In the absence of adaptive immunity, NK cells polarize M1 macrophages to facilitate cancer immunoediting.
Abstract
Cancer immunoediting is the process whereby immune cells protect against cancer formation by sculpting the immunogenicity of developing tumors. Although the full process depends on innate and adaptive immunity, it remains unclear whether innate immunity alone is capable of immunoediting. To determine whether the innate immune system can edit tumor cells in the absence of adaptive immunity, we compared the incidence and immunogenicity of 3′methylcholanthrene-induced sarcomas in syngeneic wild-type, RAG2−/−, and RAG2−/−x γc−/− mice. We found that innate immune cells could manifest cancer immunoediting activity in the absence of adaptive immunity. This activity required natural killer (NK) cells and interferon γ (IFN-γ), which mediated the induction of M1 macrophages. M1 macrophages could be elicited by administration of CD40 agonists, thereby restoring editing activity in RAG2−/−x γc−/− mice. Our results suggest that in the absence of adaptive immunity, NK cell production of IFN-γ induces M1 macrophages, which act as important effectors during cancer immunoediting.
Immune cells can infiltrate a developing tumor mass and either promote or inhibit tumorigenesis (Balkwill and Coussens, 2004; Ben-Neriah and Karin, 2011; Schreiber et al., 2011). Cancer immunoediting describes the process whereby the interaction between immune cells and tumor cells either eliminates the developing tumor, holds it in a state of growth dormancy, or generates a tumor cell repertoire that is capable of survival in immune-competent hosts (Shankaran et al., 2001; Dunn et al., 2004b; Vesely et al., 2011). Several studies have revealed the contribution of adaptive and innate immunity in cancer immunoediting (Shankaran et al., 2001; Dunn et al., 2004a; Smyth et al., 2006; Dunn et al., 2005; Smyth et al., 2005; Street et al., 2004; Crowe et al., 2002; Takeda et al., 2002; Smyth et al. 2001), but it is not clear whether the unmanipulated innate immune system can suppress tumor formation without adaptive immunity.
In this study, we examined the ability of the innate immune system to control tumor formation in the absence of adaptive immunity. It has been shown that natural killer cells (NK; Smyth et al., 2002; Raulet and Guerra, 2009) and classically activated M1 macrophages (Sica et al., 2008; Lewis and Pollard, 2006) support a Th1 response that can ultimately lead to tumor rejection in the presence of adaptive immunity, but it is not clear whether these cells interact in the absence of adaptive immunity to suppress tumor formation in primary tumor models. In contrast, other studies have found that the innate immune system can promote tumor formation via alternatively activated M2 macrophages (Gordon and Taylor, 2005) that augment angiogenesis and promote tissue invasion. M2 macrophages also inhibit the formation of antitumor adaptive immunity, and therefore it is possible that innate immunity would promote tumor formation in the absence of adaptive immunity.
Using the 3′methylcholanthrene (MCA) model of sarcomagenesis, we previously found that the immune system in WT mice could edit tumors more effectively than the immune system in RAG2−/− mice (which lack adaptive immunity; Shankaran et al., 2001; Takahashi and Yamanaka, 2006), but we did not assess whether tumors from RAG2−/− mice were edited by the innate immune system. Because RAG2−/− mice and other immunodeficient mice such as nude and SCID mice are routinely used as “immunodeficient” models for xenotransplantation and preclinical studies, it is critical to assess whether the innate immune system in these mice could have an impact, positive or negative, on tumor growth. Toward this end, we set out to quantitate tumor editing in WT versus RAG2−/− versus RAG2−/−x γc−/− mice.
RAG2−/−x γc−/− mice lack all lymphocytes, including NK, NK-T, γδ-T, classical CD4+,and CD8+ αβ-T cells and B cells, and thus show deficits in both innate and adaptive immunity. If cells of the innate immune system could hinder tumor growth, then we would expect RAG2−/−x γc−/− mice to demonstrate increased tumor incidence and decreased tumor editing compared with RAG2−/− mice. Indeed, when we compared MCA-induced sarcoma incidence and tumor cell immunogenicity between the groups of mice, we found both increased incidence and immunogenicity of MCA-induced sarcomas in RAG2−/−x γc−/− mice compared with RAG2−/− mice, which, consistent with previous results (Shankaran et al., 2001), had increased incidence and immunogenicity of tumors compared with WT mice. When transplanted into RAG2−/− recipients, RAG2−/−x γc−/− regressor sarcoma cell lines formed tumors that became heavily infiltrated with M1 macrophages. The infiltration of M1 macrophages was associated with tumor editing and required host γc and IFN-γ activity. In contrast, in the absence of γc and IFN-γ function, RAG2−/−x γc−/− regressors were infiltrated with more M2 macrophages, which can promote tumor formation (Sica et al., 2008). We also found that M1 macrophages can be elicited by CD40 agonistic antibodies to restore the editing capacity of RAG2−/−x γc−/− mice. These studies document that components of the innate immune system present in RAG2−/− mice can manifest certain types of cancer immunoediting capacity in the absence of adaptive immunity and point, specifically, to M1 macrophages as important effectors in this process.
RESULTS
MCA-induced sarcoma incidence is increased in RAG2−/−x γc−/− mice compared with syngeneic RAG2−/− and WT mice
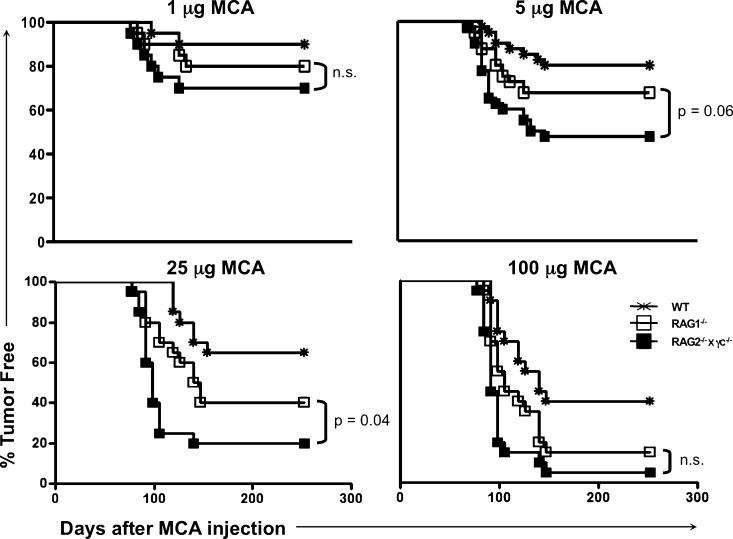
To determine whether the innate immune system of RAG2−/− mice was capable of tumor immunosurveillance, we compared the incidence of MCA-induced sarcomas in immunologically intact WT C57BL/6 mice to that of C57BL/6 mice with defects in either adaptive immunity only (RAG2−/− mice) or in both adaptive and innate immunity (RAG2−/−x γc−/− mice). Fig. 1 shows that the incidence of sarcomas was higher in RAG2−/−x γc−/− mice compared with RAG2−/− mice at all doses tested. For example, at a dose of 25 µg MCA, the incidence of sarcomas in WT, RAG2−/−, and RAG2−/−x γc−/− mice was 35, 60, and 80%, respectively. In addition, at MCA doses of 25 or 100 µg, RAG2−/−x γc−/− mice developed sarcomas slightly faster than RAG2−/− mice, indicating that the innate immune system in RAG2−/− mice controlled MCA-induced tumor outgrowth to some extent. The difference in tumor incidence between cohorts of mice were not caused by inherent strain differences, as heterozygote RAG+/− littermates did not show differences compared with WT mice (unpublished data).
Figure 1.
RAG2−/−x γc−/− mice are more susceptible to MCA-induced sarcomas than syngeneic RAG2−/− and WT mice. The indicated dose of MCA was injected into the subcutaneous space of mice, and sarcoma formation was monitored over time. All cohorts consisted of 20 mice. Tumor-positive mice were defined as those that harbored a progressively growing mass ≥25 mm2. Similar results were found in a repeat experiment that included the 5 and 25 µg doses.
Growth of MCA-induced sarcoma cell lines derived from RAG2−/−x γc−/− mice is inhibited when transplanted into syngeneic WT mice
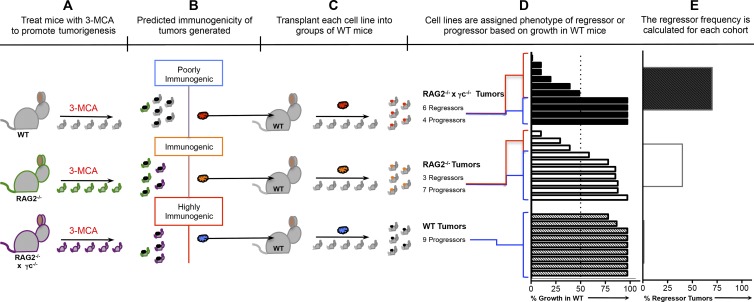
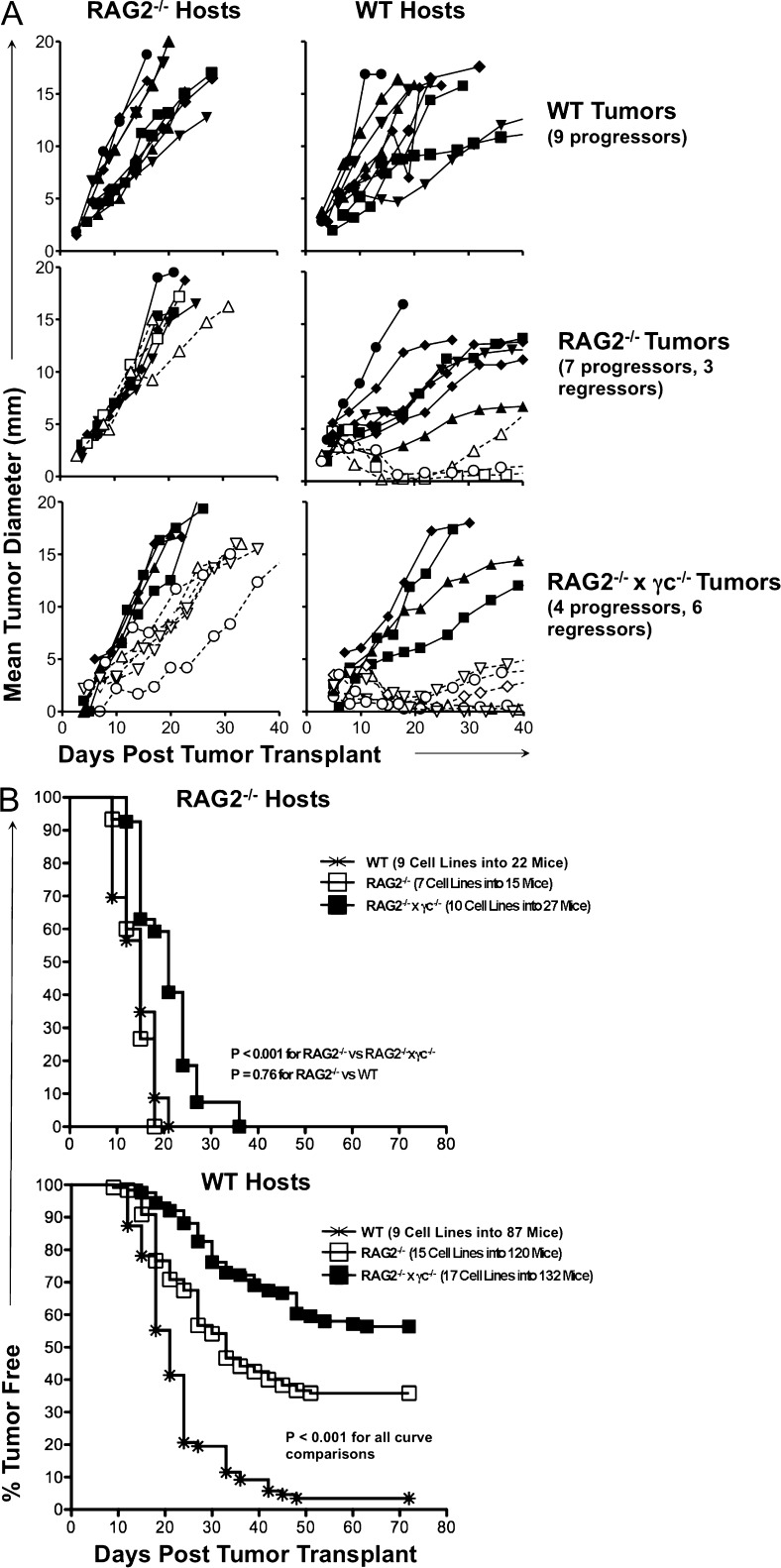
To study tumor editing, low-passage cell lines were derived from primary MCA tumor masses generated in C57BL/6 WT, RAG2−/−, and RAG2−/−x γc−/− mice, and the immunogenicity of each cell line was assessed by transplanting them into naive WT syngeneic mice and monitoring their growth (Fig. 2). As described previously (Kripke, 1974; Boon and Kellermann, 1977; Flood et al., 1987; Shankaran et al., 2001; Dunn et al., 2005), we observed two divergent growth phenotypes among the transplanted sarcomas: a regressor phenotype, defined by a failure to form a mass of >9 mm in diameter in >50% of transplantations into syngeneic WT mice, and a progressor phenotype, defined by the formation of masses >9 mm in >50% of transplantations into WT mice. When we examined groups of MCA-induced sarcoma cell lines generated from WT, RAG2−/−, and RAG2−/−x γc−/− mice, we found that the proportion of regressor MCA-induced sarcoma cell lines was 0/9 WT, 3/10 RAG2−/−, and 6/10 RAG2−/−x γc−/− (Fig. 3 A, right). All cell lines grew when transplanted into RAG2−/− mice (Fig. 3 A, left), indicating that their rejection was caused by the adaptive immune system and was not simply a failure to grow in vivo.
Figure 2.
Generation of MCA sarcoma cell lines of varying immunogenicities to study cancer immunoediting by innate and adaptive immunity. (A) The carcinogen MCA is administered to syngeneic mice with three levels of immune function. (n ≥ 20 mice for each cohort). (B) The immunogenicity of the MCA sarcomas is postulated to be heterogeneous. (C) MCA sarcoma cell lines are generated from each tumor mass and transplanted into syngeneic WT mice (n > 5 for each cell line) to assess growth. (D) Shown is the % growth of each cell line after transplant into WT mice. Cell lines that grow in <51% of the mice are termed regressors. Cell lines that grow in >50% of mice are termed progressors. (E) For each genotype, the percentage of tumor cell lines that displayed a regressor growth pattern is plotted. The “percent regressor tumors” is postulated to be inversely correlated with the quantity of immune pressure that is occurring during tumor formation in each mouse genotype.
Figure 3.
A majority of MCA-induced sarcoma cell lines derived from RAG2−/−x γc−/− mice cannot form tumors when transplanted into syngeneic WT mice. MCA-induced sarcoma cell lines were derived from tumors generated in syngeneic C57BL/6-strain WT, RAG2−/−, and RAG2−/−x γc−/− mice. These cell lines were transplanted into syngeneic RAG2−/− (n ≥ 2 for each cell line) or WT (n ≥ 5 for each cell line) hosts, and tumor growth was measured over time. (A) The mean growth for each cell line is shown (open symbols = regressor cell lines; closed symbols = progressor cell lines). (B) The percentage of WT mice that developed tumors is shown for group of cell lines. Tumor-free mice were defined as having a nonenlarging mass <9 mm in average diameter. The number of cell lines and mice are indicated in the figure.
To determine the overall immunogenicity of each group of tumors, we examined the tumor-free survival of large cohorts of WT and RAG2−/− mice challenged with panels of tumor cell lines derived from WT, RAG−/−, or RAG2−/−x γc−/− mice (Fig. 3 B). All MCA-induced sarcoma cell lines formed tumors in RAG2−/− mice by 36 d after tumor cell transplant (Fig. 3 B, top). In contrast, the kinetics and frequency of tumor formation in WT recipients was dependent on the level of immune function of the original source from which the tumor cells were derived. Specifically, when 17 tumor cell lines derived from RAG2−/−x γc−/− mice were transplanted into a total of 132 naive, syngeneic WT mice, only 46% of the mice formed tumors by 70 d after transplant (Fig. 3 B, bottom; P < 0.001 for all comparisons). Over a similar time course, MCA-induced sarcoma cell lines from 15 RAG2−/− and 9 WT mice formed tumors in 64 and 97% of WT recipients, respectively. These results were reproduced in an independent MCA induction experiment, and the combined results of these two experiments, encompassing 71 total MCA-induced sarcoma cell lines transplanted into 474 WT mice, 94 RAG2−/− mice, or 51 RAG2−/−x γc−/− mice, are shown in Table 1. Altogether, these results support the hypothesis that tumors from mice with greater immunodeficiency undergo decreased levels of immunoediting.
Table 1.
A summary of two independent MCA induction immunoediting experiments
| Tumor group | Growth in WT | Growth in RAG−/− | Growth in RAG−/−xγc−/− |
| 9 WT tumors into 87 WT or 22 RAG hosts (exp 1) | 97% (84/87) | 100% (22/22) | ND |
| 15 RAG tumors into 120 WT or 7 into 15 RAG hosts (exp 1) | 64% (77/120) | 100% (15/15) | ND |
| 17 RAGxγc tumors into 132 WT or 10 into 27 RAG hosts (exp 1) | 46% (61/132) | 100% (27/27) | ND |
| 10 WT tumors into 35 WT or 21 RAGxγc hosts(exp 2) | 100% (35/35) | ND | 100% (21/21) |
| 10 RAG tumors into 50 WT or 30 RAG hosts (exp 2) | 60% (30/50) | 100% (30/30) | ND |
| 10 RAGxγc tumors into 50 WT or 30 RAGxγc hosts (exp 2) | 30% (15/50) | ND | 100% (30/30) |
A total of 71 MCA sarcoma cell lines were generated from the indicated mice and then transplanted into 474 WT, RAG−/−, or RAG2−/−x γc−/− mice, and tumor growth was monitored. Regressor frequencies from these experiments (1 and 2) are shown in Fig. 4. ND, not determined.
Tumor cell lines generated in RAG2−/−x γc−/− mice show an increased regressor frequency compared with cell lines from WT and RAG2−/− mice
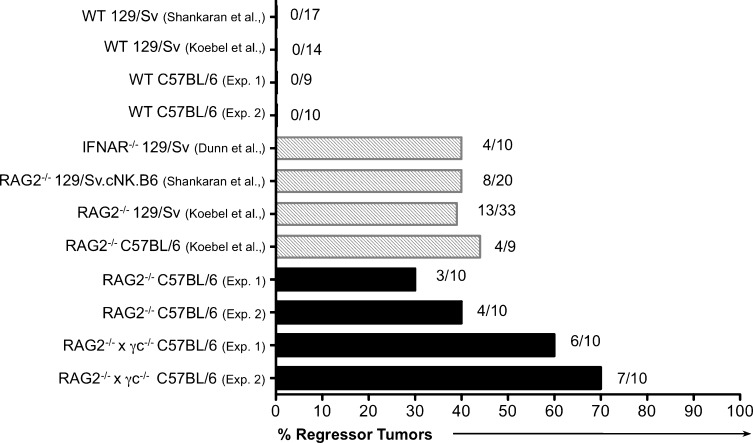
We reported previously that the percentage of regressors within a group of MCA-induced sarcoma cell lines, the “regressor frequency” (Fig. 2) was 40% when the MCA-induced sarcoma cell lines were generated in RAG2−/− mice and 0% when the cell lines were generated in WT mice (Shankaran et al., 2001). These percentages are remarkably reproducible, and have remained so even when experiments have been conducted in our three independent laboratories in La Jolla, CA, St. Louis, MO, and Melbourne, Australia (Fig. 4). Specifically, we found a consistent regressor frequency of 0% when MCA-induced sarcoma cell lines are generated in WT mice (50 cell lines from two strains and four independent experiments). Notably, MCA-induced sarcoma cell lines derived from RAG2−/− mice displayed a 30–44% regressor frequency (82 cell lines from 3 strains and 4 independent experiments). MCA-induced sarcoma cell lines derived from RAG2−/−x γc−/− mice had the highest regressor frequency (60–70%), indicating that as a group, these cell lines were the most immunogenic and least edited.
Figure 4.
The frequency of regressor cell lines is greater from tumors generated in RAG2−/−x γc−/− mice compared with WT and other immune deficient mice. A summary of two MCA-induction experiments performed in this manuscript is plotted in the context of previous MCA-induction experiments. Previously published experiments are included for comparison purposes and are from previous studies (Shankaran et al., 2001; Dunn et al., 2005; Koebel et al., 2007). Absolute numbers of regressors/total number of cell lines tested is shown next to the bar for each experiment.
RAG2−/−x γc−/− regressors undergo editing when transplanted into RAG2−/− mice
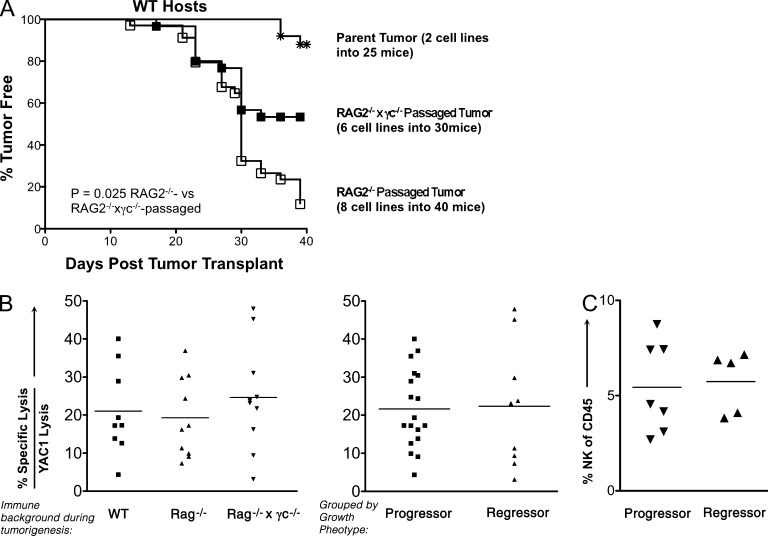
Because regressor cell lines generated from RAG2−/−x γc−/− mice displayed the highest levels of immunogenicity and, subsequently, the lowest levels of immunoediting compared with RAG2−/− and WT mice, we hypothesized that the innate immune system of RAG2−/− mice could edit these tumor cell lines in vivo. We tested this by transplanting two independent sarcoma cell lines generated from RAG2−/−x γc−/− mice into either RAG2−/− or RAG2−/−x γc−/− mice. To determine if in vivo passaging altered the immunogenicity of these cell lines, tumor masses were harvested at day 25 and converted into cell lines. When these cell lines were transplanted into WT mice, 88% of RAG2−/−-passaged tumor cell lines formed progressively growing tumor masses by day 40 compared with 46% of RAG2−/−x γc−/−–passaged and 10% of unpassaged cell lines (Fig. 5 A; P = 0.025). These results suggest a higher level of editing by the innate immune system in RAG2−/− versus RAG2−/−x γc−/− mice but also indicate that there is some level of measurable tumor sculpting in RAG2−/−x γc−/− mice, which could be caused by residual immune function or a nonimmunological editing process.
Figure 5.
RAG2−/−x γc−/− regressors are edited when transplanted into RAG2−/− mice, but are not specifically recognized by NK cells. Two independent MCA-induced sarcoma cell lines generated from RAG2−/−x γc−/− mice were transplanted into syngeneic RAG2−/−x γc−/− or RAG2−/− mice, and tumor masses were harvested at day 25 and converted into “passaged” daughter cell lines, which were transplanted into syngeneic WT mice (number of cell lines and mice are shown in the figure), and (A) the percentage of WT mice that remained tumor-free is shown for each group of cell lines. Tumor-free mice were defined to have a nonenlarging mass <9 mm in average diameter by day 40. (B) MCA sarcoma cell lines were cultured with IL-2–activated NK cells in a 5-h chromium release cytotoxicity assay and specific lysis normalized to YAC-1–specific lysis is plotted for each cell line based on immune background and phenotype. (C) Regressor and progressor cell lines were transplanted into RAG-deficient mice and analyzed for infiltrating NK cells by FACS.
NK cells do not preferentially kill regressor versus progressor tumor cells
Having shown that γc is important for the ability of innate immunity to control and edit MCA-induced sarcomas, we predicted that NK cells, dependent on γc for development (Cao et al., 1995), would participate in this editing process in vivo. To explore whether NK cells preferentially recognize regressors over progressors, we performed standard chromium release cytotoxicity assays (Bui et al., 2006) and also examined the NK cell content in regressor versus progressor tumors. We found that the overall susceptibility to NK cell killing of 10 MCA-induced sarcoma cell lines from RAG2−/−x γc−/− mice did not differ from that of 10 MCA-induced sarcoma cell lines from RAG2−/−, or 9 MCA-induced sarcomas from WT mice (Fig. 5 B). Even when all tumors were grouped based on phenotypic growth in WT mice—grouped into progressors or regressors—we observed no difference in NK cell–specific lysis (Fig. 5 B). Additionally, we did not detect a difference in NK1.1+ cell infiltration (∼5%) into any of the MCA-induced sarcomas after they were transplanted into RAG2−/− mice (Fig. 5 C).
MHC class II–positive macrophages are selectively present in regressor tumors during immunoediting
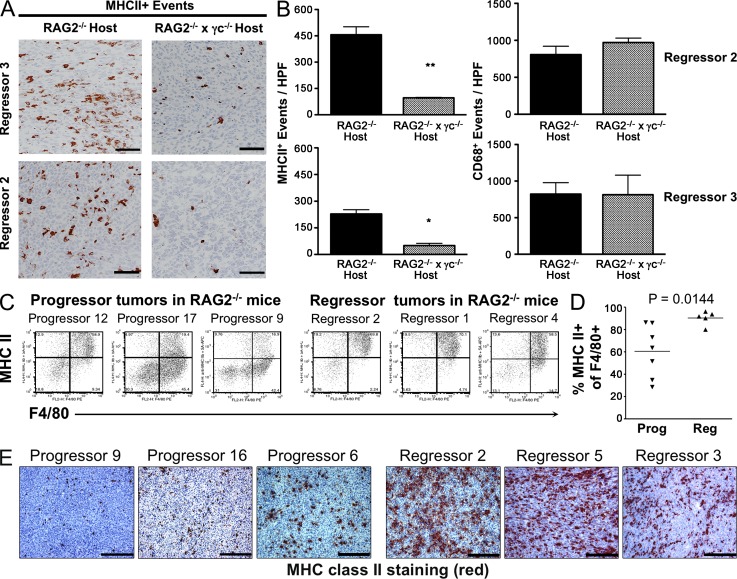
We therefore redirected our focus on myeloid cells, as they represent the major hematopoietic lineage cell type that infiltrates either rejecting or progressively growing tumors (Sica et al., 2008). To examine this issue, two RAG2−/−x γc−/− regressor cell lines were transplanted into either RAG2−/− or RAG2−/−x γc−/− hosts, and tumors were harvested at day 15 and analyzed by immunohistochemistry (IHC) to assess the number and phenotypes of infiltrating myeloid cells. Strikingly, we observed significantly higher numbers of MHC class II–positive cells in tumors growing in RAG2−/− versus RAG2−/−x γc−/− hosts (Fig. 6, A and B; P = 0.00156 and 0.0071, respectively). This was likely caused by increased MHC class II induction rather than an increase in macrophages, as no differences were detected in the total number of macrophages infiltrating tumors growing in either RAG2−/− or RAG2−/−x γc−/− hosts, as detected by the tissue macrophage marker CD68 (Fig. 6 B, right). A similar preferential accumulation of MHC class II–positive cells was also observed in unedited versus edited tumors growing in RAG2−/− mice (Fig. 6, C–E). In these studies, total monocyte-lineage cells (as marked flow cytometrically by F4/80) was similar between edited and unedited tumors, but the percentage of F4/80+ cells that expressed high levels of MHC class II was higher in unedited versus edited tumors.
Figure 6.
MHC class II+ macrophages preferentially infiltrate unedited regressors. (A) Representative images of tumor sections from RAG2−/− or RAG2−/−x γc−/− hosts stained for MHC class II. (B) Quantification of MHC class II+ events and CD68+ events in tumor sections is shown. (C–E) Regressor and progressor cell lines were transplanted into RAG2−/− mice and analyzed for activated MHC class II+ macrophages. (C) Representative FACS plots of three regressor and three progressor tumors are shown. Cells were gated on a CD45+PI− population. (D) Percentages of activated monocyte-lineage (F4/80+) cell populations are shown for regressor and progressor tumor masses. Each symbol represents a different tumor cell line transplanted into 1–3 RAG2−/− mice. (E) Frozen tumor sections of progressor and regressor tumor masses growing in RAG2−/− mice were stained for MHC class II. Nuclei were counterstained with hematoxylin. Bar, 100 μm. **, P < 0.01. Error bars are represented by ± SEM. IHC results were reproduced at least once.
Editing of regressor tumor cells from RAG2−/−x γc−/− mice and induction of MHC class II on tumor-infiltrating cells requires NK cells and IFN-γ production in vivo
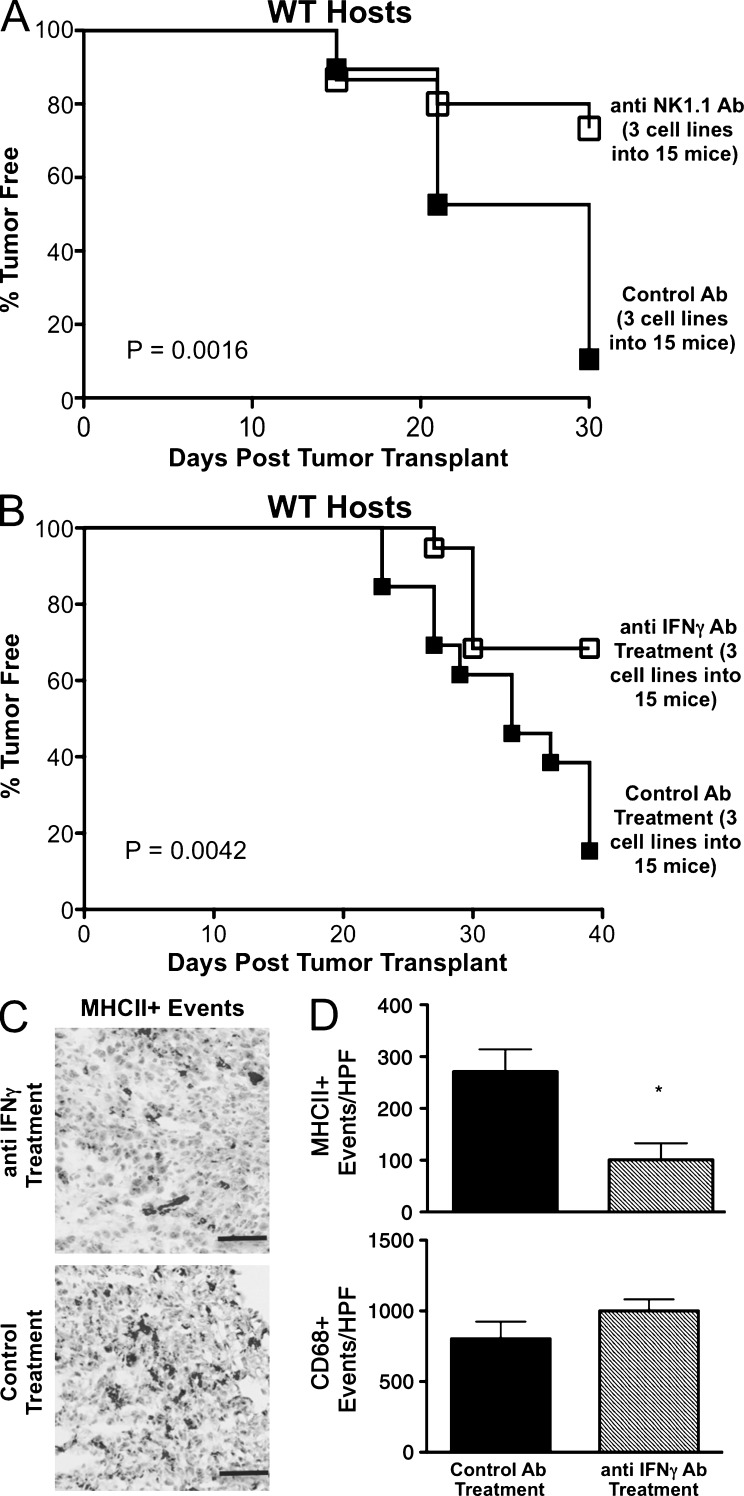
Because γc was important for editing, but NK cell–dependent tumor cell killing was not, we hypothesized that NK cell–derived IFN-γ was critical for the editing process we observed in RAG2−/− mice. We therefore transplanted a regressor cell line derived from a RAG2−/−x γc−/− mouse into RAG2−/− recipients treated either with the neutralizing H22 IFN-γ–specific monoclonal antibody (mAb), an NK1.1 specific monoclonal antibody (PK136), or a control mAb (PIP). Tumors were harvested at day 20 and converted into cell lines, which were subsequently transplanted into naive, syngeneic WT hosts to measure tumor-free survival. We observed a statistically significant increase in the survival of WT mice transplanted with MCA-induced sarcomas that had been passaged through NK cell–depleted and IFN-γ–neutralized mice versus control mice (Fig. 7, A and B; P = 0.0042 and 0.0016, respectively), indicating that NK cells and IFN-γ play critical roles in activating the editing capacity of the innate immune system in RAG2−/− mice. Analysis of tumor cross sections by IHC at day 20 showed MHC class II–positive macrophages were significantly reduced with anti–IFN-γ treatment (Fig. 7, C and D; P = 0.0432), even though total macrophage infiltration did not differ between hosts as determined by CD68+ events (Fig. 7 D). These results demonstrate that NK cells and IFN-γ may facilitate editing by activating macrophages.
Figure 7.
NK cells and IFN-γ are necessary for innate editing of a regressor tumor and M1 macrophage accumulation. Regressor cell line 2 was transplanted into RAG2−/− mice treated with anti-NK1.1, IFN-γ–blocking antibody, or control antibody, after which tumor growth was measured and passaged cell lines were generated. (A and B) The passaged cell lines were then transplanted into syngeneic WT hosts (number of cell lines and mice are indicated) and tumor-free survival was measured. Tumor-free mice were defined to have a nonenlarging mass <9 mm in average diameter by day 40. Tumor sections from RAG2−/− hosts were stained for MHC class II (C) and quantitated (D). *, P < 0.05. Error bars are represented by ± SEM. Results were reproduced at least once.
Tumor-associated macrophages (TAMs) from regressor tumors display an M1 phenotype and require NK cells and IFN-γ for polarization in vivo
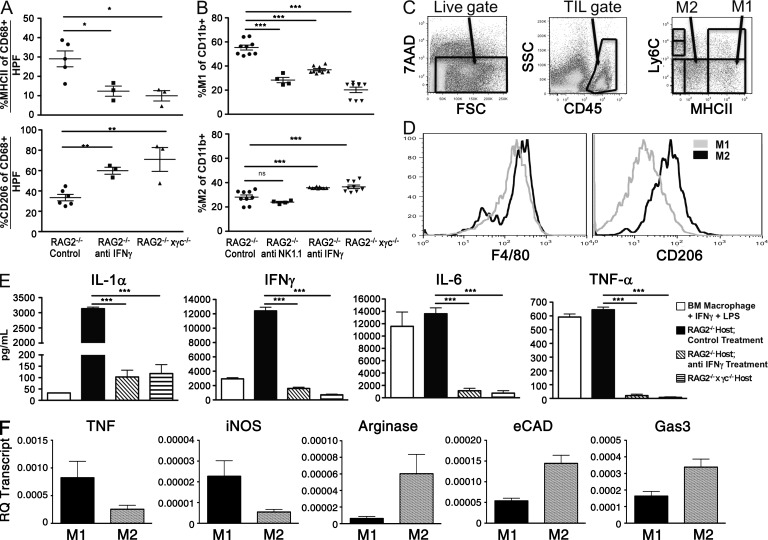
Because the MHC class II+ macrophages required IFN-γ for their accumulation, we hypothesized that these macrophages were classically activated (Gordon and Taylor, 2005) M1 macrophages and performed immunophenotyping to detect the presence of tumor-associated M1 or M2 macrophages, known to have anti- or pro-tumor functions, respectively (Lewis and Pollard, 2006; Sica et al., 2008). For this purpose, we used a combination of IHC and FACS analysis combined with defining cytokine production in freshly harvested tumors. In all cases, we analyzed no fewer than three tumors across at least two experiments. We first performed IHC analysis for the M2-type macrophage marker CD206 and compared the staining pattern to that of MHC class II (known to be up-regulated on M1 macrophages versus M2 macrophages; Fig. 8 A). We found that regressor tumors harvested from RAG2−/− mice had the highest percentage of class II high events (29%) and lowest percentage of CD206+ events (33%). In contrast, tumors harvested from both RAG2−/− mice depleted of IFN-γ or RAG2−/−x γc−/− mice had significantly lower percentages of class II events (12 and 10%, respectively) and significantly higher percentages of CD206+ events (60 and 70%, respectively; Fig. 8 A). Thus, IHC analysis suggested that M1-phenotype macrophage accumulation within tumors requires both IFN-γ and γc. We next used FACS analysis to gate on TAM subsets using combinations of CD11b, Ly6C, and MHC class II to differentiate between M1 and M2 macrophages (Fig. 8 C) as previously described (Movahedi et al., 2010). This gating strategy identified M1 macrophages as MHC class IIhi, Ly6Clo, CD206lo, F4/80hi cells and M2 macrophages as MHC class IIlo, Ly6Clo, CD206hi, F4/80hi cells (Fig. 8, C and D). This analysis showed that regressor tumors contained significantly higher percentages of M1 macrophages when isolated from RAG2−/− mice treated with control mAb PIP (56%) compared with either RAG2−/− mice treated with anti-NK1.1 mAb (28%), neutralizing IFN-γ mAb (37%), or RAG2−/−x γc−/− (20%) mice (Fig. 8 B, top; P < 0.0001 for all populations). Conversely, M2 macrophage percentages were slightly increased in tumors isolated from RAG2−/− mice treated with anti–IFN-γ (36%) and RAG2−/−x γc−/− mice (37%), but not anti-NK1.1–treated mice (27%) compared with control RAG2−/− mice (28%; Fig. 8 B, bottom; P = 0.0007 and 0.002, respectively), confirming our IHC results. Tumor cell suspensions isolated from the different groups of mice did not show differences in total numbers of CD45+ or CD11b+ cells (unpublished data), thus ruling out the possibility that the differences observed in TAM subsets were caused by differential recruitment of immune cells in mice lacking either IFN-γ or γc function.
Figure 8.
NK cells and IFN-γ are required to polarize TAMs toward an M1-type phenotype. Regressor cell line 2 was transplanted into syngeneic RAG2−/− mice (injected with isotype control, anti-NK1.1, or anti–IFN-γ monoclonal antibodies) or RAG2−/−x γc−/− mice. Tumor masses were harvested 15 d after transplantation, disaggregated into single-cell suspensions, and analyzed by IHC (A) or FACS (B) to measure the percentage of M1 and M2 macrophages as defined by MHC class II and CD206 expression of CD68+ events (for IHC) or MHC class II and Ly6C expression of CD11b+ populations (for FACS), respectively. (C and D) An example of the flow cytometry gating to quantitate M1 and M2 macrophages. M1 macrophages are 7AAD−, CD45+, Ly 6Clo, MHC class IIhi, F4/80+, CD206lo cells. M2 macrophages are 7AAD−, CD45+, Ly6Clo, MHC classIIlo, F4/80+, CD206hi cells. (E) Cultured supernatant from single-cell suspensions were assessed for production of the indicated cytokines after 24 h of culture. (F) TAMs were sorted from harvested tumors at day 15, and genes associated with classically activated M1 type genes, such as iNOS and TNF, or alternatively activated M2 type genes, such as Arginase, eCAD, and Gas3, were measured by quantitative PCR. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars are represented by ± SEM. Each symbol represents a different mouse. Results were reproduced at least once.
To test a functional marker of TAM polarization, we examined supernatant of matched tumor cell suspensions cultured in vitro. Cell suspensions from tumors growing in control RAG2−/− mice contained high levels of IL-1α and IFN-γ and produced levels of IL-6 and TNF that were similar to bone marrow–derived macrophages stimulated with LPS and IFN-γ, indicative of a classically activated M1 macrophage cytokine profile. In contrast, cell suspensions derived from tumors derived from anti-IFN-γ–treated RAG2−/− mice and RAG2−/−x γc−/− mice produced significantly lower levels of each cytokine (Fig. 8 E; P < 0.0001 for all comparisons). We did not detect IL-10, or IL-4 production in any of the cultures, indicating that the M2 TAMs are not identical to alternatively activated M2 macrophages found in certain infections. No cytokine production was observed in cultures of the tumor cell line alone (unpublished data). This result demonstrates that the cytokines that were detected in the cell suspensions derived from in vivo growing tumors can be attributed to the immune subsets that infiltrate the tumor.
To identify the phenotype of the infiltrating macrophages, cell suspensions from regressor tumors transplanted into 10 RAG2−/−x γc−/− or 10 RAG2−/− mice were harvested at day 15, and M1 and M2 TAMs were sorted and characterized by quantitative PCR, using markers known to be associated with an M1-type phenotype (TNF and inducible nitric oxide synthase [iNOS]; Martinez et al., 2009) or with an M2-type phenotype (arginase and epithelial cadherin [eCAD]), growth arrest–specific gene 3 (Gas3; Ghassabeh et al., 2006; Fig. 8 F). Macrophages sorted from tumors transplanted into RAG2−/− hosts that were identified as M1-type MHC class IIhi, Ly6Clo, CD206lo, F4/80hi displayed high levels of both TNF and iNOS transcript compared with macrophages sorted from RAG2−/−x γc−/− that were identified as M2-type MHC class IIlo, Ly6Clo, CD206hi, F4/80hi and displayed higher transcript levels of arginase, eCAD, and Gas3.
Polarization of M1 macrophages in vivo by administration of a CD40 agonist induces editing in RAG2−/−x γc−/− mice
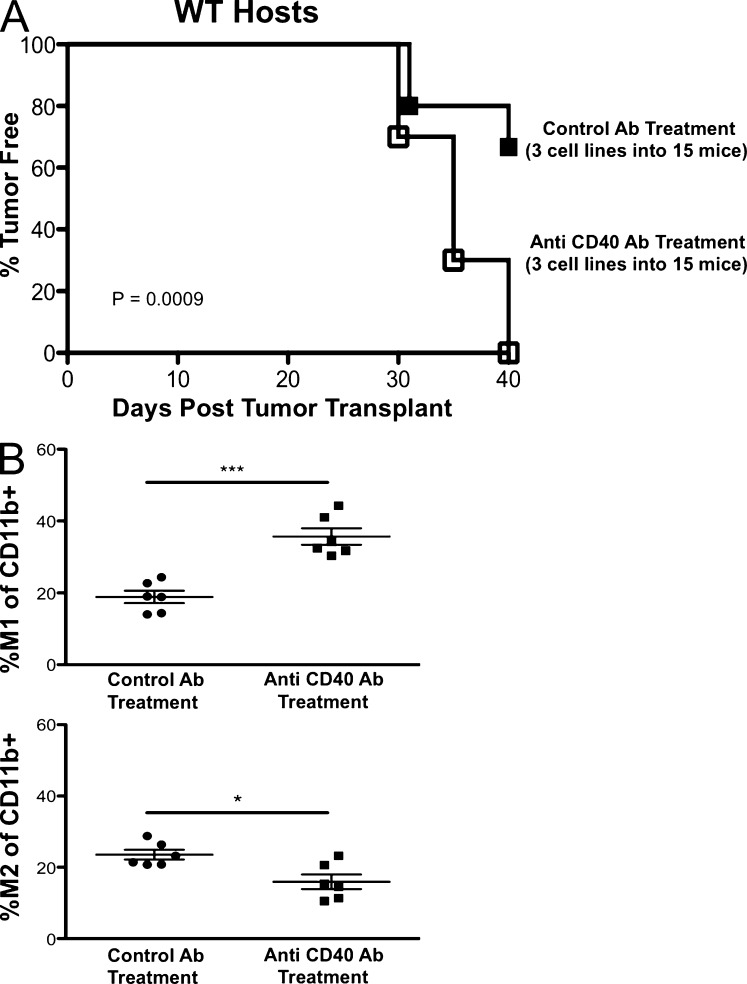
CD40 agonist administration in vivo has been shown to have antitumor properties (Buhtoiarov et al., 2005; Rakhmilevich et al., 2008) by activating TAMs to become tumoristatic through production of nitric oxide (Lum et al., 2006). We hypothesized that CD40 agonist treatment would activate macrophages in tumors growing in RAG2−/−x γc−/− mice, thereby leading to editing of cancer cells in vivo. To test this, a regressor cell line was transplanted into RAG2−/−x γc−/− mice receiving a single injection of either control IgG or anti-CD40 agonist monoclonal antibodies, tumor masses were harvested, and cell lines were generated and transplanted into WT mice. We found that cell lines from CD40 agonist-treated RAG2−/−x γc−/− mice formed tumor masses in 100% of WT recipients, whereas cell lines from isotype-treated RAG2−/−x γc−/−mice formed tumors in 33% of WT recipients in RAG2−/−x γc−/− mice (Fig. 9 A; P = 0.0009). We then analyzed the quantity of M1 macrophages in harvested tumor cell suspensions and found that M1 macrophage percentages were doubled in mice treated with CD40 agonist (36%) compared with control treatment (18%; Fig. 9 B, top; P = 0.0003). Correspondingly, M2 macrophages were decreased (23 vs. 15%) by anti-CD40 agonist treatment (Fig. 9 B, bottom; P = 0.0151). These results suggest that TAMs can be activated in RAG2−/−x γc−/− mice to effectively edit tumors in vivo.
Figure 9.
In vivo administration of CD40 agonist in RAG2−/−x γc−/− mice induces effective immunoediting and intratumoral M1 macrophages. Regressor cell line 2 was transplanted into RAG2−/−x γc−/− mice receiving a single dose of either control rat IgG or anti-CD40 agonistic monoclonal antibodies on day 5. Tumor growth was measured over time. (A) Tumor masses were converted into passaged daughter cell lines which were transplanted into syngeneic WT mice and assessed for tumor formation (number of cell lines and mice are indicated in the figure). Tumor-free mice were defined to have a nonenlarging mass <9 mm in average diameter by day 40. (B) At day 15 after transplantation, tumor masses were disaggregated into single-cell suspensions, and the percentage of M1 (top) and M2 (bottom) macrophages of CD11b+ events for each condition were quantified. *, P < 0.05; ***, P < 0.001. Error bars are represented by ± SEM. Each symbol represents a different mouse. Results were reproduced at least once.
DISCUSSION
The cancer immunoediting hypothesis predicts that tumors arising in immune-deficient individuals will be more immunogenic than tumors that develop in immune-competent individuals. Although this concept is achieving wide acceptance, the relationship between the degree of host immune deficiency and the extent of cancer immunoediting have not yet been examined. In this study, we provide evidence that the extent of host immune-deficiency directly correlates with the level of cancer immunoediting. In doing so, we document that the innate immune system present in RAG2−/− mice can mediate to some extent the immunosurveillance and immunoediting of MCA-induced sarcomas. This editing activity is associated with M1 macrophages, IFN-γ, γc, and NK cells.
Consistent with our previous studies (Shankaran et al., 2001; Koebel et al., 2007; see Fig. 4 for these data plotted in the context of our new data), we found that tumors arising in RAG-deficient mice are unedited and as a group, more immunogenic. Our evidence is based on studies of over 150 cell lines generated during a decade of experimentation performed in two separate sites, across two strains of mice, and using both RAG1- and RAG2-deficient models. A striking finding from our studies is that the regressor frequency of MCA-induced sarcoma cell lines derived from RAG-deficient mice reproducibly approximates 40%. Moreover, the regressor frequency of MCA-induced sarcomas generated in mice lacking RAG and γc is 60–70% in two independent experiments. These results suggest a quantitative nature to the immunoediting process, whereby a certain degree of basal immune function is associated with quantifiable levels of tumor sculpting, which can be measured by the regressor frequency. Because a majority of MCA cell lines generated from RAG2−/−x γc−/− mice are regressors, we speculate that the primary tumor cell repertoire consists of mostly immunogenic tumor cells that are immunologically heterogeneous (O’Sullivan et al., 2011). This heterogeneity can be partially sculpted by innate immunity in RAG2−/− mice or fully sculpted by the complete immune system in WT mice. It should be noted that we have never been able to isolate regressors from MCA-induced sarcomas that develop in WT mice (regressor frequency of 0% of 50 cell lines). These results confirm that cancer immunoediting of MCA-induced sarcomas is quite robust in WT mice and further validate that immune escape is an essential hallmark of cancer cells (Hanahan and Weinberg, 2011).
We have provided evidence that the innate immune system can edit tumors and point to M1 macrophages as participants in this process. M1 macrophages are activated classically via IFN-γ and function in the removal of intracellular pathogens (Gordon and Taylor, 2005). In the context of cancer, M1 macrophages can promote tumor elimination via activation of Th1 pathways and secretion of tumoricidal levels of nitric oxide (Sica et al., 2008). In our studies, we have defined CD45+CD11b+MHC class IIhiCD206loLy6Clo cells as M1 macrophages based not only on their phenotype but also on their classical requirement for IFN-γ for their generation. Using this definition, we found a striking correlation between the presence of M1 macrophages and productive immune responses to regressor tumors. The administration of reagents that increased M1 percentages, such as CD40 agonist, enhanced editing, whereas treatments that decreased M1 percentages, such as NK cell depletion and anti–IFN-γ mAb blocked editing.
Our findings support an anti-tumor function for macrophages that is consistent with studies performed almost 40 yr ago, when it was shown that activated macrophages from infected mice (Hibbs et al., 1971) could kill syngeneic transformed murine embryonic fibroblasts, but not primary nontransformed murine embryonic fibroblasts (Hibbs et al., 1972) in vitro. This tumoricidal activity of macrophages required cell-cell contact and was induced largely by the cytokine IFN-γ (Pace et al., 1983; Schreiber et al., 1983) in combination with additional signals such as LPS (Weinberg et al., 1978) or muramyl dipeptide (Kleinerman et al., 1983). Although we have not shown that regressor tumor cells are killed by TAMs, we have observed that regressor tumor cells can be killed effectively by IFN-γ–stimulated bone marrow macrophages in vitro (unpublished data). Our attempts to demonstrate the tumoricidal activity of regressor-associated macrophages was limited by the poor viability of sorted TAMs. Furthermore, the requirement of macrophages in immunoediting could not be tested, as treatment with clodronate-encapsulated liposomes failed to deplete macrophages in tumors, even though depletion of CD11b+ macrophages was achieved in the spleens of tumor bearing mice (unpublished data). Nevertheless, we favor the interpretation that M1 macrophages are the most likely editor in mice that lack adaptive cells, given their abundance in the tumor, their enhanced presence in response to IFN-γ and NK cell activity, and their known tumoricidal activity.
Recent studies indicate that macrophage tumoricidal activity could be enhanced in vitro and in vivo upon administration of CD40 agonistic antibodies (Buhtoiarov et al., 2005; Rakhmilevich et al., 2008; Beatty et al., 2011). Notably, Beatty et al. (2011) investigated the role of tumoricidal macrophages activated in vivo with CD40 agonist treatments in the rejection of pancreatic ductal adenocarcinoma (PDA). After demonstrating the efficacy of the anti-CD40 agonist mAb CP-870,893 in human patients, the authors used a mouse model of PDA to investigate the mechanism of tumor rejection with CD40 agonist treatment. To their surprise, the results indicated that CD40-stimulated macrophages, independent of T cell activity, are sufficient to mediate PDA rejection in vivo. Similarly, we have found that CD40 agonist treatment of RAG2−/−x γc−/− mice can induce tumor editing in the absence of adaptive immunity and NK cells, thereby suggesting that macrophages are sufficient for tumor editing. In contrast to the Beatty study, we did not see tumor rejection, suggesting that MCA sarcomas require adaptive cells for their regression. Our studies also show that unmanipulated macrophages are capable of editing through IFN-γ and NK cells without the use of CD40 agonists.
We found that the accumulation of M1 macrophages in regressor tumors required IFN-γ and NK cells. The participation of NK cells in immunosurveillance against certain types of tumors has been clearly documented in studies showing increased tumor incidences in mice lacking NK cells or molecules associated with NK cell recognition or effector function (Smyth et al., 2002, 2006; Raulet and Guerra, 2009), such as NKp46 (Gazit et al., 2006), natural killer group 2D (NKG2D; Guerra et al., 2008), DNAX accessory molecule-1 (Gilfillan et al., 2008; Iguchi-Manaka et al., 2008), perforin (van den Broek et al., 1996; Street et al., 2001), IFN-γ (Street et al., 2001), or TNF-related apoptosis-inducing ligand (Cretney et al., 2002). Therefore, we considered the possibility that there might be increased NK cell killing of MCA-induced sarcoma cells from RAG2−/−x γc−/− versus RAG2−/− or WT mice. However, we did not find major differences in the susceptibility of unedited versus edited tumors to NK cell killing. These results are consistent with recent studies showing that NKG2D, an activating receptor on NK cells that mediates tumor recognition and killing, did not play a role in the surveillance of MCA-induced sarcomas (Guerra et al., 2008). In this study, NKG2D-deficient mice had similar incidences of MCA-induced sarcomas but were more susceptible to tumor formation in prostate cancer and B lymphoma model systems, suggesting that the role of NK cells in destroying tumor cells could be dependent on the site of tumor formation. For MCA-induced sarcomas, we advocate that one role of NK cells in eliminating and/or sculpting tumors in the absence of adaptive immunity may be as a source of IFN-γ. This is based on findings that NK cells and IFN-γ are necessary for M1 macrophage polarization and subsequent editing in RAG2−/− mice. Although we cannot rule out the contribution of myeloid populations in IFN-γ production, sorted M1 and M2 TAMs do not show any IFN-γ transcript (unpublished data), suggesting that NK cells are the predominant producers of IFN-γ in the RAG2−/− host. It is not known what induces IFN-γ production by NK cells in our system, but our preliminary studies indicate that MCA-induced sarcoma cells are incapable of directly eliciting IFN-γ production from NK cells in vitro. Interestingly, IL-12p40 was shown to be required for MCA sarcoma surveillance (Smyth et al., 2000), so it is possible that local IL-12 production could stimulate NK cells to produce IFN-γ to mediate editing in the absence of adaptive immunity. It should be noted that in RAG2−/−x γc−/− mice lacking NK cells, editing could be restored with CD40 agonist treatment, suggesting that direct interaction between NK cells and tumor cells is not needed for tumor editing as long as M1 macrophages are present.
Our model is based on the postulate that immunogenic regressors, in the presence of M1 macrophages, are converted into nonimmunogenic progressors, but we have not identified the molecular basis of this phenotypic conversion. Recent studies have also found that certain tumor cells can evade macrophage killing/phagocytosis by expressing high levels of CD47 (Jaiswal et al., 2009; Majeti et al., 2009) and/or low levels of calreticulin (Chao et al., 2010). Other studies have implicated calreticulin exposure as a key initiator of innate immune responses to tumor cells, leading to antigen presentation and productive adaptive antitumor responses, and the blockade of these pathways could be a mechanism of tumor escape (Zitvogel et al., 2010). We did not find differences in the interaction between bone marrow–derived macrophages and regressor versus progressor tumor cells in vitro (unpublished data). Furthermore, our preliminary studies indicate that CD47 and calreticulin are not different between regressor and progressor cells in vitro. Future studies will compare the gene expression profiles of regressor and progressor cells to identify pathways that may mediate innate cell recognition/editing. Our matched regressor/passaged regressor cells will be critical for these experiments.
In summary, we document the generation and initial characterization of a novel set of unedited MCA-induced sarcoma cell lines that may be highly stimulatory for the innate immune system. The enhanced accumulation of M1 macrophages in these highly immunogenic tumors suggests that they can serve as models to study the early events that lead to the generation of M1 macrophages in regressing tumors. We also show that the innate immune system in RAG−/− mice contains sufficient cellular machinery to perform sculpting of MCA-induced sarcomas. This cellular machinery includes NK cells that produce IFN-γ to activate macrophages to function as innate editors. This cascade can lead to tumor elimination in the presence of adaptive immunity and/or editing in the absence of adaptive immunity. Finally, we introduce a quantitative dimension to the sculpting phase of cancer immunoediting by showing that the percentage of regressor cell lines generated from MCA-induced sarcomas is reproducible and correlates with the level of immune pressure in the tumor-bearing host.
MATERIALS AND METHODS
Experimental procedures.
All experiments involving mice were conducted under animal protocols approved by the Washington University Animal Studies Committee and the University of California, San Diego Institutional Animal Care and Use Committee (IACUC protocol #S06201) and were in accordance with ethical guidelines determined by the Peter Mac Animal Experimental Ethics Committee.
Mice and MCA induction.
Tumor induction by MCA was performed as previously described (Smyth et al., 2000; Shankaran et al., 2001). In brief, cohorts of C57BL/6-strain WT (Taconic), RAG2−/−, RAG1−/−, and RAG2−/−x γc−/− mice were injected with MCA dissolved in peanut oil at various doses. Experiment 1 was performed in St. Louis and used RAG2−/−x γc−/− mice generated by breeding IL-2Rγc−/− mice (C57/BL6 N10+1F7-strain; The Jackson Laboratory) to C57BL/6 RAG2−/− mice (Taconic). Genotyping was performed using PCR (for IL-2Rγc, following The Jackson Laboratories protocol [http://jaxmice.jax.org/strain/003174.html]) or by Southern blot for RAG2 (Shinkai et al., 1992). Genomic microsatellite analysis showed that the RAG2−/−x γc−/− mice contained C57BL/6 markers at 97% of the loci tested (3% 129/Sv markers). To control for interinstitutional breeding, minor strain differences, and housing variability, the RAG2−/− mice used in MCA experiment 1 were outcrossed from (RAG2−/−x γc−/−) x RAG2−/− breeding performed in-house. Tumors in mice were measured as previously described (Shankaran et al., 2001; Koebel et al., 2007). In experiment 1, a dose of MCA was used such that all MCA-treated mice developed tumors. Experiment 2 was performed at the Peter Mac facility in Australia and used RAG2−/−x γc−/− mice, which were provided by the Walter and Eliza Hall Institute (Bundoora), and C57BL/6 and RAG1−/− mice as previously described (Smyth et al., 2000). To rule out that RAG2−/−x γc−/− tumor cell lines were rejected based on minor strain differences, we also transplanted RAG2−/−x γc−/− regressor cell lines into F1 (C57BL/6 × 129) mice (n = 30; Taconic) and obtained identical growth patterns as in C57BL/6 mice (National Cancer Institute, Frederick Rockville, MD). For some tumor transplantation experiments, RAG2−/−x γc−/− recipient mice were purchased from Taconic. No differences in tumor growth were observed in RAG2−/−x γc−/− recipient mice purchased from Taconic Farms or bred in-house.
We discovered in the process of routine genotyping of our mice for the current study that the RAG2−/− 129/Sv mice previously obtained from Taconic Farms (RAGN12 model) and used in our 2001 publication (Shankaran et al., 2001) contained the C57BL/6 NK-C locus. Microsatellite analysis confirmed that these mice were virtually congenic at the NK-C locus and contained ∼22 cM of C57BL/6 sequence encompassing the following genes/markers: D6MIT261, D6MIT105, D6MIT018, D6MIT111, Nkrp1a, Nkrp1c, CD69, Nkg2d, Nkg2a, and Ly49a (Yokoyama and Plougastel, 2003). These mice were therefore designated 129/SvEv.cNK-C.B6 RAG2−/− mice. Because the NK-C gene locus displays allelic polymorphism and can contribute to part of the difference in NK cell activity between the C57BL/6 and 129/Sv strain (Yokoyama and Plougastel, 2003), new sets of MCA-induced sarcomas were generated using RAG2−/− mice that had been bred by Taconic Farms to be on a pure 129/SvEv background (129S6/SvEvTac-Rag2tm1Fwa). This new set of MCA-induced sarcomas was published in Koebel et al., 2007 and further studied in Fig. 3.
Cell lines.
MCA-induced and passaged daughter sarcomas were isolated and passaged in vitro as previously described (Shankaran et al., 2001; Smyth et al., 2005). In brief, tumor chunks were made into single-cell suspensions by mincing and collagenase treatment in HBSS (1 mg/ml type IA; Sigma-Aldrich), and multiple vials were frozen at passage 2. For transplantation and cytotoxicity assays, passage 2 cell lines were thawed, expanded, and studied at passage 4–8. Cell lines were maintained in RPMI 1640 (Cambrex) supplemented with 10% FCS (HyClone) as previously described (Shankaran et al., 2001; Bui et al., 2006).
Tumor transplantation.
Subconfluent tumor cell lines were harvested by trypsinization, washed 3 times with PBS, and injected at 106 cells subcutaneously into recipient C57BL/6, (129/Sv x C57BL/6) F1, RAG2−/−, or RAG2−/−x γc−/− strain mice as previously described (Shankaran et al., 2001; Smyth et al., 2005; Bui et al., 2006). RAG2−/− mice were injected i.p. with 200 µg of either control hamster IgG (PIP), anti-NK1.1 (PK136), or anti–IFN-γ (H22) on days −2 and 0 and every 4 d after until tumor harvest. RAG2−/−x γc−/− mice were injected i.p. with 200 µg either control rat IgG or anti-CD40 agonist (FGK 45.5) on day 5. Mice were monitored for tumor growth by measurement of mean tumor diameter, defined as the mean of the two maximum dimensions of the tumor mass.
Antibodies and FACS analysis.
On various days after transplantation, tumors were excised from mice, minced, and treated with 1 mg/ml type IA collagenase (Sigma-Aldrich) as previously described (Weinberg et al., 1978). Cells were vigorously resuspended, washed in FACS buffer (PBS + 1% FCS+0.05%NaN3; Sigma-Aldrich) and filtered before staining. Antibodies to CD45, F4/80, NK1.1, CD69, CD80, CD206, Ly6C, CD11b, I-A/I-E, 1A8, and streptavidin PE were obtained from BD. Staining was conducted for 15–20 min at 4° in FACS tubes containing 1–2 million total cells, 0.5–1 µl of antibody, 1 µl of anti-CD16/32, and 100 µl of FACS buffer. 7AAD (EMD Millipore) or propidium iodide (Sigma-Aldrich) was added at 1 µg/ml immediately before FACS analysis. M1-type and M2-type macrophages were gated as previously described (Movahedi et al., 2010; see Fig. 8). F4/80 and CD68 were used to detect macrophages in FACS assays or in IHC assays. F4/80 marks all monocyte lineage cells, and CD68 marks mature tissue macrophages that are in tumors (Van Ginderachter et al., 2006; Wang et al., 2012).
Immunohistochemistry.
Fresh tumor nodules were harvested, OCT-embedded, and snap frozen in cooled isopentane. Tissue blocks were cut on a cryostat into 6-µm-thick sections, mounted onto poly-l-lysine slides, air-dried overnight, and post-fixed for 10 min in acetone before staining. Purified rat anti–mouse CD16/CD32 was used to block for 20 min (BD; dilution 1:50) when appropriate. Biotin-conjugated rat anti–mouse I-A/I-E (eBioscience; dilution 1:100; 1 h at room temperature) and biotin-conjugated rat anti–mouse CD206 (BioLegend; dilution 1:100, 1 h at room temperature) staining was revealed using streptavidin-horseradish peroxidase (Vector Laboratories; 30 min at room temperature) followed by amino-ethyl-carbazole as chromogen (BD; 10-15 min at room temperature). Purified rat anti–mouse CD68 (BioLegend; dilution 1:100) staining was detected using a biotin-conjugated rabbit polyclonal anti–rat IgG, mouse adsorbed (Vector; dilution 1:200). Immunostained tissue sections were examined with a Leica DM 2500 or Nikon Eclipse E800 microscope; images were captured with a Leica DFC 420 or Nikon DXM 1200 digital camera, respectively. Quantitative analysis of MHC-II+, CD68+, CD206+ cells was obtained by counting at least 10 high power fields of tissue sections at 200× magnification.
Cell sorting.
TAMs were sorted from tumor cell suspensions on a BD FACSAria. We defined M1 macrophages as 7AAD−, CD45+, Ly 6Clo, MHC class IIhi, F4/80+, CD206lo cells, and M2 macrophages as 7AAD−, CD45+, Ly6Clo, MHC classIIlo, F4/80+, CD206hi cells.
Quantitative PCR (qPCR).
TAM RNA was isolated with a QIAGEN RNeasy Mini kit before reverse transcription with Applied Biosystems High-Capacity cDNA Reverse Transcription kit. qPCR reactions were performed on the CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories). Primers for the detection of other transcripts used are as follows: m18s forward, 5′-CGCGGTTCTATTTTGTTGGT-3′, m18s reverse, 5′-AACCTCCGACTTTCGTTCTTG-3′; mTNFα forward, 5′-CATCTTCTCAAAATTCGAGTGACAA-3′, TNF reverse, 5′-TGGGAGTAGACAAGGTACAACCC-3′; miNOS forward, 5′-CCCTTCAATGGTTGGTACATGG-3′, miNOS reverse, 5′-ACATTGATCTCCGTGACAGCC-3′; mArginase forward, 5′-GAACACGGCAGTGGCTTTAAC-3′, mArginase reverse, 5′-TGCTTAGCTCTGTCTGCTTTGC-3′; mE-cadherin forward, 5′-ACTTGGGGACAGCAACATCA-3′, mE-cadherin reverse, 5′-GGGTTTAAATCGGCCAGCAT-3′; Gas3 forward, 5′-GTAATGGACACACGACTGATC-3′, Gas3 reverse, 5′-GGAGTAGTCAGTGTTGACATG-3′.
Cytokine secretion assay.
On day 15 after transplantation, tumors were excised from mice, minced, and treated with 1 mg/ml type IA collagenase (Sigma-Aldrich) as previously described (Weinberg et al., 1978). Filtered tumor/immune cell suspensions were plated in triplicate wells at 40,000 cells/well in 100 µl for 24 h at 37°C. Supernatant was analyzed for cytokines using the mouse inflammation cytometric bead array kit from BD.
Statistical Analysis.
Statistical significance between two groups was determined by the Welch’s t test using two-tailed analysis to obtain p-values. The Log-Rank test was used to compare the survival of mice across tumor transplantation or induction conditions. Error bars are depicted using the SEM. All experiments were done at least twice.
Acknowledgments
We thank all members of the Schreiber, Smyth, and Bui Laboratories. We would like to thank Janelle Sharkey for excellent technical assistance and Jessica Archambault, Tiffany Irwin, and Michelle Stirling for animal maintenance.
J.D. Bui is supported by grants from the Cancer Research Institute, National Institutes of Health (NIH; CA128893, CA157885), the American Cancer Society (ACS-IRG #70-002), the Cancer Research Coordinating Committee (6-444951-34384), the Concern Foundation, and The Hartwell Foundation. R.D. Schreiber is supported by NIH grants CA43059 and CA107527, a grant from the Ludwig Institute for Cancer Research, and the Rhea Rosemary Finnell Clinical Investigation Grant from the Cancer Research Institute. R. Uppaluri is supported by K08 CA090403. M.J. Smyth is supported by an Australia Fellowship and a Program Grant from the National Health and Medical Research Council of Australia and a project grant from the Association of International Cancer Research. M.W.L. Teng is supported by a Cancer Council of Victoria project grant and a National Health and Medical Research Council Peter Doherty Postdoctoral Fellowship. S.F. Ngiow is supported by a Cancer Research Institute Pre-doctoral scholarship. The work cited in this publication was performed in a facility supported by National Center for Research Resources grant C06 RR012466.
The authors do not report any competing financial interests.
Footnotes
Abbreviations used:
- eCAD
- epithelial cadherin
- Gas3
- growth arrest–specific gene 3
- iNOS
- inducible nitric oxide synthase
- IHC
- immunohistochemistry
- MCA
- 3′methylcholanthrene
- NKG2D
- natural killer group 2D
- PDA
- pancreatic ductal adenocarcinoma
- qPCR
- quantitative PCR
- TAM
- tumor-associated macrophage
References
- Balkwill F., Coussens L.M. 2004. Cancer: an inflammatory link. Nature. 431:405–406 10.1038/431405a [DOI] [PubMed] [Google Scholar]
- Beatty G.L., Chiorean E.G., Fishman M.P., Saboury B., Teitelbaum U.R., Sun W., Huhn R.D., Song W., Li D., Sharp L.L., et al. 2011. CD40 agonists alter tumor stroma and show efficacy against pancreatic carcinoma in mice and humans. Science. 331:1612–1616 10.1126/science.1198443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Neriah Y., Karin M. 2011. Inflammation meets cancer, with NF-κB as the matchmaker. Nat. Immunol. 12:715–723 10.1038/ni.2060 [DOI] [PubMed] [Google Scholar]
- Boon T., Kellermann O. 1977. Rejection by syngeneic mice of cell variants obtained by mutagenesis of a malignant teratocarcinoma cell line. Proc. Natl. Acad. Sci. USA. 74:272–275 10.1073/pnas.74.1.272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhtoiarov I.N., Lum H., Berke G., Paulnock D.M., Sondel P.M., Rakhmilevich A.L. 2005. CD40 ligation activates murine macrophages via an IFN-gamma-dependent mechanism resulting in tumor cell destruction in vitro. J. Immunol. 174:6013–6022 [DOI] [PubMed] [Google Scholar]
- Bui J.D., Carayannopoulos L.N., Lanier L.L., Yokoyama W.M., Schreiber R.D. 2006. IFN-dependent down-regulation of the NKG2D ligand H60 on tumors. J. Immunol. 176:905–913 [DOI] [PubMed] [Google Scholar]
- Cao X., Shores E.W., Hu-Li J., Anver M.R., Kelsall B.L., Russell S.M., Drago J., Noguchi M., Grinberg A., Bloom E.T., et al. 1995. Defective lymphoid development in mice lacking expression of the common cytokine receptor γ chain. Immunity. 2:223–238 10.1016/1074-7613(95)90047-0 [DOI] [PubMed] [Google Scholar]
- Chao M.P., Jaiswal S., Weissman-Tsukamoto R., Alizadeh A.A., Gentles A.J., Volkmer J., Weiskopf K., Willingham S.B., Raveh T., Park C.Y., Majeti R., Weissman I.L. 2010. Calreticulin Is the Dominant Pro-Phagocytic Signal on Multiple Human Cancers and Is Counterbalanced by CD47. Science Translational Medicine. 2:63ra94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretney E., Takeda K., Yagita H., Glaccum M., Peschon J.J., Smyth M.J. 2002. Increased susceptibility to tumor initiation and metastasis in TNF-related apoptosis-inducing ligand-deficient mice. J. Immunol. 168:1356–1361 [DOI] [PubMed] [Google Scholar]
- Crowe N.Y., Smyth M.J., Godfrey D.I. 2002. A critical role for natural killer T cells in immunosurveillance of methylcholanthrene-induced sarcomas. J. Exp. Med. 196:119–127 10.1084/jem.20020092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn G.P., Old L.J., Schreiber R.D. 2004a. The immunobiology of cancer immunosurveillance and immunoediting. Immunity. 21:137–148 10.1016/j.immuni.2004.07.017 [DOI] [PubMed] [Google Scholar]
- Dunn G.P., Old L.J., Schreiber R.D. 2004b. The three Es of cancer immunoediting. Annu. Rev. Immunol. 22:329–360 10.1146/annurev.immunol.22.012703.104803 [DOI] [PubMed] [Google Scholar]
- Dunn G.P., Bruce A.T., Sheehan K.C.F., Shankaran V., Uppaluri R., Bui J.D., Diamond M.S., Koebel C.M., Arthur C., White J.M., Schreiber R.D. 2005. A critical function for type I interferons in cancer immunoediting. Nat. Immunol. 6:722–729 10.1038/ni1213 [DOI] [PubMed] [Google Scholar]
- Flood P.M., Schreiber H., Ron Y. 1987. Protective immunity to progressive tumors can be induced by antigen presented on regressor tumors. J. Immunol. 138:3573–3579 [PubMed] [Google Scholar]
- Gazit R., Gruda R., Elboim M., Arnon T.I., Katz G., Achdout H., Hanna J., Qimron U., Landau G., Greenbaum E., et al. 2006. Lethal influenza infection in the absence of the natural killer cell receptor gene Ncr1. Nat. Immunol. 7:517–523 10.1038/ni1322 [DOI] [PubMed] [Google Scholar]
- Ghassabeh G.H., De Baetselier P., Brys L., Noël W., Van Ginderachter J.A., Meerschaut S., Beschin A., Brombacher F., Raes G. 2006. Identification of a common gene signature for type II cytokine-associated myeloid cells elicited in vivo in different pathologic conditions. Blood. 108:575–583 10.1182/blood-2005-04-1485 [DOI] [PubMed] [Google Scholar]
- Gilfillan S., Chan C.J., Cella M., Haynes N.M., Rapaport A.S., Boles K.S., Andrews D.M., Smyth M.J., Colonna M. 2008. DNAM-1 promotes activation of cytotoxic lymphocytes by nonprofessional antigen-presenting cells and tumors. J. Exp. Med. 205:2965–2973 10.1084/jem.20081752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S., Taylor P.R. 2005. Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 5:953–964 10.1038/nri1733 [DOI] [PubMed] [Google Scholar]
- Guerra N., Tan Y.X., Joncker N.T., Choy A., Gallardo F., Xiong N., Knoblaugh S., Cado D., Greenberg N.M., Raulet D.H. 2008. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity. 28:571–580 10.1016/j.immuni.2008.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. 2011. Hallmarks of cancer: the next generation. Cell. 144:646–674 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Hibbs J.B., Jr, Lambert L.H., Jr, Remington J.S. 1971. Resistance to murine tumors conferred by chronic infection with intracellular protozoa, Toxoplasma gondii and Besnoitia jellisoni. J. Infect. Dis. 124:587–592 10.1093/infdis/124.6.587 [DOI] [PubMed] [Google Scholar]
- Hibbs J.B., Jr, Lambert L.H., Jr, Remington J.S. 1972. Control of carcinogenesis: a possible role for the activated macrophage. Science. 177:998–1000 10.1126/science.177.4053.998 [DOI] [PubMed] [Google Scholar]
- Iguchi-Manaka A., Kai H., Yamashita Y., Shibata K., Tahara-Hanaoka S., Honda S.I., Yasui T., Kikutani H., Shibuya K., Shibuya A. 2008. Accelerated tumor growth in mice deficient in DNAM-1 receptor. J. Exp. Med. 205:2959–2964 10.1084/jem.20081611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S., Jamieson C.H.M., Pang W.W., Park C.Y., Chao M.P., Majeti R., Traver D., van Rooijen N., Weissman I.L. 2009. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell. 138:271–285 10.1016/j.cell.2009.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinerman E.S., Erickson K.L., Schroit A.J., Fogler W.E., Fidler I.J. 1983. Activation of tumoricidal properties in human blood monocytes by liposomes containing lipophilic muramyl tripeptide. Cancer Res. 43:2010–2014 [PubMed] [Google Scholar]
- Koebel C.M., Vermi W., Swann J.B., Zerafa N., Rodig S.J., Old L.J., Smyth M.J., Schreiber R.D. 2007. Adaptive immunity maintains occult cancer in an equilibrium state. Nature. 450:903–907 10.1038/nature06309 [DOI] [PubMed] [Google Scholar]
- Kripke M.L. 1974. Antigenicity of murine skin tumors induced by ultraviolet light. J. Natl. Cancer Inst. 53:1333–1336 [DOI] [PubMed] [Google Scholar]
- Lewis C.E., Pollard J.W. 2006. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 66:605–612 10.1158/0008-5472.CAN-05-4005 [DOI] [PubMed] [Google Scholar]
- Lum H.D., Buhtoiarov I.N., Schmidt B.E., Berke G., Paulnock D.M., Sondel P.M., Rakhmilevich A.L. 2006. Tumoristatic effects of anti-CD40 mAb-activated macrophages involve nitric oxide and tumour necrosis factor-alpha. Immunology. 118:261–270 10.1111/j.1365-2567.2006.02366.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeti R., Chao M.P., Alizadeh A.A., Pang W.W., Jaiswal S., Gibbs K.D., Jr, van Rooijen N., Weissman I.L. 2009. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 138:286–299 10.1016/j.cell.2009.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez F.O., Helming L., Gordon S. 2009. Alternative activation of macrophages: an immunologic functional perspective. Annu. Rev. Immunol. 27:451–483 10.1146/annurev.immunol.021908.132532 [DOI] [PubMed] [Google Scholar]
- Movahedi K., Laoui D., Gysemans C., Baeten M., Stangé G., Van den Bossche J., Mack M., Pipeleers D., In’t Veld P., De Baetselier P., Van Ginderachter J.A. 2010. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res. 70:5728–5739 10.1158/0008-5472.CAN-09-4672 [DOI] [PubMed] [Google Scholar]
- O’Sullivan T., Dunn G.P., Lacoursiere D.Y., Schreiber R.D., Bui J.D. 2011. Cancer immunoediting of the NK group 2D ligand H60a. J. Immunol. 187:3538–3545 10.4049/jimmunol.1100413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace J.L., Russell S.W., Schreiber R.D., Altman A., Katz D.H. 1983. Macrophage activation: priming activity from a T-cell hybridoma is attributable to interferon-gamma. Proc. Natl. Acad. Sci. USA. 80:3782–3786 10.1073/pnas.80.12.3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakhmilevich A.L., Buhtoiarov I.N., Malkovsky M., Sondel P.M. 2008. CD40 ligation in vivo can induce T cell independent antitumor effects even against immunogenic tumors. Cancer Immunology. Immunotherapeutics. 57:1151–1160 10.1007/s00262-007-0447-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raulet D.H., Guerra N. 2009. Oncogenic stress sensed by the immune system: role of natural killer cell receptors. Nat. Rev. Immunol. 9:568–580 10.1038/nri2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber R.D., Pace J.L., Russell S.W., Altman A., Katz D.H. 1983. Macrophage-activating factor produced by a T cell hybridoma: physiochemical and biosynthetic resemblance to gamma-interferon. J. Immunol. 131:826–832 [PubMed] [Google Scholar]
- Schreiber R.D., Old L.J., Smyth M.J. 2011. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 331:1565–1570 10.1126/science.1203486 [DOI] [PubMed] [Google Scholar]
- Shankaran V., Ikeda H., Bruce A.T., White J.M., Swanson P.E., Old L.J., Schreiber R.D. 2001. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 410:1107–1111 10.1038/35074122 [DOI] [PubMed] [Google Scholar]
- Shinkai Y., Rathbun G., Lam K.P., Oltz E.M., Stewart V., Mendelsohn M., Charron J., Datta M., Young F., Stall A.M., et al. 1992. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 68:855–867 10.1016/0092-8674(92)90029-C [DOI] [PubMed] [Google Scholar]
- Sica A., Larghi P., Mancino A., Rubino L., Porta C., Totaro M.G., Rimoldi M., Biswas S.K., Allavena P., Mantovani A. 2008. Macrophage polarization in tumour progression. Semin. Cancer Biol. 18:349–355 10.1016/j.semcancer.2008.03.004 [DOI] [PubMed] [Google Scholar]
- Smyth M.J., Thia K.Y., Street S.E., Cretney E., Trapani J.A., Taniguchi M., Kawano T., Pelikan S.B., Crowe N.Y., Godfrey D.I. 2000. Differential tumor surveillance by natural killer (NK) and NKT cells. J. Exp. Med. 191:661–668 10.1084/jem.191.4.661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth M.J., Crowe N.Y., Godfrey D.I. 2001. NK cells and NKT cells collaborate in host protection from methylcholanthrene-induced fibrosarcoma. Int. Immunol. 13:459–463 10.1093/intimm/13.4.459 [DOI] [PubMed] [Google Scholar]
- Smyth M.J., Hayakawa Y., Takeda K., Yagita H. 2002. New aspects of natural-killer-cell surveillance and therapy of cancer. Nat. Rev. Cancer. 2:850–861 10.1038/nrc928 [DOI] [PubMed] [Google Scholar]
- Smyth M.J., Swann J., Cretney E., Zerafa N., Yokoyama W.M., Hayakawa Y. 2005. NKG2D function protects the host from tumor initiation. J. Exp. Med. 202:583–588 10.1084/jem.20050994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth M.J., Dunn G.P., Schreiber R.D. 2006. Cancer immunosurveillance and immunoediting: the roles of immunity in suppressing tumor development and shaping tumor immunogenicity. Adv. Immunol. 90:1–50 10.1016/S0065-2776(06)90001-7 [DOI] [PubMed] [Google Scholar]
- Street S.E.A., Cretney E., Smyth M.J. 2001. Perforin and interferon-gamma activities independently control tumor initiation, growth, and metastasis. Blood. 97:192–197 10.1182/blood.V97.1.192 [DOI] [PubMed] [Google Scholar]
- Street S.E.A., Hayakawa Y., Zhan Y., Lew A.M., MacGregor D., Jamieson A.M., Diefenbach A., Yagita H., Godfrey D.I., Smyth M.J. 2004. Innate immune surveillance of spontaneous B cell lymphomas by natural killer cells and gammadelta T cells. J. Exp. Med. 199:879–884 10.1084/jem.20031981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 126:663–676 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Takeda K., Smyth M.J., Cretney E., Hayakawa Y., Kayagaki N., Yagita H., Okumura K. 2002. Critical role for tumor necrosis factor-related apoptosis-inducing ligand in immune surveillance against tumor development. J. Exp. Med. 195:161–169 10.1084/jem.20011171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Broek M.E., Kägi D., Ossendorp F., Toes R., Vamvakas S., Lutz W.K., Melief C.J., Zinkernagel R.M., Hengartner H. 1996. Decreased tumor surveillance in perforin-deficient mice. J. Exp. Med. 184:1781–1790 10.1084/jem.184.5.1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ginderachter J.A., Meerschaut S., Liu Y., Brys L., De Groeve K., Hassanzadeh Ghassabeh G., Raes G., De Baetselier P. 2006. Peroxisome proliferator-activated receptor gamma (PPARgamma) ligands reverse CTL suppression by alternatively activated (M2) macrophages in cancer. Blood. 108:525–535 10.1182/blood-2005-09-3777 [DOI] [PubMed] [Google Scholar]
- Vesely M.D., Kershaw M.H., Schreiber R.D., Smyth M.J. 2011. Natural innate and adaptive immunity to cancer. Annu. Rev. Immunol. 29:235–271 10.1146/annurev-immunol-031210-101324 [DOI] [PubMed] [Google Scholar]
- Wang S.-C., Hong J.-H., Hsueh C., Chiang C.-S. 2012. Tumor-secreted SDF-1 promotes glioma invasiveness and TAM tropism toward hypoxia in a murine astrocytoma model. Lab. Invest. 92:151–162 10.1038/labinvest.2011.128 [DOI] [PubMed] [Google Scholar]
- Weinberg J.B., Chapman H.A., Jr, Hibbs J.B., Jr 1978. Characterization of the effects of endotoxin on macrophage tumor cell killing. J. Immunol. 121:72–80 [PubMed] [Google Scholar]
- Yokoyama W.M., Plougastel B.F.M. 2003. Immune functions encoded by the natural killer gene complex. Nat. Rev. Immunol. 3:304–316 10.1038/nri1055 [DOI] [PubMed] [Google Scholar]
- Zitvogel L., Kepp O., Senovilla L., Menger L., Chaput N., Kroemer G. 2010. Immunogenic tumor cell death for optimal anticancer therapy: the calreticulin exposure pathway. Clin. Cancer Res. 16:3100–3104 10.1158/1078-0432.CCR-09-2891 [DOI] [PubMed] [Google Scholar]