Abstract
Endogenous retroviruses provide molecular fossils for studying the ancient evolutionary history of retroviruses. Here, we report our independent discovery and analysis of endogenous lentiviral insertions (Mustelidae endogenous lentivirus [MELV]) within the genomes of weasel family (Mustelidae). Genome-scale screening identified MELV elements in the domestic ferret (Mustela putorius furo) genome (MELVmpf). MELVmpf exhibits a typical lentiviral genomic organization. Phylogenetic analyses position MELVmpf basal to either primate lentiviruses or feline immunodeficiency virus. Moreover, we verified the presence of MELV insertions in the genomes of several species of the Lutrinae and Mustelinae subfamilies but not the Martinae subfamily, suggesting that the invasion of MELV into the Mustelidae genomes likely took place between 8.8 and 11.8 Ma. The discovery of MELV in weasel genomes extends the host range of lentiviruses to the Caniformia (order Carnivora) and provides important insights into the prehistoric diversity of lentiviruses.
Keywords: weasel, lentivirus, endogenous retrovirus
The long-term evolutionary mode of lentiviruses remains elusive. Retroviruses can integrate into germline genomes, providing important insights into the deep history of these viruses (Johnson and Coffin 1999). However, genome invasion by lentiviruses appears to be very rare (Katzourakis et al. 2007). The short list includes those of the lemurs (pSIV) and the European rabbit (RELIK) (Katzourakis et al. 2007; Gifford et al. 2008; Gilbert et al. 2009). Recently, Cui and Holmes (2012) discovered lentiviral insertions in the genome of the domestic ferret (Mustela putorius furo) (ELVmpf). Here, we report on our independent discovery of endogenous lentiviral elements in the weasel family, both in the domestic ferret and also several wild species. We suggest the weasel family lentiviral elements be designated “Mustelidae endogenous lentivirus” (MELV).
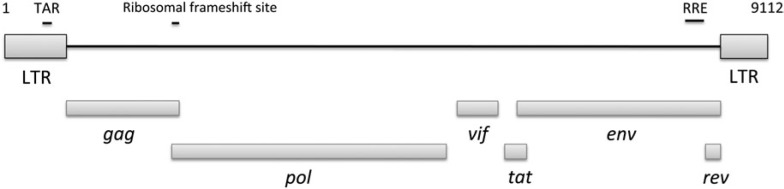
We screened all Whole Genome Shotgun (WGS) sequences from GenBank using TBLASTN and various lentivirus proteins (supplementary table S1, Supplementary Material online) and identified six full or partial lentiviral insertions and 13 solo LTRs (supplementary table S2, Supplementary Material online) in the domestic ferret genome (MELVmpf). The consensus sequence exhibits a typical lentiviral organization, encoding three long open reading frames, gag, pol, and env, and three putative accessory genes, vif, tat, and rev (fig. 1 and supplementary figs. S1 and S2, Supplementary Material online). MELVmpf also encodes three putative RNA secondary structure elements: a ribosomal frameshift site, a transactivation responsive region, and a Rev responsive element (supplementary fig. S3, Supplementary Material online). A maximum-likelihood (ML) tree of MELVmpf and exogenous retroviruses shows that MELVmpf groups within the lentiviruses with robust support (supplementary fig. S3, Supplementary Material online), confirming it is an endogenous lentivirus.
FIG. 1.
MELVmpf genomic organization. LTR, long-terminal repeat; TAR, transactivation responsive element; RRE, Rev responsive element.
We also discovered a previously overlooked putative vif gene in another endogenous lentivirus, RELIK (supplementary fig. S2, Supplementary Material online). Putative vif genes in both RELIK and MELVmpf genomes suggest vif may be a very ancient feature of lentivirus genomes; all members of the lentiviral family except equine infectious anemia virus possess a putative vif gene.
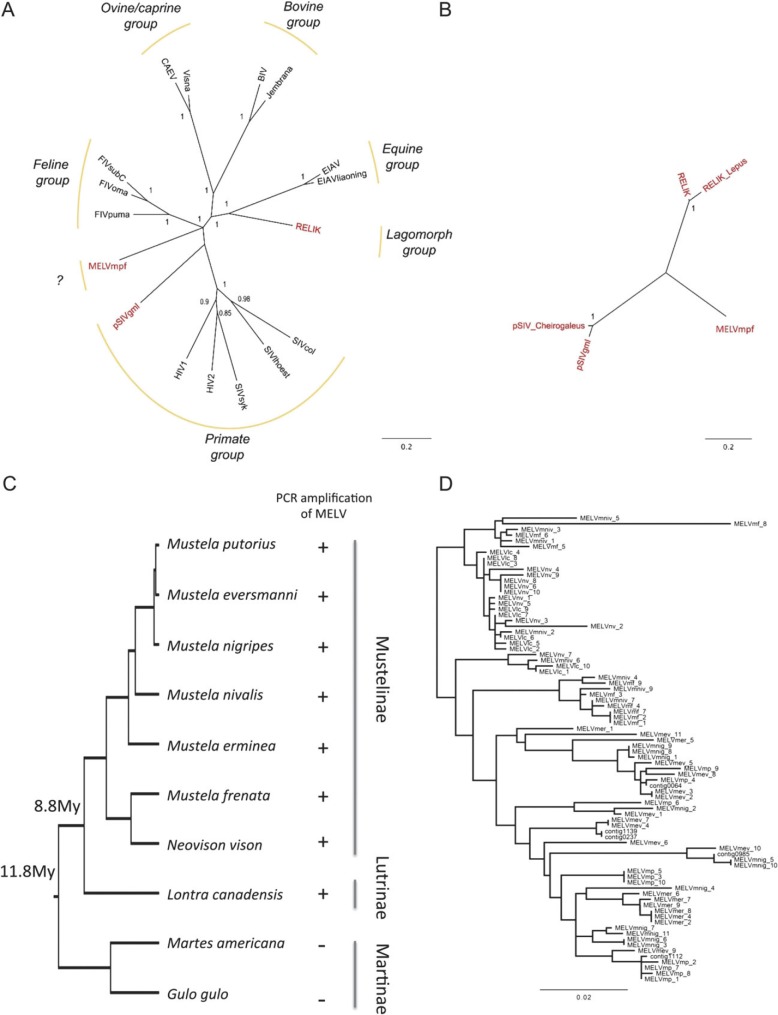
To determine the relationship between MELVmpf and other lentiviruses, we analyzed conserved regions of the pol amino acid sequences using a Bayesian phylogenetic approach, finding a similar overall topology as previous phylogenies (Katzourakis et al. 2007; Gifford et al. 2008) (fig. 2A). There was considerable posterior support for placing MELVmpf basal to either primate lentiviruses or feline immunodeficiency virus (FIV), leaving the precise phylogenetic position for MELVmpf unresolved. The order Carnivora is composed of the suborders Caniformia and Feliformia (Flynn et al. 2005). The Mustelidae family belongs to the Caniformia suborder. FIV is endemic in the Feliformia (Troyer et al. 2005). The phylogenetic results suggest MELV might either be a novel lentiviral subgroup or might group with FIV in a carnivore lentivirus lineage.
FIG. 2.
(A) Tree of MELVmpf and both endogenous and exogenous lentiviruses. (B) Tree of endogenous lentiviruses only. Both are 50% majority-rule consensus trees. Node labels are posterior probabilities; branch lengths are in expected changes per site (mean values in the MCMC sample). The phylogenies are drawn to the same scale. (C) Phylogeny and timeline of Mustelidae based on Koepfli et al. (2008). The “+” and “−” indicate the presence or absence of MELV by PCR. (D) ML phylogeny of MELV gag sequences: lc, Lontra canadensis; mf, M. frenata; mp, M. putorius; mer, M. erminea; mniv, M. nivalis; nv, Neovison vison, mnig, M. nigripes; and mev, M. eversmanni. Sequences beginning with “contig” are derived from domestic ferret WGS contigs.
To evaluate the diversity of ancient lentiviruses, we undertook a separate Bayesian phylogenetic analysis using the conserved amino acid residues used above but including only the five known endogenous lentiviruses. After eliminating the effects of postendogenization evolution, the maximum distance between endogenous viruses was 0.72 (between the common ancestors of pSIV and RELIK), while the maximum distance between any pair of taxa on the combined endogenous and exogenous lentiviral phylogeny was not much higher: 1.28 (between SIVsyk and bovine immunodeficiency virus). This suggests that endogenous lentiviruses already exhibited, several Ma genetic divergence comparable with that of modern exogenous lentiviruses (fig. 2A and B). This is a remarkable illustration of long-term evolutionary stasis in viruses that evolve extremely rapidly over short time scales.
To date the genomic invasion of MELV, we employed two approaches: calculating the divergence between 5′ LTR and 3′ LTR and determining the taxonomic distribution of MELV. The 5′ LTR and 3′ LTR of endogenous retroviruses may diverge at a neutral rate after endogenization (Johnson and Coffin 1999; Katzourakis et al. 2007). Based on 17 intron loci from six Mustela species, we estimated a neutral substitution rate for Mustela of 4.9 (95% highest posterior density [HPD]: 3.6 − 6.5) × 10−9 substitutions per site per year. We identified a complete lentiviral insertion within contig098598 of the M. putorius genome (supplementary table S2, Supplementary Material online) with a 5′LTR to 3′ LTR distance of 5.9%. Using the intron-based rate, the invasion time estimate is 6.0 (95% HPD: 4.5–8.2) Ma. This estimate, however, should be taken with caution, as the LTRs are short (295 bp) and may not have evolved at a neutral evolutionary rate.
We verified the presence of MELV insertions in the genomes of the Lutrinae and Mustelinae subfamilies but not in Martinae via PCR amplification with degenerate primers designed for a conserved region of the gag gene (fig. 2C and D). Lutrinae and Mustelinae diverged from each other ∼8.8 Ma and Martinae diverged from Lutrinae and Mustelinae around 11.8 Ma (Koepfli et al. 2008). Our findings thus suggest an MELV invasion into Mustelidae genomes between 8.8 and 11.8 Ma, close to the upper bound estimated with 5′ LTR and 3′ LTR, and close to the estimate by Cui and Holmes (2012). However, these analyses come with two caveats. First, MELV gag sequences could have arisen from multiple endogenization events in some species, as with pSIV insertions in lemurs (Gilbert et al. 2009). Second, the PCR results for the Martinae might be false negatives if mutations have occurred in primer binding sites.
Fadel et al. (2012) recently reported that engineered domestic ferret cells support productive replication of HIV-1. This discovery, along with ours and that of Cui and Holmes (2012), suggests there may be a role for advancing lentiviral biology using this animal model. The ferret's amenability to experimental research and its physiological and immunological similarities to humans have been amply demonstrated with other important diseases (Ball 2006).
Materials and Methods
Ethanol preserved tissue samples of 10 Mustelidae species (fig. 2C) were obtained in November 2011 from the Museum of Southwestern Biology, University of New Mexico, and the Museum of Vertebrate Zoology, University of California, Berkeley. Genomic DNA was extracted using the DNeasy tissue kit (QIAGEN, MD). PCR amplification of an ∼860-bp gag gene fragment was performed with the degenerate primer pair, FeEL-2R (5′-GCCTTGCArTCCTCATTwGC-3′) and FeEL-2F (5′-GTrTCTGGGCCTTGrAGAyA-3′). Purified PCR products were cloned into the pGEM-T Easy vector (Promega, WI). Cloned products were sequenced by the University of Arizona Genetics Core with an Applied Biosystems 3730XL DNA Analyzer. The sequences have been deposited in GenBank (accession numbers JQ700124–JQ700200).
Protein sequences were aligned using Clustal Omega (Sievers et al. 2011). Gblocks 0.91b was used to eliminate ambiguous regions (Talavera and Castresana 2007). An ML approach was used to reconstruct the retrovirus phylogenetic trees using PHYML 3.0 (Guindon et al. 2010) with the rtREV model (Dimmic et al. 2002) and 500 bootstrap replicates. To further evaluate the relationship and divergence of lentiviruses, two data sets were used: the 426 conserved pol amino acid residues from both exogenous and endogenous representative lentiviruses and a separate alignment including only endogenous lentiviruses. These analyses were performed with MrBayes 3.1.2 (Ronquist and Huelsenbeck 2003) and the rtREV amino acid substitution model (Dimmic et al. 2002). The ML phylogenetic tree of MELV elements, based on the gag sequences we recovered from museum specimens, was reconstructed using PHYML 3.0 (Guindon et al. 2010) with 500 bootstrap replicates and the TrN + I + Γ4 substitution model determined using jModelTest (Posada 2008). MELV nucleotide sequences were aligned using MUSCLE (Edgar 2004).
We used an alignment of 17 nuclear intron loci from six Mustela species to estimate the neutral evolutionary rate (Yu et al. 2011), with a normal prior (mean = 5.3 Ma, standard deviation = 0.5) on the TMRCA of Mustela according to fossil records (Koepfli et al. 2008). An uncorrelated lognormal relaxed clock model (Drummond et al. 2006) and Yule model of speciation was used. The BEAST package of programs (http://beast.bio.ed.ac.uk, v1.6.1) was employed for these Bayesian MCMC analyses. MCMC chains were run 100 million steps to achieve adequate mixing for all parameters (effective sample size > 200). Tracer v1.5 was used to summarize and analyze the resulting posterior sample. All alignments and input files used in the phylogenetic analyses are available from the authors upon request.
Supplementary Material
Supplementary figures S1–S3 and tables S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank the Museum of Southwestern Biology, University of New Mexico, and the Museum of Vertebrate Zoology, University of California, Berkeley, for providing tissue samples of the Mustelidae species, Dr Li Yu for providing the Mustela sequence alignment, and two reviewers for extremely constructive suggestions. This research was supported by National Institute of Allergy and Infectious Diseases grant R01 AI084691 and the David and Lucile Packard Foundation.
Associate Editor's note: This work was initially received at MBE on 13 Jan 2012, concurrent with the online publication of Cui and Holmes (2012).
References
- Ball RS. Issues to consider for preparing ferrets as research subjects in the laboratory. ILAR J. 2006;47:348–357. doi: 10.1093/ilar.47.4.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Holmes EC. Endogenous lentiviruses in the ferret genome. J Virol. 2012;86:3383–3385. doi: 10.1128/JVI.06652-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmic MW, Rest JS, Mindell DP, Goldstein D. rtREV: an amino acid substitution matrix for inference of retrovirus and reverse transcriptase phylogeny. J Mol Evol. 2002;55:65–73. doi: 10.1007/s00239-001-2304-y. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadel HJ, Saenz DT, Guevara R, von Messling V, Peretz M, Poeschla EM. Productive replication and evolution of HIV-1 in Ferret cells. J Virol. 2012;86:2312–2322. doi: 10.1128/JVI.06035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JJ, Finarelli JA, Zehr S, Hsu J, Nedbal MA. Molecular phylogeny of the carnivora (mammalia): assessing the impact of increased sampling on resolving enigmatic relationships. Syst Biol. 2005;54:317–337. doi: 10.1080/10635150590923326. [DOI] [PubMed] [Google Scholar]
- Gifford RJ, Katzourakis A, Tristem M, Pybus OG, Winters M, Shafer RW. A transitional endogenous lentivirus from the genome of a basal primate and implications for lentivirus evolution. Proc Natl Acad Sci U S A. 2008;105:20362–20367. doi: 10.1073/pnas.0807873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Maxfield DG, Goodman SM, Feschotte C. Parallel germline infiltration of a lentivirus in two Malagasy lemurs. PLoS Genet. 2009;5:e1000425. doi: 10.1371/journal.pgen.1000425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Coffin JM. Constructing primate phylogenies from ancient retrovirus sequences. Proc Natl Acad Sci U S A. 1999;96:10254–10260. doi: 10.1073/pnas.96.18.10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A, Tristem M, Pybus OG, Gifford RJ. Discovery and analysis of the first endogenous lentivirus. Proc Natl Acad Sci U S A. 2007;104:6261–6265. doi: 10.1073/pnas.0700471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepfli KP, Deere KA, Slater GJ, Begg C, Begg K, Grassman L, Lucherini M, Veron G, Wayne RK. Multigene phylogeny of the Mustelidae: resolving relationships, tempo and biogeographic history of a mammalian adaptive radiation. BMC Biol. 2008;6:10. doi: 10.1186/1741-7007-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, et al. (12 co-authors) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Troyer JL, Pecon-Slattery J, Roelke ME, et al. (16 co-authors) Seroprevalence and genomic divergence of circulating strains of feline immunodeficiency virus among Felidae and Hyaenidae species. J Virol. 2005;79:8282–8294. doi: 10.1128/JVI.79.13.8282-8294.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Peng D, Liu J, Luan P, Liang L, Lee H, Lee M, Ryder OA, Zhang Y. On the phylogeny of Mustelidae subfamilies: analysis of seventeen nuclear non-coding loci and mitochondrial complete genomes. BMC Evol Biol. 2011;11:92. doi: 10.1186/1471-2148-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.