+Nrp1+ and Foxp3+Nrp1− cells in vivo by i.v. transfer into Thy1 congenic recipients. Virtually all (97 ± 2%) of Foxp3+Nrp1+ nT reg cells retained the expression of these proteins (Fig. 4 b). The majority of Foxp3+Nrp1− cells were also stable, with 84.5 ± 3% of the cells remaining Nrp1− negative. Nrp1− cells that acquired Nrp1 expression did so early after transfer. Nrp1 expression in these converted cells did not reach the fluorescence intensity displayed by the originally sorted Nrp1+ cell population. There was very little loss of Foxp3 expression under these lymphosufficient conditions. We conclude that, under the in vivo conditions tested, both Nrp1+ and Nrp1− T reg cells are stable.
We next tested the stability of Nrp1 expression of nT reg and iT reg cells after in vitro activation. Foxp3+Nrp1+ and Foxp3+Nrp1− cells were purified from Thy1 congenic WT Foxp3GFP mice, labeled with the cell proliferation dye eFluor670, and co-cultured. Under these conditions, Nrp1+ and Nrp1− T reg cells proliferated to the same extent (Fig. 4 c). Analysis of Nrp1 expression over time showed that Nrp1+ cells did not change their levels of Nrp1. Nrp1− cells in the co-cultures behaved somewhat differently, up-regulating Nrp1 expression at day 5, but without reaching the fluorescence intensity of Nrp1+ cells (Fig. 4 c).
Next, we tested the kinetics of Nrp1 expression during in vivo and in vitro differentiation of iT reg cells. Oral OVA induced Foxp3 expression, whereas Nrp1 expression remained low throughout the course of the induction and after oral OVA treatment was stopped at day 7 (Fig. 4 d, left). When the in vitro induction protocol was applied (Chen et al., 2003), the results were different, with a transient up-regulation of Nrp1. Nrp1 surface expression peaked at day 3, when the cells were placed into wells containing only IL-2. Subsequently, Nrp1 levels decreased, becoming negative at day 9 (Fig. 4 d, right).
We sought to compare different in vivo methods of iT reg generation to establish a relationship between proliferation and Nrp1 expression by iT reg cells in vivo.
Naive D011.10 RAG−/− Foxp3GFP− cells labeled with the proliferation dye eFluor 670 were injected into Thy1 congenic hosts, and treated either with oral or i.v. OVA. Although many T reg cells from oral OVA-treated mice proliferated during their acquisition of Foxp3, the expression of Nrp1 remained low in the proliferating and nonproliferating cells. Similarly, Nrp1 expression was low in i.v.-induced T reg cells, which did not proliferate during Foxp3 acquisition (Fig. 4 e).
To further establish whether Nrp1 expression was related to the degree of proliferation during the generation of iT reg cells in vivo, we used a method in which homeostatic proliferation is essential to the development of iT reg cells (Curotto de Lafaille et al., 2004). We injected eFluor 670-labeled Foxp3− T cells into RAG−/− and TCRα−/−β−/− mice and analyzed in vivo–generated iT reg cells after 1 wk. Even in this highly proliferative environment, the vast majority of iT reg cells were Nrp1− (79.8 ± 4%; representative example in Fig. 4 f, top). We also analyzed Dapl1 mRNA expression in these iT reg cells. Consistent with the selective expression of Dapl1 mRNA in iT reg cells, we found that Nrp1− iT reg cells, and even the small fraction of Nrp1+ iT reg cells, expressed Dapl1 mRNA (Fig. 4 f, bottom).
Nrp1 expression is controlled by TGF-β
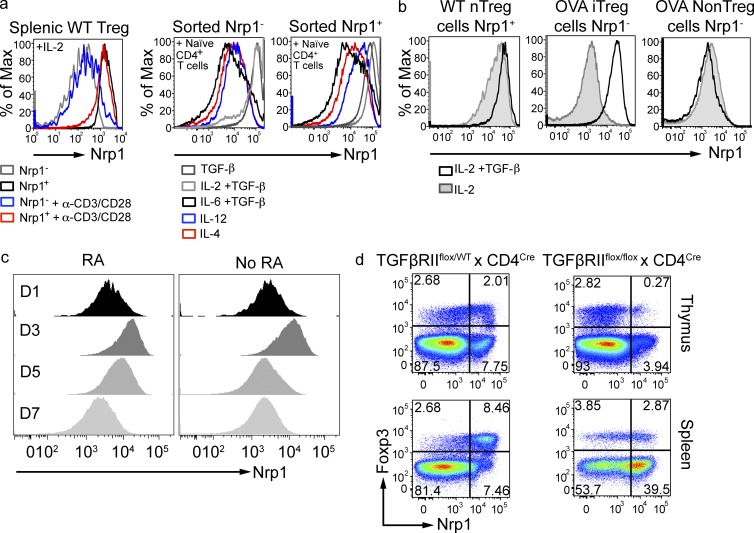
We sought to understand the fluctuation of Nrp1 expression under in vitro culture conditions, reasoning that the cytokines used for in vitro polarization could have played a role. First, we confirmed that, in the absence of added TGF-β1, sorted WT Nrp1+ cells did not change Nrp1 levels, whereas sorted Nrp1− T reg cells displayed a modest increase in Nrp1 expression (Fig. 5 a, left). We then co-cultured T reg cells with naive T cells in the presence of different cytokines. On day 4, Nrp1− T reg cells cultured in the presence of TGF-β1 highly up-regulated Nrp1 expression, whereas other cytokines did not affect Nrp1 expression (Fig. 5 a, middle). The effect of TGF-β1 in boosting Nrp1 expression was also noticeable on Nrp1+ T reg cells (Fig. 5, a [right] and b [left]). We also evaluated the impact of TGF-β on OVA-specific iT reg cells generated through oral OVA treatment. OVA-specific iT reg and conventional T (T conv) cells were sorted from OVA-fed TBmc mice and cultured in the presence of TGF-β1 and IL-2. Strikingly, TGF-β increased Nrp1 expression of OVA-specific iT reg cells, but not of OVA-specific T conv cells (Fig. 5 b, middle and right).
Figure 5.
Nrp1 expression is controlled by TGF-β. (a) Splenic Nrp1+ and Nrp1− T reg cells were FACS-sorted from WT mice and cultured in the presence or absence of plate-bound anti-CD3 and soluble anti-CD28 and IL-2 (left). Nrp1+ and Nrp1− T reg cells were also cultured together with naive T cells in the presence of the indicated cytokines (middle and right). Stability of Nrp1 expression was assessed 4 d after culture. FACS plots are gated on CD4+ Foxp3GFP+ cells. Histograms are representative of one out of two independent experiments performed in duplicate. (b) T reg cells were sorted from WT or OVA-fed TBmc mice and cultured for 4 d in the presence of plate-bound anti-CD3 and soluble anti-CD28 and IL-2 with or without TGF-β. Foxp3− Nrp1− CD4+ T cells were also sorted from TBmc mice treated with OVA and Nrp1 expression analyzed 4 d after culture. FACS plots are gated on CD4+ Foxp3GFP+ cells. Histograms are representative of one out of two independent experiments performed in duplicate. (c) In vitro iT reg cells were generated as in Fig. 4 d, with the addition of 10 nM of retinoic acid. FACS plots are gated on CD4+ Foxp3GFP+ cells. Data are representative of two experiments run in duplicate. (d) Thymi and spleens of 2.5-wk-old TGF-β receptor II–deficient mice and heterozygous littermates were harvested, and Nrp1 expression by CD4+ T cells were cells analyzed. FACS plots are gated on CD4+ CD8− cells. Data are representative of 3 independent experiments (n = 2 mice/group).
The TGF-β–mediated up-regulation of Nrp1 expression in the aforementioned in vitro experiments presents an apparent contradiction with our in vivo data, which shows that oral OVA-induced iT reg cells are Nrp1− and are not generated in the presence of blocking anti–TGF-β antibodies (Mucida et al., 2005). We reasoned that, in vivo, other factors would counterbalance the effect of TGF-β on Nrp1 expression. It has been shown that retinoic acid (RA) is a cofactor involved in gut iT reg generation. We therefore tested whether or not RA could play a role in Nrp1 expression. Although RA increased the percentage of Foxp3+ cells, it did not prevent Nrp1 up-regulation (Fig. 5 c). However, we found that IL-6 prevented up-regulation of Nrp1 on Nrp1− T reg cells treated with TGF-β1 (Fig. 5 a, middle). IL-6 is but an example of many factors that could influence Nrp1 expression in vivo. We surmise that the low expression of Nrp1 in mucosa-induced iT reg cells reflects the opposing effects of several factors present in the environment.
To provide definitive evidence that TGF-β controls the levels of Nrp1, we analyzed thymi and spleens of 2.5-wk-old TGF-β receptor II–conditional knock-out mice and heterozygous littermates. Nrp1 expression was almost absent in thymic T reg cells, and greatly reduced in splenic T reg cells lacking TGF-β receptor II (Fig. 5 d). The presence of a small population of Nrp1+ T reg cells is likely caused by the incomplete Cre-mediated deletion of TGFβRII in these cells, as we could show the presence of residual floxed alleles in T cells of TGFβRII conditional knockout mice (unpublished data). In contrast to T reg cells, a higher fraction of splenic T conv cells expressed Nrp1 in TGF-β receptor II–deficient mice (Fig. 5 d). These results are consistent with the in vitro data (Fig. 5, a and b) and indicate that TGF-β selectively control Nrp1 expression on T reg cells but not on T conv cells.
Both Nrp1+ and Nrp1− T reg cells from WT mice can suppress immune responses in vitro and in vivo
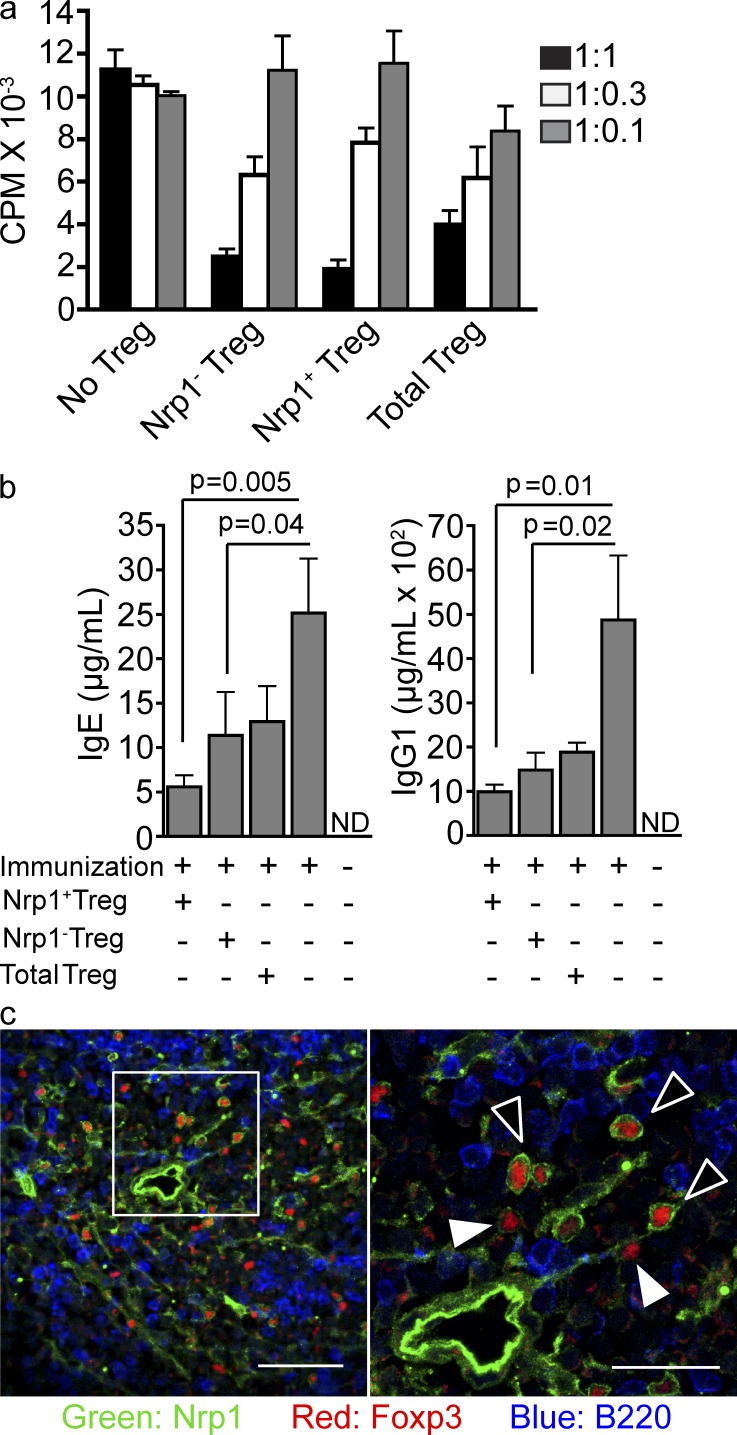
It was important to determine the functionality of Nrp1+ and Nrp1− Foxp3+ T reg cells in different situations. We therefore compared the effectiveness of Nrp1+ and Nrp1− T reg cells in the in vitro suppression assay (Takahashi et al., 1998; Thornton and Shevach, 1998). At three different ratios, nT reg and iT reg cells displayed equivalent in vitro suppressive capacity (Fig. 6 a).
Figure 6.
Both Nrp1+ and Nrp1− T reg cells from WT mice can suppress immune responses in vitro and in vivo. (a) In vitro suppression assay. Naive T cells from WT mice were co-cultured in a 1:1, 1:0.3, or 1:0.1 ratio (Naive T: T reg) with the indicated T reg populations and with APCs in the presence of anti-CD3. As a control, naive T cells were cultured in the absence of T reg cells, but in the presence of additional naive T cells at the indicated ratios. Cell proliferation was measured by [3H]-thymidine incorporation. Graph is representative of two independent experiments run in triplicate. (b) Nrp1+ and Nrp1− T reg cells in suppression of Th2 response. TBmc mice were transferred or not with the indicated T reg population from the Foxp3GFP+ KJ1.26+ population of TBmc RAG+ mice (2 × 105 cells/mouse), and 24 h later, immunized with OVA-HA adsorbed in alum. IgE and IgG1 serum levels were analyzed by ELISA 2 wk after immunization (n = 3 mice/group, 2 independent experiments). Significance was determined by unpaired Student’s t test. (c) Nrp1+ and Nrp1− T reg cells distribution in the spleen under steady state. Confocal images of spleen sections showing the distribution of Nrp1+ and Nrp1− T reg cells in the white pulp. Sections were stained with anti-Nrp1, Foxp3 and B220. Bar, 50 µm. 60× magnification. (right) 2.5× digital zoom of inset from left pane; bar, 25 µm. Black arrowheads point to Nrp1+ T reg cells, and white arrowheads point to Nrp1− T reg cells. Images are representative of at least three different mice.
We also tested the capacity of OVA-specific Nrp1+ and Nrp1− T reg cells, sorted from D011.10 RAG+ mice, to suppress an antibody response in vivo. TBmc mice lack nT reg cells, and thus respond to a single immunization by producing very high levels of IgE and IgG1 antibodies (Curotto de Lafaille et al., 2001). When we compared the ability of Nrp1+ and Nrp1− T reg cells to reduce the antibody responses, we found that both subsets were effective (Fig. 6 b). Foxp3+Nrp1+ cells were slightly more effective than Foxp3+Nrp1− cells, without reaching statistical significance.
We next determined the location of Foxp3+Nrp1+ and Foxp3+Nrp1− T reg cells in secondary lymphoid organs. Confocal microscopy images of WT spleens indicated that both Nrp1− and Nrp1+ T reg cells colocalized in the periarteriolar lymphoid sheath area of the white pulp (Fig. 6 c), with few Foxp3+ cells in the red pulp. Thus, even though a division of labor between nT reg and iT reg cells may exist, there is no segregation of Foxp3+ T reg cell types in the WT spleen.
Tumor-infiltrating T reg cells are enriched in iT reg cells
It has long been noted that many tumors harbor Foxp3+ T reg cells (Cao, 2010; Nishikawa and Sakaguchi, 2010; Wilke et al., 2010). The iT reg or nT reg origin of tumor-infiltrating lymphocytes (TILs) is controversial. Some lines of experimentation support the in situ generation of iT reg cells (Valzasina et al., 2006; Liu et al., 2007), whereas others favor a differential migration and expansion of nT reg cells (Curiel et al., 2004; Hindley et al., 2011). In fact, both possibilities are not mutually exclusive (Zhou and Levitsky, 2007). We therefore assessed the surface expression of Nrp1 on tumor-infiltrating Foxp3+ T reg cells.
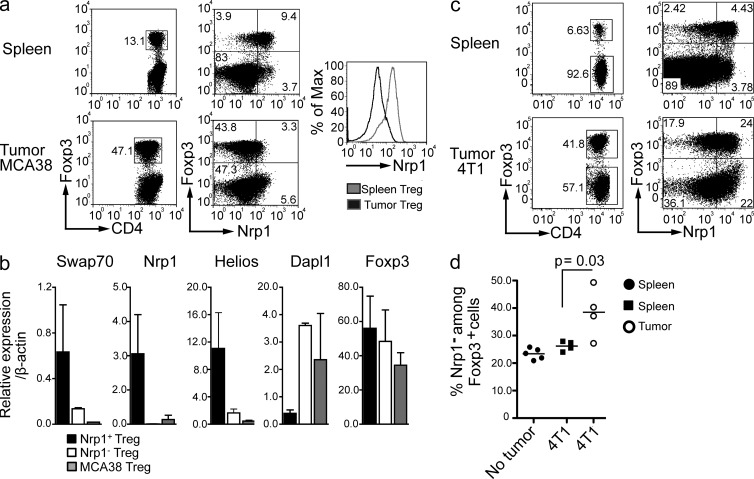
First, we injected mice with the MCA-38 colon adenocarcinoma cell line i.p., and the CD4+ TILs were analyzed by flow cytometry. MCA-38 tumors had extensive infiltration by Foxp3+ cells, which constituted up to 50% of the CD4+ TIL. Strikingly, the vast majority of these Foxp3+ cells were Nrp1−, suggesting that they were iT reg cells (Fig. 7 a). In contrast, splenic Foxp3+ cells in the same mice were predominantly Nrp1+. To confirm that the Foxp3+ cells from the tumor were iT reg cells, as opposed to nT reg cells that lost Nrp1 expression in the tumor environment, we analyzed by qPCR the expression of the typical iT reg and nT reg genes described in Fig. 1. The Foxp3+Nrp1− TIL expressed low levels of Nrp1, Helios, and SWAP70 and high levels of Dapl1, indicating that they were, indeed, iT reg cells (Fig. 7 b).
Figure 7.
Tumor-infiltrating T reg cells are enriched in iT reg cells. (a) C57BL/6 were injected i.p. with 2 × 105 MCA38 cells. After 2 wk, tumor and spleen were harvested and Nrp1 expression on CD4+ T cells was analyzed by FACS. FACS plots are gated on CD4+ CD45+ cells. Dot plots are representative of 2 different experiments (n = 5 mice/group). (b) Real-time PCR analysis of sorted CD4+ Foxp3+ T reg cells from MCA38 tumor-infiltrating T reg cells. n = 2 biological replicates run in duplicate. P < 0.05 for Nrp1, Swap 70, Helios, and Dapl1; P = 0.85 for Foxp3. Significance was determined by non parametric Mann-Whitney U test. (c) BALB/c mice were injected subcutaneously on the right flank with 5 × 104 4T1 cells. After 2 wk, tumor and spleen were harvested and Nrp1 expression on CD4+ T cells was analyzed by FACS. FACS plots are gated on CD4+ CD45+ CD3+ cells. Dot plots are representative of 2 different experiments (n = 4 mice/group). (d) Statistical analysis of the percentage of Nrp1− cells contained within each Foxp3+ population from mice bearing or not 4T1 tumors. Graph is representative of one experiment. Significance was determined by unpaired Student’s t test.
Next, we injected s.c. the 4T1 breast cancer cell line into BALB/c mice. Compared with the MCA-38 tumors, there was a slightly lower frequency of Foxp3+ cells among CD4+ TIL. (Fig. 7 c). In addition, there was a lower proportion of Nrp1− cells among Foxp3+ cells in the 4T1 model. We also noticed an elevation of Nrp1 surface expression in CD4+ Foxp3− conventional T cells infiltrating the 4T1 tumor (Fig. 7 c). Foxp3− TIL expressed slightly higher Nrp1 levels than the corresponding spleen cells, Foxp3+ TIL had a lower expression of Nrp1 than splenic Foxp3+ cells. Thus, the propensity toward a lower Nrp1 expression in the tumor T reg cells than in the spleen is conserved between MCA-38 and 4T1 tumors (Fig. 7 d).
We conclude that iT reg cells are predominant among tumor-resident T reg cells in some tumor models, but not in others. It will therefore be important to analyze the T reg population in different types of tumors to uncover the rules that govern the migration and/or generation of T reg cells in each tumor type.
In inflammatory environments, iT reg cells express Nrp1
Myelin basic protein (MBP)–specific TCR transgenic mice crossed with RAG−/− mice, TCRα−/−β−/− mice, or into homozygosity, lack nT reg cells and develop EAE spontaneously with 100% penetrance (Lafaille et al., 1994; Olivares-Villagómez et al., 1998; Furtado et al., 2001). The phases of disease are: a preclinical phase in which the mice are still healthy, a subclinical phase with a rapidly developing inflammation in the CNS but no neurological signs of disease, an acute phase with a clinically progressing disease, and a long chronic phase in which mice remain relatively stable (Furtado et al., 2008).
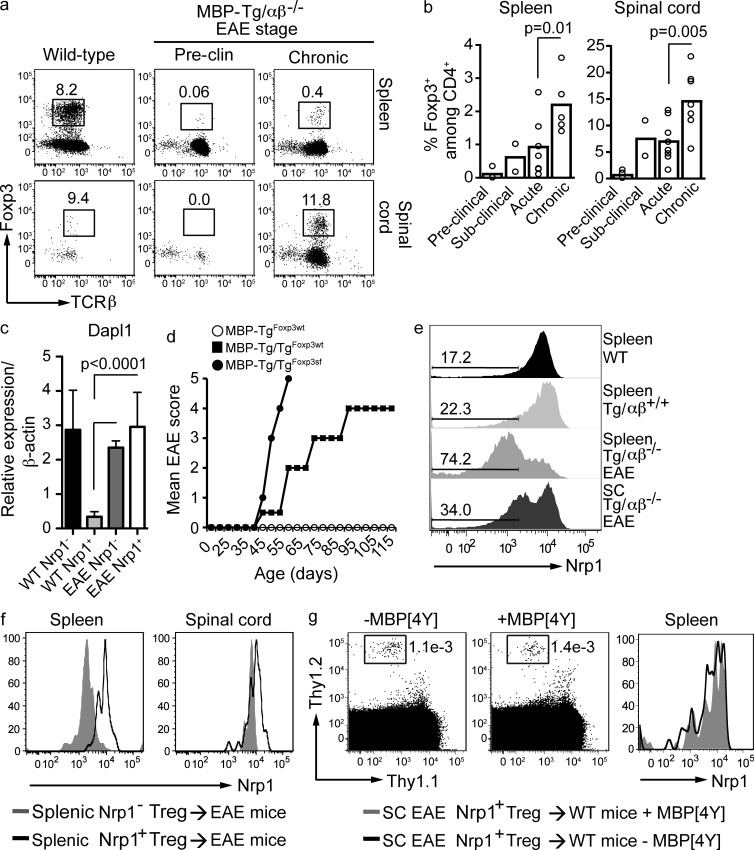
A population of Foxp3+ iT reg cells (range of 10–30% of CD4+ cells) was readily detected in the central nervous system (CNS) and secondary lymphoid organs of mice with chronic spontaneous EAE, whereas little or no Foxp3+ cells were detected during the preclinical stage (Fig. 8 a). Note that healthy mice have very few CNS-infiltrating CD4+ cells, and their characterization is therefore unreliable. The appearance of iT reg cells in the CNS could be detected as soon as mice entered the subclinical EAE stage (Fig. 8 b), but not before. The percentage of iT reg cells in the spleen was consistently much lower than in the CNS. In summary, iT reg cells were not observed in any organ before disease onset but became detectable shortly after disease onset. To strengthen the notion that T reg cells in the spinal cords of mice with spontaneous EAE were iT reg cells, we determined expression of Dapl1 mRNA, which is elevated in iT reg cells. As anticipated, Foxp3+ cells from the highly inflamed spinal cords of mice with EAE expressed high levels of Dapl1 mRNA, even those that were Nrp1+ (Fig. 8 c).
Figure 8.
iT reg cells from inflamed central nervous system express Nrp1. (a and b) The frequency of T reg cells in the spleen and spinal cords of MBP-Tg/TCRαβ−/− mice during the different stages of spontaneous EAE was analyzed by flow cytometry. Dot-plots show frequency of Foxp3+ cells among total CD4+ CD45High cells. Pre-clinical, before onset of EAE (4–5 wk old); sub-clinical, 1 d after weight loss, but before onset of EAE; acute, 3–7 d after onset of EAE; chronic, 20–30 d after onset of EAE. Dot plots are representative of 1 out of 2–8 mice analyzed/group during a 4–8-wk period. Graph shows all mice analyzed. Unpaired Student’s t test was used to determine significance. (c) qPCR analysis of Dapl1 expression by the indicated T reg cell populations (n = 2 biological replicates run in duplicate) sorted from either spleen of healthy WT mice or spinal cords of mice afflicted by EAE. Significance was determined by nonparametric Mann-Whitney U test. (d) EAE was monitored in homozygous MBP-Tg/Tg Foxp3-sufficient mice (MBP-Tg/TgFoxp3wt, n = 30), or Foxp3-deficient scurfy mice (MBP-Tg/TgFoxp3sf, n = 10), and hemizygous MBP-Tg and Foxp3-sufficient mice (MBP-TgFoxp3wt, n = 10) that harbor nT reg cells. Mice that died during the follow-up period were scored as level 5, and were included in calculation of the mean score. (e) Expression of Nrp1 by T reg of MBP-Tg/TCRαβ−/− mice with EAE, compared with hemizygous MBP-Tg TCRαβ-sufficient mice and to WT mice. Numbers on the left indicate the percentage of Nrp1− cells among CD4+ Foxp3+ cells. SC, spinal cord. FACS plots are gated on CD4+ CD45High cells. Data are representative of at least three independent experiments with at least two mice per group. (f) MBP-Tg/Tg mice afflicted by acute EAE were transferred with T reg cells (Nrp1+ or Nrp1− as indicated) sorted from spleens of healthy MBP-Tg Foxp3-GFP mice. Nrp1 levels among transferred T reg cells were analyzed 48 h after transfer. FACS plots are gated on CD4+ CD45+ cells. Histograms are representative of two independent experiments (two mice per group). (g) Thy1.2+ Nrp1+ T reg cells were sorted from spinal cords (SC) of MBP-Tg/Tg Foxp3-GFP mice afflicted by chronic EAE and transferred into Thy1.1 congenic recipients. On the following day, mice were injected i.p. with 10 µM MBP[4Y] peptide. Nrp1 levels on the transferred T reg cells were analyzed 4 d after peptide injection. FACS plots are gated on CD4+ CD45+ cells. Representative of two mice per group.
To assess the impact of iT reg cells in spontaneous EAE, we generated MBP TCR transgenic (Tg) homozygous mice with Foxp3-deficient scurfy mice (Foxp3sf). The results were striking. All Tg/Tg Foxp3sf mice died within 1 wk of EAE onset, whereas the majority of mice capable of generating Foxp3+ iT reg cells (Foxp3WT) did not die (Fig. 8 d). These mice remained in a chronic stage with permanent disability caused by myelin damage and secondary axonal losses, but otherwise gained weight and displayed healthy behavior. Thus, in the EAE inflammatory environment, in which iT reg cells are generated too late to prevent disease, iT reg cells nevertheless exert a beneficial effect by allowing the establishment of a chronic stage of disease.
We then analyzed Nrp1 expression on Foxp3+ T cells from the spleen and CNS of mice afflicted with spontaneous EAE. The majority of splenic iT reg cells were Nrp1−, as expected for iT reg cells. However, in the CNS of the same mice, the majority of iT reg cells were Nrp1+ (Fig. 8 e). Foxp3+ cells from spleens of MBP-specific hemizygous TCR Tg TCRαβ+/+ mice and WT mice were predominantly Nrp1+, as expected for mice in which nT reg cells constitute the main T reg cell population (Fig. 8 e). There were little or no T cells in the CNS of the latter two mouse strains.
One possible explanation for the presence of Nrp1+ iT reg cells in spinal cord of mice with EAE is the abundance of antigen and cytokines within the inflamed CNS. In that environment, the balance of factors favors Nrp1 expression, similar to the in vitro differentiation experiment shown in Fig. 4 d.
To investigate the hypothesis that the inflammatory environment within the CNS of mice afflicted by EAE influenced the levels of Nrp1 on iT reg cells, we performed T reg cell transfer experiments into healthy and EAE-affected mice. First, we transferred Nrp1+ or Nrp1− T reg cells sorted from spleens of healthy TCR Tg TCRαβ+/+ Foxp3GFP mice into MBP-Tg/Tg mice during acute spontaneous EAE. Nrp1 levels on transferred T reg cells were analyzed after 48 h. Whereas in the spleen Nrp1− T reg cells remained Nrp1− (and Nrp1+ remained Nrp1+), in the inflamed spinal cord, Nrp1− T reg cells up-regulated Nrp1 levels (Fig. 8 f). When Nrp1+ T reg cells were sorted from inflamed spinal cords and transferred into Thy1 congenic healthy recipients, they remained Nrp1+, even when the cognate antigen was not added (Fig. 8 g). These results indicate that, in sharp contrast to iT reg cells generated in tolerogenic environments, many iT reg cells generated during EAE expressed Nrp1.
We hypothesized that Nrp1 expression in iT reg cells in EAE reflected the Th1/Th17 inflammatory environment in which they were generated. We therefore tested Nrp1 expression in iT reg cells generated in a different inflammatory environment, one caused by a Th2 response. Chronic lung inflammation was induced in TBmc by repeated airway antigen exposure. Foxp3+ iT reg cells were induced and found at its’ highest percentage in the lungs (Fig. 9 a). Similarly to the Th1/Th17 environment of EAE, the majority of iT reg cells isolated from chronically inflamed lungs expressed high levels of Nrp1 (Fig. 9 b). The spleens of the same mice harbored mostly Nrp1− cells, as we observed in the EAE model. Thus, inflammatory iT reg cells can be distinguished from iT reg cells generated under tolerogenic conditions by the higher percentage of cells expressing Nrp1 in the former group.
Figure 9.
iT reg cells from inflamed lungs are Nrp1+. (a) TBmc mice were immunized i.p. with OVA cross-linked to hemagglutinin peptide adsorbed in alum and treated intranasally twice a week with OVA-HA. After 8 wk of OVA-HA intranasal administration, lungs, mediastinal LN, and spleen were harvested and stained for CD4 and Foxp3. WT BALB/c mouse was used as control. FACS plots are gated on CD4+ CD45+ cells. (b) Nrp1 expression by Foxp3+ T reg cells shown in panel a. FACS plots are gated on CD4+ CD45+ Foxp3+ cells. Data are representative of 2 experiments (n = 2 mice/experiment).
Foxp3−Nrp1+ cells are activated T cells that are not related to the T reg cell lineage
A population of ∼1% Foxp3−Nrp1+ was noticeable in the WT thymus (Fig. 3 a). These cells could be nT reg progenitors if they would up-regulate Foxp3 expression at a later time. To determine whether Foxp3−Nrp1+ CD4SP cells could give rise to Foxp3+Nrp1+ cells we injected those cells intrathymically. Clearly, Foxp3−Nrp1+ cells did not become Foxp3+Nrp1+ cells (Fig. 10 a).
Figure 10.
Foxp3−Nrp1+ cells are activated T cells not related to the T reg lineage. (a) Thymic Thy1.2+ CD4SP Foxp3GFP− Nrp1+ T cells were intrathymically injected into Thy1.1+ congenic mice using ultrasound guidance. Nrp1 and Foxp3 expression by splenic CD4+ cells was analyzed 1–2 wk later. Cells are gated on Thy1.2+ Thy1.1− CD4SP T cells. Dot plots are representative of 2 independent experiments (n = 2 mice/group). (b) Nrp1+ CD4+ T cells have a memory/activated phenotype. Spleens of steady-state Foxp3GFP mice were harvested and the indicated activation markers were analyzed by flow cytometry. Dotted line indicates separation of Nrp1+ and Nrp1− populations. All plots are gated on CD4+ CD8− cells. Histograms and dot plots are representative of one out of three different mice.
We also observed a population of Foxp3−Nrp1+ in secondary lymphoid organs and inflamed tissues. To further characterize this population, spleens of healthy WT Foxp3GFP mice were harvested and the expression of different activation/memory markers was analyzed. Most Foxp3−Nrp1+ cells have an activated/memory phenotype, largely CD62Llow, CD45RBlow, and CD44high (Fig. 10 b). This is consistent with the fact that TGF-β RII–deficient mice, which undergo an autoinflammatory disease from an early age, have a greatly increased population of Foxp3−Nrp1+ cells (Fig. 5 d). In contrast to Foxp3− cells, Foxp3+Nrp1+ T reg cells were CD44low and CD62Lhigh (Fig. 10 b). Thus, for T conv cells, Nrp1 expression reflects an activated/memory stage, and expression of Nrp1 in these cells occurs in the absence of TGF-β signaling. This result suggests that the rules that govern Nrp1 expression on T reg and T conv cells are different.
Our data indicates that, in unmanipulated mice, Foxp3+Nrp1+ cells are stable, thymic-derived nT reg cells, whereas Foxp3+Nrp1− T reg cells are iT reg cells derived at sites such as the mucosa. Foxp3−Nrp1+ cells are not related to T reg cells.
DISCUSSION
We compared iT reg cells generated through oral antigen administration with WT T reg cells and found that low surface Nrp1 expression on Foxp3+ cells could be used to identify iT reg cells. iT reg cells generated by other tolerogenic methods, such as i.v. injection and homeostatic proliferation, were also Nrp1−. Some of the microbiota components that are responsible for iT reg cell induction have been recently identified (Round and Mazmanian, 2010; Atarashi et al., 2011). A substantial proportion of Nrp1− iT reg cells are generated via the mucosal route in response to microbiota, as they are greatly reduced in colons of germ-free mice. Our studies with germ-free mice confirmed, by surface Nrp1 staining, the previous finding that iT reg cells in the intestine are partially induced by the commensal microbiota (Atarashi et al., 2011), and further corroborated the usefulness of Nrp1 as a marker.
In the thymus, all Foxp3high cells were Nrp1+. In secondary lymphoid organs of unimmunized SPF mice, the majority of Foxp3+ cells (70–80%) were also Nrp1+, with a well-defined minor peak of Nrp1− cells. This latter population was reduced in CNS1-deficient mice, which have defective iT reg generation, but normal thymic generation of nT reg cells. Thus, in the steady state, bona fide iT reg cells are Nrp1− and bona fide nT reg cells are Nrp1+.
Nrp1 is, however, not a perfectly specific marker. In unimmunized mice, Nrp1 distinguishes well, although not absolutely, nT reg from iT reg cells. Even in mice undergoing severe inflammatory reactions, the vast majority of iT reg cells in secondary lymphoid organs express low levels of Nrp1, and this trend also applies to LN draining the inflammatory sites. However, most iT reg cells at the sites of inflammation express Nrp1. So there is a “blind spot” in which Nrp1+ cells collected at inflammatory sites from WT mice cannot be ascribed as having nT reg or iT reg origin. We found that, in these cases, the expression of Dapl1 mRNA helps to trace the origin of the T reg cells, as Dapl1 mRNA is expressed by iT reg cells regardless of Nrp1 surface expression. Dapl1 (death-associated protein-like 1) is a 107-aa-long protein with a predicted molecular weight of ∼11.8 KD. It is homologous to 102-aa-long Dap1 (death-associated protein 1; Deiss et al., 1995). There are parallelisms between the use of Nrp1 as an nT reg cell–specific marker and the use of CD25 as a T reg cell–specific marker. Although not an absolute T reg cell marker, CD25 (Sakaguchi et al., 1995) was very effective in moving the field forward for a long time.
Nrp1+ nT reg cells are very stable in regard to Foxp3 expression, and they have a zero methylation score of the TSDR. The TSDR colocalizes with conserved non-coding sequence-2 (CNS2), a region shown to be involved in the maintenance of Foxp3 expression (Zheng et al., 2010). There is a debate about the stability of the T reg phenotype in general, and of Foxp3 expression in particular (Duarte et al., 2009; Oldenhove et al., 2009; Zhou et al., 2009; Rubtsov et al., 2010; Sharma et al., 2010; Bailey-Bucktrout and Bluestone, 2011). Our work shows that Nrp1 can be used in addition to Foxp3 to isolate nT reg cells that are stable in regard to Foxp3 expression, and fail to acquire effector functions. Thus, it appears that nT reg cells are stable, and the observed instability is likely derived from iT reg cells or, more generally, more immature T reg cells. In that sense, Nrp1 may compare favorably with a population of CD25high cells shown to also be highly stable (Komatsu et al., 2009; Miyao et al., 2012).
The gene expression studies provided interesting results. Although mucosa-induced iT reg cells are fully functional in vitro and in vivo (Mucida et al., 2005; Curotto de Lafaille et al., 2008), they retain the expression of some genes that are more typical of conventional T cells than nT reg cells, such as Igfbp4 and Dapl1. These two genes, for instance, are not part of the “T reg signature” established in previous publications (Hill et al., 2007; Lin et al., 2007). It is clear that the oral administration of antigen changes the expression of many genes that are required for T reg cell function (Foxp3 for example), but some noncritical T conv genes are retained by the iT reg cells; in combination with Foxp3, expression of these genes could be used to distinguish iT reg from nT reg cells. We did not find any gene up-regulated in iT reg cells relative to nT reg cells that was not also expressed by conventional T cells.
We have shown that in several instances the Nrp1 surface expression correlates with known or expected iT reg versus nT reg origin of the Foxp3+ cells. However, the complete gene expression pattern of oral antigen–induced Foxp3+ iT reg cells in mice lacking nT reg cells is not entirely overlapping with the pattern of Foxp3+Nrp1− cells in WT mice. Therefore, it would be of interest to address the meaning of the differences in future studies. The differences could indicate that mucosa-generated iT reg cells are only one of the contributors to the splenic iT reg cell pool in WT mice, probably an important contributor, but not the only one. It would be interesting to determine the gene expression profile of iT reg cells generated at other locations. For example, it is to be expected that in the MCA-38 model there is generation of Nrp1− iT reg cells, and these tumor iT reg cells are unlikely to have a fully overlapping gene expression pattern with mucosa-generated or other iT reg cells.
iT reg cells in spleens of WT BALB/c mice represented ∼30% of Foxp3+ cells, whereas in WT C57BL/10.PL mice housed in the same animal facility, splenic iT reg cells constituted <20% of total T reg cells. Differences in the genetic makeup could explain why oral tolerance is easier to induce in some mouse strains than in others.
Nrp1 was proposed as a T reg cell marker in 2004 (Bruder et al., 2004); here, we propose its use a marker of nT reg cells. However, the role of Nrp1 in T reg cell biology is unclear. A study on a conditional Nrp1 knockout mouse strain driven by lck-Cre showed no defect in T reg cell generation; more importantly, there was no report of any disease in mice harboring Nrp1–deficient T cells (Corbel et al., 2007). Had T reg cell function been compromised, evidence of autoimmune disease would not have been missed. Despite the knockout data, Nrp1 could still play a role in nT reg cell function. For example, it was proposed that Nrp1 strengthened the contacts between T reg cells and APC (Sarris et al., 2008). Because iT reg cells express little or no Nrp1, they would have to use a different mechanism to exert their suppressive function.
We have recently described that iT reg cells can be divided into two groups based on their origin in tolerogenic or inflammatory environments (Bilate and Lafaille, 2012). We found that iT reg cells generated in tolerogenic environments were largely Nrp1−. In contrast, iT reg generated under inflammatory conditions displayed a high proportion (>50%) of Nrp1+ cells, as we exemplified in EAE and chronic lung inflammation models. These results are consistent with those reported by Haribhai et al. (2011). In their microarray analysis, these authors found Nrp1 mRNA expression in iT reg cells. These iT reg cells were generated in vivo in Foxp3-deficient mice, therefore a highly inflammatory environment.
We also observed that Nrp1 expression was transiently up-regulated during in vitro iT reg cell induction in the presence of TGF-β1, but the same phenomenon was not observed upon iT reg induction in vivo.
Intriguingly, TGF-β1 appears to be an important driver of Nrp1 expression in T reg cells, but not on T conv cells. This applies to nT reg and iT reg cells. TGF-β signaling deficiency on T cells dramatically reduced Nrp1 expression by thymic Foxp3+ cells. It has been shown that blockade of TGF-β signaling abolishes iT reg generation and the acquisition of oral tolerance, which is mediated by iT reg cells (Cobbold et al., 2004; Mucida et al., 2005). However, in vivo, the TGF-β–induced Nrp1 expression on iT reg cells is countered by other opposing factors, such that, in balance, iT reg cells generated under tolerogenic conditions are Nrp1−, whereas iT reg cells generated under inflammatory conditions are predominantly Nrp1+. It has been said that Nrp1 acts as a co-receptor for TGF-β, as Nrp1+ cells have enhanced TGF-β signaling compared with Nrp1− cells (Glinka et al., 2011). This may create a positive loop, in which Nrp1+ T reg cells display higher TGF-β signaling, which further supports Nrp1 expression. On the other hand, Nrp1− T reg cells lack the positive loop mechanism, and respond to TGF-β without the enhancement.
There was an apparent discrepancy between the role of IL-6 in vitro and in vivo. During in vitro polarization of T cells, IL-6 blocked TGF-β1–induced up-regulation of Nrp1 expression on Nrp1− T reg cells. However, in vivo, proinflammatory environments, which are known to contain IL-6, (e.g., the CNS of mice afflicted with EAE), did not result in Nrp1 down-modulation on iT reg cells. In all likelihood, these results reflect the complexity of the in vivo inflammatory microenvironment, with multiple factors influencing Nrp1 expression on T reg cells in opposing ways. The net result in EAE and chronic lung inflammation is that Nrp1 expression is up-regulated on iT reg cells.
A previous study used an EAE model induced by MOG immunization with CFA and pertussis toxin; in this study, conversion of naive T cells into iT reg cells was not observed (Korn et al., 2007). There are key differences between the aforementioned work and our current study. First, in the former work the mice already had a normal complement of nT reg and iT reg cells before immunization. Second, EAE induction with CFA and Pertussis toxin does not favor the generation of iT reg cells; instead, antigen/CFA emulsions generate T reg cells in the thymus (Zelenay et al., 2010). Therefore, the situation is quite different in the spontaneous EAE model, in which there is no nT reg cell presence at any point before or during disease. However, unlike the CFA-created environment, the inflamed CNS milieu of spontaneous EAE favors the generation of iT reg cells, as it probably occurs in other inflammatory conditions such as asthma. It is noteworthy that in mice with severe spontaneous EAE or chronic lung inflammation there was little up-regulation of Nrp1 in iT reg cells outside of the target tissues.
We also observed an elevated percentage of iT reg cells in one tumor model, and a smaller elevation in another. Future studies will indicate which proportion of tumors are infiltrated by large numbers of iT reg cells, and what are the rules that determine the predominant type of T reg cells in the different tumors. For those tumors with high frequency of iT reg cells, our results open the possibility of a selective manipulation of nT reg and iT reg cells. It would be of great importance to assess whether one could, for example, improve anticancer immune response by targeting iT reg cells, while, by keeping nT reg cells untouched, not augmenting substantially the likelihood of autoimmune diseases.
MATERIALS AND METHODS
Mice.
TBmc and MBP Ac1-11–specific TCR transgenic mice have been previously described (Lafaille et al., 1994; Curotto de Lafaille et al., 2001). MBP-Tg/Tg mice are mice in which the TCR transgenes have been bred to homozygosity. MBP-Tg mice refer to mice hemizygous for the TCR α/β integration. MBP-Tg/αβKO mice are MBP-Tg mice bred to TCRα−/−β−/− mice. Foxp3GFP mice (Bettelli et al., 2006) were backcrossed to the TBmc mice and BALB/c mice for at least five generations and into the MBP-Tg/Tg C57BL/10.PL background for at least two generations to incorporate the MHC H-2u/u restriction. C57BL/6 mice were obtained from The Jackson Laboratory. Swiss Webster germ-free and SPF mice were obtained from Taconic laboratories. Foxp3 CNS1 knockout mice (Foxp3ΔCNS1−gfp) were generated as previously described (Zheng et al., 2010). T cell–specific TGF-βRII–deficient mice (Tgfbr2−/−) were generated by crossing Tgfbr2-floxed mice with the CD4-Cre transgene as previously described (Li et al., 2006).
As indicated in the text, some mice were treated with an antibiotic combination of 1 g/liter each of Ampicillin (sodium salt; Sigma-Aldrich), Neomycin sulfate (Thermo Fisher Scientific), and Metronidazole (Thermo Fisher Scientific) and 0.5 g/liter Vancomycin hydrochloride (Thermo Fisher Scientific) in drinking water.
Animals were housed at the NYU Medical Center Skirball Animal Facility under SPF conditions or at Memorial Sloan-Kettering Cancer Center facilities (MSKCC). All procedures were approved by MSKCC and New York University School of Medicine Institutional Animal Care Use Committee.
Antigen treatment.
Oral and airway tolerance to chicken OVA were induced as previously described (Mucida et al., 2005). Alternatively, when indicated, WT mice were transferred with 106 naive DO11.10 CD4+ T cells and treated as described in the previous section. For immunization, TBmc mice were injected with 100 µg OVA-HA in 1 mg of alum by the intraperitoneal route as previously described (Curotto de Lafaille et al., 2008) and either transferred or not with Nrp1+, Nrp1−, or total Foxp3+ T reg cells sorted from TBmc RAG+/− Foxp3GFP+ KJ1.26int. Lung inflammation (asthma model) was induced as previously described (Curotto de Lafaille et al., 2008).
To assess the stability of Nrp1 expression on iT reg cells isolated from spinal cords of mice with EAE, recipient mice were injected with an MHC I-Au–stable derivative of MBP peptide Ac1-11, named MBP[4Y].
EAE evaluation.
EAE was scored as previously described (Olivares-Villagómez et al., 1998): level 1, limp tail; level 2, hind leg weakness or partial paralysis; level 3, total hind leg paralysis; level 4, hind leg paralysis and front leg weakness or partial paralysis; level 5, moribund.
Flow cytometry and cell purification.
Cells were stained with the following conjugated antibodies: CD4 (RM4-5), CD45RB (C363.16A), Thy 1.1 (HIS51), Thy 1.2 (53–2.1), Foxp3 (FJK-16s), rNrp1 biotinylated (BAF566; R&D Systems) and unconjugated (AF566; R&D Systems), fluorophore-conjugated rabbit anti–goat IgG (H+L; A-11078 and A-21446; Invitrogen), fluorophore-conjugated Streptavidin (eBioscience), anti-TCRβ (H57-597; BD), Helios (22F6; BioLegend), CD45 (30-F11), and DO11.10 TCR (KJ1.26). Intranuclear staining was performed using the eBioscience kit according to the manufacture’s instructions.
Stained cells were analyzed on a FACSCalibur or a LSRII flow cytometer (BD) and data processed using FlowJo (Tree Star). For cell isolation, CD4+ T cells were enriched from total splenic or LN cell suspensions by negative selection against CD8- and B220-expressing cells using MACS (Miltenyi Biotec) and further sorted using an Aria II cell sorter (BD) or MoFlo sorter (Dako) taking the clearly negative and clearly positive Nrp1 populations.
For sorting of cell populations from the thymus, CD4+ CD8− T cells were enriched by negative selection against CD8-expressing cells using MACS and further sorted using an Aria II cell sorter.
Microarray analysis.
Microarray analyses were performed in triplicate or duplicate. Foxp3GFP+ cells were FACS-sorted from mLN of OVA-fed TBmc mice or from BALB/c mice. Splenic CD4+ Foxp3+ Nrp+ T reg and Nrp− T reg cells were isolated from Foxp3GFP BALB/c mice. The purity was always >95%. RNA was then extracted using TRIzol Reagent (Invitrogen) according to manufacture’s instructions. Total RNA was reverse transcribed, amplified, labeled, and hybridized to Mouse Genome 430 2.0 arrays (Affymetrix) at New York University NYU Cancer Institute Genomics Facility or Genomics Core Laboratory of the MSKCC. Microarray data were analyzed using Agilent Genespring GX 11 software. Raw data for the microarray analyses performed in this study are available under GEO accession no. GSE40488).
In vitro suppression assay.
FACS-sorted naive 4 × 104 CD4+ CD45RBhigh Foxp3GFP− T cells were co-cultured with T reg populations at different ratios, in the presence of 8 × 104 mitomycin C-treated splenocytes isolated from TCRαβ KO mice and of soluble low endotoxin, azide-free anti-CD3 (17A2) at 0.5 ng/ml in a 96-well plate. Proliferation was determined by adding [3H]-thymidine (1 µCi/well; Du Pont) on the third day of culture, and analyzing incorporation 6 h later using a Wallac 4500 Microbeta Plus scintillation counter.
ELISA.
Total IgE and IgG1 antibodies were quantified by ELISA as previously described (Curotto de Lafaille et al., 2001). Antibodies used were as follows: anti-IgG1 (clone A85-3; BD), anti-IgG1 HRP conjugated (clone LO-MG1-2; Invitrogen), anti-IgE (clone LO-ME-3; Invitrogen), and anti-IgE HRP conjugated (clone 23G3; Southern Biotech).
In vitro activation and in vitro iT reg generation.
For T reg activation, FACS-sorted cells were placed in flat-bottom, 96-well plates with plate-bound low endotoxin/azide–free anti-CD28 (37.51) and anti-CD3 (17A2; BioLegend) at 5 ng/ml and 100 U/ml rhIL2 (PeproTech). At day 3, cells were transferred to new wells with 100 U/ml IL-2. For iT reg generation, sorted naive CD4+ CD45RBhigh Foxp3GFP− cells were cultured with plate-bound anti-CD28 and anti-CD3 with 100 U/ml rhIL2 in addition to 20 ng/ml active TGF-β1 (PeproTech). Retinoic acid was used at 10 nM. Proliferation was measured by dilution of eFluor 670 (eBioscience) according to the manufacturer’s instructions.
Isolation of lamina propria mononuclear cells.
Small and large intestines were harvested, Peyer’s patches were removed, and tissues were digested with a mixture of collagenase type VIII (Sigma-Aldrich) and DNase I (Roche). Mononuclear cells were enriched using a 40:80% Percoll (Sigma-Aldrich) gradient as previously described (Ivanov et al., 2009)
BrdU incorporation.
WT BALB/c mice were injected i.p. with 1 mg of BrdU and sacrificed 4 h later. Spleens, LNs, and large intestine were harvested, surface-stained, and intranuclearly stained for BrdU using the BD BrdU proliferation kit, followed by Foxp3 staining using the eBioscience Foxp3 Staining Buffer kit according to the manufacture’s instructions. eBioscience fixable viability dye-eFluor 780 was used to gate dead cells out.
Isolation of TILs.
2 × 105 MCA38 cells were injected i.p. into C57BL/6 WT or C57BL/6 Foxp3GFP mice. 5 × 104 4T1 tumor cells were injected subcutaneously into BALB/c Foxp3GFP mice on the right flank. After 12 d, tumor-infiltrating cells were obtained as previously described (Radoja et al., 2001). In brief, tumors were excised, minced, and digested twice with a mixture of enzymes. CD4+ lymphocytes were enriched using magnetic beads (Miltenyi Biotec). Single-cell suspensions were then stained for flow cytometry analysis.
Isolation of mononuclear cells from spinal cords.
Mice were anesthetized with a mixture containing 12.5mg/ml ketamine, 2.5mg/ml xylazine, and 25 mg/ml acepromazine and intracardially perfused with PBS with 5 mM EDTA. Spinal cords were dissected, torn into small pieces and digested in 80 U/ml of collagenase D (Roche) at 37°C for 35 min. Samples were incubated for 5 min in 10 mM EDTA to disrupt DC–T cell complexes. Mononuclear cells were enriched using a 38% Percoll (Sigma-Aldrich) gradient and centrifuged at 1,000 g for 30 min.
Ultrasound-guided intrathymic injection.
4–8-wk-old mice were anesthetized with 4% Isoflurane (Aerrane) in medical air and maintained under anesthesia using a nose-cone with 1.5% Isoflurane. Hair was removed from the thorax using with depilatory cream, and animals were placed on a heat pad set at 37°C during the injection procedure. Thymus was visualized with a 30-MHz 707B ultrasound probe (VisualSonics). 1–2 × 105 sorted cells of the indicated populations were resuspended in 20 µl of PBS and injected (10 µl in each lobe) using a Hamilton syringe and a 30-gauge needle with the aid of a three-dimensional micromanipulator. After the indicated time points, thymus and/or pooled spleen and LNs (axillary, inguinal, and mesenteric) of injected mice were harvested and depleted of CD8+ (thymus) and B220+ cells (spleen and LN) using magnetic beads (Miltenyi Biotec). Single-cell suspensions were stained for the indicated markers and analyzed by flow cytometry.
Methylation analysis.
105 cells of each type were FACS-sorted to >95% purity, and then genomic DNA was purified using DNEasy blood and tissue gDNA purification kit (QIAGEN) according to the manufacturer’s instructions. Bisulfite sequencing was performed as previously described (Floess et al., 2007).
Immunofluorescence and confocal microscopy.
BALB/c mice were perfused with 1% PFA in PBS, spleens were harvested and further fixed with 4% PFA for 30 min to 1 h at room temperature and cryoprotected with 30% sucrose for 18 h. Fixed spleens were then embedded in optimum cutting temperature (OCT) compound and snap frozen at −80°C until use. 8–10-µm-thick cryostat sections of spleen were fixed with cold acetone for 10 min at −20°C and blocked/permeabilized with 0.2% Triton X-100 in PBS with 5% BSA and 10% FCS for 10 min at room temperature. Endogenous biotin and avidin were blocked with an avidin and biotin blocking kit (Vector Laboratories) according to the manufacture’s instructions. Sections were stained using the following antibodies diluted in PBS 5% BSA, 10% FCS, and 10% horse and donkey serum for 30 min to 1 h at room temperature under 40-rpm agitation: biotinylated anti-Foxp3 (FJK-16s; eBioscience), unconjugated goat anti–rat Nrp-1 (R&D Systems), Alexa Fluor 647–conjugated anti-B220 (RA3-6B2; eBioscience), and Alexa Fluor 488–conjugated rabbit anti–goat IgG (Invitrogen). Streptavidin–Alexa Fluor 555 was obtained from Invitrogen. Confocal images were acquired on an Fluoview BX50W1 microscope (Olympus) and a Radiance confocal system with 488-, 568-, and 637-nm excitation lines, 560- and 650-nm detectors and HQ515/30 and HQ600/40 filters (Bio-Rad Laboratories). Images were obtained with a Plan-Apochromat 20× (NA 0.8) and 60× (NA 1.4) oil immersion objectives. Images were exported to ImageJ 1.43u (National Institutes of Health) for final processing. Maximal intensity Z projections were generated, and contrast was adjusted linearly.
Real-time PCR analysis.
RNA from FACS-sorted cells was extracted by the TRIzol method according to the manufacture’s instructions (Invitrogen) and treated with RNase-free DNase I (Invitrogen) to eliminate contaminating genomic DNA. Reverse transcription was performed using Superscript II reverse-transcriptase and oligo(dT)12–18 primer (Invitrogen). Quantitative real-time PCR was done with SYBR Green with the primers described in Table S1. An iCycler real-time qPCR system (Bio-Rad Laboratories) was used for all reactions and detection. Expression values were normalized to β-actin.
Statistical analysis.
Mean and SD values were calculated with GraphPad Prism (GraphPad Software). Unpaired Student’s t test or nonparametric Mann-Whitney U test were used to compare two variables, as indicated in each figure legend. P-values < 0.05 were considered significant.
Online supplemental material.
Table S1 lists percent methylation for all positions tested in CNS2. Table S2 lists the PCR primers used for qPCR in Figs. 1, 3, 4, 7, and 8. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20120914/DC1.
Supplementary Material
Acknowledgments
We thank the Memorial Sloan Kettering Cancer Center and New York University School of Medicine Genomics Facility for performing the microarray experiments; all cell sorting experiments were performed by Peter Lopez and Michael Gregory at the NYUCI Flow Cytometry and Cell Sorting Center, which is supported by grant National Institutes of Health/National Cancer Institute 5 P30CA16087-31. DNA methylation experiments were developed and performed by Udo Baron and Sven Olek at Epiontis GmbH. We also thank Drs. V. Kuchroo and M. Oukka for the Foxp3GFP mice, and Orlando Aristizábal for advice with the ultrasound-guided injections, and Sang V. Kim and Maria Ciofani for their invaluable advice.
A.M. Bilate was the recipient of a postdoctoral fellowship from the Pew Latin American Fellows Program. Work by S. Floess and J. Huehn was supported by the German Research Foundation (SFB621). Work in the Lafaille laboratory is supported by the National Institutes of Health/National Institute of Allergy and Infectious Diseases, the National Multiple Sclerosis Society, the Crohn’s and Colitis Foundation of America, and The Leona M. and Harry B. Helmsley Charitable Trust.
The authors have no conflicting financial interest.
Footnotes
Abbreviations used:
- CNS
- central nervous system
- EAE
- experimental autoimmune encephalomyelitis
- MBP
- myelin basic protein
- mLN
- mesenteric LN
- Nrp1
- Neuropilin 1
- SPF
- specific pathogen–free
- TBmc
- T cell/B cell monoclonal
- TIL
- tumor-infiltrating lymphocyte
- TSDR
- T reg–specific demethylated region
References
- Apostolou I., von Boehmer H. 2004. In vivo instruction of suppressor commitment in naive T cells. J. Exp. Med. 199:1401–1408 10.1084/jem.20040249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K., Tanoue T., Shima T., Imaoka A., Kuwahara T., Momose Y., Cheng G., Yamasaki S., Saito T., Ohba Y., et al. 2011. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 331:337–341 10.1126/science.1198469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Bucktrout S.L., Bluestone J.A. 2011. Regulatory T cells: stability revisited. Trends Immunol. 32:301–306 10.1016/j.it.2011.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett C.L., Christie J., Ramsdell F., Brunkow M.E., Ferguson P.J., Whitesell L., Kelly T.E., Saulsbury F.T., Chance P.F., Ochs H.D. 2001. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 27:20–21 10.1038/83713 [DOI] [PubMed] [Google Scholar]
- Bettelli E., Carrier Y., Gao W., Korn T., Strom T.B., Oukka M., Weiner H.L., Kuchroo V.K. 2006. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 441:235–238 10.1038/nature04753 [DOI] [PubMed] [Google Scholar]
- Bilate A.B., Lafaille J.J. 2011. It takes two to tango. Immunity. 35:6–8 10.1016/j.immuni.2011.07.003 [DOI] [PubMed] [Google Scholar]
- Bilate A.M., Lafaille J.J. 2012. Induced CD4+Foxp3+ regulatory T cells in immune tolerance. Annu. Rev. Immunol. 30:733–758 10.1146/annurev-immunol-020711-075043 [DOI] [PubMed] [Google Scholar]
- Bluestone J.A., Abbas A.K. 2003. Natural versus adaptive regulatory T cells. Nat. Rev. Immunol. 3:253–257 10.1038/nri1032 [DOI] [PubMed] [Google Scholar]
- Bruder D., Probst-Kepper M., Westendorf A.M., Geffers R., Beissert S., Loser K., von Boehmer H., Buer J., Hansen W. 2004. Neuropilin-1: a surface marker of regulatory T cells. Eur. J. Immunol. 34:623–630 10.1002/eji.200324799 [DOI] [PubMed] [Google Scholar]
- Brunkow M.E., Jeffery E.W., Hjerrild K.A., Paeper B., Clark L.B., Yasayko S.A., Wilkinson J.E., Galas D., Ziegler S.F., Ramsdell F. 2001. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 27:68–73 10.1038/83784 [DOI] [PubMed] [Google Scholar]
- Cao X. 2010. Regulatory T cells and immune tolerance to tumors. Immunol. Res. 46:79–93 10.1007/s12026-009-8124-7 [DOI] [PubMed] [Google Scholar]
- Chen W., Jin W., Hardegen N., Lei K.J., Li L., Marinos N., McGrady G., Wahl S.M. 2003. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J. Exp. Med. 198:1875–1886 10.1084/jem.20030152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbold S.P., Castejon R., Adams E., Zelenika D., Graca L., Humm S., Waldmann H. 2004. Induction of foxP3+ regulatory T cells in the periphery of T cell receptor transgenic mice tolerized to transplants. J. Immunol. 172:6003–6010 [DOI] [PubMed] [Google Scholar]
- Coombes J.L., Siddiqui K.R., Arancibia-Cárcamo C.V., Hall J., Sun C.M., Belkaid Y., Powrie F. 2007. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J. Exp. Med. 204:1757–1764 10.1084/jem.20070590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbel C., Lemarchandel V., Thomas-Vaslin V., Pelus A.S., Agboton C., Roméo P.H. 2007. Neuropilin 1 and CD25 co-regulation during early murine thymic differentiation. Dev. Comp. Immunol. 31:1082–1094 10.1016/j.dci.2007.01.009 [DOI] [PubMed] [Google Scholar]
- Curiel T.J., Coukos G., Zou L., Alvarez X., Cheng P., Mottram P., Evdemon-Hogan M., Conejo-Garcia J.R., Zhang L., Burow M., et al. 2004. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med. 10:942–949 10.1038/nm1093 [DOI] [PubMed] [Google Scholar]
- Curotto de Lafaille M.A., Lafaille J.J. 2009. Natural and adaptive foxp3+ regulatory T cells: more of the same or a division of labor? Immunity. 30:626–635 10.1016/j.immuni.2009.05.002 [DOI] [PubMed] [Google Scholar]
- Curotto de Lafaille M.A., Muriglan S., Sunshine M.J., Lei Y., Kutchukhidze N., Furtado G.C., Wensky A.K., Olivares-Villagómez D., Lafaille J.J. 2001. Hyper immunoglobulin E response in mice with monoclonal populations of B and T lymphocytes. J. Exp. Med. 194:1349–1359 10.1084/jem.194.9.1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curotto de Lafaille M.A., Lino A.C., Kutchukhidze N., Lafaille J.J. 2004. CD25- T cells generate CD25+Foxp3+ regulatory T cells by peripheral expansion. J. Immunol. 173:7259–7268 [DOI] [PubMed] [Google Scholar]
- Curotto de Lafaille M.A., Kutchukhidze N., Shen S., Ding Y., Yee H., Lafaille J.J. 2008. Adaptive Foxp3+ regulatory T cell-dependent and -independent control of allergic inflammation. Immunity. 29:114–126 10.1016/j.immuni.2008.05.010 [DOI] [PubMed] [Google Scholar]
- Deiss L.P., Feinstein E., Berissi H., Cohen O., Kimchi A. 1995. Identification of a novel serine/threonine kinase and a novel 15-kD protein as potential mediators of the gamma interferon-induced cell death. Genes Dev. 9:15–30 10.1101/gad.9.1.15 [DOI] [PubMed] [Google Scholar]
- Duarte J.H., Zelenay S., Bergman M.L., Martins A.C., Demengeot J. 2009. Natural Treg cells spontaneously differentiate into pathogenic helper cells in lymphopenic conditions. Eur. J. Immunol. 39:948–955 10.1002/eji.200839196 [DOI] [PubMed] [Google Scholar]
- Faria A.M., Weiner H.L. 2005. Oral tolerance. Immunol. Rev. 206:232–259 10.1111/j.0105-2896.2005.00280.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floess S., Freyer J., Siewert C., Baron U., Olek S., Polansky J., Schlawe K., Chang H.D., Bopp T., Schmitt E., et al. 2007. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 5:e38 10.1371/journal.pbio.0050038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furtado G.C., Olivares-Villagómez D., Curotto de Lafaille M.A., Wensky A.K., Latkowski J.A., Lafaille J.J. 2001. Regulatory T cells in spontaneous autoimmune encephalomyelitis. Immunol. Rev. 182:122–134 10.1034/j.1600-065X.2001.1820110.x [DOI] [PubMed] [Google Scholar]
- Furtado G.C., Marcondes M.C., Latkowski J.A., Tsai J., Wensky A., Lafaille J.J. 2008. Swift entry of myelin-specific T lymphocytes into the central nervous system in spontaneous autoimmune encephalomyelitis. J. Immunol. 181:4648–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinka Y., Stoilova S., Mohammed N., Prud’homme G.J. 2011. Neuropilin-1 exerts co-receptor function for TGF-beta-1 on the membrane of cancer cells and enhances responses to both latent and active TGF-beta. Carcinogenesis. 32:613–621 10.1093/carcin/bgq281 [DOI] [PubMed] [Google Scholar]
- Hadis U., Wahl B., Schulz O., Hardtke-Wolenski M., Schippers A., Wagner N., Müller W., Sparwasser T., Förster R., Pabst O. 2011. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity. 34:237–246 10.1016/j.immuni.2011.01.016 [DOI] [PubMed] [Google Scholar]
- Haribhai D., Lin W., Edwards B., Ziegelbauer J., Salzman N.H., Carlson M.R., Li S.H., Simpson P.M., Chatila T.A., Williams C.B. 2009. A central role for induced regulatory T cells in tolerance induction in experimental colitis. J. Immunol. 182:3461–3468 10.4049/jimmunol.0802535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haribhai D., Williams J.B., Jia S., Nickerson D., Schmitt E.G., Edwards B., Ziegelbauer J., Yassai M., Li S.H., Relland L.M., et al. 2011. A requisite role for induced regulatory T cells in tolerance based on expanding antigen receptor diversity. Immunity. 35:109–122 10.1016/j.immuni.2011.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J.A., Feuerer M., Tash K., Haxhinasto S., Perez J., Melamed R., Mathis D., Benoist C. 2007. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 27:786–800 10.1016/j.immuni.2007.09.010 [DOI] [PubMed] [Google Scholar]
- Hindley J.P., Ferreira C., Jones E., Lauder S.N., Ladell K., Wynn K.K., Betts G.J., Singh Y., Price D.A., Godkin A.J., et al. 2011. Analysis of the T-cell receptor repertoires of tumor-infiltrating conventional and regulatory T cells reveals no evidence for conversion in carcinogen-induced tumors. Cancer Res. 71:736–746 10.1158/0008-5472.CAN-10-1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huehn J., Polansky J.K., Hamann A. 2009. Epigenetic control of FOXP3 expression: the key to a stable regulatory T-cell lineage? Nat. Rev. Immunol. 9:83–89 10.1038/nri2474 [DOI] [PubMed] [Google Scholar]
- Ivanov I.I., Atarashi K., Manel N., Brodie E.L., Shima T., Karaoz U., Wei D., Goldfarb K.C., Santee C.A., Lynch S.V., et al. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 139:485–498 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz S.Z., Niec R.E., Kim H.Y., Treuting P., Chinen T., Zheng Y., Umetsu D.T., Rudensky A.Y. 2012. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 482:395–399 10.1038/nature10772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.M., Rasmussen J.P., Rudensky A.Y. 2007. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 8:191–197 10.1038/ni1428 [DOI] [PubMed] [Google Scholar]
- Komatsu N., Mariotti-Ferrandiz M.E., Wang Y., Malissen B., Waldmann H., Hori S. 2009. Heterogeneity of natural Foxp3+ T cells: a committed regulatory T-cell lineage and an uncommitted minor population retaining plasticity. Proc. Natl. Acad. Sci. USA. 106:1903–1908 10.1073/pnas.0811556106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn T., Reddy J., Gao W., Bettelli E., Awasthi A., Petersen T.R., Bäckström B.T., Sobel R.A., Wucherpfennig K.W., Strom T.B., et al. 2007. Myelin-specific regulatory T cells accumulate in the CNS but fail to control autoimmune inflammation. Nat. Med. 13:423–431 10.1038/nm1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer K., Apostolou I., Hawiger D., Khazaie K., Nussenzweig M.C., von Boehmer H. 2005. Inducing and expanding regulatory T cell populations by foreign antigen. Nat. Immunol. 6:1219–1227 10.1038/ni1265 [DOI] [PubMed] [Google Scholar]
- Lafaille J.J., Nagashima K., Katsuki M., Tonegawa S. 1994. High incidence of spontaneous autoimmune encephalomyelitis in immunodeficient anti-myelin basic protein T cell receptor transgenic mice. Cell. 78:399–408 10.1016/0092-8674(94)90419-7 [DOI] [PubMed] [Google Scholar]
- Lathrop S.K., Bloom S.M., Rao S.M., Nutsch K., Lio C.W., Santacruz N., Peterson D.A., Stappenbeck T.S., Hsieh C.S. 2011. Peripheral education of the immune system by colonic commensal microbiota. Nature. 478:250–254 10.1038/nature10434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.O., Sanjabi S., Flavell R.A. 2006. Transforming growth factor-beta controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity. 25:455–471 10.1016/j.immuni.2006.07.011 [DOI] [PubMed] [Google Scholar]
- Lin W., Haribhai D., Relland L.M., Truong N., Carlson M.R., Williams C.B., Chatila T.A. 2007. Regulatory T cell development in the absence of functional Foxp3. Nat. Immunol. 8:359–368 10.1038/ni1445 [DOI] [PubMed] [Google Scholar]
- Liu V.C., Wong L.Y., Jang T., Shah A.H., Park I., Yang X., Zhang Q., Lonning S., Teicher B.A., Lee C. 2007. Tumor evasion of the immune system by converting CD4+CD25- T cells into CD4+CD25+ T regulatory cells: role of tumor-derived TGF-beta. J. Immunol. 178:2883–2892 [DOI] [PubMed] [Google Scholar]
- Miyao T., Floess S., Setoguchi R., Luche H., Fehling H.J., Waldmann H., Huehn J., Hori S. 2012. Plasticity of Foxp3(+) T cells reflects promiscuous Foxp3 expression in conventional T cells but not reprogramming of regulatory T cells. Immunity. 36:262–275 10.1016/j.immuni.2011.12.012 [DOI] [PubMed] [Google Scholar]
- Mucida D., Kutchukhidze N., Erazo A., Russo M., Lafaille J.J., Curotto de Lafaille M.A. 2005. Oral tolerance in the absence of naturally occurring Tregs. J. Clin. Invest. 115:1923–1933 10.1172/JCI24487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucida D., Park Y., Kim G., Turovskaya O., Scott I., Kronenberg M., Cheroutre H. 2007. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 317:256–260 10.1126/science.1145697 [DOI] [PubMed] [Google Scholar]
- Nishikawa H., Sakaguchi S. 2010. Regulatory T cells in tumor immunity. Int. J. Cancer. 127:759–767 [DOI] [PubMed] [Google Scholar]
- Oldenhove G., Bouladoux N., Wohlfert E.A., Hall J.A., Chou D., Dos Santos L., O’Brien S., Blank R., Lamb E., Natarajan S., et al. 2009. Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity. 31:772–786 10.1016/j.immuni.2009.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares-Villagómez D., Wang Y., Lafaille J.J. 1998. Regulatory CD4(+) T cells expressing endogenous T cell receptor chains protect myelin basic protein-specific transgenic mice from spontaneous autoimmune encephalomyelitis. J. Exp. Med. 188:1883–1894 10.1084/jem.188.10.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polansky J.K., Kretschmer K., Freyer J., Floess S., Garbe A., Baron U., Olek S., Hamann A., von Boehmer H., Huehn J. 2008. DNA methylation controls Foxp3 gene expression. Eur. J. Immunol. 38:1654–1663 10.1002/eji.200838105 [DOI] [PubMed] [Google Scholar]
- Radoja S., Saio M., Frey A.B. 2001. CD8+ tumor-infiltrating lymphocytes are primed for Fas-mediated activation-induced cell death but are not apoptotic in situ. J. Immunol. 166:6074–6083 [DOI] [PubMed] [Google Scholar]
- Round J.L., Mazmanian S.K. 2010. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. USA. 107:12204–12209 10.1073/pnas.0909122107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov Y.P., Niec R.E., Josefowicz S., Li L., Darce J., Mathis D., Benoist C., Rudensky A.Y. 2010. Stability of the regulatory T cell lineage in vivo. Science. 329:1667–1671 10.1126/science.1191996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S., Sakaguchi N., Asano M., Itoh M., Toda M. 1995. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J. Immunol. 155:1151–1164 [PubMed] [Google Scholar]
- Sarris M., Andersen K.G., Randow F., Mayr L., Betz A.G. 2008. Neuropilin-1 expression on regulatory T cells enhances their interactions with dendritic cells during antigen recognition. Immunity. 28:402–413 10.1016/j.immuni.2008.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M.D., Hou D.Y., Baban B., Koni P.A., He Y., Chandler P.R., Blazar B.R., Mellor A.L., Munn D.H. 2010. Reprogrammed foxp3(+) regulatory T cells provide essential help to support cross-presentation and CD8(+) T cell priming in naive mice. Immunity. 33:942–954 10.1016/j.immuni.2010.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C.M., Hall J.A., Blank R.B., Bouladoux N., Oukka M., Mora J.R., Belkaid Y. 2007. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J. Exp. Med. 204:1775–1785 10.1084/jem.20070602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Kuniyasu Y., Toda M., Sakaguchi N., Itoh M., Iwata M., Shimizu J., Sakaguchi S. 1998. Immunologic self-tolerance maintained by CD25+CD4+ naturally anergic and suppressive T cells: induction of autoimmune disease by breaking their anergic/suppressive state. Int. Immunol. 10:1969–1980 10.1093/intimm/10.12.1969 [DOI] [PubMed] [Google Scholar]
- Thornton A.M., Shevach E.M. 1998. CD4+CD25+ immunoregulatory T cells suppress polyclonal T cell activation in vitro by inhibiting interleukin 2 production. J. Exp. Med. 188:287–296 10.1084/jem.188.2.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton A.M., Korty P.E., Tran D.Q., Wohlfert E.A., Murray P.E., Belkaid Y., Shevach E.M. 2010. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J. Immunol. 184:3433–3441 10.4049/jimmunol.0904028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valzasina B., Piconese S., Guiducci C., Colombo M.P. 2006. Tumor-induced expansion of regulatory T cells by conversion of CD4+CD25- lymphocytes is thymus and proliferation independent. Cancer Res. 66:4488–4495 10.1158/0008-5472.CAN-05-4217 [DOI] [PubMed] [Google Scholar]
- Wildin R.S., Ramsdell F., Peake J., Faravelli F., Casanova J.L., Buist N., Levy-Lahad E., Mazzella M., Goulet O., Perroni L., et al. 2001. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat. Genet. 27:18–20 10.1038/83707 [DOI] [PubMed] [Google Scholar]
- Wilke C.M., Wu K., Zhao E., Wang G., Zou W. 2010. Prognostic significance of regulatory T cells in tumor. Int. J. Cancer. 127:748–758 [DOI] [PubMed] [Google Scholar]
- Zelenay S., Bergman M.L., Paiva R.S., Lino A.C., Martins A.C., Duarte J.H., Moraes-Fontes M.F., Bilate A.M., Lafaille J.J., Demengeot J. 2010. Cutting edge: Intrathymic differentiation of adaptive Foxp3+ regulatory T cells upon peripheral proinflammatory immunization. J. Immunol. 185:3829–3833 10.4049/jimmunol.1001281 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Josefowicz S., Chaudhry A., Peng X.P., Forbush K., Rudensky A.Y. 2010. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 463:808–812 10.1038/nature08750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G., Levitsky H.I. 2007. Natural regulatory T cells and de novo-induced regulatory T cells contribute independently to tumor-specific tolerance. J. Immunol. 178:2155–2162 [DOI] [PubMed] [Google Scholar]
- Zhou L., Lopes J.E., Chong M.M., Ivanov I.I., Min R., Victora G.D., Shen Y., Du J., Rubtsov Y.P., Rudensky A.Y., et al. 2008. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 453:236–240 10.1038/nature06878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Bailey-Bucktrout S.L., Jeker L.T., Penaranda C., Martínez-Llordella M., Ashby M., Nakayama M., Rosenthal W., Bluestone J.A. 2009. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat. Immunol. 10:1000–1007 10.1038/ni.1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.