Abstract
The matrix metalloproteinase (MMP) family of proteins degrades extracellular matrix (ECM) components as well as processes cytokines and growth factors. MMPs are involved in regulating ECM homeostasis in both normal physiology and disease pathophysiology. Here, we report the critical roles of mmp23b in normal zebrafish liver development. Mmp23b was initially identified as a gene linked to the genomic locus of an enhancer trap transgenic zebrafish line in which GFP expression was restricted to the developing liver. Follow-up analysis of mmp23b mRNA expression confirmed its liver-specific expression pattern. Morpholino (MO) knockdown of mmp23b resulted in defective hepatocyte proliferation, causing a reduction in liver size while maintaining relatively normal pancreas and gut development. Genetically, we showed that mmp23b functions through the tumor necrosis factor (TNF) signaling pathway. Antisense knockdown of tnfa or tnfb in zebrafish caused similar reductions of liver size whereas overexpression of tnfa or tnfb rescued liver defects in mmp23b morphants but not vice versa. Biochemically, MMP23B, the human ortholog of Mmp23b, directly interacts with TNF and mediates its release from the cell membrane in a cell culture system. Since mmp23b/MMP23B is highly conserved, our findings in zebrafish warrant further investigation of its role in regulating liver development in mammals.
Keywords: mmp23b, TNF, liver development, hepatocyte proliferation, zebrafish
Introduction
Matrix metalloproteinases (MMPs) are a family of proteinase that degrade the extracellular matrix (ECM) by substrate cleavage (1), regulating physiological processes during embryonic development, tissue remodeling, and disease processes such as metastasis, arthritis, and liver fibrosis (2, 3). Hepatic fibrosis is characterized by increased deposition, altered composition of the ECM, and regenerative nodule formation (4). In response to injury, increased proinflammatory cytokines induce MMP expression in hepatocytes including hepatic stellate cells (HSC) (4). For example, levels of MMP-13, MMP-2, MMP-9, MT1-MMP, MMP-3 and MMP-10 are all increased during liver injury and fibrosis (5). MMPs degrade normal ECM in the space of Disse, which in turn activates HSC trans-differentiation into myofibroblasts producing fibrillar, contractile ECM while the remaining HSC population undergoes apoptosis (4).
In addition to being responsible for the turnover and degradation of connective-tissue proteins, MMPs regulate the activity of cytokines and chemokines by substrate cleavage (6, 7). One MMP-regulated pro-inflammatory cytokine is tumor necrosis factor (TNF) (8). Hepatic TNF is mainly derived from Kupffer cells and regulates liver homeostasis by modulating hepatocyte proliferation as well as cell death (9, 10). TNF has also been implicated in the pathogenesis of alcoholic and nonalcoholic fatty liver injury as well as inflammation response during liver fibrosis in human and lower vertebrates (11, 12). Systemic administration of TNF induces hepatocyte proliferation (13). In TNF-R1-deficient animal models, hepatocyte DNA synthesis is severely impaired, establishing that TNF can initiate hepatocyte proliferation in response to tissue loss (14). TNF is produced as a 26-kDa membrane bound protein (proTNF) that is cleaved into a soluble 17-kDa cytokine by TNF-converting enzyme (TACE/ADAM17) (15, 16), a member of the disintegrin family of metalloproteinases (ADAMs). MMPs have also been shown to be capable of processing TNF to its soluble form (17–19), but no known MMPs can cleave proTNF as effectively as TACE (20).
Compared to their role in liver fibrosis, MMPs’ functions during normal liver development are not well known. The regulation of ECM remodeling during morphogenesis is poorly understood because of the lack of good in vivo models for the study of MMP regulation during embryonic development. Improved understanding of ECM remodeling may also improve the understanding of the pathological mechanisms underlying the misregulation of ECM activity in tumors (21). A recent study by Waytt et al. developed several elaborate techniques for studying MMPs in zebrafish (Danio rerio), demonstrating the zebrafish embryo as a more tractable experimental organism to study the regulation of MMP activity during development (21).
Zebrafish have a conserved liver developmental process that shares striking similarity to mammals (22). The optical clarity during embryogenesis allows visualization of the entire liver developmental process and morphology if a transgenic green or red fluorescent protein (GFP or RFP) reporter is expressed in the liver. We performed a large-scale Tol2 based enhancer trap screen for transgenic zebrafish exhibiting tissue specific GFP expression and mapped 15 lines with liver expression patterns to specific trapped genes. One of such lines, mp255c, trapped a gene designated as matrix metalloproteinase 23b (mmp23b); mmp23b is orthologous to mammalian MMP23B, which has no described function. A recent microarray study using a zebrafish liver cancer model showed that mmp23b is significantly down-regulated in liver cancers while other MMPs are increased (23), implying the uniqueness of this MMP member in liver biology. We show that endogenous mmp23b is expressed in hepatocytes during development and its proper function is required for hepatocyte proliferation. Genetically, mmp23b functions through tumor necrotic factors (TNF). Evidence in vitro suggests an MMP23B-mediated protein shedding mechanism might be involved in regulating TNF activity. Since mmp23b/MMP23B is highly conserved, its role in liver development in mammals should be further studied.
Materials and methods
More reagents and procedures used in this report were described in detail as supplemental materials and methods.
Zebrafish husbandry
Zebrafish were raised and kept under standard laboratory conditions at approximately 28 °C. Embryos were staged according to Kimmel et al (24). Mmp23b:GFP fish was generated from a previous Tol2 based enhancer trap screen. The wild-type line used was AB.
RNA whole-mount in situ hybridization (WISH)
WISH was performed essentially as previously described (25). NBT/BCIP (50 mg/ml; Promega) were used as alkaline phosphatase substrates. The following probes were used: mmp23b, ceruloplasmin, hhex, gata6, insulin, and carboxypeptidase A.
RNA synthesis and injection
Mmp23b, tnfa and tnfb full-length coding sequences were obtained from Open Biosystems and subcloned into a pCS2+ vector. mRNAs were synthesized using T7 or SP6 mMessage mMachine kit (Ambion). mRNA injection was performed as described at the one-cell stage (26). Sterile water was used for the control experiments. 100 pg of mmp23b, tnfa and tnfb mRNA was used for all experiments.
Antisense morpholino oligonucleotides
MOs designed against mmp23b targeting either the ATG (5’-AGCACACGAACAAACCCAGAACATC-3’) or 5’-UTR (5’-TGAAATCACAACTTTCCTCACGGAT-3’) was obtained from Gene Tools. Tnfa-MO1 (5’-GGCAGGATTTTCACCTTATGGAGCG-3’) and tnfb-MO1 (5’-AATTTCAGTCTTACCATCACATGCC-3’) were obtained from Open Biosystems. A standard MO control oligo was obtained from Gene Tools. All MOs were resuspended in sterile water at 2 ng/nl and injected into one- to two-cell stage embryos. More sequence information is provided in supplemental materials.
Whole-mount immunostaining and immunohischemistry
Whole-mount immunostaining with phospho-histone H3 (pH3) antibody was performed as described (27). Mmp23b:GFP transgenic fish were used for double staining. Primary antibodies were used as follows: rabbit anti-pH3 Ser 10 (Santa Cruz) at 1:400, mouse anti-GFP (Invitrogen) at 1:200. Alexa Fluor 488-conjugated anti-mouse IgG and Alexa Fluor 568-conjugated goat anti-rabbit IgG were used as secondary antibodies (Invitrogen). The whole embryos were then observed with confocal microscopy. Preparation of paraffin sections and cryosections for immunohischemistry was described in supplemental materials.
Cell culture and transfection
Mtace (gift from Dr. R. Black) and HMMP23B (Open Biosystems) full-length coding sequences were subcloned into pcDNA3.1+ (Invitrogen) vector. Murine TACE−/− fibroblasts were gifts from Dr. R. Black and were cultured as described before (28). Transfection and samples collection were described in supplemental materials.
Western blots and ELISA detection
Western blotting was performed using a standard protocol; anti-FLAG was obtained from Sigma; anti-tubulin (Invitrogen) was used as a loading control. Cell lysates were measured for TNF content by ELISA as previously described (29) and normalized to total protein using a protein assay (Pierce).
Results
Tol2 transposon-based enhancer trap screen
We have performed a large-scale enhancer trap screen in zebrafish using Tol2 transposon-mediated transgenic approaches (30). The Tol2GT2MP constructs are modified Tol2 mobile elements created for the purpose of enhancer trapping in zebrafish. They carry a minimal promoter fragment from the zebrafish gata2 gene (31) fused upstream of the EGFP coding sequence (Fig.1A). To generate founder fish, circular DNA constructs of Tol2GT2MP plasmid and the Tol2-transposase mRNA synthesized in vitro were co-injected into zebrafish embryos. The injected fish were raised to adulthood and mated. GFP-positive embryos were identified under a fluorescence microscope at 24–48hpf. A total of 1,738 individual F1 fish lines were isolated, and fluorescent protein reporter expression patterns of these lines exhibited a broad range of tissue specificities, including brain, spinal cord, floor plate, sensory organs, neural crest cells, notochord, blood vessels, gut, pancreas, skin, and primordial germ cells (data unpublished). To detect the insertion sites, a modified linker-mediated PCR procedure based on previous reports (32, 33) was performed. Genomic flanking sequences of the trap insertions were identified and cloned for sequencing. These sequences were used in BlastN searches in the zebrafish genomic database Ensembl (http://www.ensembl.org/). A sequence was regarded as a match to the zebrafish genome sequence only when they were over 95% identity.
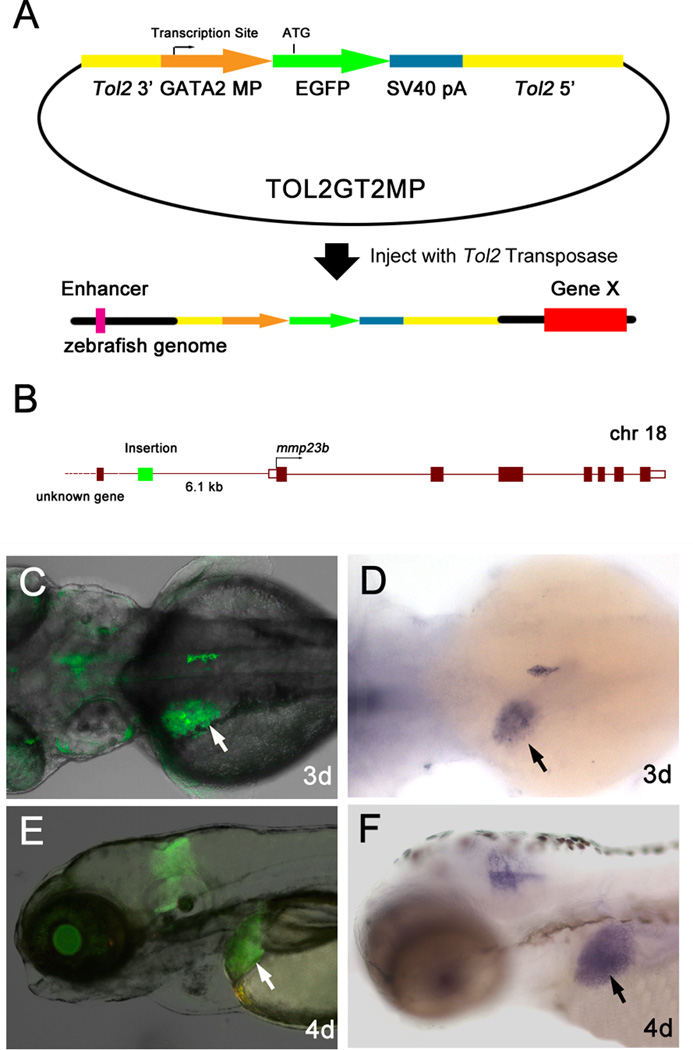
Figure 1. Tol2 transposon based enhancer trap screen.
(A) The enhancer trap constructs contain Tol2 transposon elements and a minimal promoter of the zebrafish gata2 gene fused to EGFP reporter gene. When they are inserted in the operating area of an enhancer element, the expression of the reporter gene can be induced and observed. (B) Tol2 insertion location of mp255c fish line. Tol2 elements were inserted at chromosome 18, 6.1 kb upstream of gene mmp23b. (C and E) mmp23b:GFP expression pattern. (C) Dorsal view at 3dpf. Note liver expression (white arrow). (E) Lateral view at 4dpf. (D and F) WISH using an mmp23b probe. (D) Dorsal view at 3dpf (100%, n=30). Note tissue specific expression in the developing liver (black arrow). (F) Lateral view at 4dpf (100%, n=30).
Among these transgenic fish lines, a liver-specific screen was performed at later developmental stages (3dpf, 5dpf). A total of 15 fish lines with liver-specific GFP expression were generated from the screen. Genomic flanking sequences were cloned and insertion sites were mapped in the zebrafish genome. One of such lines, mp255c, trapped the gene designated as matrix metalloproteinase 23b (mmp23b, GeneID: 550589), which is orthologous to mammalian MMP23B (GeneID: 8510). Sequence analysis of the insertion site by linker-mediated PCR revealed that mmp23b was approximately 6.1kb downstream of the GFP insertion (Fig.1B).
Mmp23b is specifically expressed in the developing zebrafish liver
The enhancer trap line, mp255c, exhibits liver- and brain-specific GFP expression patterns (Fig.1C and E). RNA WISH showed that mmp23b expression is mostly specific to the liver but has a minor domain in the brain (Fig.1D and F). Overall, GFP expression in mp255c faithfully recapitulated the endogenous mmp23b expression in developing zebrafish embryos, including the liver and brain. Mmp23b was expressed throughout all developmental stages, starting at 1dpf in the brain and peaking at 2–4dpf after liver development began (Fig.S1C). Protein alignment and phylogenic tree analysis showed that zebrafish mmp23b shares significant homology with mouse MMP23 and human MMP23B (Fig.S1A and B). RT-PCR analysis revealed that Mmp23 is also expressed in mouse liver (Fig.S1D) and brain (34). These studies indicate that mmp23b/Mmp23/MMP23B is highly conserved from zebrafish to human.
Knockdown of mmp23b specifically affects liver development
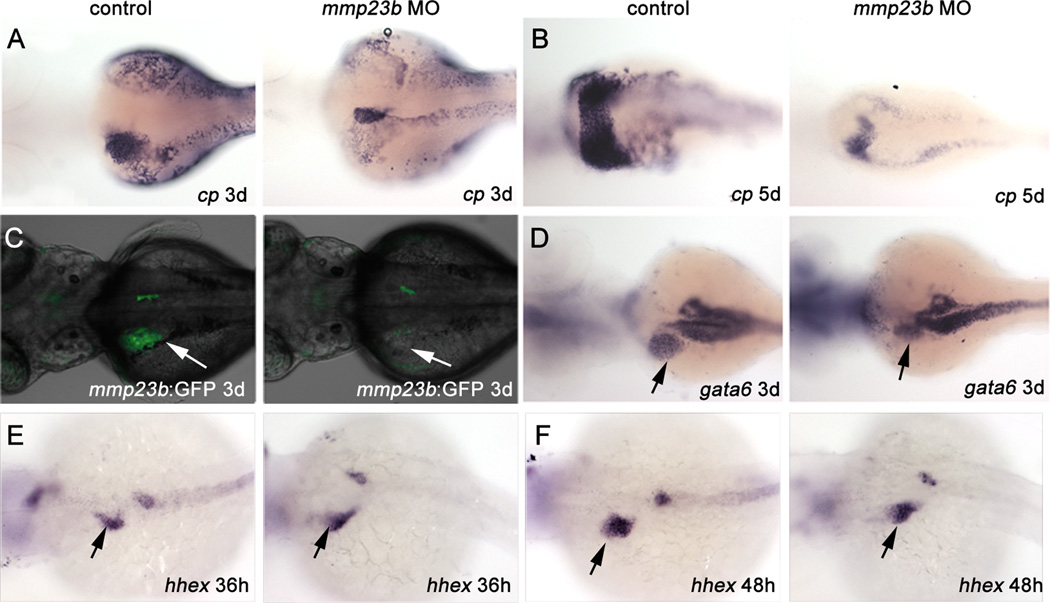
To examine mmp23b function, we generated two MOs against mmp23b, blocking the translation start site (MO1) and the 5’ UTR (MO2), respectively. Injection of either MO caused no apparent morphological changes (Fig.S3B and F). Further analysis by WISH using a liver marker, ceruloplasmin (cp), revealed significant liver mass reduction in mmp23b morphants (Fig.2A–B). We observed a similar reduction in liver GFP expression when the mmp23b:GFP line was injected with MO1 (Fig.2C). To determine whether liver size reduction was tissue specific, we analyzed gata6 expression, which is present in the liver, pancreas, and gut. Pancreatic and gut expression of gata6 in the mmp23b morphants was similar to that of control embryos (Fig.2D), whereas the liver expression was significantly reduced (Fig.2D, arrow). In addition, pancreatic expression of endocrine cell marker insulin (ins) and exocrine cell marker carboxypeptidase A (cpa) remained normal in mmp23b morphants (Fig.S2A–B). Furthermore, liver size reduction can be rescued with mmp23b mRNA co-injection with the MO (Fig.S4C).
Figure 2. Mmp23b knockdown phenotype.
(A–B) WISH using a ceruloplasmin (cp) probe. (A) Control and mmp23b morphant (88%, n=25) embryos at 3dpf. Note the dramatically reduced liver in the mmp23b morpholino injected embryo (black arrow) compared with that in the control embryo. (B) Control and mmp23b morphant (91.7%, n=24) embryos at 5dpf. The liver in the morphant remains underdeveloped. (C) mmp23b:GFP expression at 3dpf. Note the severe reduction of GFP expression in the developing liver area in the mmp23b morphant (white arrow, 86.4%, n=22,). (D) WISH using a gata6 probe. Control embryo and mmp23b morphant at 3dpf. Note only the liver expression of gata6 is reduced (black arrow, 73.9%, n=23) whereas pancreatic and gut expression of gata6 remain unaffected. (E–F) WISH using a hhex probe. (E) Control and mmp23b morphant embryos (96.2%, n=26) at 36hpf. (F) Control and mmp23b morphant embryos (84%, n=25) at 48hpf. All embryos are mounted as dosal view with anterior to the left.
To examine whether size reduction of the developing liver was due to a defect of progenitor cell specification, we performed WISH using a probe against hhex, one of the earliest liver markers in zebrafish. Endodermal expression of hhex at 36hpf and 48hpf (Fig.2E–F) in zebrafish appears normal in the developing liver and pancreas of mmp23al morphants. These results suggest that mmp23al deficiency leads to an organ-specific size reduction in the developing liver that is independent of early hepatic specification.
Endothelial cells have been shown to be required for liver morphogenesis in a mouse model (35). The MMP family is also known to be involved in blood vessel formation and remodeling (36). We thus tested capillary morphogenesis in the zebrafish liver with a flk:RFP/mmp23b:GFP double transgenic fish line. However, there was no significant change of liver capillaries in mmp23b morphants at 3dpf (Fig.S6).
Knockdown of mmp23b causes defect of liver cell proliferation
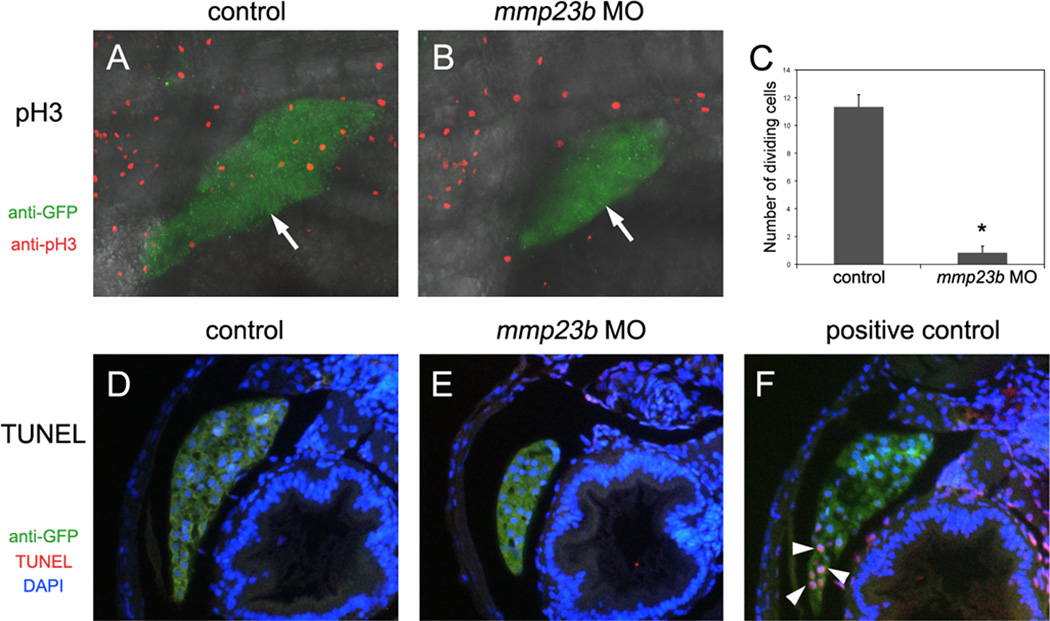
To investigate the underlying mechanism of liver size reduction in mmp23b morphants, we performed immunostaining analysis of phospho-histone H3 (pH3), a cell proliferation marker. In control embryos, we detected an average of 11.3±0.9 (n=6) pH3-positive cells vs. 0.8±0.5 (n=6, p<0.0005) in mmp23b morphant livers (Fig.3 A–C). To assess whether cell death contributes to liver size reduction in mmp23b morphants, we conducted a TUNEL assay. Embryonic hepatocytes presented a low apoptotic index at 4dpf, which was unchanged in mmp23b morphants (Fig.3 D–F). Therefore, reduction of liver size appears to be due to defective cell proliferation in the developing liver of mmp23b morphants.
Figure 3. Cell division defect in mmp23b morphants.
(A and B) Anti-phospho Histone H3 (pH3) staining for control and mmp23b morphant embryos at 3dpf. Red: pH3 staining. Green: GFP staining. pH3 staining in the liver (B, white arrow) of mmp23b morphant is rarely observed. (C) Quantification of dividing cells by pH3 staining. *P<0.00001. (D–F) TUNEL analysis at 3dpf. Red: TUNEL staining. Green: GFP staining. Blue: DAPI staining. (D) Control embryo. (E) Mmp23b morphant. No apoptotic cells were observed in the developing liver of control or mmp23b morphant (L). (F) Positive control. Embryos were treated with DNaseI before TUNEL assay. Note apoptotic cells in the developing liver (white arrowheads).
Knockdown of tnfa or tnfb induces liver phenotypes similar to that of mmp23b deficiency
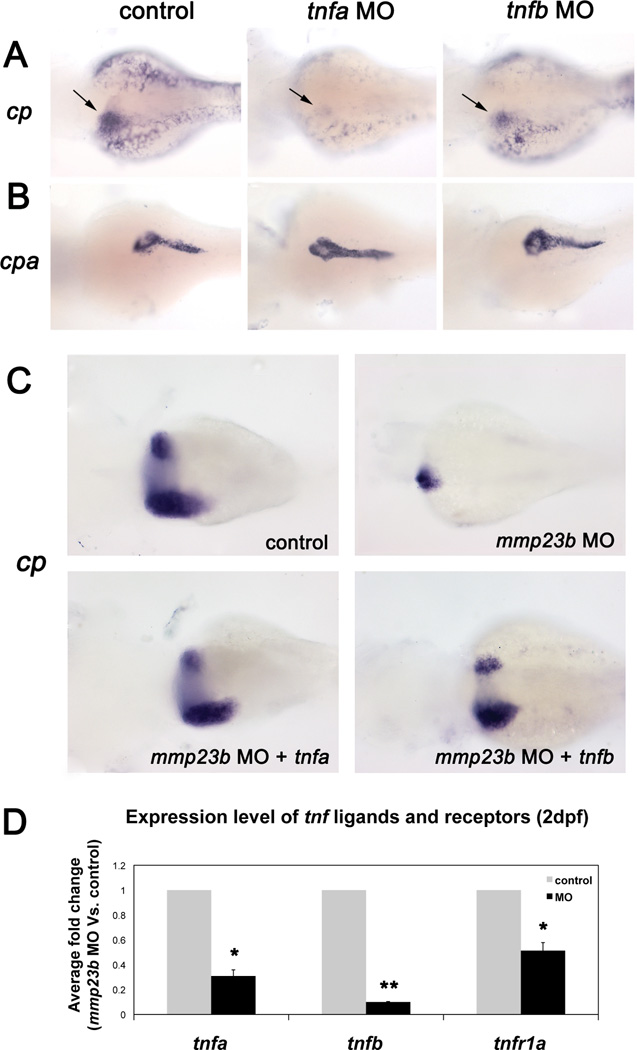
The proinflammatory cytokine TNF is a key regulator of liver homeostasis in mammals (37). There are two TNF isoforms in zebrafish, tnfa (GeneID: 405785) and tnfb (GeneID: 554167). To examine whether TNFs are required for liver development in zebrafish, we generated antisense MOs to knockdown tnfa or tnfb, respectively. Embryos injected with either tnfa MO or tnfb MO exhibited a severely underdeveloped liver at 3dpf (Fig.4A). In contrast, pancreas development was unaffected as indicated by normal cpa expression in both tnfa or tnfb morphants (Fig.4B). Therefore, tnfa and tnfb knockdown produces hepatic phenotypes similar to that of mmp23b morphants.
Figure 4. tnfa and tnfb function downstream of mmp23b.
(A) WISH using a cp probe at 3dpf. Control embryo and tnfa/tnfb morphant embryos (tnfa, 85.2%, n=27; tnfb, 68.2%, n=22). Note dramatically reduced expression of cp (black arrow). (B) WISH using a cpa probe. Note the similar expression pattern of cpa in all embryos (tnfa, 95.8%, n=24; tnfb, 95% n=20). (C) WISH using a cp probe at 4dpf. Control embryo and mmp23b morphant (100%, n=18). Note the severely reduced liver size (arrow). Mmp23b morphant injected with tnfa/tnfb mRNA at 1-cell stage (I, 63.4%, n=33; J, 41.4% n=29). Note the restoration of liver development. (D) Fold changes of tnf ligands and receptor in control and mmp23b morphant embryos at 2 dpf. The expression levels of each marker were measured by quantitative RT-PCR and fold changes were calculated by comparing morphants expression level to that of the control embryos. β-actin serves as the reference gene. Error bar represents the standard deviation. *P<0.005, **P<0.0005.
Forced expression of tnfa or tnfb restores liver development in mmp23b morphants
Since tnfa and tnfb morphants exhibit similarly underdeveloped livers as in mmp23b morphants, we tested whether mmp23b and TNF interact genetically. We analyzed expression levels of tnfa, tnfb, tnfr1a (tnf receptor1a) in mmp23b morphants by quantitative RT-PCR. Knockdown of mmp23b decreased the expression level of both tnf ligands (tnfa 0.31±0.05, p<0.005; tnfb 0.10±0.003, p<0.0005) and receptor (tnfr1a 0.51±0.07, p<0.005; Fig.4D). Then we co-injected mmp23b MO with tnfa or tnfb mRNA and analyzed liver development by cp WISH. At 5dpf, injection of mmp23b MO alone resulted in a dramatically reduced liver size (Fig.4C). However, co-injection of tnfa mRNA with mmp23b MO restored the liver size (Fig.4C) comparable to that of control embryos. Co-injection of tnfb mRNA with mmp23b MO also rescued liver development (Fig.4C), although to a lesser extent. Conversely, forced expression of mmp23b mRNA in tnfa or tnfb morphants failed to restore liver development (Fig.S4D–F). These data suggest that mmp23b acts upstream of tnf in liver development.
Mmp23b/MMP23B is involved in TNF shedding in a cell culture system
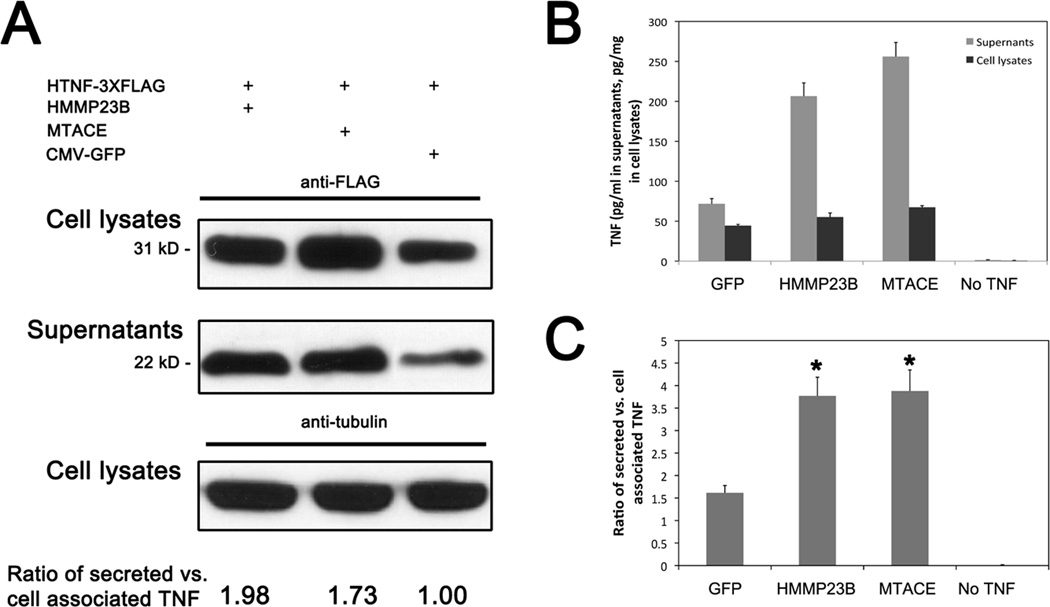
TNF is expressed on the cell surface as a transmembrane pro-protein and requires processing by TACE/ADAM17 to become active, although membrane bound pro-TNF has some capacity as an inflammatory activator (15). The mmp23b gene encodes a 377 amino acids protein that contains a catalytic domain similar to that of TACE/ADAM17. This led us to postulate that Mmp23b is involved in TNF shedding. To test this hypothesis, we took advantage of the null mutant TACE−/− fibroblasts, which has minimal endogenous TNF converting ability (15). In the control cells with co-transfection of HTNF-3xFLAG (human TNF) and CMV-GFP constructs, membrane bound TNF (from cell lysates, 31 kD with FLAG,) and soluble TNF (from supernatants, 22 kD with FLAG) were detected by western blot using an anti-FLAG antibody (Fig.5A). As expected, co-transfection of HMMP23B (human MMP23B) and TNF caused a significant increase of soluble TNF comparable to that of MTACE (mouse TACE) co-transfection (Fig.5A). Western blots of secreted TNF and membrane associated TNF showed both HMMP23B and MTACE could lead to an increase of soluble TNF. ELISA analysis with anti-HTNF was also performed separately to quantify the TNF release efficiency by HMMP23B (Fig.5B). Compared to the control group, HMMP23B increased shedding of TNF by 2.32 fold whereas MTACE increased by 2.36 fold (Fig.5C). These data suggest that Mmp23b/MMP23B might be involved in TNF shedding.
Figure 5. Mmp23b/MMP23B is involved in TNF shedding.
(A) Western blot of supernatants and cell lysates using an anti-FLAG antibody. Co-transfection with HTNF-3xFLAG and GFP serves as negative control. Co-transfection with HTNF-3xFLAG and either HMMP23B or MTACE lead to a significant increase of soluble TNF level in the supernatants. Normalization of the ration of secreted TNF vs. cell associated TNF is listed. Anti-tubuling was used as a loading control. (B) ELISA analysis of TNF levels in supernatants and cell lysates. Grey: TNF levels in supernatants. Black: TNF levels in cell lysates. Co-transfection of HMMP23B increased the supernatants TNF level to an extent comparable to that of MTACE co-transfection. (C) Ratio of secreted vs. cell associated TNF. Ratios were calculated from supernatants TNF divided by cell lysates TNF. Both HMMP23B and MTACE led to a significant increase of TNF release from cell membrane comparing to that of control cells (GFP and TNF co-transfection). Error bars represent standard deviation. *P<0.05.
To test if HMMP23B interacts with TNF, we performed an immunoprecipitation experiment. TACE−/− cells were transfected with CMV-MYC or HMMP23B-MYC, along with HTNF-3xFLAG. Protein lysates were immunoprecipitated with anti-MYC antibody and blotted with anti-FLAG antibody. We found that HMMP23B has a physical interaction with HTNF (Fig.S7), leading us to speculate that HMMP23B might be directly involved in the shedding process, either as a proteinase or as a co-factor.
Discussion
Several MMPs have been previously implicated in liver injury in response to increased proinflammatory cytokines (5). However, none of them are expressed specifically in the liver under physiological conditions. Mmp23b is the first MMP member that mainly has liver-specific expression. In zebrafish, it is initially expressed at 24hpf and continues to be highly expressed in the developing liver (Fig.1C–F, Fig.S1C). Preliminary analysis showed that the mammalian homolog of mmp23b is also expressed in normal liver cells (Fig.S1D), which is in contrast with other MMP family members activated primarily by liver injury and disease processes. Mmp23b is also the first MMP member that has been demonstrated to have a clear biological function in liver development. Knockdown of mmp23b resulted in a severe reduction of liver size without affecting early liver development marker hhex (Fig.2E–F), suggesting that mmp23b is required for hepatocyte proliferation after initial hepatoblast determination and specification. The liver is the only organ in mammals capable of natural regeneration of lost tissue and as little as 25% of the liver can regenerate into a whole liver due to hepatocyte re-enterance into the cell cycle. It is known that hepatocyte proliferation is controlled by the neurotrophin receptor p75NTR, which instructs HSCs to differentiate and then make growth factors and ECM to support hepatocyte proliferation (38). P75NTR is a TNF receptor superfamily member and it will be interesting to see if mmp23b is essential for liver cell proliferation during regeneration in response to p75NTR signaling.
There are twenty-five vertebrate MMPs including two subfamilies, membrane-bounded and non-membrane bounded (1). Mmp23b/MMP23B contains a type II transmembrane domain at the amino-terminal that likely enables it membrane bounded. Due to the unavailability of a zebrafish hepatocyte in vitro cell culture system, we took advantage of the broadly used TACE −/− fibroblast cell line system. We provide evidence that Mmp23b/MMP23B is also involved in TNF shedding using the cell culture system, suggesting membrane bound MMP family members can process cytokines in a tissue-specific manner. Epistatic studies in zebrafish place TNF downstream of mmp23b in liver development, which is consistent with biochemical data showing that TNFs are related to Mmp23b/MMP23B activity. Although our in vitro immunoprecipitation experiment suggests that there are direct interactions between HMMP23B and HTNF, HMMP23B’s in vivo biochemical partners in regulating TNF releasing remains to be determined. Previous immunoprecipitation and Western blotting analysis have also shown that TACE could interact with the proform of TNF (39). It is likely that other MMPs including MMP23A and closely related MMP19 may share the similar specificity. Further study about the mechanism of the interaction is necessary to understand of the roles of MMPs in TNF activation.
The MMP family is involved in blood vessel formation and remodeling, which is necessary for liver morphogenesis in mice. In this study, no capillaries appear affected by mmp23b knockdown. Therefore, the function of mmp23b in liver proliferation is independent of capillary morphogenesis. This finding is consistent with previous studies in zebrafish since liver and biliary tree form normally in the absence of blood vessels in cloche mutants (40, 41).
Like other MMPs, mmp23b may have more than one target contributing to inhibited cell proliferation. Although we show here MMP23B interacts with TNFs, it may have other cytokine targets such as transforming growth factor-β1 (TGF-β1), which is also an important indicator of liver fibrosis (42). TGF-β1 is released by cells but with its cleaved pro-domain still latently associated. Several mechanisms, including MMP proteolysis (MMP3, MMP9, and MMP14), have been proposed to release the active cytokine from this complex (8). It is possible that MMP23B can also act in the TGF-β1 releasing process in vivo. Recently, Wyatt et al. have developed two novel assays that allow the detection and characterization of active MMPs in vivo (differential in vivo zymography and activity-based protease profiling) (21). These techniques should be useful for further detection of in vivo MMP23B activities and its potential substrates. Activity-based protease profiling allows detection of biologically activity of MMP23B during embryonic development. Fluorogenic TNF or TGF-β1 as well as some other candidates could be injected into a living zebrafish embryo, and then visualized for the fluorescent breakdown products by confocal microscopy. The fluorescent breakdown products of observed in liver could be a candidate substrate of MMP23B.
Hepatic fibrosis represents the final common pathway of chronic injuries associated with viral hepatitis, alcohol abuse, autoimmune diseases, drugs, and metabolic syndromes (43). In nonalcoholic fatty liver disease, the disease spectrum ranges from early hepatic steatosis to end-stage liver disease, with fibrosis and cirrhosis accounting for the most common cause of abnormal liver function tests and approximately 14% of liver transplants in the United States. Several MMP family members, which normally have relatively low expression in the liver, have been shown to be abnormally activated in fibrosis (4). On the other hand, mmp23b is highly expressed in normal livers, appearing to be different from those MMP family members involved in fibrosis. We did some preliminary experiments to investigate whether partial mmp23b deficiency might be related to liver fibrosis. Several markers of gene expression for fibrosis were changed in a way that reflects the ontogeny of fibrosis, as detected by quantitative RT-PCR (Fig.S8). These findings, although preliminary, suggest that deficiency of mmp23b is likely a marker or a cause of early liver fibrosis. However, further studies, including the generation of mmp23b genetic mutations, are needed to link this gene to liver fibrosis. We failed to detect collagen deposition by trichrome staining in mmp23b-deficient zebrafish embryos at 6dpf, possibly due to the short half life of antisense MO. Mmp23b is also unique in relation to liver carcinogenesis as it has been shown by gene expression profiling of zebrafish liver cancer samples that mmp23b is significantly decreased while other MMPs are increased (23). Again, it will be interesting to determine if mmp23b/MMP23B deficiency represents one of the causes leading to liver cancer whereas the increases of other MMPs are a consequence of hepatic carcinogenesis.
Supplementary Material
ACKNOWLEDGMENTS
We thank numerous members of the S. Lin and B. Zhang laboratories for excellent fish care, technical assistance, and helpful discussions; R. Black for sharing TACE−/− cell line; J. Kolls for sharing HTNF-3xFLAG plasmid. This work was supported in part by grants from the National Institutes of Health to SL (RR13227, DK054508, HD061570), from NSFC (No. 30721064 and 30730056) as well as 973 Program from MOST of P. R. China (2005CB522504, 2006CB943801, 2007CB914502 and 2009CB941203). FQ was supported in part by China Scholarship Council, Ministry of Education, China.
Reference
- 1.Sternlicht MD, Werb Z. How matrix metalloproteinases regulate cell behavior. Annu Rev Cell Dev Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pinzani M, Rombouts K, Colagrande S. Fibrosis in chronic liver diseases: diagnosis and management. J Hepatol. 2005;42(Suppl):S22–S36. doi: 10.1016/j.jhep.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 3.Kisseleva T, Brenner DA. Mechanisms of fibrogenesis. Exp Biol Med (Maywood) 2008;233:109–122. doi: 10.3181/0707-MR-190. [DOI] [PubMed] [Google Scholar]
- 4.Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134:1655–1669. doi: 10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knittel T, Mehde M, Grundmann A, Saile B, Scharf JG, Ramadori G. Expression of matrix metalloproteinases and their inhibitors during hepatic tissue repair in the rat. Histochem Cell Biol. 2000;113:443–453. doi: 10.1007/s004180000150. [DOI] [PubMed] [Google Scholar]
- 6.Levi E, Fridman R, Miao HQ, Ma YS, Yayon A, Vlodavsky I. Matrix metalloproteinase 2 releases active soluble ectodomain of fibroblast growth factor receptor 1. Proc Natl Acad Sci U S A. 1996;93:7069–7074. doi: 10.1073/pnas.93.14.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelissen I, Martens E, Van den Steen PE, Proost P, Ronsse I, Opdenakker G. Gelatinase B/matrix metalloproteinase-9 cleaves interferon-beta and is a target for immunotherapy. Brain. 2003;126:1371–1381. doi: 10.1093/brain/awg129. [DOI] [PubMed] [Google Scholar]
- 8.Parks WC, Wilson CL, Lopez-Boado YS. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat Rev Immunol. 2004;4:617–629. doi: 10.1038/nri1418. [DOI] [PubMed] [Google Scholar]
- 9.Schwabe RF, Brenner DA. Mechanisms of Liver Injury. I. TNF-alpha-induced liver injury: role of IKK, JNK, and ROS pathways. Am J Physiol Gastrointest Liver Physiol. 2006;290:G583–G589. doi: 10.1152/ajpgi.00422.2005. [DOI] [PubMed] [Google Scholar]
- 10.Wullaert A, van Loo G, Heyninck K, Beyaert R. Hepatic tumor necrosis factor signaling and nuclear factor-kappaB: effects on liver homeostasis and beyond. Endocr Rev. 2007;28:365–386. doi: 10.1210/er.2006-0031. [DOI] [PubMed] [Google Scholar]
- 11.Nagata K, Suzuki H, Sakaguchi S. Common pathogenic mechanism in development progression of liver injury caused by non-alcoholic or alcoholic steatohepatitis. J Toxicol Sci. 2007;32:453–468. doi: 10.2131/jts.32.453. [DOI] [PubMed] [Google Scholar]
- 12.Connolly MK, Bedrosian AS, Mallen-St Clair J, Mitchell AP, Ibrahim J, Stroud A, Pachter HL, et al. In liver fibrosis, dendritic cells govern hepatic inflammation in mice via TNF-alpha. J Clin Invest. 2009;119:3213–3225. doi: 10.1172/JCI37581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feingold KR, Soued M, Grunfeld C. Tumor necrosis factor stimulates DNA synthesis in the liver of intact rats. Biochem Biophys Res Commun. 1988;153:576–582. doi: 10.1016/s0006-291x(88)81134-3. [DOI] [PubMed] [Google Scholar]
- 14.Yamada Y, Kirillova I, Peschon JJ, Fausto N. Initiation of liver growth by tumor necrosis factor: deficient liver regeneration in mice lacking type I tumor necrosis factor receptor. Proc Natl Acad Sci U S A. 1997;94:1441–1446. doi: 10.1073/pnas.94.4.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, et al. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature. 1997;385:729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- 16.Moss ML, Jin SL, Milla ME, Bickett DM, Burkhart W, Carter HL, Chen WJ, et al. Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-alpha. Nature. 1997;385:733–736. doi: 10.1038/385733a0. [DOI] [PubMed] [Google Scholar]
- 17.Gearing AJ, Beckett P, Christodoulou M, Churchill M, Clements J, Davidson AH, Drummond AH, et al. Processing of tumour necrosis factor-alpha precursor by metalloproteinases. Nature. 1994;370:555–557. doi: 10.1038/370555a0. [DOI] [PubMed] [Google Scholar]
- 18.English WR, Puente XS, Freije JM, Knauper V, Amour A, Merryweather A, Lopez-Otin C, et al. Membrane type 4 matrix metalloproteinase (MMP17) has tumor necrosis factor-alpha convertase activity but does not activate pro-MMP2. J Biol Chem. 2000;275:14046–14055. doi: 10.1074/jbc.275.19.14046. [DOI] [PubMed] [Google Scholar]
- 19.Haro H, Crawford HC, Fingleton B, Shinomiya K, Spengler DM, Matrisian LM. Matrix metalloproteinase-7-dependent release of tumor necrosis factor-alpha in a model of herniated disc resorption. J Clin Invest. 2000;105:143–150. doi: 10.1172/JCI7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohan MJ, Seaton T, Mitchell J, Howe A, Blackburn K, Burkhart W, Moyer M, et al. The tumor necrosis factor-alpha converting enzyme (TACE): a unique metalloproteinase with highly defined substrate selectivity. Biochemistry. 2002;41:9462–9469. doi: 10.1021/bi0260132. [DOI] [PubMed] [Google Scholar]
- 21.Wyatt RA, Keow JY, Harris ND, Hache CA, Li DH, Crawford BD. The zebrafish embryo: a powerful model system for investigating matrix remodeling. Zebrafish. 2009;6:347–354. doi: 10.1089/zeb.2009.0609. [DOI] [PubMed] [Google Scholar]
- 22.Chu J, Sadler KC. New school in liver development: lessons from zebrafish. Hepatology. 2009;50:1656–1663. doi: 10.1002/hep.23157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lam SH, Gong Z. Modeling liver cancer using zebrafish: a comparative oncogenomics approach. Cell Cycle. 2006;5:573–577. doi: 10.4161/cc.5.6.2550. [DOI] [PubMed] [Google Scholar]
- 24.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 25.Thisse C, Thisse B, Schilling TF, Postlethwait JH. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- 26.Hyatt TM, Ekker SC. Vectors and techniques for ectopic gene expression in zebrafish. Methods Cell Biol. 1999;59:117–126. doi: 10.1016/s0091-679x(08)61823-3. [DOI] [PubMed] [Google Scholar]
- 27.Shepard JL, Stern HM, Pfaff KL, Amatruda JF. Analysis of the cell cycle in zebrafish embryos. Methods Cell Biol. 2004;76:109–125. doi: 10.1016/s0091-679x(04)76007-0. [DOI] [PubMed] [Google Scholar]
- 28.Zhao XJ, Marrero L, Song K, Oliver P, Chin SY, Simon H, Schurr JR, et al. Acute alcohol inhibits TNF-alpha processing in human monocytes by inhibiting TNF/TNF-alpha-converting enzyme interactions in the cell membrane. J Immunol. 2003;170:2923–2931. doi: 10.4049/jimmunol.170.6.2923. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Z, Cork J, Ye P, Lei D, Schwarzenberger PO, Summer WR, Shellito JE, et al. Inhibition of TNF-alpha processing and TACE-mediated ectodomain shedding by ethanol. J Leukoc Biol. 2000;67:856–862. doi: 10.1002/jlb.67.6.856. [DOI] [PubMed] [Google Scholar]
- 30.Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 31.Meng A, Tang H, Ong BA, Farrell MJ, Lin S. Promoter analysis in living zebrafish embryos identifies a cis-acting motif required for neuronal expression of GATA-2. Proc Natl Acad Sci U S A. 1997;94:6267–6272. doi: 10.1073/pnas.94.12.6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 33.Wang D, Jao LE, Zheng N, Dolan K, Ivey J, Zonies S, Wu X, et al. Efficient genome-wide mutagenesis of zebrafish genes by retroviral insertions. Proc Natl Acad Sci U S A. 2007;104:12428–12433. doi: 10.1073/pnas.0705502104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gajecka M, Yu W, Ballif BC, Glotzbach CD, Bailey KA, Shaw CA, Kashork CD, et al. Delineation of mechanisms and regions of dosage imbalance in complex rearrangements of 1p36 leads to a putative gene for regulation of cranial suture closure. Eur J Hum Genet. 2005;13:139–149. doi: 10.1038/sj.ejhg.5201302. [DOI] [PubMed] [Google Scholar]
- 35.Matsumoto K, Yoshitomi H, Rossant J, Zaret KS. Liver organogenesis promoted by endothelial cells prior to vascular function. Science. 2001;294:559–563. doi: 10.1126/science.1063889. [DOI] [PubMed] [Google Scholar]
- 36.Ghajar CM, George SC, Putnam AJ. Matrix metalloproteinase control of capillary morphogenesis. Crit Rev Eukaryot Gene Expr. 2008;18:251–278. doi: 10.1615/critreveukargeneexpr.v18.i3.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wajant H, Pfizenmaier K, Scheurich P. Tumor necrosis factor signaling. Cell Death Differ. 2003;10:45–65. doi: 10.1038/sj.cdd.4401189. [DOI] [PubMed] [Google Scholar]
- 38.Passino MA, Adams RA, Sikorski SL, Akassoglou K. Regulation of hepatic stellate cell differentiation by the neurotrophin receptor p75NTR. Science. 2007;315:1853–1856. doi: 10.1126/science.1137603. [DOI] [PubMed] [Google Scholar]
- 39.Itai T, Tanaka M, Nagata S. Processing of tumor necrosis factor by the membrane-bound TNF-alpha-converting enzyme, but not its truncated soluble form. Eur J Biochem. 2001;268:2074–2082. doi: 10.1046/j.1432-1327.2001.02085.x. [DOI] [PubMed] [Google Scholar]
- 40.Lorent K, Yeo SY, Oda T, Chandrasekharappa S, Chitnis A, Matthews RP, Pack M. Inhibition of Jagged-mediated Notch signaling disrupts zebrafish biliary development and generates multi-organ defects compatible with an Alagille syndrome phenocopy. Development. 2004;131:5753–5766. doi: 10.1242/dev.01411. [DOI] [PubMed] [Google Scholar]
- 41.Sakaguchi TF, Sadler KC, Crosnier C, Stainier DY. Endothelial signals modulate hepatocyte apicobasal polarization in zebrafish. Curr Biol. 2008;18:1565–1571. doi: 10.1016/j.cub.2008.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parsons CJ, Takashima M, Rippe RA. Molecular mechanisms of hepatic fibrogenesis. J Gastroenterol Hepatol. 2007;22(Suppl 1):S79–S84. doi: 10.1111/j.1440-1746.2006.04659.x. [DOI] [PubMed] [Google Scholar]
- 43.Guo J, Friedman SL. Hepatic fibrogenesis. Semin Liver Dis. 2007;27:413–426. doi: 10.1055/s-2007-991517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.