Abstract
Estimates of the proportion of amino acid substitutions that have been fixed by selection (α) vary widely among taxa, ranging from zero in humans to over 50% in Drosophila. This wide range may reflect differences in the efficacy of selection due to differences in the effective population size (Ne). However, most comparisons have been made among distantly related organisms that differ not only in Ne but also in many other aspects of their biology. Here, we estimate α in three closely related lineages of house mice that have a similar ecology but differ widely in Ne: Mus musculus musculus (Ne ∼ 25,000–120,000), M. m. domesticus (Ne ∼ 58,000–200,000), and M. m. castaneus (Ne ∼ 200,000–733,000). Mice were genotyped using a high-density single nucleotide polymorphism array, and the proportions of replacement and silent mutations within subspecies were compared with those fixed between each subspecies and an outgroup, Mus spretus. There was significant evidence of positive selection in M. m. castaneus, the lineage with the largest Ne, with α estimated to be approximately 40%. In contrast, estimates of α for M. m. domesticus (α = 13%) and for M. m. musculus (α = 12 %) were much smaller. Interestingly, the higher estimate of α for M. m. castaneus appears to reflect not only more adaptive fixations but also more effective purifying selection. These results support the hypothesis that differences in Ne contribute to differences among species in the efficacy of selection.
Keywords: substitution, adaptation, evolution, effective population size, house mouse, Mus musculus
Introduction
Resolving the relative contributions of stochastic and adaptive processes to amino acid substitutions remains a fundamental goal of evolutionary biology. Several methods have been developed to empirically estimate the proportion of amino acid substitutions fixed by positive selection termed α (e.g., Bustamante et al. 2001; Smith and Eyre-Walker 2002; Eyre-Walker and Keightley 2009). These methods follow from the neutral expectation that the ratio of synonymous to nonsynonymous mutations should be the same within and between species (McDonald and Kreitman 1991). Assuming that synonymous variation is not under selection, deviations from this expectation may be used to estimate α (e.g., Smith and Eyre-Walker 2002; Fay et al. 2002).
The fraction of substitutions fixed by selection is a function of Nes, where Ne is the effective population size and s is the selection coefficient (Kimura 1983). Therefore, differences in estimates of α among taxa may reflect differences in s, differences in Ne, or both. Ne is expected to influence the contribution of positive selection to amino acid substitution in two ways. First, given an equal rate of mutation, larger populations will have a greater input of new mutations simply due to the greater number of individuals contributing mutations. Second, drift can overwhelm selection in small populations such that mutations with s on the order of 1/Ne or smaller will behave as if effectively neutral (e.g., Ohta 1973; Kimura 1983). Consequently, α is predicted to be larger in larger populations. Indeed, empirical studies provide support for a correlation between Ne and rates of adaptive substitution. Taxa with the highest estimates of α tend to have high Ne. In Drosophila, estimates of α are consistently above 50% (Smith and Eyre-Walker 2002; Andolfatto 2007; Begun et al. 2007; Maside and Charlesworth 2007; Shapiro et al. 2007; Bachtrog 2008; Eyre-Walker and Keightley 2009; for review, see Sella et al. 2009). Estimates of α in the European Aspen, Populus tremula (Ne ∼ 105), and the outcrossing crucifer, Capsella grandiflora (Ne ∼ 5 × 105), are also large and in the range of 30–40% (Ingvarsson 2010; Slotte et al. 2010). Among mammals, there is evidence that α is large (up to 57%; Halligan et al. 2010) in the house mouse, Mus musculus castaneus for which Ne estimates range from 200,000 to over 700,000 (Geraldes et al. 2008, 2011; Halligan et al. 2010). On the other hand, in Arabidopsis (Foxe et al. 2008), yeast (Doniger et al. 2008), and humans (Mikkelsen et al. 2005; Zhang and Li 2005; Boyko et al. 2008), taxa with low Ne, adaptive substitutions appear to represent only a small fraction of amino acid divergence. However, these comparisons are between taxa that differ not only in Ne but also in many aspects of their biology. To separate the effects of s and Ne, one would ideally like to compare closely related taxa that differ in Ne but are otherwise similar. Two recent studies have taken this approach, one comparing two fruit fly species separated by ∼2 my (Jensen and Bachtrog 2011) and the other comparing multiple species of sunflowers diverging roughly 8 Ma (Strasburg et al. 2011). Although the taxa compared are still separated by millions of years, they are more closely related than any previous comparisons and, in both studies, the species with larger Ne showed more evidence of adaptive fixations.
Here, we build on these results by comparing ratios of synonymous to nonsynonymous mutations within and between subspecies of house mice. These three subspecies are estimated to have diverged ∼500,000 years ago (Geraldes et al. 2008, 2011; Duvaux et al. 2011) and show some evidence of reproductive isolation (e.g., Britton-Davidian et al. 2005; Good et al. 2008). All three subspecies are commensal with humans and thus occupy similar ecological niches. Although estimates of Ne are large for M. m. castaneus (see above), they are much smaller for M. m. domesticus (∼58,000–200,000) and M. m. musculus (∼25,000–120,000) (Salcedo et al. 2007; Geraldes et al. 2008, 2011; Halligan et al. 2010; see supplementary table 1, Supplementary Material online). We find significant evidence of positive selection only in the subspecies with the highest Ne, M. m. castaneus. However, higher estimates of α for M. m. castaneus appear to result not only from more adaptive fixations but also from more effective purifying selection.
Materials and Methods
Samples
All mice were wild-caught or from wild-derived inbred strains (supplementary table 2, Supplementary Material online). We first conducted analyses using only wild-caught individuals, including 10 M. m. castaneus from India, 8 M. m. domesticus from Western Europe, and 14 M. m. musculus from Eastern Europe. To provide a more evenly distributed geographical sample, we then expanded our analysis to include several wild-derived inbred lines from each of the three subspecies. Results from these two different sampling strategies were broadly concordant. A wild-derived laboratory strain of Mus spretus was included as an outgroup in all analyses (supplementary fig. 1, Supplementary Material online; Tucker et al. 2005). Mus spretus diverged from M. musculus approximately 1 Ma (She et al. 1990; Suzuki et al. 2004).
Genotyping
The McDonald–Kreitman (MK) framework is based on a comparison of synonymous and nonsynonymous mutations (McDonald and Kreitman 1991). Data for this comparison typically come from DNA sequences of individual genes or sets of genes (e.g., Halligan et al. 2010). Here, we take advantage of a newly developed Affymetrix genotyping array that interrogates over 600,000 single nucleotide polymorphisms (SNPs) across the genome to assess patterns of nonsynonymous and synonymous variation (Yang et al. 2009, 2011). In this study, we focus only on those SNPs found in autosomal coding regions, a set of over 6,200 SNPs from more than 4,100 genes. The SNPs cover all of the autosomes at an average density of 2.5 SNPs/Mb (standard deviation = 0.75). DNA samples were prepared at the University of Arizona or at the University of North Carolina, and genotyping was performed at the Jackson Laboratory. Details on base calling can be found in Yang et al. (2011).
The use of a genotyping array rather than DNA sequences of individual genes has both advantages and disadvantages. SNP genotyping has the principal advantage of being a relatively low cost, high throughput method, enabling a survey of coding regions throughout the genome. Therefore, SNP genotyping reduces the biases that might be introduced by sampling a limited set of genes, biases that may be significant for traditional sequencing studies. The chief disadvantage of this approach lies in the potential ascertainment bias of SNPs interrogated by the array. First, although SNPs included are likely to represent variation segregating at higher frequencies, this bias, in and of itself, does not preclude the use of the MK framework. In addition, low-frequency variants are often removed from MK analyses (Fay et al. 2001, 2002) because they contain disproportionately more weakly deleterious mutations. Second, bias can result if a chip developed solely from data from one taxon is applied to others. SNPs on the Affymetrix chip were ascertained in a sample of house mice that included all three subspecies, but there is some evidence of ascertainment bias in SNPs used in this study (Yang et al. 2011). The average minor allele frequency (MAF) was slightly higher for M. m. domesticus, the subspecies in which the majority of SNPs on the Affymetrix chip were ascertained, than for M. m. castaneus or M. m. musculus ( (standard error): M. m. domesticus, synonymous = 0.242 (0.003), nonsynonymous = 0.234 (0.007); M. m. castaneus, synonymous = 0.229 (0.004), nonsynonymous = 0.214 (0.008); M. m. musculus, synonymous = 0.203 (0.005), nonsynonymous = 0.203 (0.008)). However, the average MAF for the other two subspecies was also high, and all three subspecies have appreciable numbers of sites segregating at intermediate frequencies (supplementary fig. 2, Supplementary Material online). Finally, because the MK framework compares ratios of synonymous and nonsynonymous mutations, bias in the results due to the manner in which SNPs were chosen would require not only that nonsynonymous or synonymous SNPs were preferentially included on the array but also that this was done differently for the three subspecies, which seems unlikely. Empirical evidence that the conclusions are not strongly biased comes from the comparison of our results for one subspecies (M. m. castaneus) to those of Halligan et al. (2010) who conducted analyses based on DNA sequences sampled from the same populations and obtained similar values of α (see Discussion). One limitation of the use of ascertained SNPs is that estimates of polymorphism are not comparable to traditional estimates of diversity from sequence studies and do not reflect the true site frequency spectra (SFSs). As a result, we cannot use our data to estimate Ne nor can we use methods based on the SFS to estimate α. Instead, we refer to previously published estimates of Ne (supplementary table 1, Supplementary Material online).
Estimates of α
We compared polymorphism within each of the three subspecies of M. musculus to fixed differences between M. musculus and M. spretus. Sites with missing data for a given pairwise comparison were excluded. We also excluded sites at which any of the sampled alleles were designated as VINOs (variable intensity oligonucleotides; for details, see Yang et al. 2011; Didion et al. 2012). Variation at each coding site was classified as either synonymous or nonsynonymous based on Ensembl notation (Build 63) of base position in the Mus musculus reference sequence (Build 37). If a site had conflicting overlapping designations, fell within noncoding regions in any transcript or fell within a coding region with no stop or start codon, it was excluded from the analysis. To mitigate the effects of nearly neutral mutations that may contribute proportionally more to polymorphism than to divergence, MK tests are often repeated, excluding polymorphisms that fall below a given threshold (Fay et al. 2001, 2002). Although such an approach is arbitrary, it does address the potential underestimation of α and can be applied to SNP data. We repeated all analyses, counting polymorphic sites only when the MAF exceeded 10% or 20%, levels chosen to represent meaningful differences given the number of alleles sampled. Significance levels were calculated using χ2 tests. We then calculated the neutrality index (NI; Rand and Kann 1996),
where Pns is the number of polymorphic nonsynonymous sites, Fns is the number of fixed nonsynonymous sites, Ps is the number of polymorphic synonymous sites, and Fs is the number of fixed synonymous sites. Neutrality indices less than one are consistent with positive selection, whereas values greater than one are consistent with weak purifying selection. From NI, we estimated α (Smith and Eyre-Walker 2002):
Confidence intervals (CI) were determined using 1,000 bootstrap replicates of sites. These calculations were completed using a combination of custom PERL and R scripts.
The Impact of Population Structure and Changes in Ne
Hidden population structure and admixture can result in biased estimates of α. In addition, changes in Ne may bias estimates of α. For example, consider a population that has contracted at some time in the past and then expanded to its current size. Weakly deleterious amino acid mutations may have fixed when the population was smaller, inflating estimates of α (McDonald and Kreitman 1991). Reductions in Ne may produce the opposite result, underestimating α (Eyre-Walker and Keightley 2009). However, the effects of changes in Ne on estimates of α are complex, dependent on the number of mutations that are nearly neutral, and thus difficult to predict (Charlesworth and Eyre-Walker 2006; Eyre-Walker and Keightley 2009). In this study, all subspecies are believed to have expanded in the recent past as human commensals (Rajabi-Maham et al. 2008) and in M. m. domesticus and M. m. musculus, there is evidence that those expansions were preceded by major contractions (Rajabi-Maham et al. 2008; Duvaux et al. 2011). The ancestral Ne of these subspecies has been estimated at ∼400,000–500,000 (Geraldes et al. 2011) and is closest to the estimated current Ne of M. m. castaneus. Therefore, changes in Ne may lead to overestimates of α in M. m. domesticus and M. m. musculus and underestimates of α in M. m. castaneus (see Discussion). Finally, we note that the wild-caught M. m. castaneus were sampled from the presumed ancestral range of house mice, whereas wild-caught M. m. musculus and M. m. domesticus were sampled from derived populations.
We addressed the potential effects of these demographic phenomena in three ways. First, as stated above, we repeated our analyses with different thresholds for polymorphism. Second, we used STRUCTURE to test for evidence of admixture between subspecies and for evidence of further subdivision within each subspecies (Pritchard et al. 2000; see supplementary analyses, Supplementary Material online). Using these results, we repeated our analyses with subsamples from each subspecies that showed little evidence of subdivision. Third, to make sampling more consistent between subspecies, we expanded our study to include wild-derived inbred lines (6 in M. m. castaneus, 20 in M. m. domesticus, and 8 in M. m. musculus; supplementary table 2, Supplementary Material online). By doing so, we increased both the number of alleles sampled and the geographic range of the samples, including alleles from the ancestral and derived ranges of each subspecies. Using this expanded sample, we again repeated the estimation of α (see supplementary analyses, Supplementary Material online). As seen below, results were similar for all analyses.
Results
Estimates of α
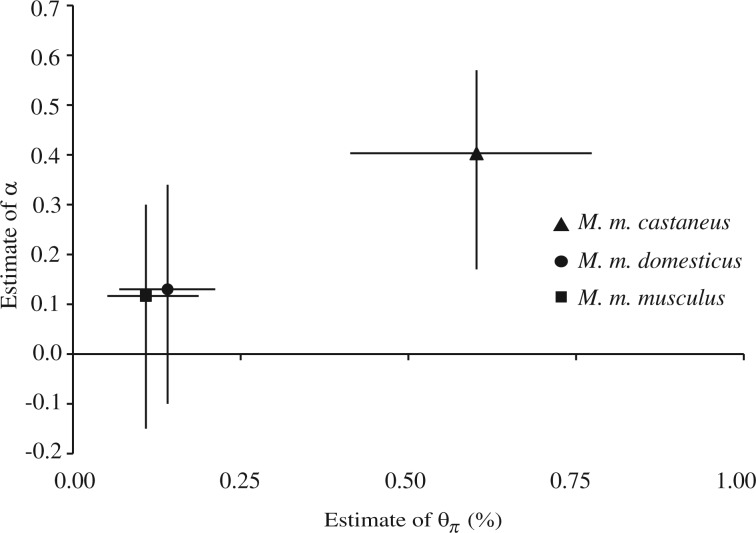
Estimates of the NI and of α for each of the three subspecies of M. musculus are given in table 1. Whether low-frequency polymorphisms were excluded or not, M. m. castaneus had the largest values of α (0.37–0.43), whereas estimates for M. m. domesticus and M. m. musculus were much smaller (0.10–0.16 and 0.10–0.13, respectively). Notably, it is only in M. m. castaneus that CIs for α did not include zero. Estimates of Ne were taken from the literature and are based on data from resequencing studies (supplementary table 1, Supplementary Material online). The wide range of values for Ne for each subspecies reflects uncertainty in the generation time and associated estimates of mutation rate as well as differences in the methods employed. Nonetheless, there is broad agreement between estimates of Ne and α; estimates of α and Ne are largest in M. m. castaneus, whereas estimates of both Ne and α for M. m. musculus and M. m. domesticus are smaller. To account for the variation in Ne introduced by differences in assumptions regarding generation time and mutation rate, we plotted estimates of α against published estimates of nucleotide diversity (supplementary table 1, Supplementary Material online) as a proxy for Ne (fig. 1).
Table 1.
Estimates of the Neutrality Index (NI) and the Proportion of Amino Acid Substitutions Fixed by Positive Selection (α) Derived from the Ratios of Replacement and Silent Polymorphisms and Fixed Differences.
| Subspecies | Polymorphism Cutoffa | Replacement Polymorphism | Replacement Fixed | Silent Polymorphism | Silent Fixed | Replacement Polymorphism/Silent Polymorphism | Replacement Fixed/Silent Fixed | NI | αˆ | CIb | Pearson’s χ2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| M. m. castaneus | >0 | 339 | 89 | 1,332 | 222 | 0.25 | 0.40 | 0.64 | 0.36 | 0.17–0.51 | 10.75* |
| M. m. castaneus | >0.10 | 207 | 89 | 903 | 222 | 0.23 | 0.40 | 0.57 | 0.43 | 0.24–0.57 | 14.64** |
| M. m. castaneus | >0.20 | 151 | 89 | 636 | 222 | 0.24 | 0.40 | 0.59 | 0.41 | 0.21–0.56 | 11.61** |
| M. m. domesticus | >0 | 442 | 152 | 1,509 | 467 | 0.29 | 0.33 | 0.90 | 0.10 | −0.10 to 0.27 | 0.96 |
| M. m. domesticus | >0.10 | 352 | 152 | 1,243 | 467 | 0.28 | 0.33 | 0.87 | 0.13 | −0.09 to 0.30 | 1.57 |
| M. m. domesticus | >0.20 | 216 | 152 | 786 | 467 | 0.27 | 0.33 | 0.84 | 0.16 | −0.07 to 0.34 | 1.96 |
| M. m. musculus | >0 | 314 | 185 | 969 | 501 | 0.32 | 0.37 | 0.88 | 0.12 | −0.09 to 0.30 | 1.47 |
| M. m. musculus | >0.10 | 216 | 185 | 652 | 501 | 0.33 | 0.37 | 0.90 | 0.10 | −0.15 to 0.28 | 0.87 |
| M. m. musculus | >0.20 | 142 | 185 | 444 | 501 | 0.32 | 0.37 | 0.87 | 0.13 | −0.11 to 0.33 | 1.24 |
The polymorphism cutoff is given in terms of the MAF.
CIs for αˆ were derived via 1,000 bootstrap replicates. α cannot be less than 0. However, negative estimates of α do occur when the ratio of replacement polymorphism to divergence exceeds the ratio of silent polymorphism to divergence.
*P < 0.05, **P < 0.001.
FIG. 1.
Average estimate of α for each subspecies of Mus musculus versus average estimate of nucleotide diversity, θπ. Vertical error bars reflect the range of CIs for α estimates derived using different polymorphism cutoffs. Horizontal error bars reflect the range of reported estimates of θπ.
It is important to note that although α is defined as the contribution of positive selection to amino acid substitution, estimates of α are influenced not only by the ratio of replacement to silent fixed differences but also by the ratio of replacement to silent polymorphism. Therefore, more effective positive selection can be confounded with more effective purifying selection. A simple comparison of the ratios of replacement to silent polymorphisms and ratios of replacement to silent fixations among subspecies in our study suggests that both purifying selection and positive selection are more effective in M. m. castaneus, contributing to higher overall estimates of α (table 1). The ratio of replacement to silent polymorphism, which reflects purifying selection, is lower in M. m. castaneus than in either M. m. musculus or M. m. domesticus, whereas the ratio of replacement to silent fixed differences, which reflects adaptive fixation, is higher in M. m. castaneus than in either M. m. musculus or M. m. domesticus.
The use of a closely related outgroup can result in biased estimates of α when diversity is high relative to divergence (Keightley and Eyre-Walker 2012). One potential source of bias is the misattribution of polymorphism to divergence, particularly when a single allele is sampled from either the focal taxon or the outgroup. Neutral polymorphisms misattributed to divergence tend to dilute estimates of α, whereas slightly deleterious mutations tend to inflate estimates of α. Bias may also result from the contribution of ancestral polymorphism to divergence and from the differences in rates of fixation of neutral and advantageous alleles (Keightley and Eyre-Walker 2012). In this study, we sampled multiple individuals of the focal taxon and a single allele from the outgroup because it allowed us to survey more sites. However, we addressed the potential misattribution of polymorphism to divergence by repeating the analysis using genotype data for five additional inbred lines of M. spretus only counting fixed differences when there was no variation segregating in M. spretus. Results were similar (supplementary table 3, Supplementary Material online).
The Impact of Population Structure and Demography
Population structure and changes in population size can result in biased estimates of α. We found no evidence for significant admixture among subspecies in our wild-caught samples, a result that is in agreement with previous analysis (Yang et al. 2011). We also found no evidence for hidden structure within M. m. domesticus (see supplementary analyses, Supplementary Material online). There was evidence for subdivision within our samples of M. m. castaneus and M. m. musculus (see supplementary analyses, Supplementary Material online). However, we estimated α again for both subspecies using the largest sample for which there was no evidence of subdivision (eight individuals for M. m. castaneus and six for M. m. musculus) and found very similar results (supplementary http://www.mbe.oxfordjournals.org/lookup/suppl/doi:10.1093/molbev/mss105/-/DC1table 4, Supplementary Material online). Estimates of α for M. m. castaneus ranged from 0.39 to 0.43 and for M. m. musculus from 0.07 to 0.09. Changes in Ne could also affect the interpretation of our results. However, the fact that similar patterns were obtained using all the data and when low-frequency polymorphisms were excluded (table 1) suggests that the bias introduced by changes in Ne may not be great. In addition, we expanded our sample to include wild-derived inbred lines from a larger geographic range for each subspecies (see supplementary analyses, Supplementary Material online). Although this analysis does not address the impact of changes in Ne directly, it does mitigate the potential impact of our sampling on the effects of such changes. Estimates of α for the expanded sample were lower in M. m. castaneus (0.26–0.36) and for M. m. domesticus (−0.09 to 0.04) and slightly higher for M. m. musculus (0.09–0.14, supplementary table 5, Supplementary Material online). Nonetheless, consistent with the wild-caught samples, there was significant evidence for positive selection only in M. m. castaneus.
Discussion
Ne and the Contribution of Positive Selection to Amino Acid Divergence
Empirical estimates of α have varied widely among taxa (e.g., Smith and Eyre-Walker 2002; Mikkelsen et al. 2005; Andolfatto 2007; Boyko et al. 2008; Doniger et al. 2008; Foxe et al. 2008; Halligan et al. 2010; Ingvarsson 2010; Slotte et al. 2010). The contribution of positive selection to amino acid divergence is expected to depend on both selective and demographic processes. However, previous comparisons of α among taxa have largely been restricted to groups that are relatively distantly related, varying in aspects of ecology and life history and in Ne (but see Jensen and Bachtrog 2011; Strasburg et al. 2011). In this study, we demonstrate that three closely related taxa with similar ecology and life history show marked variation in α. Furthermore, as reported for fruit flies and sunflowers (Jensen and Bachtrog 2011; Strasburg et al. 2011), observed variation in estimates of α among house mouse subspecies is consistent with differences in estimates of Ne.
Our analyses were based on polymorphism and divergence in ascertained SNPs rather than in DNA sequence data, allowing us to survey the genome broadly. Ascertainment bias in a form that could affect our results seems unlikely (see Materials and Methods), and comparison of levels of variation among polymorphic sites in the three subspecies provides evidence that ascertainment was not dramatically different among subspecies. In addition, our results for M. m. castaneus are consistent with previously published estimates of α based on DNA sequence analysis. Halligan et al. (2010) used resequencing data from 77 autosomal loci to estimate α for M. m. castaneus collected from the same geographic region surveyed in our study. Using a variety of different methods, their estimates of α based on comparisons among 0-fold and 4-fold degenerate sites ranged from 0.22 to 0.57. Despite using different outgroups, their estimates are in good agreement with ours (0.37–0.43). However, it is worth considering how ascertainment bias could affect our results. If the SNPs were primarily discovered in M. m. domesticus, then they may be expected to be at higher frequencies in M. m. domesticus than in M. m. castaneus. Since deleterious replacement mutations will mostly be in excess on the low-frequency end, ascertainment bias could result in a higher ratio of replacement to silent polymorphism in the subspecies in which the SNPs were not ascertained. In this case, we observed the opposite pattern: M. m. castaneus has the lowest rates of replacement to silent polymorphism.
Population Structure, Changes in Ne, and Estimates of α
Population structure and changes in Ne are predicted to affect estimates of α. In this study, the potential impact of demography is particularly relevant because of the close relationship between the subspecies. Exploring the relationship between Ne and the contribution of positive selection to amino acid substitution using recently diverged taxa has the advantage of minimizing the effects of differing biology. However, recently diverged taxa necessarily share evolutionary history, having evolved together away from the outgroup until the time of their divergence. In this case, the three subspecies of M. musculus diverged from each other ∼500,000 years ago, whereas they diverged from M. spretus ∼ 1 Ma. Therefore, the time during which they experienced differences in Ne is relatively short. As a result, estimates of α may be particularly influenced by the effects of changes in Ne reflected in patterns of polymorphism rather than differences in positive selection reflected in patterns of divergence (Keightley and Eyre-Walker 2012).
Ideally, α could be estimated using a lineage specific approach. Because there is limited power for such an approach with this data set, it is important to consider the potential impact of shared evolutionary history and changes in Ne on the interpretation of results. Estimates of the ancestral Ne range from ∼400,000 to ∼500,000 (Geraldes et al. 2011), slightly smaller than estimates of Ne for M. m. castaneus and substantially larger than estimates of Ne for M. m. musculus or M. m. domesticus. Although increases in Ne in M. m. castaneus are predicted to lead to underestimates of α, population contractions in M. m. domesticus and M. m. musculus may have resulted in overestimates of α due to the fixation or near fixation of slightly deleterious mutations segregating prior to the bottlenecks. Both of these potential biases suggest that the inference of more positive selection in M. m. castaneus relative to the other two subspecies is conservative. Nevertheless, specific biases are difficult to predict without more detailed information on the demographic history of the subspecies. The nature of ascertained SNPs prevents the application of methods based on the SFS that have been developed to better account for complex demographic scenarios (Keightley and Eyre-Walker 2007; Eyre-Walker and Keightley 2009). However, we have attempted to test the robustness of our result repeating our analysis with different thresholds for polymorphism, with subsamples to reduce population subdivision within subspecies, and with additional sampling of wild-derived inbred lines to cover a broader geographic range across the subspecies. We consistently found evidence of a significant contribution of positive selection to amino acid substitution for the subspecies with the largest Ne, M. m. castaneus, whereas finding little evidence of positive selection in the other two subspecies.
Importantly, we also found evidence that purifying selection is more effective in the species with the largest Ne, M. m. castaneus. Estimates of α depend on both the ratio of replacement to silent fixed differences, which reflects positive selection, and the ratio of replacement to silent polymorphisms, which reflects purifying selection. Our results underscore that dependence and caution against the strict interpretation of differences in α among taxa as evidence for differences in adaptive substitution.
This study is among the first genome-wide attempts to compare estimates of α among close relatives that occupy similar niches but differ in Ne. The marked and consistent differences in estimates of α among the subspecies observed in this study suggest that this system is well suited for further investigation of the impact of Ne on the efficacy of selection. The likely complex demographic histories of the subspecies add to that potential, providing an opportunity to test theoretical predictions in wild populations.
Supplementary Material
Supplementary analyses, tables 1–9, figures 1 and 2, and data table 1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the National Institutes of Health (GM074245 to M.W.N. and GM076468 to G.A.C.) and the Czech Science Foundation (206/08/0640 to J.P.). We thank F. Pardo-Manuel de Villena for help with sample preparation and F. Pardo-Manuel de Villena, J. Didion, and H. Yang for assistance with the genotyping data used in this study. We thank M. Bomhoff and P. Degnan for assistance with PERL programming. We also thank P. Campbell, B. Harr, F. Mendez, F. Pardo-Manuel de Villena, J.B. Walsh, and reviewers for their thoughtful comments.
References
- Andolfatto P. Hitchhiking effects of recurrent beneficial amino acid substitutions in the Drosophila melanogaster genome. Genome Res. 2007;17:1755–1762. doi: 10.1101/gr.6691007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D. Similar rates of protein adaptation in Drosophila miranda and D. melanogaster, two species with different current effective population sizes. BMC Evol Biol. 2008;8:334. doi: 10.1186/1471-2148-8-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Holloway AK, Stevens K, et al. (13 co-authors) Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko AR, Williamson SH, Indap AR, et al. (14 co-authors) Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 2008;4:e1000083. doi: 10.1371/journal.pgen.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton-Davidian J, Fel-Clair F, Lopez J, Alibert P, Boursot P. Postzygotic isolation between the two European subspecies of the house mouse: estimates from fertility patterns in wild and laboratory-bred hybrids. Biol J Linn Soc. 2005;84:379–393. [Google Scholar]
- Bustamante CD, Wakeley J, Sawyer S, Hartl DL. Directional selection and the site-frequency spectrum. Genetics. 2001;159:1779–1788. doi: 10.1093/genetics/159.4.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth J, Eyre-Walker A. The rate of adaptive evolution in enteric bacteria. Mol Biol Evol. 2006;23:1348–1356. doi: 10.1093/molbev/msk025. [DOI] [PubMed] [Google Scholar]
- Didion JP, Yang H, Sheppard K, Fu C-P, McMillan L, Pardo-Manuel de Villena F, Churchill GA. Discovery of novel variants in genotyping arrays improves genotype retention and reduces ascertainment bias. BMC Genomics. 2012;13 doi: 10.1186/1471-2164-13-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniger SW, Kim HS, Swain D, Corcuera D, Williams M, Yang SP, Fay JC. A catalog of neutral and deleterious polymorphism in yeast. PLoS Genet. 2008;4:e1000183. doi: 10.1371/journal.pgen.1000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvaux L, Belkhir K, Boulesteix M, Boursot P. Isolation and gene flow: inferring the speciation history of European house mice. Mol Ecol. 2011;20:5248–5264. doi: 10.1111/j.1365-294X.2011.05343.x. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD. Estimating the rate of adaptive molecular evolution in the presence of slightly deleterious mutations and population size change. Mol Biol Evol. 2009;26:2097–2108. doi: 10.1093/molbev/msp119. [DOI] [PubMed] [Google Scholar]
- Fay JC, Wyckoff GJ, Wu CI. Positive and negative selection on the human genome. Genetics. 2001;158:1227–1234. doi: 10.1093/genetics/158.3.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wyckoff GJ, Wu CI. Testing the neutral theory of molecular evolution with genomic data from Drosophila. Nature. 2002;415:1024–1026. doi: 10.1038/4151024a. [DOI] [PubMed] [Google Scholar]
- Foxe JP, Dar VUN, Zheng H, Nordborg M, Gaut BS, Wright SI. Selection on amino acid substitutions in Arabidopsis. Mol Biol Evol. 2008;25:1375–1383. doi: 10.1093/molbev/msn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldes A, Basset P, Gibson B, Smith KL, Harr B, Yu HT, Bulatova N, Ziv Y, Nachman MW. Inferring the history of speciation in house mice from autosomal, X-linked, Y-linked and mitochondrial genes. Mol Ecol. 2008;17:5349–5363. doi: 10.1111/j.1365-294X.2008.04005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldes A, Basset P, Smith KL, Nachman MW. Higher differentiation among subspecies of the house mouse (Mus musculus) in genomic regions with low recombination. Mol Ecol. 2011;20:4722–4736. doi: 10.1111/j.1365-294X.2011.05285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good JM, Handel MA, Nachman MW. Asymmetry and polymorphism of hybrid male sterility during the early stages of speciation in house mice. Evolution. 2008;62:50–65. doi: 10.1111/j.1558-5646.2007.00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan DL, Oliver F, Eyre-Walker A, Harr B, Keightley PD. Evidence for pervasive adaptive protein evolution in wild mice. PLoS Genet. 2010;6:e1000825. doi: 10.1371/journal.pgen.1000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson PK. Natural selection on synonymous and nonsynonymous mutations shapes patterns of polymorphism in Populus tremula. Mol Biol Evol. 2010;27:650–660. doi: 10.1093/molbev/msp255. [DOI] [PubMed] [Google Scholar]
- Jensen J, Bachtrog D. Characterizing the influence of effective population size on the rate of adaptation: Gillespie's Darwin Domain. Genome Biol Evol. 2011;3:687–701. doi: 10.1093/gbe/evr063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Eyre-Walker A. Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies. Genetics. 2007;177:2251–2261. doi: 10.1534/genetics.107.080663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Eyre-Walker A. Estimating the rate of adaptive molecular evolution when the evolutionary divergence between species is small. J Mol Evol. 2012;74:61–68. doi: 10.1007/s00239-012-9488-1. [DOI] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge (United Kingdom): Cambridge University Press; 1983. [Google Scholar]
- Maside X, Charlesworth B. Patterns of molecular variation and evolution in Drosophila americana and its relatives. Genetics. 2007;176:2293–2305. doi: 10.1534/genetics.107.071191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Hillier LW, Eichler EE, et al. (67 co-authors) Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Ohta T. Slightly deleterious mutant substitutions in evolution. Nature. 1973;246:96–98. doi: 10.1038/246096a0. [DOI] [PubMed] [Google Scholar]
- Pritchard J, Stephens M, Donelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajabi-Maham H, Orth A, Bonhomme F. Phylogeography and postglacial expansion of Mus musculus domesticus inferred from mitochondrial DNA coalescent, from Iran to Europe. Mol Ecol. 2008;17:627–641. doi: 10.1111/j.1365-294X.2007.03601.x. [DOI] [PubMed] [Google Scholar]
- Rand DM, Kann LM. Excess amino acid polymorphism in mitochondrial DNA: contrasts among genes from Drosophila, mice, and humans. Mol Biol Evol. 1996;13:735–748. doi: 10.1093/oxfordjournals.molbev.a025634. [DOI] [PubMed] [Google Scholar]
- Salcedo T, Geraldes A, Nachman MW. Nucleotide variation in wild and inbred mice. Genetics. 2007;177:2277–2291. doi: 10.1534/genetics.107.079988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sella G, Petrov DA, Przeworski M, Andolfatto P. Pervasive natural selection in the Drosophila genome? PLoS Genet. 2009;5:e1000495. doi: 10.1371/journal.pgen.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro JA, Huang W, Zhang CH, et al. (12 co-authors) Adaptive genic evolution in the Drosophila genomes. Proc Natl Acad Sci U S A. 2007;104:2271–2276. doi: 10.1073/pnas.0610385104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She JX, Bonhomme F, Boursot P, Thaler L, Catzeflis F. Molecular phylogenies in the genus Mus—comparative analysis of electrophoretic, scDNA hybridization, and mtDNA RFLP data. Biol J Linn Soc. 1990;41:83–103. [Google Scholar]
- Slotte T, Foxe JP, Hazzouri KM, Wright SI. Genome-wide evidence for efficient positive and purifying selection in Capsella grandiflora, a plant species with a large effective population size. Mol Biol Evol. 2010;27:1813–1821. doi: 10.1093/molbev/msq062. [DOI] [PubMed] [Google Scholar]
- Smith NGC, Eyre-Walker A. Adaptive protein evolution in Drosophila. Nature. 2002;415:1022–1024. doi: 10.1038/4151022a. [DOI] [PubMed] [Google Scholar]
- Strasburg JL, Kane N, Raduski A, Bonin A, Michelmore R, Rieseberg L. Effective population size is positively correlated with levels of adaptive divergence among annual sunflowers. Mol Biol Evol. 2011;28:1569–1580. doi: 10.1093/molbev/msq270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Shimada T, Terashima M, Tsuchiya K, Aplin K. Temporal, spatial, and ecological modes of evolution of Eurasian Mus based on mitochondrial and nuclear gene sequences. Mol Phylogenet Evol. 2004;33:626–646. doi: 10.1016/j.ympev.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Tucker P, Sandstedt S, Lundrigan B. Phylogenetic relationships in the subgenus Mus (genus Mus, family Muridae, subfamily Murinae): examining gene trees and species trees. Biol J Linn Soc. 2005;84:653–662. [Google Scholar]
- Yang H, Ding Y, Hutchins LN, Szatkiewicz J, Bell T, Paigen BJ, Graber JH, Pardo-Manuel de Villena F, Churchill GA. A customized and versatile high-density genotyping array for the mouse. Nat Methods. 2009;6:663–666. doi: 10.1038/nmeth.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang JR, Didion JP, Buus RJ, Bell TA, Welsh CE, Bonhomme F, Yu HT, Nachman MW, Pialek J, et al. (12 co-authors) Subspecific origin and haplotype diversity in the laboratory mouse. Nat Genet. 2011;43:648–655. doi: 10.1038/ng.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LQ, Li WH. Human SNPs reveal no evidence of frequent positive selection. Mol Biol Evol. 2005;22:2504–2507. doi: 10.1093/molbev/msi240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.