Abstract
Although both genotypes with elevated mutation rate (mutators) and mobilization of insertion sequence (IS) elements have substantial impact on genome diversification, their potential interactions are unknown. Moreover, the evolutionary forces driving gradual accumulation of these elements are unclear: Do these elements spread in an initially transposon-free bacterial genome as they enable rapid adaptive evolution? To address these issues, we inserted an active IS1 element into a reduced Escherichia coli genome devoid of all other mobile DNA. Evolutionary laboratory experiments revealed that IS elements increase mutational supply and occasionally generate variants with especially large phenotypic effects. However, their impact on adaptive evolution is small compared with mismatch repair mutator alleles, and hence, the latter impede the spread of IS-carrying strains. Given their ubiquity in natural populations, such mutator alleles could limit early phase of IS element evolution in a new bacterial host. More generally, our work demonstrates the existence of an evolutionary conflict between mutation-promoting mechanisms.
Keywords: evolution, IS elements, mutation rate
Introduction
Microbes have a broad range of molecular toolkits that promote adaptation under stressful conditions (Aertsen and Michiels 2005). For example, genotypes with increased mutation rates are considered to have an important role during microbial evolution and are frequently found in evolving natural (Matic et al. 1997; Oliver et al. 2000) and experimental populations (Sniegowski et al. 1997). Most repair deficient mutators in nature result from mutations in the methyl-directed mismatch repair (MMR) system (Matic et al. 1997; Oliver et al. 2000). Various forms of stress-induced mutagenesis also have substantial impact on adaptation (Tenaillon et al. 2004; Foster 2007).
In sharp contrast, the evolutionary role of transposable genetic elements, including bacterial insertion sequences (ISs), is still a matter of intense debate (Orgel and Crick 1980; Kidwell and Lisch 2000; Wagner 2009). IS elements are the smallest autonomously replicating mobile genetic elements widely distributed in bacterial genomes (Siguier et al. 2006). Generally, these elements carry only the genes related to their own transposition and its regulation (Siguier et al. 2006). It is well established that insertions of these elements frequently cause alteration of gene expression of adjacent genes and can induce complex genomic rearrangements (Charlier et al. 1982; Prentki et al. 1986; Schneider et al. 2000; Schneider and Lenski 2004). Given their ubiquity and the diversity of mutational events they generate, it is hardly surprising that a fraction of these mutations have beneficial effects. However, the fact that they occasionally play important roles in adaptive evolution (Cooper et al. 2001; Schneider and Lenski 2004; Chou et al. 2009) does not imply that they have been directly selected to enhance the rate of evolutionary adaptation (Lynch 2007).
Specifically, the selective forces driving gradual accumulation of these elements in nascent bacterial genomes are largely unknown. The first steps of this process are especially problematic: it is unclear whether one or few elements in an initially transposon-free bacterial genome can produce sufficient number of beneficial mutations to be favored by selection. Indeed, bacterial transposable genetic elements are only one of the many molecular mechanisms that can potentially promote evolutionary adaptation (Aertsen and Michiels 2005). As mentioned above, bacteria with an elevated genome-wide mutation rate are frequently found in natural populations and are involved in adaptation to stressful conditions (Aertsen and Michiels 2005). However, no prior study has investigated the relative impacts of mutator genotypes and IS elements during evolutionary adaptation.
To examine the potential interactions of IS elements and other mutator alleles on evolvability, we concentrate on Escherichia coli MDS42 (Pósfai et al. 2006), a derivative of E. coli K12. With the use of synthetic biology tools, this strain was specifically designed for elimination of nonessential genes including all mobile DNA and cryptic virulence genes while preserving good growth profiles and protein production (Pósfai et al. 2006). Thus, compared with previous efforts (Chao et al. 1983), E. coli MDS42 is an ideal model system for investigating the early spread of IS elements in a bacterial genome.
We investigated the evolution of IS-carrying and IS-free strains in a medium that contains salicin (a β-glucoside) as the sole carbon source. Normally, E. coli is unable to utilize salicin because the genetic system that encodes the functions for catabolism, the bgl operon, is silent and uninducible (Hall 1998; Madan et al. 2005). Thus, population size declines unless mutants that can exploit salicin arise. Mutations that activate the operon arise spontaneously at an extremely low, but detectable frequency (Moorthy and Mahadevan 2002): Transposition of DNA insertion elements IS1 and IS5 within the regulatory region of the promoter constitutes one class of activating mutations (Schnetz and Rak 1992) (fig. 1). This selection pressure is expected to favor invasion of IS-carrying cells. More generally, our experimental setup mimics two key aspects of bacterial physiology: 1) due to severe nutrient limitations, bacteria are generally starving and are in stationary phase (Morita 1997) and 2) very rare mutations allow growth on nutrients which are otherwise inaccessible. These may be common features of many natural bacterial habitats.
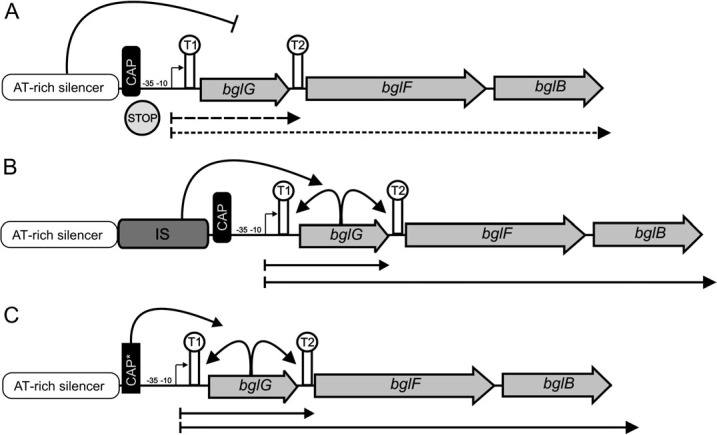
FIG. 1.
Activation modes of the Escherichia coli cryptic bgl operon. Genes are indicated by gray arrows. (A) In wild-type cells, transcription of the bgl operon is silenced by multiple mechanisms. First, the promoter region (−35 and −10 region) is directly silenced by a nearby AT-rich DNA silencer element. Second, two rho-independent transcriptional terminators (T1 and T2) interrupt ongoing transcription (at the beginning and at the end of bglG). (B) IS insertion (dark gray box) can displace the silencer element from the bgl promoter region (−35 and −10) and allows transcription of the BglG antiterminator protein. Due to efficient antitermination by BglG in the presence of salicin, the operon is transcribed (filled, black arrows). (C) Mutations changing the CAP-binding site (starred black box) allow strong catabolic activation during carbon starvation. In addition, there are also numerous known point mutations that can activate the bgl operon. For more details, see Hall (1998) and Moorthy and Mahadevan (2002).
We demonstrate that when initially rare in a given bacterial genome, a highly active family of IS elements (IS1) produces a relatively low number of beneficial mutations compared with MMR mutator alleles, and hence, the latter can take over in evolving bacterial populations. Our work highlights the importance of studying interactions between mechanisms that promote evolvability. Investigating various mutational mechanisms in isolation can lead to a false conclusion on their role in evolution.
Materials and Methods
Bacterial Strains
This study uses series of competition experiments among IS-free (MDS42), single IS1-harboring (MDS42-IS1), and double IS1-harboring (MDS42-2IS1) E. coli strains to investigate the evolutionary role of IS1 elements. E. coli strain MDS42, lacking all mobile DNA elements from its genome, has been described previously (Pósfai et al. 2006). A single copy of IS1 was transferred from MG1655 to MDS42 by P1 transduction (Perna et al. 2001) into position of the yeaJ gene (Pósfai et al. 2006), leading to MDS42-IS1. To allow selection during P1 transduction, a suicide plasmid was inserted in the near vicinity of the yeaJ gene, as described earlier (Pósfai et al. 1999), which was removed after the transduction process. Another copy of IS1 interrupting the crl gene was transferred in a similar manner to make MDS42-2IS1. To eliminate potential fitness effects of crl and yeaJ gene interruptions during competitions, these genes were also deleted in the corresponding competing partners (supplementary methods and table 1, Supplementary Material online). The ΔmutS versions of all strains were generated using the suicide plasmid method (Pósfai et al. 1999).
To allow automatic readout of competition experiments, competing strains were labeled with yellow (YFP) or cyan (CFP) fluorescent protein. Genes of the fluorescent proteins, inserted into the genome, were under strict transcriptional regulation and were expressed only in the “readout phase” at the very end of competition experiments, that is when 66 ng/ml anhydotetracycline (AhTc) was added to the medium (see supplementary methods, Supplementary Material online). Thus, the outcome of competition experiments is not biased by the potential fitness cost of fluorescent protein expression or differences in initial fitness (supplementary figs. 1 and 2, Supplementary Material online). Fluorescence emissions of MDS42-CFP and MDS42-YFP strains were also tested using confocal fluorescence microscopy (supplementary fig. 3, Supplementary Material online), and mixtures of labeled strains were used for calibration (supplementary fig. 4, Supplementary Material online).
The presence of IS insertions and all gene deletions were verified using polymerase chain reaction (PCR). For more details, see supplementary methods and table 1 (Supplementary Material online).
Competition Experiments
In the first type of competition experiments, experimental populations with starting 1:1 ratio of IS+/IS− genotypes were allowed to adapt to mineral salts minimal (MS-minimal) medium (Hall 1998) that contains salicin (salicin-minimal) as the sole carbon source (fig. 2B). Pairwise competitions were carried out by incubating ∼109 cells in a 1:1 initial ratio in 0.4% salicin-minimal medium in at least 480 parallel cultures. Readout of the experiments was performed after 7 days by transferring a small aliquot of the mixture into another microplate containing fresh salicin-minimal medium and the inducer of the fluorescent proteins (fig. 2B). The genetic background of each bgl+ population was inferred from YFP and CFP fluorescent intensities measured in a microplate reader (Hegreness et al. 2006). We concentrated on the most clear-cut cases, that is when 1) the population shows evidence for exploitation of salicin and 2) one of the two genotypes has reached at least 90% frequency in the population. We confirmed that induction of fluorescent labeling has no measurable effects on the outcome of competition during long-term culturing in MS-minimal supplemented with 0.4% glucose (supplementary fig. 1, Supplementary Material online). Furthermore, addition of the inducer did not cause differential fitness effects across differently labeled strains (supplementary fig. 2, Supplementary Material online).
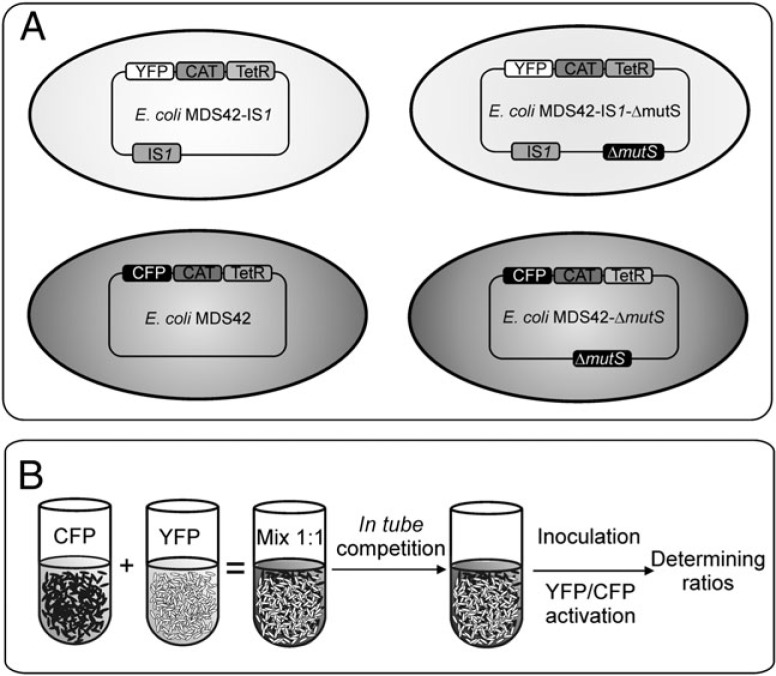
FIG. 2.
Competing genotypes and experimental setup. (A) This study uses a series of competition experiments among IS-free and IS1-harboring Escherichia coli MDS42 strains to investigate the impact of IS elements on evolutionary adaptation. E. coli strain MDS42 lacks all mobile DNA elements from its genome. An active single copy of IS1 was inserted to MDS42 by P1 transduction. Mutator versions of the above strains were generated by deleting the MMR gene mutS. Competing strains were also labeled with inducible yellow (YFP) or cyan (CFP) fluorescent proteins by inserting the corresponding genes into the genome (for a complete list of strains, see supplementary table 1, Supplementary Material online). The constitutively expressed TetR repressor enables transcriptional control of fluorescent protein–coding genes. An additional chloramphenicol resistance marker (CAT) ensures the stability of fluorescent markers. (B) Competition experiments were carried out with a starting 1:1 ratio of two strains carrying the YFP or CFP genes, respectively. Unless otherwise indicated in the main text, the first phase of the competition experiments involved long-term culturing of the mixed population in a medium that contains salicin as the sole carbon source. Small amounts of evolved cultures were transferred into fresh salicin-minimal medium containing inducer of fluorescent protein expression. The ratios of the two strains were inferred from the measured fluorescence intensities.
The second type of competition experiments was carried out in mixed glucose/salicin medium. One hundred and sixty-four populations of MDS42 (CFP) and MDS42-IS1 (YFP) cells were mixed 1:1 and were grown in two 96-well microtiter plates. Each well contained 100 μl of 0.04% glucose and 0.4% salicin. These populations were allowed to evolve for 22 days. Every 48 h, ∼2 μl of the grown-up cultures were transferred to fresh medium using a 96-pin replicator (VP407). As above, all plates were shaken at 37 °C, 300–350 rpm, and 5 cm shaking diameter (Duetz 2007). None of the bacteria-free wells were contaminated in the course of the entire experiment (data not shown).
To reliably infer the ratios of YFP- to CFP-labeled cells based on measured fluorescent intensities, we trained a linear regression model using fluorescent data from a reference set with known ratios of labeled cells (see supplementary methods, Supplementary Material online). Estimated ratios of differentially labeled fluorescent cells were confirmed by fluorescent microscopy (supplementary fig. 4, Supplementary Material online).
Estimating Growth Parameters of bgl+ Genotypes
Cells from bgl+ MDS42 and MDS42-IS1 populations were isolated and purified to single colonies. Identification of the mutation type that led to the bgl+ phenotype was carried out by colony PCR analysis using the bglR1/bglR2 primer pair (Schnetz and Rak 1992). Overnight cultures (1–0.6 μl) of the isolated 15–23 non-IS and IS+ mutants (all from independent source populations) were inoculated into 384-well shallow plates (eight replicates per mutants). Each well contained 50 μl salicin-minimal medium. Growth curves were recorded by measuring every 7 min for 72 h at 37 °C using a Biotek automated plate reader. Growth parameters (length of lag phase, growth rate, optical density increment) were calculated from the obtained growth curves following a reported procedure (Warringer and Blomberg 2003; Warringer et al. 2003).
Determining the Rate of Adaptive Mutations
Mutation rate was calculated using fluctuation analysis based on the fraction of bgl+ cells (salicin exploiters) as observed in 20 parallel cultures (see supplementary material, Supplementary Material online). Briefly, fully grown bacterial cultures were plated on salicin-minimal plates, and colonies were counted after 4 days. Fluctuation analyses were performed on the colony numbers using the maximum likelihood method (Sarkar et al. 1992) to determine the number of mutations that occurred in a culture (mutation/culture). Mutation rates were obtained by dividing the mutation/culture values by the total cell counts, determined by plating appropriate dilutions from three randomly chosen tubes onto Luria-Bertani plates.
Results
A Single IS Element Provides Evolutionary Advantage to the Bacterial Host in the Laboratory
We inserted a single IS1 element in the genome of E. coli MDS42. The IS1 family is one of the smallest highly active mobile elements of E. coli and relatives (Sawyer et al. 1987; Mahillon and Chandler 1998) and is frequently involved in reactivation and silencing of bacterial host genes (Mahillon and Chandler 1998). The fluorescent reporter constructs YFP (yellow) and CFP (cyan) with inducible promoters were inserted into the genome of IS1 bearing (IS+) and IS1-free (IS−) strains, respectively (fig. 2A). Importantly, using isogenic IS-free strains, we could confirm that induction of these markers was neutral with respect to bacterial fitness (supplementary figs. 1 and 2, Supplementary Material online).
After only 1 week of adaptation, we found that the ratio of IS+/IS− strains was 9:1 among bgl+ populations, that is those that are capable of growth on salicin medium (fig. 3 and supplementary table 2, Supplementary Material online). After an additional 2 weeks of culturing in the same salicin-containing medium, we found no increase in the number of bgl+ populations (data not shown). Emerging mutations at the bgl locus were investigated by streaking samples onto salicin-minimal plates, followed by colony PCR amplification and sequencing (supplementary methods, Supplementary Material online). The analysis confirmed an inserted IS1 element in the bgl regulatory locus in ∼90% of the evolved IS+ populations (supplementary table 3, Supplementary Material online). In agreement with known molecular mechanisms of transposition (Mahillon and Chandler 1998), PCR analysis also confirmed the presence of an IS1 copy at the original genomic location (data not shown).
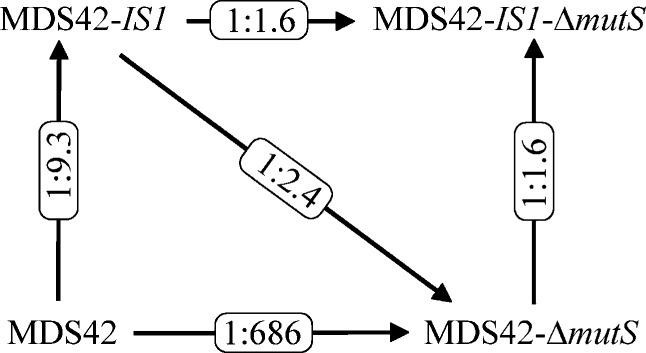
FIG. 3.
Outcome of competition experiments. Summary of competition experiments. Each arrow indicates the result of a competition experiment. Arrows point toward the genotypes with a higher probability of fixation in the populations, while the numbers above the arrows represent the ratios of the outcomes of parallel evolved populations. As can be seen on the figure, both IS carrying (IS+) and mutator (ΔmutS) genotypes outcompete MDS42 (χ2 = 39.8 degrees of freedom [df] = 1, P < 10−4 and χ2 = 454 df = 1, P << 10−4, respectively). Moreover, direct competition between IS+ and ΔmutS showed a higher selection of the latter (χ2 = 7.8 df = 1, P = 0.005). Remarkably, when competing genotypes have high underlying mutation rates (ΔmutS), selection of IS+ over IS− is significantly reduced (χ2 = 32.6 df = 1, P << 10−4): genotypes can fix in ∼61% of the adapted populations. Similarly, when both genotypes contain an IS element (ΔmutS IS+ vs. IS+), spread of ΔmutS mutator in the population is significantly lower compared with that in populations with IS-free genotypes (χ2 = 271 df = 1, P << 10−4). For more details on the outcome of competition experiments, see supplementary table 2 (Supplementary Material online).
As mutational processes (including IS transposition) differ substantially under starvation from those occurring during logarithmic growth phase (Hall 1998; Bjedov et al. 2003), it is important to confirm the advantage of IS+ strains under different culturing conditions. Equal ratios of IS+/IS− cells were allowed to evolve for 22 days through serial transfers (Materials and Methods). One hundred and sixty-four parallel populations adapted to a glucose-limited minimal medium that also contained salicin. Thus, populations can only grow at a low rate unless they acquire the capacity to exploit salicin. After 22 days, 89 of the evolved populations acquired the capacity to exploit salicin, and IS+ genotype became fixed in 84% of these populations, a significant departure from random expectation (χ2 = 41.2 degrees of freedom = 1, P << 10−4). We conclude that a single IS1 element can accelerate host adaptation to salicin irrespective of the details of culturing conditions. Therefore, all further experiments were performed under the first condition, where salicin was the sole carbon source.
IS Element–Induced Mutations Allow More Efficient Growth
To estimate the impact of IS1 elements on mutagenesis, we first measured the rate of mutations conferring growth on salicin-minimal plates. A single copy of IS1 in the genome causes an approximately 10-fold increase in the rate of such mutations over an IS-free host (fig. 4A). Next, we asked whether IS element-generated mutations are phenotypically different from mutations found in the absence of IS elements. We isolated 24 IS+ and 13 IS− independent bgl+ mutant strains and measured their growth characteristics in liquid salicin medium (see Materials and Methods). High-resolution growth curves were measured over 72 h and three relevant growth variables—length of lag phase, rate (slope), and efficiency (increment in optical density)—were extracted. We failed to find any advantage of IS+ mutants over IS− ones when lag phase length and rate were investigated (P = 0.65 and P = 0.58, respectively, using Wilcoxon rank tests). In sharp contrast, we observed a large variation in final optical densities of IS+, but not that of IS− mutants. Remarkably, at least 7 of the 24 investigated IS+ mutants showed an especially high OD increment (fig. 4B, P < 0.001 for the difference between IS+ and IS−). Taken together, these results indicate that, at least in our environmental settings, IS1 has the capacity to create mutations with exceptional phenotypic effects (but see Stoebel and Dorman 2010).
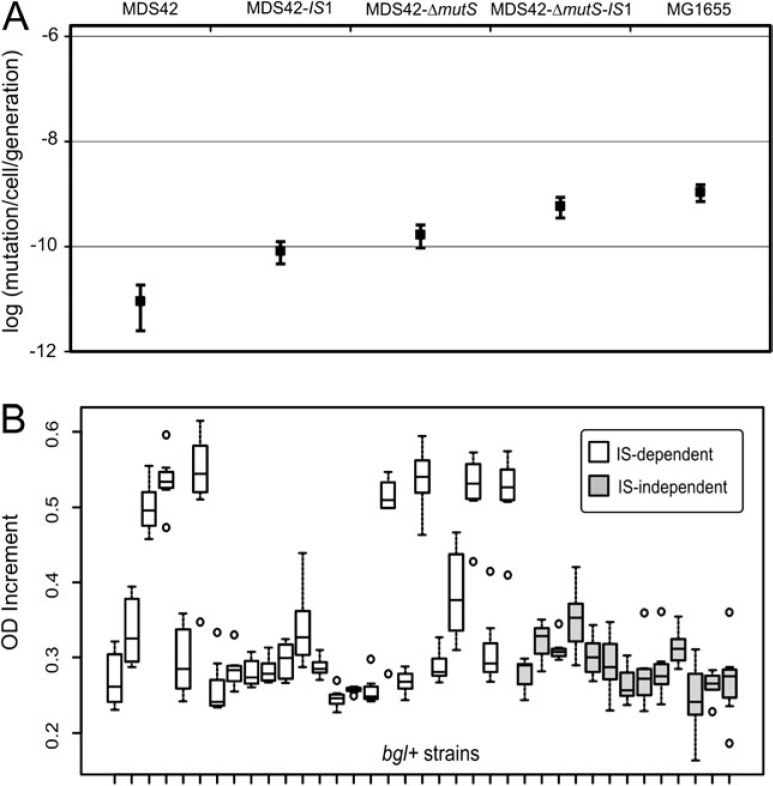
FIG. 4.
Mutation rate and fitness measurements. (A) The figure shows mutation rates calculated from the numbers of colonies growing on salicin-minimal plates using fluctuation analyses. Bars indicate 95% confidence intervals. (B) Fitness of salicin-exploiting genotypes. We compared the growth characteristics of 37 strains where growth data were available and OD increment showed a coefficient of variation less than 0.2. Horizontal lines of the boxplots correspond to the medians, the bottoms and tops of the boxes show the 25th and 75th percentiles, respectively. Whiskers show either the maximum (minimum) value or 1.5 times the interquartile range of the data, whichever is smaller (higher). Points more than 1.5 times the interquartile range above the third quartile or below the first quartile are plotted individually as outliers. Difference of bgl+ IS-mutants compared with point mutants was highly significant (Wilcoxon rank-sum test, P < 10−4).
IS-Carrying Strains Are Outcompeted by MMR Mutators
Natural populations typically carry a proportion of hypermutating cells with deficient repair enzymes (Matic et al. 1997; Oliver et al. 2000). Might the presence of such mutants interfere with the spread of IS elements? The relative impact of repair-mutators versus IS1 elements on adaptive evolution was addressed by constructing a mutS deletion in starting IS+ and IS− MDS42 genotypes (fig. 2A). It has been previously shown that deletion of this MMR gene confers an approximately 100-fold increase in genome-wide mutation rates (Rewinski and Marinus 1987). First, we confirmed that when mutS mutant (ΔmutS) and nonmutator (mutS+) were competed in the absence of IS elements, mutators had a massive selective advantage (fig. 3), and they became fixed in nearly all bgl+ populations. In isolation, both IS- and repair-deficient mutators function as boosters of mutability and hence of evolvability. Additionally, direct competition between IS+/mutS+ and IS−/ΔmutS strains revealed that ΔmutS mutators generally outcompeted IS+/mutS+ strains (fig. 3).
Does this conclusion remain valid when more than one copy of IS1 resides initially in the genome? Competition experiments between strains with one and two IS1 elements revealed a substantial advantage of the latter genotype (supplementary table 2, Supplementary Material online). However, ΔmutS mutator strains can generally outcompete genotypes with two IS elements as well (supplementary table 2, Supplementary Material online). Taken together, our results indicate that rare IS1 elements produce sufficient numbers of mutations to be favored by selection. Some of these IS-generated mutations deliver exceptional high-fitness phenotypes. However, the impact of ΔmutS mutators on evolvability is higher, and hence, they are favored over IS-carrying strains in adapting bacterial populations.
The above assays consider what happens when an IS element arrives in a repair-proficient background. What would be the fate of strains carrying both mutation-promoting mechanisms? First, we established that lack of MMR activity in ΔmutS genetic background has no negative effect on the frequency of transposition (supplementary table 3, Supplementary Material online). Next, we repeated the competition of mutator (ΔmutS) and non-mutator strains, but this time, both genotypes had a fixed IS1 element in the genome. Remarkably, in contrast to the outcome of the competition with IS1-free genotypes where non-mutators were always outcompeted, in this case, the ratio of IS1+/ΔmutS:IS1+/mutS+ was 1.6:1 among bgl+ populations (fig. 3). This result suggests that presence of a single IS1 in the bacterial genome can alter the evolutionary trajectory of repair deficiency mutator alleles.
Last, we examined the other side of the coin: How frequently IS+ genotypes can outcompete IS− when both genotypes have high underlying genomic mutation rates (ΔmutS)? Results showed that in ΔmutS mutator genetic background, fixation probability of the IS-carrying strain is substantially reduced: genotypes can spread in 61% of the competitions (fig. 3 and supplementary table 3, Supplementary Material online), whereas 50% would be expected in the case of a neutral allele. Thus, in the presence of one mutation promoting mechanism (IS or MMR mutator allele) in the genome, the other can still invade the population, albeit much less frequently. Thus, IS elements and mutators slightly but significantly reduce second-order selection of the other.
Discussion
In this work, through a series of competition experiments, we demonstrated that a single IS element can accelerate host adaptation under a specific environmental condition. However, even in such an environment, MMR mutators have higher impact on evolvability and can invade IS-carrying populations. Although we do not wish to conclude that IS elements never cause adaptive mutations, our work highlights a problem with the hypothesis that transposable elements spread as they provide host genomes with the ability to enhance their own evolution. Once present in many copies across the genome, they generate numerous beneficial (and detrimental) genomic rearrangements and insertions. However, when only one or few copies exist in a given bacterial genome, their impact on adaptive evolution is expected to be small compared with other mutation-generating pathways, such as MMR deficiency. Given the ubiquity of repair-deficient strains in wild populations, it is unclear how often IS elements spread owing to their mutagenic effects. Indeed, the E. coli K12 genome harbors 7 IS1 and 12 IS5 elements (Blattner et al. 1997). Consequently, due to their combined effects, the rate of bgl+ mutations in this strain is nearly 118-fold higher compared with that of MDS42 (fig. 4A). However, the same comparison between MDS42 ΔmutS and E. coli K12 reveals a mere 6-fold increase (fig. 4A).
Although we considered in detail adaptation associated with just one gene, we think our results might yet be quite general, not least because the IS1 element is one of the most highly active bacterial elements, frequently involved in genomic changes (Mahillon and Chandler 1998; Schneider et al. 2000; Fehér et al. 2006). Moreover, although the profile of the forms of mutations (insertions, point mutations, inversions etc.) generated by mutS and IS1 are likely to be different, we were conservative in choosing a system where IS1 is a priori more likely to be the source of adaptive mutations. To address this issue further, we investigated the contribution of IS and non-IS generated mutations in growing populations of bacteria by selection of mutants resistant to d-cycloserine, a cell wall synthesis inhibitor antibiotic. In MG1655, IS transpositions (including IS1, see Fehér et al. 2006) accounted for 24.2% of the resistance conferring mutations (Pósfai et al. 2006). Remarkably, a single IS element in MDS42 has no significant impact on the rate of resistance mutations, whereas the same figure in MDS42 ΔmutS shows a 17-fold increase (for more details, see supplementary fig. 5, Supplementary Material online). These preliminary results support the general conclusion that MMR mutators generate more adaptive mutations than a single IS element. Nonetheless, it is at least conceivable that adaptation to certain environments might be only achievable through IS elements, in which case our conclusions would not hold. Clearly, future works should examine how robust our conclusions are across other classes of IS elements, environmental conditions, and bacterial species.
We note, however, that IS elements are frequently activated during times of stress (Siguier et al. 2006), via the SOS response (Levy et al. 1993). Thus, constitutive mutators may accumulate more deleterious mutations under no stress conditions, conferring a potential long-term advantage to IS-carrying populations. In a similar vein, under certain environmental conditions, intermediate mutation frequency provides the highest fitness benefit at the population level (Denamur et al. 2005).
More generally, our results also highlight the evolutionary interplay between different mutational systems, whereby the presence of one variability-generating mechanism can influence the fate of others (Tenaillon et al. 2000; Bjedov et al. 2003). We speculate that such competition may also hold among different classes of IS elements as long as they share some potential target genes subject to selection.
For studies of evolvability, interactions between different mechanisms are likely to be important. For example, sex can promote adaptive evolution of bacterial populations (Cooper 2007). The impact of IS elements on host evolution may be more pronounced in sexual populations. Theoretical studies indicate that even rare genetic exchanges can prevent mutator alleles from spreading in the population, as mutators no longer remain associated with the rare favorable mutations they generate (Tenaillon et al. 2000). Given that mobile element insertions generally affect neighboring genes, they are more likely to be genetically linked to the mutation they generate (Chao et al. 1983; Zeyl et al. 1996). Future works should reveal the evolutionary interactions between diverse recombinational repair pathways and mobile genetic elements.
Supplementary Material
Supplementary methods, figures 1–6, and tables 1–4 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We would like to thank the reviewers for their insightful comments on the manuscript. Plasmids were kindly provided by Natalie Balaban. This work was supported by grants from European Research Council (202591), Welcome Trust and EMBO Young Investigator Programme (C.P.), the ‘Lendület Program’ of the Hungarian Academy of Sciences (B.P.), the International Human Frontier Science Program Organization, and the Hungarian Scientific Research Fund (PD 75261 [B.P.] and PD 72719 [T.F.]).
References
- Aertsen A, Michiels CW. Diversify or die: generation of diversity in response to stress. Crit Rev Microbiol. 2005;31:69–78. doi: 10.1080/10408410590921718. [DOI] [PubMed] [Google Scholar]
- Bjedov I, Tenaillon O, Gerard B, Souza V, Denamur E, Radman M, Taddei F, Matic I. Stress-induced mutagenesis in bacteria. Science. 2003;300:1404–1409. doi: 10.1126/science.1082240. [DOI] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, 3rd, Bloch CA, et al. (17 co-authors) The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Chao L, Vargas C, Spear BB, Cox EC. Transposable elements as mutator genes in evolution. Nature. 1983;303:633–635. doi: 10.1038/303633a0. [DOI] [PubMed] [Google Scholar]
- Charlier D, Piette J, Glansdorff N. IS3 can function as a mobile promoter in E. coli. Nucleic Acids Res. 1982;10:5935–5948. doi: 10.1093/nar/10.19.5935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HH, Berthet J, Marx CJ. Fast growth increases the selective advantage of a mutation arising recurrently during evolution under metal limitation. PLoS Genet. 2009;5:e1000652. doi: 10.1371/journal.pgen.1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TF. Recombination speeds adaptation by reducing competition between beneficial mutations in populations of Escherichia coli. PLoS Biol. 2007;5:e225. doi: 10.1371/journal.pbio.0050225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper VS, Schneider D, Blot M, Lenski RE. Mechanisms causing rapid and parallel losses of ribose catabolism in evolving populations of Escherichia coli B. J Bacteriol. 2001;183:2834–2841. doi: 10.1128/JB.183.9.2834-2841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denamur E, Tenaillon O, Deschamps C, Skurnik D, Ronco E, Gaillard JL, Picard B, Branger C, Matic I. Intermediate mutation frequencies favor evolution of multidrug resistance in Escherichia coli. Genetics. 2005;171:825–827. doi: 10.1534/genetics.105.045526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duetz WA. Microtiter plates as mini-bioreactors: miniaturization of fermentation methods. Trends Microbiol. 2007;15:469–475. doi: 10.1016/j.tim.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Fehér T, Cseh B, Umenhoffer K, Karcagi I, Posfai G. Characterization of cycA mutants of Escherichia coli. An assay for measuring in vivo mutation rates. Mutat Res. 2006;595:184–190. doi: 10.1016/j.mrfmmm.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Foster PL. Stress-induced mutagenesis in bacteria. Crit Rev Biochem Mol Biol. 2007;42:373–397. doi: 10.1080/10409230701648494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BG. Activation of the bgl operon by adaptive mutation. Mol Biol Evol. 1998;15:1–5. doi: 10.1093/oxfordjournals.molbev.a025842. [DOI] [PubMed] [Google Scholar]
- Hegreness M, Shoresh N, Hartl D, Kishony R. An equivalence principle for the incorporation of favorable mutations in asexual populations. Science. 2006;311:1615–1617. doi: 10.1126/science.1122469. [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Lisch DR. Transposable elements and host genome evolution. Trends Ecol Evol. 2000;15:95–99. doi: 10.1016/s0169-5347(99)01817-0. [DOI] [PubMed] [Google Scholar]
- Levy MS, Balbinder E, Nagel R. Effect of mutations in SOS genes on UV-induced precise excision of Tn10 in Escherichia coli. Mutat Res. 1993;293:241–247. doi: 10.1016/0921-8777(93)90075-r. [DOI] [PubMed] [Google Scholar]
- Lynch M. The origins of genome architecture. Sunderland (MA): Sinauer Press; 2007. [Google Scholar]
- Madan R, Kolter R, Mahadevan S. Mutations that activate the silent bgl operon of Escherichia coli confer a growth advantage in stationary phase. J Bacteriol. 2005;187:7912–7917. doi: 10.1128/JB.187.23.7912-7917.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matic I, Radman M, Taddei F, Picard B, Doit C, Bingen E, Denamur E, Elion J. Highly variable mutation rates in commensal and pathogenic Escherichia coli. Science. 1997;277:1833–1834. doi: 10.1126/science.277.5333.1833. [DOI] [PubMed] [Google Scholar]
- Moorthy S, Mahadevan S. Differential spectrum of mutations that activate the Escherichia coli bgl operon in an rpoS genetic background. J Bacteriol. 2002;184:4033–4038. doi: 10.1128/JB.184.14.4033-4038.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita RY. Bacteria in oligotrophic environments: starvation-survival lifestyle. New York: Chapman & Hall; 1997. [Google Scholar]
- Oliver A, Canton R, Campo P, Baquero F, Blazquez J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science. 2000;288:1251–1254. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- Orgel LE, Crick FH. Selfish DNA: the ultimate parasite. Nature. 1980;284:604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- Perna NT, Plunkett G, 3rd, Burland V, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- Pósfai G, Kolisnychenko V, Bereczki Z, Blattner FR. Markerless gene replacement in Escherichia coli stimulated by a double-strand break in the chromosome. Nucleic Acids Res. 1999;27:4409–4415. doi: 10.1093/nar/27.22.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pósfai G, Plunkett G, 3rd, Feher T, et al. Emergent properties of reduced-genome Escherichia coli. Science. 2006;312:1044–1046. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- Prentki P, Teter B, Chandler M, Galas DJ. Functional promoters created by the insertion of transposable element IS1. J Mol Biol. 1986;191:383–393. doi: 10.1016/0022-2836(86)90134-8. [DOI] [PubMed] [Google Scholar]
- Rewinski C, Marinus MG. Mutation spectrum in Escherichia coli DNA mismatch repair deficient (mutH) strain. Nucleic Acids Res. 1987;15:8205–8215. doi: 10.1093/nar/15.20.8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Ma WT, Sandri GH. On fluctuation analysis: a new, simple and efficient method for computing the expected number of mutants. Genetica. 1992;85:173–179. doi: 10.1007/BF00120324. [DOI] [PubMed] [Google Scholar]
- Sawyer SA, Dykhuizen DE, DuBose RF, Green L, Mutangadura-Mhlanga T, Wolczyk DF, Hartl DL. Distribution and abundance of insertion sequences among natural isolates of Escherichia coli. Genetics. 1987;115:51–63. doi: 10.1093/genetics/115.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D, Duperchy E, Coursange E, Lenski RE, Blot M. Long-term experimental evolution in Escherichia coli. IX. Characterization of insertion sequence-mediated mutations and rearrangements. Genetics. 2000;156:477–488. doi: 10.1093/genetics/156.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D, Lenski RE. Dynamics of insertion sequence elements during experimental evolution of bacteria. Res Microbiol. 2004;155:319–327. doi: 10.1016/j.resmic.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Schnetz K, Rak B. IS5: a mobile enhancer of transcription in Escherichia coli. Proc Natl Acad Sci U S A. 1992;89:1244–1248. doi: 10.1073/pnas.89.4.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguier P, Filee J, Chandler M. Insertion sequences in prokaryotic genomes. Curr Opin Microbiol. 2006;9:526–531. doi: 10.1016/j.mib.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Sniegowski PD, Gerrish PJ, Lenski RE. Evolution of high mutation rates in experimental populations of E. coli. Nature. 1997;387:703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- Stoebel DM, Dorman CJ. The effect of mobile element IS10 on experimental regulatory evolution in Escherichia coli. Mol Biol Evol. 2010;27:2105–2112. doi: 10.1093/molbev/msq101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O, Denamur E, Matic I. Evolutionary significance of stress-induced mutagenesis in bacteria. Trends Microbiol. 2004;12:264–270. doi: 10.1016/j.tim.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Tenaillon O, Le Nagard H, Godelle B, Taddei F. Mutators and sex in bacteria: conflict between adaptive strategies. Proc Natl Acad Sci U S A. 2000;97:10465–10470. doi: 10.1073/pnas.180063397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. Transposable elements as genomic diseases. Mol Biosyst. 2009;5:32–35. doi: 10.1039/b814624c. [DOI] [PubMed] [Google Scholar]
- Warringer J, Blomberg A. Automated screening in environmental arrays allows analysis of quantitative phenotypic profiles in Saccharomyces cerevisiae. Yeast. 2003;20:53–67. doi: 10.1002/yea.931. [DOI] [PubMed] [Google Scholar]
- Warringer J, Ericson E, Fernandez L, Nerman O, Blomberg A. High-resolution yeast phenomics resolves different physiological features in the saline response. Proc Natl Acad Sci U S A. 2003;100:15724–15729. doi: 10.1073/pnas.2435976100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeyl C, Bell G, Green DM. Sex and the spread of retrotransposon Ty3 in experimental populations of Saccharomyces cerevisiae. Genetics. 1996;143:1567–1577. doi: 10.1093/genetics/143.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.