Summary
Mutations in ribosomal proteins are associated with a congenital syndrome, Diamond–Blackfan anaemia (DBA), manifested by red blood cell aplasia, developmental abnormalities and increased risk of malignancy. Recent studies suggest the involvement of p53 activation in DBA. However, which pathways are involved and how they contribute to the DBA phenotype remains unknown. Here we show that a zebrafish mutant for the rpl11 gene had defects both in the development of haematopoietic stem cells (HSCs) and maintenance of erythroid cells. The molecular signature of the mutant included upregulation of p53 target genes and global changes in metabolism. The changes in several pathways may affect haematopoiesis including upregulation of pro-apoptotic and cell cycle arrest genes, suppression of glycolysis, downregulation of biosynthesis and dysregulation of cytoskeleton. Each of these pathways has been individually implicated in haematological diseases. Inhibition of p53 partially rescued haematopoiesis in the mutant. Altogether, we propose that the unique phenotype of DBA is a sum of several abnormally regulated molecular pathways, mediated by the p53 protein family and p53-independent, which have synergistic impact on haematological and other cellular pathways affected in DBA. Our results provide new insights into the pathogenesis of DBA and point to the potential avenues for therapeutic intervention.
Keywords: haematopoiesis, HSC, p53, Diamond–Blackfan anaemia
Diamond–Blackfan anaemia (DBA) is a bone marrow failure syndrome also characterized by developmental malformations and cancer (Lipton & Ellis, 2010). DBA is associated with mutations in ribosomal protein (RP) genes, most often in RPS19, RPL5 and RPL11, while mutations in several other RP genes are found at lower frequencies (Draptchinskaia et al, 1999; Lipton & Ellis, 2010). The acquired bone marrow failure in 5q-syndrome is also associated with the loss of a ribosomal protein, in this case RPS14 (Ebert et al, 2008). Deficiency in many RPs leads to the impairment of ribosome biogenesis (Flygare et al, 2007). Ribosome biogenesis is also compromised in several other bone marrow failure syndromes with clinical features that overlap with DBA, such as dyskeratosis congenita, Shwachman–Diamond syndrome and cartilage-hair hypoplasia (Ganapathi & Shimamura, 2008).
Previously, we demonstrated that developmental and haematopoietic defects in rps19-deficient zebrafish were mediated by p53; which was also upregulated in zebrafish mutants for rps8, rps11 and rps18 (Danilova et al, 2008). These results are consistent with earlier reports that haploinsufficiency of rps6 leads to the activation of a p53-dependent checkpoint during gastrulation (Panic et al, 2006) and RPL23 knockout in human cell lines leads to p53 accumulation (Jin et al, 2004). Also, McGowan et al (2008) reported that in mice, mutations in Rps20 and Rps19 cause p53-mediated decrease in the number of erythrocytes and skin darkening. Knockdown of zebrafish rpl11 by a morpholino also resulted in p53 upregulation, although no blood defects were reported (Chakraborty et al, 2009). Macrocytic anaemia in a mouse model of 5q-syndrome is also accompanied by upregulation of p53 (Barlow et al, 2010). Both in zebrafish and in mice, suppression of p53 rescues the abnormal phenotype (Danilova et al, 2008; McGowan et al, 2008; Chakraborty et al, 2009; Barlow et al, 2010). Deficiency of other proteins involved in ribosome biogenesis, such as nucleolar protein Bop1, cause p53-mediated cell cycle arrest (Pestov et al, 2001). These results suggest that p53 upregulation may be a common response to ribosomal stress and is involved in the pathogenesis of DBA and related bone marrow failure syndromes.
The p53 protein family is involved in development, differentiation and cell response to stress (Murray-Zmijewski et al, 2006). Full-length proteins (TAp63, TAp73 and p53) induce cell cycle arrest, apoptosis, senescence, or differentiation depending on the cellular context, while isoforms expressed from the internal promoters (ΔN) inhibit the activity of full-length proteins and promote proliferation. Stress signals activate TA isoforms whereas growth factors activate ΔN isoforms. Thus, the p53 family works as a cellular processor, integrating signals from the cell’s external and internal environment to determine cell fate.
To further understand molecular pathways associated with ribosomal insufficiency, we studied a zebrafish mutant for rpl11 gene (Amsterdam et al, 2004a). The most severe defects in the mutant were confined to the development and maintenance of red blood cells. The mutant had decreased numbers of haematopoietic stem cells (HSCs) that could be rescued by p53 inhibition. The shortened life span of erythroid cells in the mutant was also rescued by p53 inhibition. The mutant had altered expression of hundreds of genes and, despite the changes in expression of some erythroid-specific genes, the changes most detrimental to erythrocytes may be common to all cell types. Upregulation of genes involved in cell cycle arrest and apoptosis are examples of such changes. Specific physiology of red blood cells makes them selectively vulnerable to these changes. A distinctive feature of the mutant phenotype was abnormal regulation of metabolic pathways that involved a shift from glycolysis to aerobic respiration, suppressed biosynthesis, activated catabolism and increased levels of insulin mRNA and glucose.
Approximately 40% of DBA patients have congenital malformations pointing to a global dysregulation of development. Accordingly, we have found that expression of many genes affecting development was altered in the rpl11 mutant. Hormonal and immune dysregulation were also apparent. Remarkably, mitogenic factors were over expressed in the mutant on the background of the increased cell death. Overall, the analysis of rpl11 mutant suggests that ribosomal deficiency leads to a systemic disease – a sum of multiple defects that probably have a synergistic effect on development and haematopoiesis.
Methods
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
RNA was prepared using Trizol (Invitrogen, Carlsbad, CA, USA) from pool of 30–40 embryos. cDNA was synthesized by reverse transcription of 2 µg of RNA with the random hexamer primers. Quantitative PCR (qPCR) was performed using iQ SYBR Green Super Mix and a MyiQ Single-Color PCR thermal cycler (Biorad, Hercules, CA, USA). Each experiment was performed in triplicate. Levels of mRNA expression in mutants relative to sibling controls were normalized to bactin1 and calculated according to the Cτ method. Primers are shown in Table SVII.
Microarray analysis
The assay was performed at the UCLA microarray core. Two µg of total RNA were amplified and labelled with biotin using GeneChip Expression 3′-Amplification Reagents (Affymetrix, Santa Clara, CA, USA). Twelve microgram of biotin-labelled fragmented cRNA was hybridized with GeneChip zebrafish Genome Array and scanned using GeneChip Scanner 3000. Cell intensity calculation and scaling was performed using GENECHIP Operating Software v 1.4. The data were submitted to ArrayExpress, Acc: E-MEXP-2381.
Morpholinos
p53 MO inhibitor of translation, 5′-gcgccattgctttgcaagaattg (Langheinrich et al, 2002) (Gene Tools, Corvallis, OR, USA). The 3 ng of morpholino was injected at the one cell stage.
Staining of erythroid cells
Six milligram of o-dianizidine was dissolved in 50 ml acetic acid, the volume brought to 6 ml with water and neutralized to pH 4·5 by NaOH following by addition of 4 ml of ethanol and 130 ml of 50% H2O2. Embryos were treated in the dark in this solution and washed with water.
Staining of proliferating cells
Embryos were incubated overnight with rabbit antibodies to H3 histone phosphorylated at serine 10 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and, after washing, for 2 h at room temperature with goat anti rabbit, (Fab)2 fragments labelled with A594 fluorophore (Invitrogen) as the secondary antibodies.
In situ hybridization
Whole mount in situ hybridization was carried out as described (Thisse et al, 1993) with runx1, cmyb, kdrl (flk1), efnb2a, mpx and rag1 riboprobes.
Western blot
Thirty-five embryos were lysed with lysis buffer and protein concentrations were determined by bicinchoninic acid (BCA) Protein Assay kit (Thermo Scientific, Rockford, IL, USA). 10 mg of total protein was separated on 12% sodium dodecyl sulphate polyacrylamide gel electophoresis. The proteins were transferred onto a nitrocellulose membrane and probed with rabbit anti-RPL11 antibody, ab79352, 1:1000 (Abcam, Cambridge, MA, USA) followed by horseradish peroxidase–conjugated anti-rabbit antibody (Santa Cruz Biotechnology). The membrane was stripped and reprobed with anti-mouse alpha-tubulin antibody (Sigma, Saint Louis, MI, USA) followed by peroxidase-conjugated anti-mouse antibody (Santa Cruz Biotechnology).
Glucose levels
The blood from adult fish was obtained by tail cutting. To measure glucose levels in body fluid of zebrafish embryos, 80 embryos were placed on the tube cup with a mesh, water was removed by brief centrifugation, embryos were homogenized and centrifuged. The body liquid or blood was applied to a test stripe of Accu-Chek Compact Plus blood glucose meter (Roche Diagnostics, Indianapolis, IN, USA).
Results
rpl11 mutant has defective haematopoiesis
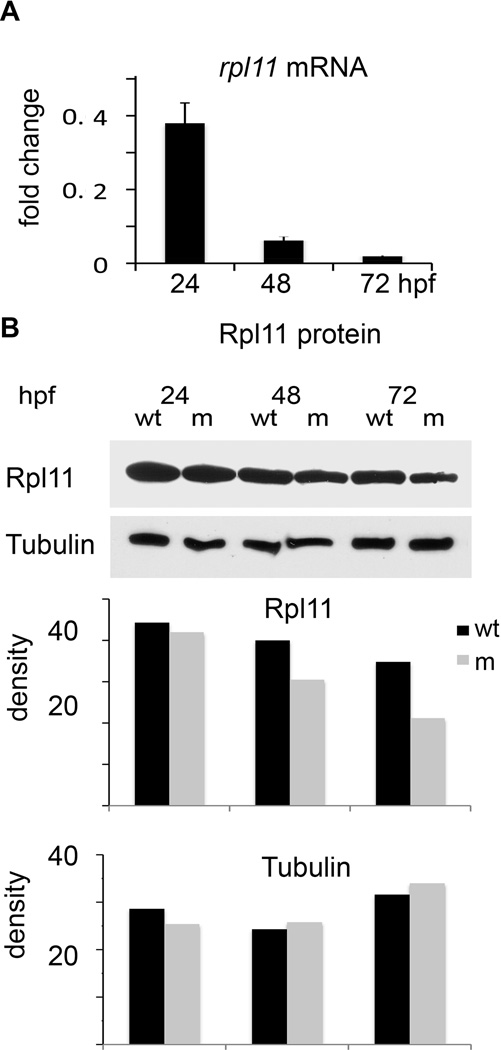
A zebrafish mutant for rpl11 was identified in a mutagenic screen performed by the Center for Cancer Research and Department of Biology, Massachusetts Institute of Technology (Cambridge, MA, USA) (Amsterdam et al, 2004a). The mutant contains a retroviral insertion in the first rpl11 intron, which precludes normal mRNA splicing resulting in reduction of rpl11 expression (Fig 1A). Rpl11 is an essential RP and the mutation is homozygously lethal (Amsterdam et al, 2004a). However, embryos are supplied with maternal ribosomes and mRNA for RPs that let mutants proceed throughout the initial developmental stages even in the absence of synthesis of their own Rpl11. The level of Rpl11 protein in mutants was comparable to that in wild type embryos during the first day of development and it gradually decreased thereafter (Fig 1B). With exhaustion of maternal supplies, mutants eventually died around day 5–7. Rpl11-deficient embryos could be distinguished from siblings started from 24-h post-fertilization (hpf) by signs of brain apoptosis. At day two, they had smaller head and eyes and underdeveloped liver and gut (Amsterdam et al, 2004a); although the number of primitive erythrocytes was not reduced and circulation was established (Fig 2A, B). The maturation of erythrocytes was, however, delayed in the mutants as evidenced by 2·6-fold elevated gata1a expression at 48 hpf (Fig S1). gata1a is highly expressed in developing erythrocytes, and the expression decreases when cells mature. In wild-type zebrafish, gata1a expression is very low at 48 hpf.
Fig 1.
In the rpl11 mutant, the levels of rpl11 mRNA and protein decreased with different dynamics. (A) Embryos were supplied with maternal ribosomes and mRNA for rpl11 and progressed through initial developmental stages using these resources. At 24 hpf the level of rpl11 mRNA was only 0·35-fold lower from wild-type embryos but sharply decreased thereafter when wild-type embryos started to transcribe more of their own rpl11 mRNA. qPCR, RNA pooled from 30 embryos. (B) At 24 hpf, the level of Rpl11 protein in the mutant was comparable to that in wild type fish. It declined gradually with exhaustion of maternal supplies. The intensity of staining relative to background was measured using IMIGEJ program. Representative of two independent experiments is shown.
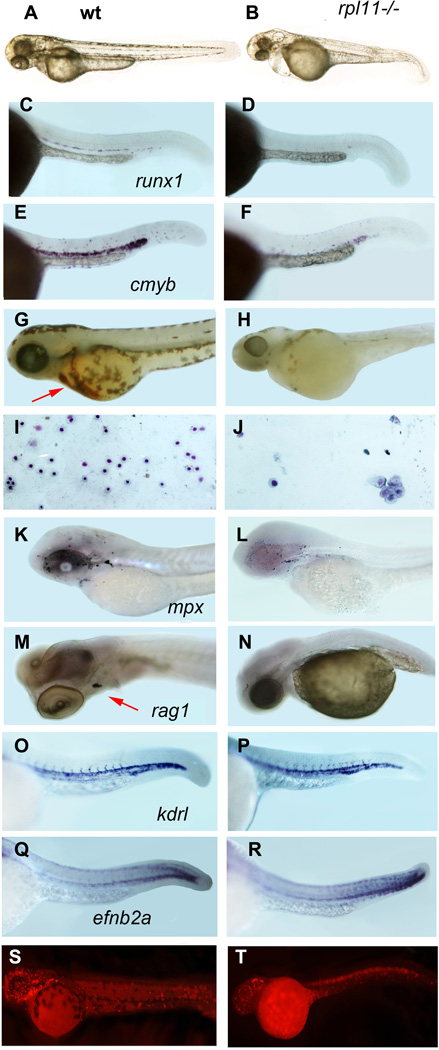
Fig 2.
The rpl11 mutant has developmental and haematopoietic defects. (A, B) At 48 hpf, rpl11 mutants had smaller heads and eyes, underdeveloped liver/gut and occasionally pericardial oedema. (C–F) The number of HSCs marked by expression of runx1 (C,D) and cmyb (E,F) is reduced in the mutant, in situ hybridization, 30 hpf, 30 embryos per group (G,H) Only few red blood cells remained in the mutant after day 3·5. O-dianizidine staining, 84 hpf, 30 embryos per group. Arrow points to erythrocytes in the wild-type embryo (I,J) Some erythroid cells from the blood of a mutant embryo are macrocytic. Blood smears were prepared from individual embryos; the results are representative of 6–10 embryos. Giemsa stain. (K,L) The number of granulocytes in the mutant was only slightly decreased. 72 hpf, in situ hybridization, mpx probe. (M,N) The expression of rag1 in the thymus was reduced in the mutant. Day 4, in situ hybridization. (O–R) The expression of the vascular marker kdrl and arterial marker efnb2a was not significantly changed in the mutant, 30 hpf (S,T) Proliferation is decreased in the brain and blood of in the mutant embryo but cells in some other tissues continue to proliferate. 24 hpf. Antibodies to phosphorylated histone H3.
Similar to other vertebrate species, zebrafish have two waves of haematopoiesis, primitive and definitive. Primitive erythrocytes and macrophages develop at c. 18–23 hpf directly from the lateral plate mesoderm (Davidson & Zon, 2004). They stay in circulation for 5–10 d until replaced by definitive cells. The latter are generated in the posterior blood island (Bertrand et al, 2007) and from haematopoietic stem cells (HSCs) developing in the region of the dorsal aorta starting from 26 hpf (Murayama et al, 2006). HSCs can be visualized by expression of runx1 and cmyb. In mouse and zebrafish, Runx1 was shown to be necessary for the formation of HSCs (Kalev-Zylinska et al, 2002). Cmyb controls the proliferation and differentiation of HSC and progenitor cells (Sandberg et al, 2005). Staining with these probes suggested that in the Rpl11-deficient zebrafish, the number of HSCs was decreased (Fig 2C–F).
The circulation of primitive erythrocytes in the mutant did not initially differ from that in wild type fish. However, beginning at day 2·5, mutant erythrocytes started to pool in the yolk in large aggregates and the number of cells in circulation started to decrease. One day later, the aggregated cells were cleared, presumably by macrophages, and the rpl11 mutant became almost completely devoid of erythrocytes (Fig 2G–H); the few cells that remained in circulation were blast-like or macrocytic (Fig 2I,J). These data suggest that two mechanisms contributed to the anaemia in the Rpl11-deficient zebrafish: (i) decreased production of new cells and (ii) increased destruction of existing cells.
To determine if the development of definitive blood lineages other than erythrocytes was affected we analysed the expression of mpx, which is a marker for granulocytes, and rag1, which is a marker for lymphoid cells. At day 3, the number of granulocytes in the Rpl11-deficient zebrafish was only slightly lower than in wild type (Fig 2K,L). At day 4, the expression of rag1 was significantly reduced in the mutant (Fig 2M,N). However, at this age, the level of Rpl11 protein dropped below 50% of the wild-type level, making the deficiency more severe than in the heterozygous DBA patients. Nevertheless these data suggest that all definitive lineages can be affected to various degrees by RP mutation. DBA patients also occasionally show defects in blood lineages other than erythrocytes (Halperin & Freedman, 1989; Giri et al, 2000).
Previous studies have suggested the existence of a common progenitor for haematopoietic and endothelial cells with Runx1 acting at the endothelial to haematopoietic fate transition. A zebrafish mutant for rps29, which lacks HSCs, underwent vasculogenesis normally but failed to establish artery identity (Burns et al, 2009). We therefore analysed expression of kdrl specific to all vasculature and efnb2a, which is necessary for artery specification. rpl11 mutants had normal levels of flk1 expression in major and intersomitic vessels and a normal pattern of efnb2a expression at 30 hpf (Fig 2O,P). Therefore, in the rpl11 mutant, deficiency of blood production does not appear to depend on endothelial lineage defects but is associated with defects in development/maintenance of HSCs.
To further characterize the effects of rpl11 mutation on cell proliferation, we compared the number of proliferating cells using antibodies to phosphorylated histone H3. Cell proliferation in the brain and blood cells was significantly reduced compared to other tissues pointing to different sensitivity of various cell types to rpl11 deficiency (Fig 2S,T).
p53 network is activated in the rpl11 mutant
To determine which molecular pathways are affected in the mutant, we performed gene expression profiling of 48 hpf mutants and siblings using Affimetrix chips representing 14 900 transcripts. Expression of selected genes was verified by qPCR. Expression of additional genes of interest not represented in the array was examined by qPCR alone. From genes represented in the array, at least 400 genes were upregulated more than twofold and more than 300 were downregulated more than twofold. Less substantial changes were observed in expression of many more genes, some of those nevertheless might be functionally important. Among upregulated genes with known function the largest fraction comprised genes involved in ribosome biogenesis, RNA processing, translation and mitochondria (Tables SI–VI) (Fig S2). Among the downregulated genes, the largest fraction comprised neural genes followed by genes involved in energy production, transport, biosynthesis, cytoskeleton and extracellular matrix.
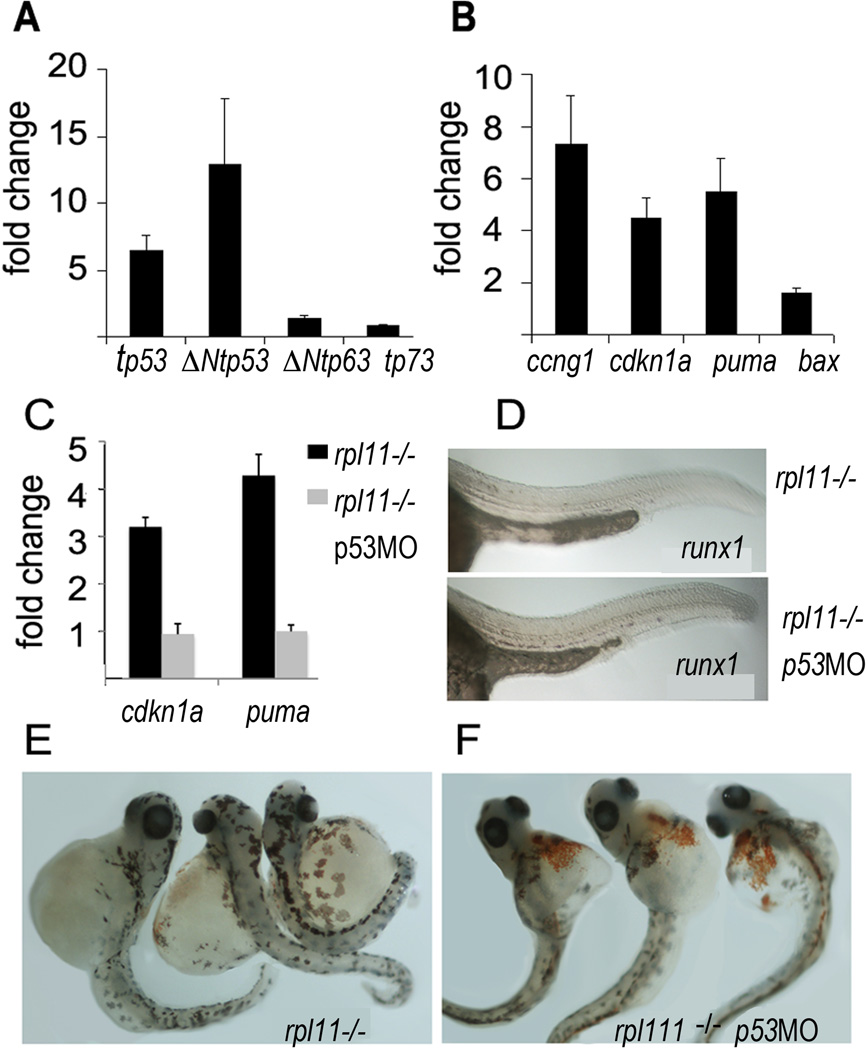
Previously, we found transcriptional activation of both full-length and N-truncated p53 isoforms in several RP mutants and in rps19 knockdown (Danilova et al, 2008). We also observed upregulation of ΔNtp63, although to a lesser degree. Similarly in the rpl11 mutant, we observed transcriptional upregulation of tp53 (p53), Δ113tp53 and ΔNtp63 (Fig 3A). Many alterations in gene expression in the mutant corresponded to known p53 family targets (Riley et al, 2008). The most important changes included upregulation of genes responsible for cell cycle arrest such as cdkn1a (p21) and ccng1 (cyclin G1) and for apoptosis such as puma, bax (Fig 3B), gadd45ab and gadd45ba (Table SI).
Fig 3.
p53-dependent pathways were dysregulated in the rpl11 mutant. (A) Expression of total tp53 and Δ113Ntp53 was increased, ΔNtp63 expression was slightly (1·5-fold) increased, tp73 expression was not changed, qPCR, 48 hpf. In A-C, RNA was pooled from 30 mutant or sibling embryos (B) cdkn1a and ccng1 responsible for cell cycle suppression and pro-apoptotic puma and bax were upregulated in the rpl11 mutant. (C) After p53 was inhibited by a morpholino, expression of p53 targets returned to the wild-type level. (D) Expression of runx1 was rescued in the rpl11 mutant by p53 inhibition. (E,F) The rpl11 mutant embryos with suppressed p53 had more red blood cells at day 3·5. In A–C, representative of two and in D–F, three independent experiments are shown.
To determine the effect of the elevated expression of p53 and its targets on haematopoiesis in the rpl11 mutant, we inhibited tp53 translation using morpholinos. p53 inhibition resulted in downregulation of genes controlling cell cycle and apoptosis and rescue of runx1 expression in the mutant (Fig 3C, D). Cell loss was also reduced. As a result, at day 3·5, rpl11 mutants with suppressed p53 had more erythroid cells than mutants with intact p53 (Fig 3E, F). Therefore both the life span of erythrocytes and development of new cells depended to a large extent on p53 target genes.
Upregulation of p53 targets mediating cell cycle arrest and apoptosis are probably the major factors responsible for haematopoietic defects in the rpl11 mutant. In addition, p53 controls many other cellular processes including energy production, general metabolism, cytoskeleton, extracellular matrix, transport, etc. (Riley et al, 2008; Vousden & Ryan, 2009). Accordingly, our data show alteration in many correspondent pathways in the mutant.
Metabolic changes in the rpl11 mutant
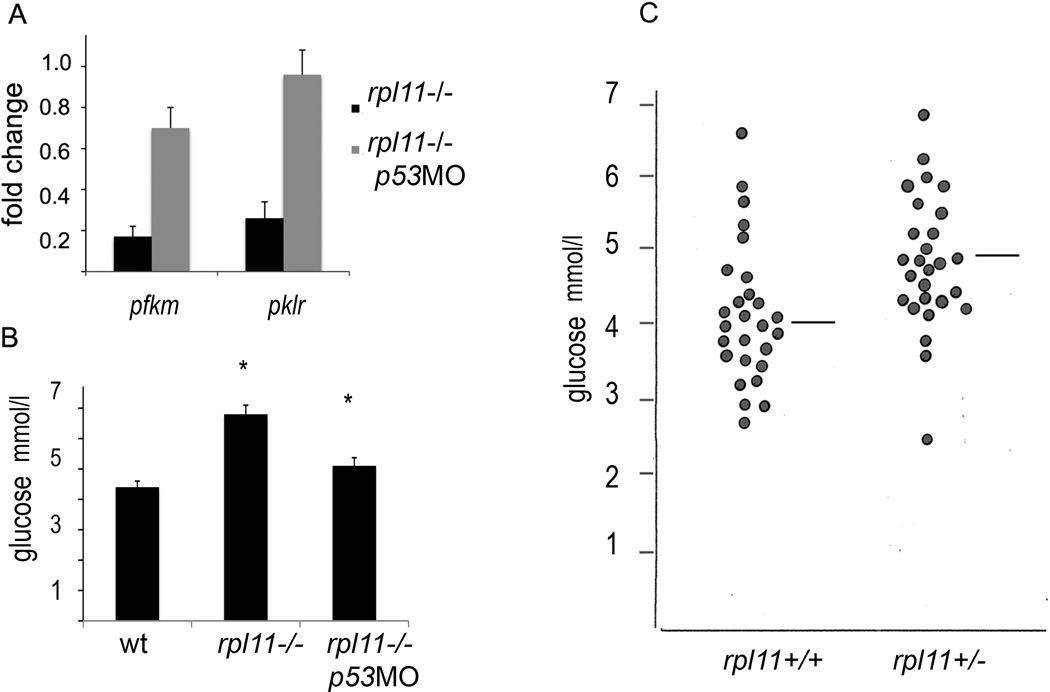
A distinct feature of the rpl11 mutant was a shift from glycolysis to aerobic respiration. Genes encoding glycolytic enzymes were down regulated approximately twofold including phosphorylase glycogen muscle b(pygmb) and liver (pygl), glucose-6 phosphate isomerase b(gpib), 6-phosphofructokinase muscle (pfkm), triosephosphate isomerase 1a(tpi1a), phosphoglycerate mutase 2 (pgam2), aldolase and pyruvate kinase liver and RBC (pklr) (Table SII). At the same time, enzymes involved in aerobic respiration were upregulated. These changes were consistent with observations in cell lines, where the relative fraction of ATP derived from glycolysis versus aerobic respiration is inversely proportional to tp53 gene dosage (Matoba et al, 2006). Accordingly, in the rpl11 mutant, inhibition of p53 rescued the expression of glycolytic enzymes (Fig 4A).
Fig 4.
The rpl11 mutant had major p53-dependent changes in metabolism. (A) Expression of glycolytic enzymes was downregulated and was rescued by p53 inhibition. Adult zebrafish heterozygous for rpl11 mutations were mated and eggs were divided into two groups. One group was injected with 3 ng of p53 morpholino at one cell stage, the other group was left as a control. At 60 hpf, mutants were collected in both groups based on their morphology and RNA was prepared from 30 mutant and corresponding siblings. Fold change in expression of 6-phosphofructokinase, muscle (pfkm) and pyruvate kinase, liver and RBC (pklr) was determined by qPCR in control and p53 morpholino-treated mutants relative to their siblings. (B) The level of glucose was elevated in the rpl11 mutant and p53 inhibition led to its decrease. Control and p53 morpholino-treated rpl11 mutants were grown until 72 hpf and glucose measurement was performed in pools of 80 embryos. Shown is an average of four repeats (P < 0·001 Student’s t-test). (C) The median levels of blood glucose in adult fish were higher in rpl11 heterozygous fish in comparison to their wild-type siblings. The blood glucose was measured after 12 h of fasting. The median level for wild-type fish was 4·01 mmol/l, for mutants, 4·62 mmol/l (P < 0·005 Student’s t-test, n = 27).
To determine how changes observed in zebrafish relate to mammalian models we analysed expression of glycolytic enzymes in mouse fetal liver cells with downregulated Rps19 (Sieff et al, 2010). Similar to zebrafish embryos, mouse fetal liver cells respond to RP deficiency by transcriptional upregulation of p53 and its targets. The glycolytic enzymes were downregulated in RPS19-deficient fetal liver cells even stronger than in the rpl11 mutant zebrafish (Fig S3).
Erythrocytes differ from other cells in that they lack mitochondria and derive most of their energy from glycolysis. In lower vertebrates such as zebrafish, young red blood cells have mitochondria but lose them with ageing (Moyes et al, 2002). Therefore suppression of glycolysis would selectively affect erythrocytes. Deficiency of glycolytic enzymes is a known cause of hereditary nonspherocytic haemolytic anaemia (HNHA) (Steiner & Gallagher, 2007). Haemolysis of erythrocytes, however, has not been often reported in DBA. Contrary to HNHA, where the failure of one enzyme disrupts the whole pathway, in RP deficiency, the glycolytic pathway is functional but down regulated, which may lead to less severe defects.
Other genes involved in energy production and utilization were also downregulated in the rpl11 mutant including citric acid cycle, transporters, ATP synthases and creatine kinases. Downregulation of creatine kinases has been noted in DBA (Gazda et al, 2006). Another important source of energy, beta-oxidation of fatty acids, was also compromised in the mutant (Table SII). In humans, such deficiency causes HELLP (haemolysis, elevated liver enzymes, low platelets) syndrome. In general, lack of energy due to shortage of ATP leads to destabilization of the red cell membrane.
An increase in aerobic respiration in the mutant was accompanied by the decreased expression of detoxifying enzymes, such as superoxide dismutase, which converts superoxide radicals into hydrogen peroxide and catalase, which destroys hydrogen peroxide. The downregulation of catalase was rescued by p53 inhibition (Fig S4). Members of the aldehyde dehydrogenase family were also downregulated. These enzymes protect cells from the toxic effects of aldehydes by oxidizing them to acids. Oxidative stress resulting from the decreased expression of all types of detoxifying factors may decrease the life span of erythrocytes and affect HSC maintenance.
Glycolysis and the citric acid cycle provide precursors for synthesis of biomolecules. Therefore, suppressed glycolysis would compromise biosynthesis. In addition, many genes involved in biosynthesis were directly down regulated, especially those involved in the energy consuming processes such as synthesis of cholesterol and fatty acids (Table SII). Biosynthesis of some erythroid-specific genes including alas2 (aminolevulinate, delta-, synthetase 2), which catalyze the first step in haem biosynthesis, was also suppressed (Table SVI). Defects in this gene cause sideroblastic anaemia characterized by hypochromic erythrocytes and accumulation of iron (Harigae et al, 2003). Another important group of genes downregulated in the rpl11 mutant comprised genes involved in vitamin A absorption, transport and utilization.
In contrast to biosynthesis, salvage pathways were activated, consistent with p53 upregulation (Vousden & Ryan, 2009). One example is adenosine deaminase (ADA), which participates in nucleotide metabolism (1·7-fold) and its upregulation is commonly associated with DBA. The gene cyp24a1, encoding enzyme catabolizing vitamin D, was 10-fold upregulated, which would negatively impact many cellular processes (Table SII).
Another feature of the rpl11 mutant’s metabolism was the increased expression of genes involved in gluconeogenesis and upregulation of the preproinsulin mRNA (Table SII). To determine the outcome of these changes, we measured the levels of glucose in body fluids of the mutant embryos and found that they were elevated in the mutants in comparison to siblings (Fig 4B). Next, we measured glucose levels after overnight fasting in adult rpl11+/− fish and their wild type siblings. The median blood glucose levels were higher in rpl11+/− fish (Fig 4C). A combination of high insulin levels with elevated glucose would lead to a condition resembling human diabetes type II. With p53 downregulation, the levels of glucose returned to normal indicating that these changes were p53-dependent (Fig 4B).
Cytoskeleton and extracellular matrix
Many genes of the cytoskeleton and extracellular matrix are p53 targets (Riley et al, 2008). In the rpl11 mutant, various structural genes showed downregulation (Table SIII). Several collagens were among the most downregulated genes. Cellular membranes are stabilized through protein linkages, such as Ankyrin1, with the underlying cytoskeleton. There are two Ankyrin1-like proteins in the zebrafish. One of them (NM_001110783) was ubiquitously expressed while the other (XM_001919434) was more restricted to erythroid cells (data not shown). The latter was twofold downregulated in the mutant. Several other components of the skeletal-membrane linkages were also downregulated (Table SIII). Consistent with findings in zebrafish rpl11 mutant, mouse fetal liver cells with suppressed Rps19 also had downregulated Ank1 (Fig S3).
The quality of cytoskeleton and membranes is important for all cells but especially for the erythrocytes. They need to withstand circulation and exposure to oxidative stress in lungs/gills. Defects in linkage proteins, such as Ankyrin 1, Band 3 and Spectrins, lead to hereditary spherocytosis characterized by loss of membrane, reduced deformability and eventual trapping of such cells by macrophages (Steiner & Gallagher, 2007).
Stress-response and immune genes
Altogether, our data indicate that the allostatic changes in the mutant included cell cycle arrest, increased apoptosis, shift from glycolysis to aerobic respiration, decreased biosynthesis and increased catabolism, decreased expression of detoxifying factors, decreased expression of structural, membrane and extracellular matrix components. Not surprisingly, stress response pathways became activated. Upregulation of acute phase and heat shock proteins was prominent. The fourfold upregulation of prolactin may constitute a component of the endocrine stress response to metabolic imbalance.
There is an evolutionary conserved connection between the metabolic and immune systems. Metabolic stress can activate interferon and complement systems and activation of the autoimmune response can modify insulin signalling. In the rpl11 mutant, genes involved with the interferon and complement systems were upregulated along with some pro-inflammatory factors (Table SVI). Increased expression of the complement system may enhance the elimination of erythrocytes from circulation by macrophages. Inappropriate activation of complement can lead to damage of other tissues and, because the complement system controls B cell response, it may lead to activation of antibody-mediated autoimmunity. Upregulation of interferons and other proinflammatory factors activates macrophages and directs T helper cells development to pro-inflammatory Th1 pathway (Hu & Ivashkiv, 2009). Interferon response is thought to contribute to the apoptosis of haematopoietic progenitors induced by cytotoxic T cells in acquired aplastic anaemia (Young et al, 2008). Increased proportion of cytotoxic T cells (decreased T4/T8 ratio) was reported in DBA and may also be connected to the interferon activation (Halperin & Freedman, 1989). At the same time, certain protective immune mechanisms were downregulated in the rpl11 mutant, such as antimicrobial peptides defensins and lysozyme. These data suggest that RP mutations may result in weakened immunity and increased risk for inflammation and autoimmunity.
Increased expression of growth-promoting factors
Along with increased expression of factors promoting apoptosis, the rpl11 mutant had increased expression of factors promoting proliferation including myca (c-myc), fos and jun families, cdc25, ccnd1 (cyclin D1) and pcna (Fig 5 and Table SI). The activation of both death and growth-promoting pathways is a feature predisposing to oncogenic transformation. In addition, many factors promoting the survival and spread of cancer cells were upregulated including antiapoptotic proteins, ΔNtp63, heat shock proteins, matrix metalloproteinases and vegfaa (Tables SI–VI).
Fig 5.
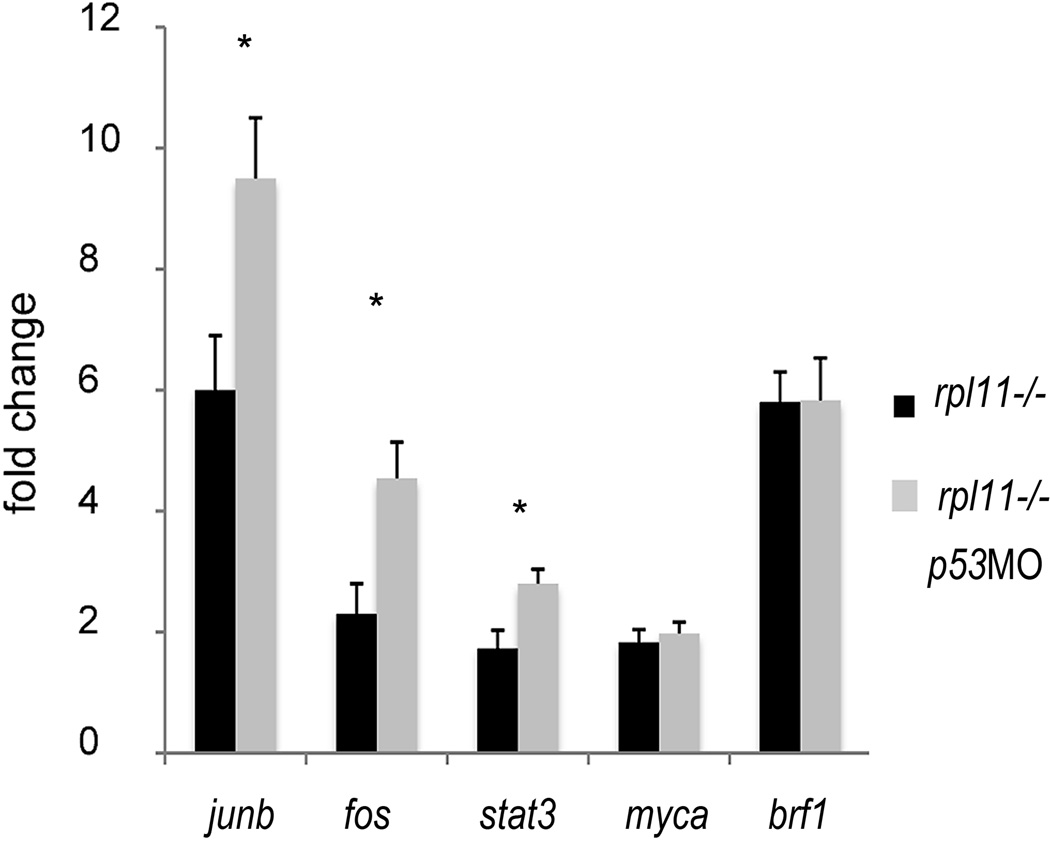
The rpl11 mutant upregulated some factors promoting proliferation. Early response genes such as fos, junb and stat3 showed higher expression when p53 was inhibited by a morpholino (junb P < 0·05, fos and stat3 P < 0·01, Student’s t-test). Upregulation of genes involved in synthesis of non-coding RNA and ribosome biogenesis did not depend on p53. Example is brf1, involved in tRNA biosynthesis. qPCR, 48 hpf, RNA was pooled from 30 embryos. Representative of two independent experiments is shown.
p53 inhibition had a different impact on growth-promoting factors (Fig 5). Our study suggests that upregulation of genes involved in ribosome biosynthesis, synthesis of tRNAs and translation were p53-independent. For example, brf1, subunit of RNA polymerase III transcription initiation factor IIIB, was among the most upregulated genes in the mutant and its expression did not change after p53 inhibition (Fig 5). AP-1 factor genes and stat3, involved in acute phase response, were also among the most upregulated genes. p53 inhibition in the mutant led to the increase in expression of some of these factors (Fig 5) indicating that their upregulation was p53 independent and that p53 opposed their upregulation.
Drugs currently used for DBA treatment may act through the suppression of p53
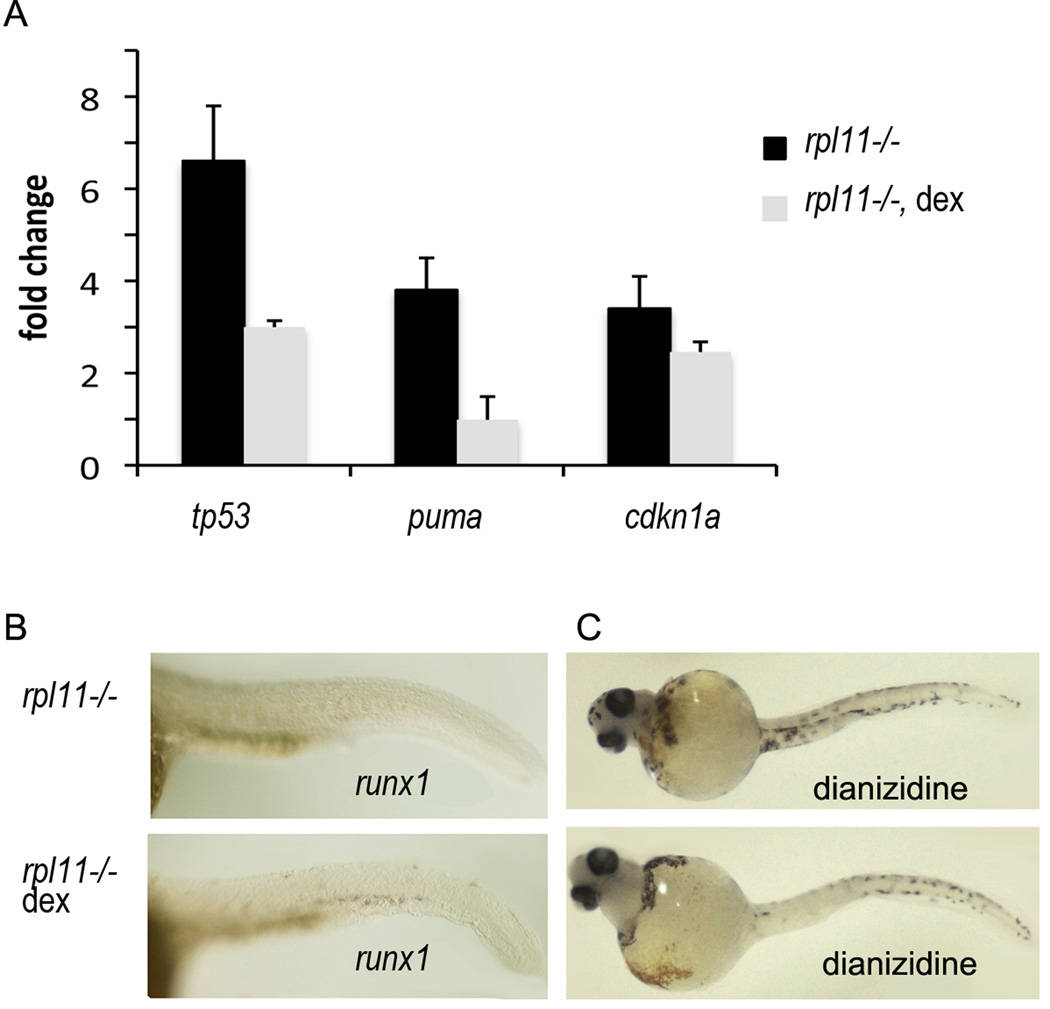
The dependence of many changes of gene expression in the rpl11 mutant on p53 suggests modulation of p53-dependent pathways as a possible therapeutic approach in DBA. Currently, corticosteroids are the most effective form of treatment for DBA. How they achieve their beneficial effects is, however, unclear. The analysis of RPS19-deficient CD34(+) cells treated with dexamethasone revealed that dexamethasone did not alter expression of RPS19 but activated haematopoietic genes such as RUNX1(Ebert et al, 2005). As runx1 expression was p53-dependent in our model, we decided to analyse the effect of dexamethasone on the p53 network in conditions of Rpl11 deficiency. Our results demonstrated that treatment of Rpl11-deficient zebrafish with pharmacological concentrations of dexamethasone led to a decreased expression of tp53 and its downstream targets cdkn1a and puma (Fig 6A). This suggests that dexamethasone acts, at least partially, by inhibiting p53 and in this way reduces suppression of HSC development imposed by p53. The decreased expression of runx1 and anaemia in the mutant were also rescued by dexamethasone treatment (Fig 6B,C). Dexamethasone may also relieve RP-deficient conditions by suppressing autoimmune reactions.
Fig 6.
Corticosteroids used for DBA treatment may act by downregulating p53 and its targets and in this way shifting cellular homeostasis to more normal conditions. Adult zebrafish heterozygous for rpl11 mutation were mated and the embryos were treated with 100 ng/ml dexamethasone from 2 hpf. The embryos were collected at 32–84 hpf and divided into mutant and sibling groups. (A) Expression of tp53, cdkn1a and puma mRNA was downregulated in mutants treated with dexamethasone. qPCR, fold change is shown as ratio to sibling controls. Each group had at least 30 embryos. (B) runx1 expression is downregulated in mutants and its expression was rescued by dexamethasone treatment. 32 hpf, In situ hybridization. (C) rpl11 mutants have few red blood cells at day 3·5 as estimated by staining with dianizidine. Treatment with dexamethasone led to the increase in number of red blood cells. Representative of two independent experiments are shown.
Discussion
The goal of this study was to understand the molecular mechanisms underlying ribosomal protein insufficiency and the specific phenotypes associated with DBA. A convenient feature of zebrafish is the ease of studying early development that enables the analysis of rpl11 mutant embryos during early embryogenesis and organogenesis when mutants develop on supplies of maternal Rpl11. The mutant used in this study was a severe hypomorph exhibiting increasing deficiency in Rpl11 from the second day of development; therefore it serves as a useful model for studying the pathogenesis of DBA.
The wave of primitive blood cells forms normally in the mutant, later however, these cells are eliminated from the circulation and the development of HSC and definitive red blood cells is suppressed. Analysis of the gene expression profile of the mutant suggests that the p53 family is responsible for many alterations. Despite the universal nature of the changes, the blood cells are the most susceptible to RP deficiency. Previous studies demonstrated that p53 targets are critical for haematopoesis. p53-deficient mice have twice as many HSCs with greater repopulating capacity (TeKippe et al, 2003). Upregulation of p53 targets suppressing cell cycle affects haematopoiesis also at later stages by limiting the expansive capacity of multipotent progenitors (Akala et al, 2008). Human CD34+ cells with downregulated RPS19 arrest at the G0 phase of the cell cycle (Kuramitsu et al, 2008). DBA erythrocytes with upregulated pro-apoptotic genes are more sensitive to apoptotic stimuli; in addition such cells expose phosphatidylserine on the cell surface, signalling to macrophages to eliminate them (Perdahl et al, 1994; Gazda et al, 2006; Miyake et al, 2008).
We hypothesize that the increased sensitivity of erythroid cells to ribosomal stress depends on two factors: (i) high proliferation rate of erythroid progenitors and (ii) specific physiology of erythroid cells. The response of a particular cell to stress depends on the ratio of growth promoting and stress signals. Demands for proliferation are higher in the fetal environment and in highly propagating cells such as blood progenitors. It may explain differences between the published findings. p53 accumulation at the level of mRNA and protein is easily detected in response to RP deficiency in a fetal background. In contrast, in DBA patients, only accumulation of p53 targets has been detected. Arginyl tRNA synthetase is upregulated in DBA blood progenitors (Gazda et al, 2006), similar to zebrafish rpl11 mutant. In contrast, tRNA synthetases are downregulated in DBA fibroblasts (Avondo et al, 2009), which agrees with the lower proliferation rate of these cells. Enhanced delivery of growth factors has been demonstrated to induce stalling and collapse of DNA replication forks, leading to the formation of DNA double stranded breaks; Ras, Mycs, Cyclin E, Mos, Cdc25a and E2f1 have similar effects (Halazonetis et al, 2008). This could lead to p53 activation through the ATM pathway as well as to the increased chance of oncogenic transformation. The exact mechanism of p53 activation is however unknown. One mechanism could be through free ribosomal proteins because RP deficiency leads to their misbalance and increased amounts of ribosomal proteins not incorporated into ribosome. Several ribosomal proteins can activate p53 by binding Mdm2 (Zhang & Lu, 2009). Transcriptional upregulation of p53 in the fetal environment suggest pathways leading to stabilization of tp53 mRNA or transactivation of tp53 promoter may be involved. Early response genes such as myca and AP-1 factor group are induced by growth factors and tp53 promoter was reported to be upregulated by these factors (Kirch et al, 1999). Disturbances in nucleolus organization/assembly may also provide stress signal leading to p53 upregulation.
Red blood cells are distinct from other tissues not only due to high proliferation during their development. They have a distinct physiology, which may make them selectively vulnerable to alterations in several common pathways. An example is the shift from glycolysis to aerobic respiration, which takes place in zebrafish rpl11 mutant. This shift happens in all cells but only erythrocytes rely almost exclusively on glycolysis and are, therefore, selectively affected by this change. Accordingly, erythrocytes would have more severe deficit of energy in comparison to other cells. Defects in membranes and cytoskeleton, although common for all cells, are more important for erythrocytes because of especially high requirements for quality of their structure, which has to be both strong and deformable. We suggest that the increased elimination of erythroid cells from the circulation in rpl11 mutant is a cumulative effect of upregulation of pro-apoptotic genes, downregulation of detoxifying enzymes, defects in membranes and cytoskeleton and upregulation of autoimmunity (Fig 7).
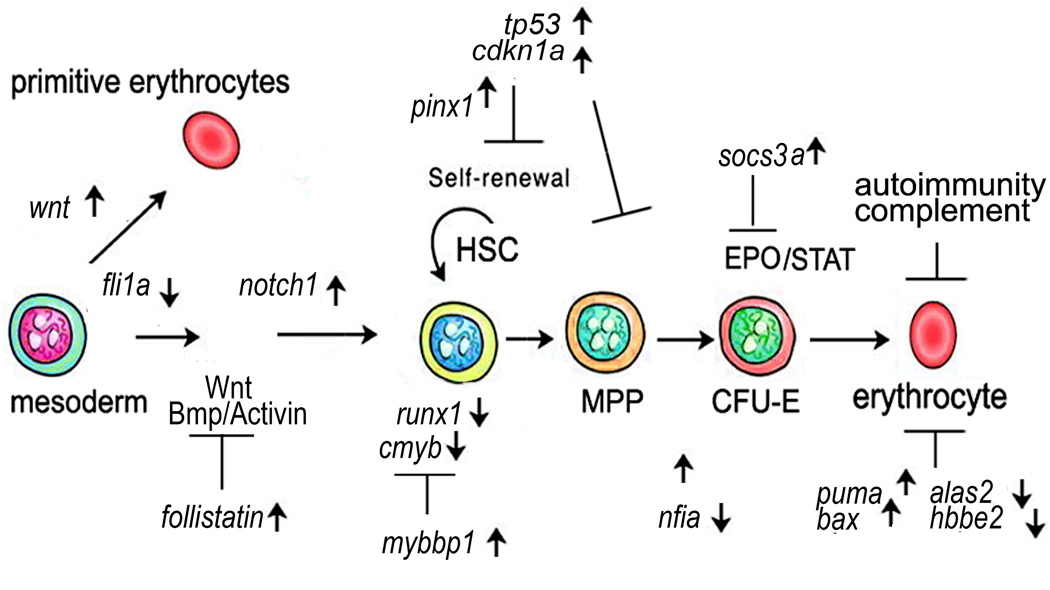
Fig 7.
Development and function of red blood cells may be compromised in multiple ways in the rpl11 mutant. Several factors that are important for the development of HSCs such as runx1 and cmyb are downregulated in the rpl11 mutant. Self-renewal of HSCs may be compromised by upregulation of factors responsible for cell cycle arrest such as cdkn1a (p21). Upregulation of cdkn1a may also negatively affect proliferation of erythroblasts. Suppression of EPO/STAT signaling by socs3a may also contribute to decreased proliferation of erythroid progenitors. The function of mature erythroid cells may be compromised in a number of ways. They include increased expression of pro-apoptotic genes, decreased haem production (Alas2 enzyme), dysregulation of haemoglobin synthesis, decreased glycolysis, decrease of enzymes detoxifying ROS, defects in cytoskeleton and membranes. Defective red blood cells would be a subject for enhanced trapping and removal by immune cells.
The failure of new erythrocytes to develop may be a result of inhibited HSC formation and self-renewal and inhibition of cell cycle progression in progenitors among other causes. Also, erythroid cells mature in association with macrophages and as RP-deficient erythrocytes are inherently defective, many of them can be eliminated at this stage. Some other factors may impair the function of mature RP-deficient erythroid cells, such as deficit of energy, decreased haem production and dysregulation of splicing and translational machineries.
Our analysis underscored the importance of known regulators of haematopoiesis and reveals new players. Shift in energy production ruled by p53 may be an important contributor to the increased vulnerability of erythroid cells to stresses. The global changes in metabolism in the mutant agrees with recent data that point to the important role p53 plays in normal and stressed metabolism (Vousden & Ryan, 2009). The increased levels of glucose and insulin in the rpl11 mutant and heterozygous rpl11+/− zebrafish suggest that DBA patients may have similar defects. This is an issue worthy of investigation.
Changes in gene expression observed in the rpl11 mutant affect development and function of not only blood cells but other tissues as well. It is likely that virtually any defect observed in DBA patients cannot be ascribed to dysregulation of a single gene. For example, in DBA, translation is decreased (Cmejlova et al, 2006). It is regulated by mTOR pathway integrating growth-promoting and stress signals. Increased levels of insulin and mitogens would lead to PI3K-Akt pathway activation and stimulation of translation; they are opposed by AMPK, p53 and PTEN. The rate-limiting step of translation is initiation, controlled by the eukaryotic translation initiation factors (eIFs), whose transcription is stimulated by growth signals. On the other hand, stress signals lead to translational silencing through eIFs post-translational modifications, such as eIF2a phosphorylation. In the rpl11 mutant, expression of 21 eIFs from 23 present on the chip as well as rps6 kinase was upregulated. Therefore, in the mutant, translation initiation factors are probably produced in excess but are suppressed by posttranslational mechanisms preventing optimal regulation of the translational machinery.
The cancer predisposition noted in DBA patients may also be due to cumulative effects of multiple alterations. Upregulation of mitogens leads to an enhanced transformation capability of RP-deficient cells. Decreased expression of detoxifying enzymes could lead to increased DNA damage and genomic instability. Upregulation of antiapoptotic factors and HSPs may support survival of cancer cells. Furthermore, upregulation of vegfaa and matrix metalloproteinases may promote cancer growth and spread. Interestingly, several zebrafish mutants for various RPs develop tumours with high frequency and the analysis of these tumours revealed decreased level of p53 protein (Amsterdam et al, 2004b; MacInnes et al, 2008). No mutations in the tp53 gene were found and the gene was transcribed. Therefore, a tumourigenesis in RPs mutants seems to involve a yet unknown regulator of p53 protein synthesis. These findings suggest that a link exists between RP deficiency and tp53 translation.
The impact of each p53-dependent pathway into the mutant phenotype is difficult to quantify due to their interdependence. Contributions from cell cycle and apoptosis are probably the main factors, however, other pathways may play an important role in a particular genetic background. From our data, the metabolic imbalance contributes to DBA pathophysiology and requires further evaluation in DBA patients.
Conclusions
Overall, our data demonstrate that systemic reprogramming takes place in RP-deficient organisms. DBA is a complex disease with multiple defects occurring at various levels: cellular, tissue and the whole organism. The developmental and haematopoietic defects in DBA and animal models are cumulative effects of deficiencies in several pathways.
These data open new avenues for drug intervention in DBA. Activation of the p53 network mediates several defects observed in RP-deficient zebrafish. Thus, finding ways to modulate these pathways without increasing the risk of cancer is one approach. When the specific molecular pathways leading to p53 activation in RP-deficient cells are determined, this information could provide new targets for therapeutic intervention. Other approaches to inhibit pathways downstream of p53 may also be beneficial, such as antioxidants. The complement system, which is upregulated in the rpl11 mutant, could also be a promising target. Finally, it is possible that the changes observed in rpl11 mutant zebrafish are shared with other human diseases with p53 upregulation that could lead to similar therapeutic interventions.
Supplementary Material
Acknowledgements
We thank Dr Colin Sieff for cDNA from mouse fetal liver with downregulated RPS19 and Xi Ren for runx1 and efnb2a probes. We thank Drs Hanna Gazda and Colin Sieff for the critical reading of the manuscript. The work was supported by Diamond–Blackfan anemia Foundation, St. Baldrick’s Foundation and NIH (NHLBI) research grant R01HL97561.
Footnotes
Authors’ contributions
N.D. designed and performed research, analysed data and wrote the paper; S.L. designed research, analysed data and wrote the paper; K.M.S. wrote the paper.
Conflict of interest
The authors have no conflict of interest to declare.
Supporting information
Additional Supporting information may be found in the online version of this article:
Fig S1. gata1 is 2·57-fold upregulated in the rpl11 mutant in comparison to siblings at 48 hpf. qPCR RNA was pooled from 30 mutant or sibling embryos.
Fig S2. rpl11 mutant had changes in expression of hundreds of genes.
Fig S3. RPS 19-deficient mouse fetal lever cells downregulated expression of a structural protein ank1 and glycolytic enzymes Tri and Pfk.
Fig S4. Inhibition of p53 resulted in normalization of expression of many dysregulated genes.
Table SI. Changes in expression of genes involved in regulation of cell cycle, transcription, proliferation and apoptosis.
Table SII. Changes in expression of genes functioning in metabolism, transport, chaperones, organelles.
Table SIII. Expression of genes functioning in cytoskeleton and extracellular matrix.
Table SIV. Changes in expression of genes involved in synthesis and processing of ribosomal RNA, other non-coding RNAs, mRNA processing and translation.
Table SV. Expression of genes affecting various aspects of development and functions of tissues and organs.
Table SVI. Expression of genes affecting function of blood and immune system.
Table SVII. Primers used in qPCR. Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Akala OO, Park IK, Qian D, Pihalja M, Becker MW, Clarke MF. Long-term haematopoietic reconstitution by Trp532/2p16Ink4a2/2p19Arf2/2multipotent progenitors. Nature. 2008;453:228–232. doi: 10.1038/nature06869. [DOI] [PubMed] [Google Scholar]
- Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proceedings of National Academy Sci of Sciences of the United States of America. 2004a;101:12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsterdam A, Sadler KC, Lai K, Farrington S, Bronson RT, Lees JA, Hopkins N. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biology. 2004b;2:E139. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avondo F, Roncaglia P, Crescenzio N, Krmac H, Garelli E, Armiraglio M, Castagnoli C, Campagnoli MF, Ramenghi U, Gustincich S, Santoro C, Dianzani I. Fibroblasts from patients with Diamond-Blackfan anaemia show abnormal expression of genes involved in protein synthesis, amino acid metabolism and cancer. BMC Genomics. 2009;10:442. doi: 10.1186/1471-2164-10-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow JL, Drynan LF, Hewett DR, Holmes LR, Lorenzo-Abalde S, Lane AL, Jolin HE, Pannell R, Middleton AJ, Wong SH, Warren AJ, Wainscoat JS, Boultwood J, McKenzie AN. A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q- syndrome. Nature Medicine. 2010;16:59–66. doi: 10.1038/nm.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand JY, Kim AD, Violette EP, Stachura DL, Cisson JL, Traver D. Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development. 2007;134:4147–4156. doi: 10.1242/dev.012385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CE, Galloway JL, Smith AC, Keefe MD, Cashman TJ, Paik EJ, Mayhall EA, Amsterdam AH, Zon LI. A genetic screen in zebrafish defines a hierarchical network of pathways required for hematopoietic stem cell emergence. Blood. 2009;113:5776–5782. doi: 10.1182/blood-2008-12-193607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty A, Uechi T, Higa S, Torihara H, Kenmochi N. Loss of ribosomal protein L11 affects zebrafish embryonic development through a p53-dependent apoptotic response. PLoS One. 2009;4:e4152. doi: 10.1371/journal.pone.0004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cmejlova J, Dolezalova L, Pospisilova D, Petrtylova K, Petrak J, Cmejla R. Translational efficiency in patients with Diamond-Blackfan anemia. Haematologica. 2006;91:1456–1464. [PubMed] [Google Scholar]
- Danilova N, Sakamoto KM, Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008;112:5228–5237. doi: 10.1182/blood-2008-01-132290. [DOI] [PubMed] [Google Scholar]
- Davidson AJ, Zon L. The ‘definitive’ (and ‘primitive’) guide to zebrafish hematopoiesis. Oncogene. 2004;23:7233–7246. doi: 10.1038/sj.onc.1207943. [DOI] [PubMed] [Google Scholar]
- Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, Tentler D, Mohandas N, Carlsson B, Dahl N. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nature Genetics. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- Ebert BL, Lee MM, Pretz JL, Subramanian A, Mak R, Golub TR, Sieff CA. An RNA interference model of RPS19 deficiency in Diamond-Blackfan anemia recapitulates defective hematopoiesis and rescue by dexamethasone: identification of dexamethasone-responsive genes by microarray. Blood. 2005;105:4620–4626. doi: 10.1182/blood-2004-08-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, Raza A, Root DE, Attar E, Ellis SR, Golub TR. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flygare J, Aspesi A, Bailey JC, Miyake K, Caffrey JM, Karlsson S, Ellis S. Human RPS19, the gene mutated in Diamond Blackfan anemia, encodes a ribosomal protein required for the maturation of 40S ribosomal subunits. Blood. 2007;109:980–986. doi: 10.1182/blood-2006-07-038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathi KA, Shimamura A. Ribosomal dysfunction and inherited marrow failure. British Journal of Haematology. 2008;141:376–387. doi: 10.1111/j.1365-2141.2008.07095.x. [DOI] [PubMed] [Google Scholar]
- Gazda HT, Kho AT, Sanoudou D, Zaucha JM, Kohane IS, Sieff CA, Beggs AH. Defective Ribosomal Protein Gene Expression Alters Transcription, Translation, Apoptosis And Oncogenic Pathways In Diamond-Blackfan Anemia. Stem Cells. 2006;24:2034–2044. doi: 10.1634/stemcells.2005-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri N, Kang E, Tisdale JF, Follman D, Rivera M, Schwartz GN, Kim S, Young NS, Rick ME, Dunbar CE. Clinical and laboratory evidence for a trilineage haematopoietic defect in patients with refractory Diamond-Blackfan anaemia. British Journal of Haematology. 2000;108:167–175. doi: 10.1046/j.1365-2141.2000.01796.x. [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Halperin DS, Freedman MH. Diamond-Blackfan anemia: etiology, pathophysiology, and treatment. American Journal of Pediatric Hematology/Oncology. 1989;11:380–394. [PubMed] [Google Scholar]
- Harigae H, Nakajima O, Suwabe N, Yokoyama H, Furuyama K, Sasaki T, Kaku M, Yamamoto M, Sassa S. Aberrant iron accumulation and oxidized status of erythroid-specific delta-aminolevulinate synthase (ALAS2)-deficient definitive erythroblasts. Blood. 2003;101:1188–1193. doi: 10.1182/blood-2002-01-0309. [DOI] [PubMed] [Google Scholar]
- Hu X, Ivashkiv LB. Cross-regulation of signaling pathways by interferon-gamma: implications for immune responses and autoimmune diseases. Immunity. 2009;31:539–550. doi: 10.1016/j.immuni.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin A, Itahana K, O’Keefe K, Zhang Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Molecular and Cellular Biology. 2004;24:7669–7680. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalev-Zylinska ML, Horsfield JA, Flores MV, Postlethwait JH, Vitas MR, Baas AM, Crosier PS, Crosier KE. Runx1 is required for zebrafish blood and vessel development and expression of a human RUNX1-CBF2T1 transgene advances a model for studies of leukemogenesis. Development. 2002;129:2015–2030. doi: 10.1242/dev.129.8.2015. [DOI] [PubMed] [Google Scholar]
- Kirch HC, Flaswinkel S, Rumpf H, Brockmann D, Esche H. Expression of human p53 requires synergistic activation of transcription from the p53 promoter by AP-1, NF-kappaB and Myc/Max. Oncogene. 1999;18:2728–2738. doi: 10.1038/sj.onc.1202626. [DOI] [PubMed] [Google Scholar]
- Kuramitsu M, Hamaguchi I, Takuo M, Masumi A, Momose H, Takizawa K, Mochizuki M, Naito S, Yamaguchi K. Deficient RPS19 protein production induces cell cycle arrest in erythroid progenitor cells. British Journal of Haematology. 2008;140:348–359. doi: 10.1111/j.1365-2141.2007.06930.x. [DOI] [PubMed] [Google Scholar]
- Langheinrich U, Hennen E, Stott G, Vacun G. Zebrafish as a model organism for the identification and characterization of drugs and genes affecting p53 signaling. Current Biology. 2002;12:2023–2028. doi: 10.1016/s0960-9822(02)01319-2. [DOI] [PubMed] [Google Scholar]
- Lipton JM, Ellis SR. Diamond Blackfan anemia 2008–2009: broadening the scope of ribosome biogenesis disorders. Current Opinion in Pediatrics. 2010;22:12–19. doi: 10.1097/MOP.0b013e328334573b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacInnes AW, Amsterdam A, Whittaker CA, Hopkins N, Lees JA. Loss of p53 synthesis in zebrafish tumors with ribosomal protein gene mutations. Proceedings of National Academy Sci of Sciences of the United States of America. 2008;105:10408–10413. doi: 10.1073/pnas.0805036105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- McGowan KA, Li JZ, Park CY, Beaudry V, Tabor HK, Sabnis AJ, Zhang W, Fuchs H, de Angelis MH, Myers RM, Attardi LD, Barsh GS. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nature Genetics. 2008;40:963–970. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake K, Utsugisawa T, Flygare J, Kiefer T, Hamaguchi I, Richter J, Karlsson S. RPS19 Deficiency Leads to Reduced Proliferation and Increased Apoptosis but Does Not Affect Terminal Erythroid Differentiation in a Cell Line Model of Diamond-Blackfan Anemia. Stem Cells. 2008;26:323–329. doi: 10.1634/stemcells.2007-0569. [DOI] [PubMed] [Google Scholar]
- Moyes CD, Sharma ML, Lyons C, Leary SC, Leon M, Petrie A, Lund SG, Tufts BL. Origins and consequences of mitochondrial decline in nucleated erythrocytes. Biochimica et Biophysica Acta. 2002;1591:11–20. doi: 10.1016/s0167-4889(02)00224-0. [DOI] [PubMed] [Google Scholar]
- Murayama E, Kissa K, Zapata A, Mordelet E, Briolat V, Lin HF, Handin RI, Herbomel P. Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity. 2006;25:963–975. doi: 10.1016/j.immuni.2006.10.015. [DOI] [PubMed] [Google Scholar]
- Murray-Zmijewski F, Lane DP, Bourdon JC. p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death and Differentiation. 2006;13:962–972. doi: 10.1038/sj.cdd.4401914. [DOI] [PubMed] [Google Scholar]
- Panic L, Tamarut S, Sticker-Jantscheff M, Barkic M, Solter D, Uzelac M, Grabusic K, Volarevic S. Ribosomal protein S6 gene haploinsufficiency is associated with activation of a p53-dependent checkpoint during gastrulation. Molecular and Cellular Biology. 2006;26:8880–8891. doi: 10.1128/MCB.00751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdahl EB, Naprstek BL, Wallace WC, Lipton JM. Erythroid failure in Diamond-Blackfan anemia is characterized by apoptosis. Blood. 1994;83:645–650. [PubMed] [Google Scholar]
- Pestov DG, Strezoska Z, Lau LF. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Molecular and Cellular Biology. 2001;21:4246–4255. doi: 10.1128/MCB.21.13.4246-4255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nature Reviews Molecular Cell Biology. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Sandberg ML, Sutton SE, Pletcher MT, Wiltshire T, Tarantino LM, Hogenesch JB, Cooke MP. c-Myb and p300 regulate hematopoietic stem cell proliferation and differentiation. Developmental Cell. 2005;8:153–166. doi: 10.1016/j.devcel.2004.12.015. [DOI] [PubMed] [Google Scholar]
- Sieff CA, Yang J, Merida-Long LB, Lodish HF. Pathogenesis of the erythroid failure in Diamond Blackfan anaemia. British Journal of Haematology. 2010;148:611–622. doi: 10.1111/j.1365-2141.2009.07993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner LA, Gallagher PG. Erythrocyte disorders in the perinatal period. Seminars in Perinatology. 2007;31:254–261. doi: 10.1053/j.semperi.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TeKippe M, Harrison DE, Chen J. Expansion of hematopoietic stem cell phenotype and activity in Trp53-null mice. Experimental Hematology. 2003;31:521–527. doi: 10.1016/s0301-472x(03)00072-9. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B, Schilling TF, Postlethwait JH. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Ryan KM. p53 and metabolism. Nature Reviews Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- Young NS, Scheinberg P, Calado RT. Aplastic anemia. Current Opinion in Hematology. 2008;15:162–168. doi: 10.1097/MOH.0b013e3282fa7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.