Abstract
Rationale
Endothelial cells are developmentally derived from angioblasts specified in the mesodermal germ cell layer. The transcription factor etsrp/etv2 is at the top of the known genetic hierarchy for angioblast development. The transcriptional events that induce etsrp expression and angioblast specification are not well understood.
Objective
We generated etsrp:gfp transgenic zebrafish and used them to identify regulatory regions and transcription factors critical for etsrp expression and angioblast specification from mesoderm.
Methods and Results
To investigate the mechanisms that initiate angioblast cell transcription during embryogenesis, we have performed promoter analysis of the etsrp locus in zebrafish. We describe three enhancer elements sufficient for endothelial gene expression when place in front of a heterologous promoter. The deletion of all three regulatory regions led to a near complete loss of endothelial expression from the etsrp promoter. One of the enhancers, located 2.3 kb upstream of etsrp contains a consensus FOX binding site that binds Foxc1a and Foxc1b in vitro by EMSA and in vivo using ChIP. Combined knockdown of foxc1a/b, using morpholinos, led to a significant decrease in etsrp expression at early developmental stages as measured by quantitative RT-PCR and in situ hybridization. Decreased expression of primitive erythrocyte genes scl and gata1 was also observed while pronephric gene pax2a was relatively normal in expression level and pattern.
Conclusions
These findings identify mesodermal foxc1a/b as a direct upstream regulator of etsrp in angioblasts. This establishes a new molecular link in the process of mesoderm specification into angioblast.
Keywords: angioblast, etsrp, foxc1a, scl, zebrafish
Introduction
Endothelial cells are developmentally derived from precursor cells termed angioblasts. These cells initially appear in the mesoderm and coalesce to form the primary vessels through a process known as vasculogenesis. From these primary vessels the rest of the vasculature spreads throughout the embryo through the process of angiogenesis. The morphological events that occur during these processes are well defined; however, the molecular mechanisms driving these processes are still unclear.
The zebrafish embryo has been a valuable tool for studying the molecular and genetic events occurring during vascular development. For example, the transcription factor Etsrp was first identified in a microarray screen for gene expression changes in the cloche mutant embryo. 1 Cloche embryos lack blood and vascular cells but have normal development of other organ systems. 2 Etsrp overexpression is sufficient to rescue expression of vascular and primitive myeloid genes in cloche embryos. 3 Additionally, overexpression of Etsrp in wild-type embryos ectopically induces the expression of hundreds of vascular and myeloid genes, whereas morpholino knockdown or mutation of Etsrp disrupts vasculogenesis as well as angiogenesis 4-6 Epistasis experiments in zebrafish embryos have demonstrated that etsrp is at the top of the angioblast transcriptional hierarchy, placing it above scl, fli1a, and kdrl. 6-8
The mammalian homolog of etsrp, Etv2 (formerly ER71 or Etsrp71), is expressed in mesodermal tissues of the early mouse embryo, including vascular and hematopoietic lineages. 9-11 Etv2 knockout mice are embryonic lethal by E11.0 with severe defects in hematopoietic and vascular development. 10, 11 In embryonic stem cells, Etv2 directly regulates Kdr (Flk1) expression and can increase the derivation of blood and endothelial cells when overexpressed. 10 Interestingly, Scl and Kdr were shown to function downstream of Etv2 in mice as was found in zebrafish. 10, 12 In fact, human or mouse Etv2 protein overexpression in zebrafish embryos was sufficient to induce the ectopic expression of scl and kdrl, 8 suggesting that etsrp and Etv2 are homologous genes that have conserved functions in vertebrate vascular development and hematopoiesis.
Although much effort has been made to study the genes downstream of etsrp/Etv2, little is known about its upstream regulators. In mouse, the transcription factor Nkx2-5 has been suggested to regulate Etv2 expression in the endocardium. 11 However, Nkx2-5 expression is limited to cardiac and endocardial lineages implying that this regulatory interaction is limited to the developing heart. 13, 14 Additionally, the zebrafish Nkx2-5 homolog nkx2.5 is expressed in the cardiac mesoderm where it is discretely segregated from the etsrp expression domain in the anterior and posterior lateral plate mesoderm, suggesting that a direct positive interaction does not occur in zebrafish. 15, 16 Combined morpholino knockdown of gata4, gata5, and gata6 can delay the expression of etsrp and other vascular and cardiac genes in the anterior lateral plate. 17 However, angioblasts in the posterior lateral plate are unaffected and a direct interaction between these factors and etsrp has not been established. The cloche mutant locus is upstream of etsrp, but the specific genetic lesion in this mutant has not been conclusively identified. Xiong et. al. suggested that the lycat gene, a predicted lipid acetyltransferase, is responsible for the cloche phenotype. 18 Although knockdown of lycat blocks the expression of etsrp18, it is unlikely that lycat directly regulates the transcription of etsrp. Therefore a significant gap exists in our knowledge of angioblast specification from mesodermal tissue at the level of the etsrp transcription factor.
To identify upstream regulators of etsrp gene expression we have studied the regulatory regions of the etsrp locus. Using transgenic zebrafish, we identify three enhancer regions that are sufficient to drive GFP expression similar to the endogenous pattern. We identify Foxc1a/b as a direct upstream regulator of etsrp and demonstrate its involvement in angioblast specification.
Methods
An expanded Methods section is available in the Online Data Supplement at http://circres.ahajournals.org.
Zebrafish embryos were maintained and staged as described. 19 The University of California, Los Angeles Animal Care and Use Committee approved all protocols used in this study. Transgene plasmids were generated using Tol2Kit plasmids20 and the Multisite Gateway System (Invitrogen). Zebrafish embryos were microinjected at the one cell stage with DNA, mRNA, or morpholinos as previously described. 21 Electrophoretic Mobility Shift Assay (EMSA) was performed using the LightShift Chemiluminescent EMSA Kit (Pierce) according to the manufacturer’s suggested protocol. Nuclear protein extracts from Porcine Aortic Endothelial (PAE) and Human Umbilical Vein Endothelial Cells (HUVEC) were isolated using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce) as recommended. In vitro synthesized Foxc1a, Foxc1b, and mCherry protein was created using the TnT in vitro transcription/translation kit (Promega). Chromatin Immunoprecipitation (ChIP) and quantitative RT-PCR methods are available in the online supplement. Whole mount in situ hybridization was performed as described22 using DIG labeled riboprobes (Roche). Images were captured on an Axioskop 2 plus microscope (Zeiss) or a Stemi2000-C (Zeiss) using 5x or 10x objectives with an AxioCam camera and Openlab 4.0 software (Improvision). Adobe Photoshop was used to adjust brightness and contrast and assemble composite images. Students t-test was used to determine significance with p<0.05 for qRT-PCR experiments.
Results
Tg(−2.3etsrp:gfp) transgene recapitulates the endogenous expression pattern of etsrp
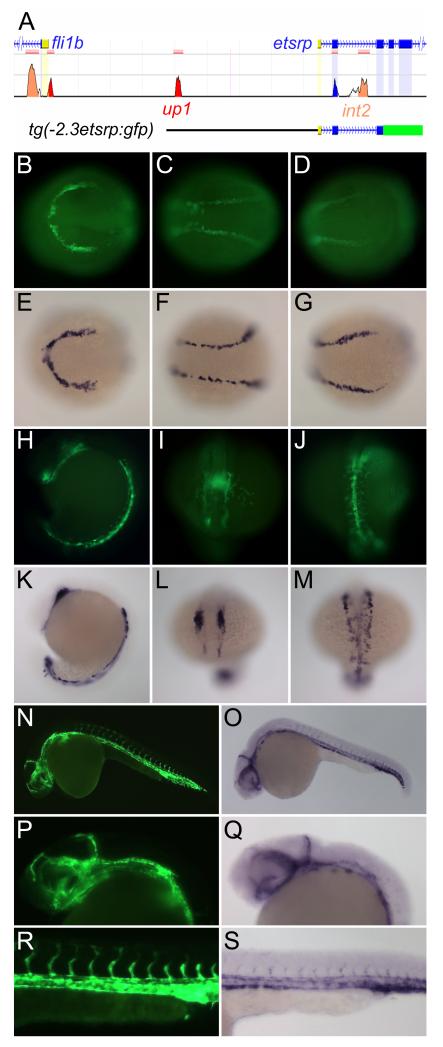
To begin dissecting the regulatory mechanisms of etsrp expression, we undertook a bioinformatic analysis of the etsrp/Etv2 locus in multiple species using Multi-Pipmaker analysis (http://pipmaker.bx.psu.edu/pipmaker) 23 and the web-based ECR Browser (http://ecrbrowser.dcode.org/).24 Comparison of approximately 200 kilobases of sequence between human, mouse, Xenopus, pufferfish, and zebrafish found very little homology outside of the exonic sequences (unpublished data). However, two peaks of conserved sequence were identified near the transcription start site of etsrp between zebrafish and pufferfish (Figure 1A). One of the conserved peaks, located 2.3 kb upstream of the etsrp transcription start site, was called up1. The second region of conservation, located in etsrp intron two, was called int2. We generated a transgene, tg(−2.3etsrp:gfp), that encompassed these two conserved sequences (Figure 1A). Transgenic embryos exhibited strong vascular specific expression at 24 hours post fertilization (hpf) suggesting that the conserved regions may be relevant to the endogenous gene’s expression.
Figure 1. Tg(−2.3etsrp:gfp) contains two evolutionarily conserved regions and faithfully recapitulates the endogenous expression pattern of etsrp.
(A) The etsrp gene locus with conserved regions up1 and int2 along with the region corresponding to tg(−2.3etsrp:gfp) highlighted. The up1 region is approximately equidistant from the transcription start sites of etsrp and the adjacent fli1b gene and int2 is located within intron 2 of etsrp. Conserved regions between zebrafish and pufferfish were identified using the ECR Browser (http://ecrbrowser.dcode.org) and the locus image is adapted from this website. (B-S) Fluorescent images of tg(−2.3etsrp:gfp) panels B-D, H-J, N, P, and R and in situ hybridization for etsrp panels E-G, K-M, O, Q, and S demonstrating near perfect correlation of expression between the transgene and endogenous gene at different developmental stages. (B-G) 10-somite stage; (H-M) 18-somite stage; (N-S) 24 hours post fertilization (hpf).
Tg(−2.3etsrp:gfp) fish exhibited GFP expression initially in the anterior (ALPM) and posterior lateral plate mesoderm (PLPM) at ~4 somite stage. This is identical in pattern to endogenous etsrp, but slightly delayed, most likely due to the time necessary for GFP to mature. By the 10 somite stage, strong GFP expression is present in the ALPM and PLPM in a pattern identical to the endogenous gene (Figure 1B-G). Similarly, at the 18 somite stage, the expression of GFP and etsrp correlate almost identically (Figure 1H-M). At 24 hpf, GFP is highly expressed in both the cranial vasculature (Figure 1N-Q) and the axial and intersomitic vessels of the trunk (Figure 1R-S). By 36 hpf, endogenous etsrp is significantly reduced in the vasculature with the exception of the aortic arches.3 Tg(−2.3etsrp:gfp) follows this pattern of expression (Online Figure I). In comparison to the well characterized tg(kdrl:gfp) 25 zebrafish, tg(−2.3etsrp:gfp) expression appears earlier in angioblasts but disappears as the vasculature matures while tg(kdrl:gfp) maintains expression throughout development and into adulthood (Online Figure I). Additionally, tg(−2.3etsrp:mcherry) co-localizes with tg(fli1a:gfp)26 in the ALPM and PLPM at 10 somite stage demonstrating the promoter drives expression in angioblasts at this early stage (Online Figure II). Overall, tg(−2.3etsrp:gfp) faithfully recapitulates the endogenous expression of etsrp both temporally and spatially.
Three enhancers drive angioblast expression in etsrp transgenic fish
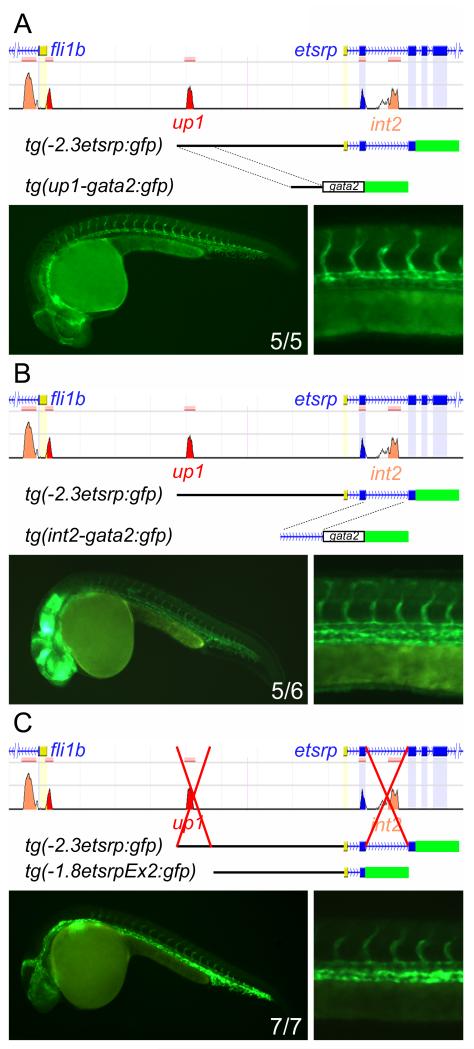
To test whether the evolutionarily conserved regions have enhancer activity, we placed them in front of the minimal gata2 promoter driving GFP. Cloning of either up1 or int2 into the reporter was sufficient to drive GFP expression in the developing vasculature both transiently and in germline transgenics (Figure 2A-B). Additionally, these regions functioned when placed in the reverse orientation, demonstrating that they are true enhancer regions and not cryptic promoters (Online Figure III). Given that we could not identify any other conserved regions within the promoter, we hypothesized that deletion of up1 and int2 would abolish expression of the etsrp transgene. However, deletion of each region separately or simultaneously did not significantly disrupt transgene expression (Figure 2C). This suggests that non-conserved regulatory sequences are present in the −1.8 kb region of the promoter.
Figure 2. Evolutionarily conserved regions up1 and int2 are sufficient but not necessary for tg(−2.3etsrp:gfp) expression in endothelial cells.
(A and B) The conserved region up1 (A) or int2 (B) was placed in front of the minimal gata2 promoter driving GFP expression and germline transgenics generated. Both regions are sufficient to drive GFP expression in the developing vasculature as demonstrated by fluorescent images of whole embryo and trunk vasculature at 24hpf. (C) Up1 and int2 are not necessary for strong vascular expression from the etsrp promoter transgene. Deletion of both up1 and int2 from the GFP reporter lines does not eliminate vascular expression as demonstrated by tg(−1.8etsrpEx2:gfp), suggesting that more regulatory elements are present. The numbers in the whole embryo image represents the number of germline transgenic fish lines expressing vascular GFP versus the total number of lines examined. Note that up1 drives stronger expression in the dorsal aorta and its deletion results in decreased relative expression in the dorsal aorta (A and C).
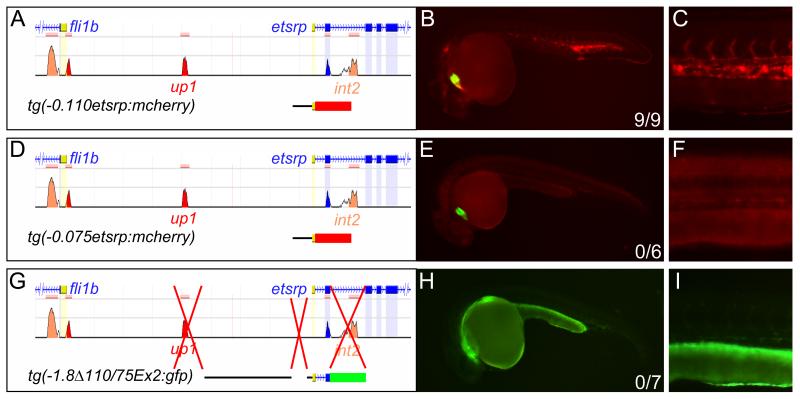
In an attempt to map the remaining regulatory elements we did a linear deletion analysis of the −1.8 kb promoter sequence (Online Figure IV). 110 base-pairs of etsrp promoter sequence were found to be sufficient for vascular expression at 24 hpf (Figure 3A-C). Deletion of 35 base-pairs from the 5′-end completely abolished vascular specific expression (Figure 3D-F). However, the remaining 75 base-pair promoter was still capable of driving non-vascular expression in several lines; presumably due to enhancer trapping effects (unpublished data). This suggests that basal promoter function had been preserved and the −0.110 kb to −0.075 kb region of the promoter was acting as an enhancer. To determine the importance of the enhancer, we deleted it along with up1 and int2 from tg(−2.3etsrp:gfp). These fish demonstrated minimal expression in the developing vasculature (Figure 3G-I) suggesting that the combination of these three enhancers drives strong etsrp expression during development.
Figure 3. An unconserved region at −110 to −75 accounts for the majority of the remaining promoter activity in endothelial cells.
(A-C) Tg(−0.110etsrp:mcherry) containing only 110 base pairs of the etsrp promoter is sufficient for vascular expression at 24hpf. (D-F) Deletion of 35 base pairs from this site tg(−0.075etsrp:mcherry) abolishes expression in the vasculature. (A-F) mCherry was used as a reporter for the element being tested and GFP was driven by a constitutive cardiac promoter in the same transgene to identify transgenic germlines independently of the element being tested. (G-I) Deletion of up1, int2, and −0.110/−0.075, tg(−1.8Δ110/75Ex2:gfp), almost completely abolishes expression from the etsrp promoter, compare expression in H and I to Figure 2C, suggesting these three regions are critical for high levels of etsrp expression in the developing vasculature.
We also noticed that some of the transgenes had stronger or weaker expression in the dorsal aorta (DA) versus the posterior cardinal vein (PCV). Specifically, the up1 and int2 enhancers exhibited stronger or equal expression in the DA versus the PCV while deletion of these enhancers resulted in PCV expression greater than DA (Figure 2A-C). A summary of the relative transgene expression in the axial vasculature for each transgene line is provided in Online Table I.
The upstream enhancer, up1, contains multiple evolutionarily conserved protein binding sites
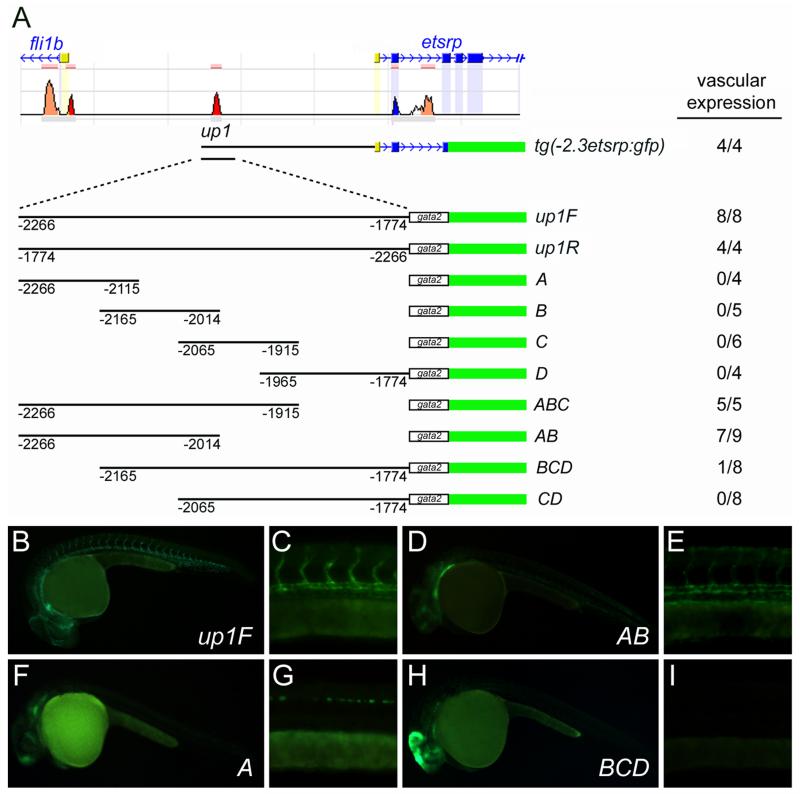
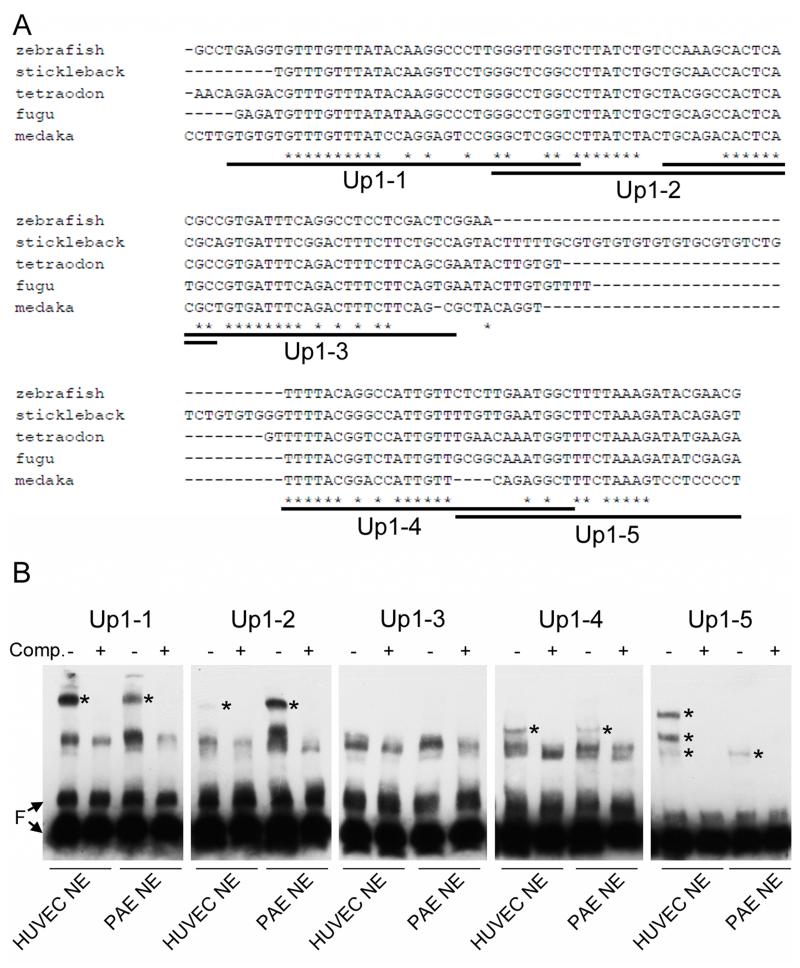
To better define the sequences necessary for enhancer activity, we studied up1 in more detail. The ~500 base-pair element was broken into four overlapping fragments of ~150 base-pairs, termed A, B, C, and D, and cloned in front of the minimal gata2 promoter transgene. Unfortunately, none of these sequences was sufficient for expression in the developing vasculature (Figure 4A, B, F, and G). Therefore, different combinations of A, B, C, and D were tested for enhancer activity. We found that the combination of A and B was sufficient for vascular expression of the reporter, while fragments C and D were dispensable (Figure 4A, and D-I). This narrowed the enhancer region to 252 base-pairs. Comparison of this sequence between fish species including zebrafish, stickleback, tetraodon, fugu, and medaka identified several regions of evolutionary conservation (Figure 5A).
Figure 4. Up1 enhancer activity is present at −2266 to −2014.
(A) Schematic of the up1 fragments tested for enhancer activity using the gata2 minimal promoter and GFP reporter. Number of lines demonstrating vascular expression out of total lines examined is noted. (B-I) Fluorescent images demonstrating that fragment AB (−2266 to −2014) is the minimal region necessary for vascular expression from the up1 enhancer. Fragments A (F and G) and BCD (H and I) exhibit some nonvascular expression presumably due to insertional enhancer trapping effects.
Figure 5. Multiple protein binding sites are present in the AB region of the up1 enhancer.
(A) Evolutionarily conserved sequence between different fish species identified using clustalW analysis at the AB region. Overlapping EMSA probe sequences are underlined and labeled Up1-(1-5). (B) EMSA using the oligonucleotide probes defined in (A) and nuclear protein extracts, NE, from HUVEC or PAE cells. Unlabeled probe competition, Comp. +/-, was used to define specific binding protein complexes denoted with an asterisk (*). All oligos bound specific, well-defined proteins except for Up1-3. F, free probe.
The AB sequence was divided into five overlapping fragments termed Up1-(1-5) that were used as probes in EMSA (Figure 5). Because no zebrafish endothelial cell lines exist and zebrafish whole embryo extract proved too complex to resolve individual binding complexes, nuclear extracts from HUVEC and PAE cells were used for this analysis. We found that specific protein binding to probes Up1-1, -2, -4, and -5 could be detected (Figure 5B). Most binding activity was present in both arterial and venous cell types. However, Up1-5 bound to three distinct protein bands in HUVEC extracts, while only the lowest band was present in PAE cells (Figure 5B). In fact, up1 drives expression more strongly in arterial cells (Figure 2A) and the difference in protein binding may be relevant to A/V specific expression levels.
Foxc1a and Foxc1b bind to up1 in vitro and in vivo
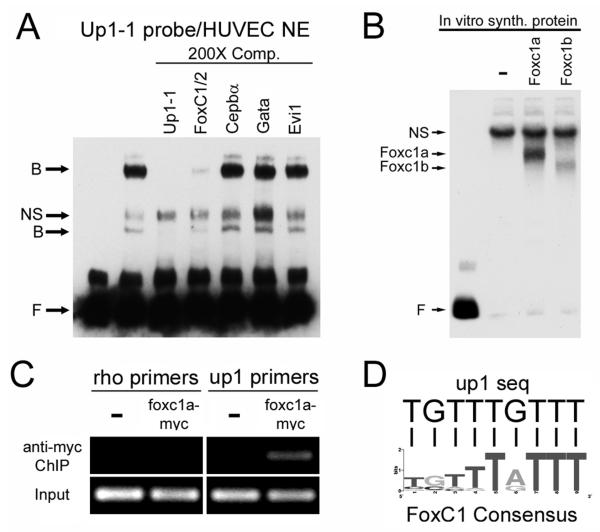
Using the TRANSFAC database27 we screened the sequence of up1 for consensus transcription factor binding sites. Four candidates were identified, FoxC1/2, Cebpα, Gata, and Evi1. To test if the binding activity identified by EMSA corresponded to any of these consensus sites, oligos for each were prepared and used as unlabeled competitors. For the Up1-1 probe, we found that the FoxC1/2 consensus site could compete for binding with HUVEC protein extracts while the others could not (Figure 6A). To determine if the zebrafish homologs of FoxC1/2, Foxc1a and Foxc1b, could bind to Up1-1, we in vitro synthesized these proteins and performed EMSA with the Up1-1 probe. Both Foxc1a and Foxc1b were able to bind to Up1-1 while a negative control protein, in vitro synthesized mCherry, could not (Figure 6B).
Figure 6. Foxc1a/b binds to up1 in vitro and in vivo.
(A) EMSA using Up1-1 probe and HUVEC nuclear protein extracts, NE, demonstrates that the FoxC1/2 consensus binding site oligo can compete for binding with the Up1-1 probe while Cepbα, Gata, and Evi1 consensus binding site oligos cannot. B, bound; NS, nonspecific; F, free probe. (B) EMSA demonstrating Up1-1 probe bound to in vitro synthesized Foxc1a and Foxc1b protein. (C) Chromatin immunoprecipitation, ChIP, from wild-type zebrafish embryos (−) or embryos expressing a foxc1a-myc. Up1 primers detect enrichment in embryos expressing foxc1a-myc while negative control rhodopsin, rho, primers do not. (D) The Up1-1 sequence, 5′-TGTTTGTTT-3′, contains a FoxC1 consensus binding site, 5′-(T/G)(G/C)(T/R)(T/Y)T(A/G)TTT-3′.
We next wanted to test if FoxC proteins were able to interact with the etsrp promoter in vivo using ChIP. Since no antibody to zebrafish FoxC proteins is available, we generated a C-terminal myc-tagged version of Foxc1a to use for immunoprecipitation. mRNA for foxc1a-myc was injected into single cell embryos resulting in ubiquitous expression of the tagged protein. Embryos were processed for ChIP at 50% epiboly due to gastulation defects and death that occurred at later stages. We could detect significant enrichment of the up1 genomic locus in foxc1a-myc mRNA injected embryos but not in uninjected control embryos (Figure 6C). The rhodopsin promoter region used as a negative control was not enriched in either uninjected or foxc1a-myc injected samples demonstrating that Foxc1a-myc binding to the up1 site is specific (Figure 6C). Comparison of the FoxC1 consensus binding site to the sequence found in Up1-1 showed perfect alignment (Figure 6D). These results suggest that forkhead transcription factors can bind to the etsrp promoter at a conserved site within a functional enhancer.
Foxc1a and foxc1b act upstream of etsrp in angioblast development
It was previously reported that foxc1a and foxc1b knockdown affects artery-vein specification and vascular morphology and integrity in developing zebrafish embryos.12, 28 These defects are reminiscent of etsrp mutants and morphants. 3, 6, 7 De Val et.al. demonstrated an epistatic relationship between foxc1a and etsrp and suggested that Etsrp and Foxc1a directly interact to activate downstream vascular genes. 12 However, an alternate explanation is that foxc1a and etsrp have a linear relationship with one factor directly regulating the other. Foxc1a and foxc1b expression initially appear in the involuting mesendoderm at the shield stage and is then maintained in paraxial mesoderm and other mesodermally derived tissues including the vasculature.12, 28, 29 This expression precedes etsrp which is first induced in the lateral plate mesoderm at the 1-2 somite stage3. We performed double fluorescent in situ hybridization to examine co-localization of foxc1a and etsrp. At 8-10 somites stage we noted co-labeling in a subset of cells in the ALPM and anterior PLPM (Online Figure V). This timing of expression suggests that foxc1a and foxc1b may act upstream to etsrp.
To determine the epistatic relationship of etsrp and foxc1a/b in vivo we performed morpholino gene knockdown studies using previously published morpholinos that had been shown to be specific and free of off-target effects.28, 30 It has been suggested that etsrp and foxc1a/b function in a complex to induce the expression of Kdr (Flk1).12 In zebrafish embryos, overexpression of etsrp is sufficient to induce robust ectopic expression of tg(kdrl:gfp).3 To determine if foxc1a/b are necessary co-factors for this induction, we blocked their expression in tg(kdrl:gfp) embryos while simultaneously overexpressing etsrp. Under these conditions, foxc1a/b are not necessary for etsrp to induce ectopic expression of tg(kdrl:gfp) (Online Figure VI). This suggests that etsrp function is independent of, or downstream to, foxc1a/b at this early developmental stage.
To determine if the etsrp promoter is regulated by foxc1a and foxc1b at the up1 enhancer, morpholinos were injected into tg(up1-gata2:gfp) fish to see if the loss of these factors affects transgene expression. By the 16 somite stage, transgene expression is visible in the axial vessels of control embryos but not foxc1a/b morphant embryos (Online Figure VII), demonstrating that the loss of foxc1a/b decreases the activity of the up1 enhancer in vivo.
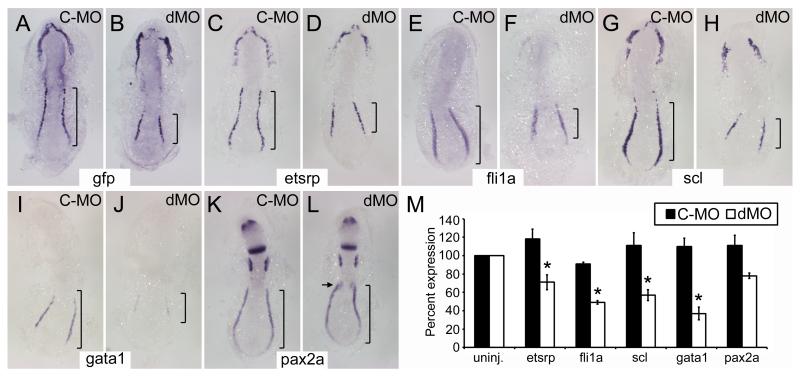
To see if foxc1a/b played a larger role in regulating etsrp, we tested the affect of their knockdown on tg(−2.3etsrp:gfp) expression and endogenous etsrp expression by in situ hybridization at the 6-8 somite stage. Morphlino knockdown of foxc1a/b significantly reduced the expression of both tg(−2.3etsrp:gfp) and etsrp, most notably in the PLPM (bracketed in Figure 7A-D). Three other PLPM markers, fli1a, scl, and gata1, were also examined. Fli1a and scl are important transcription factors that functions downstream of etsrp in vascular lineages 7; in primitive erythroid cells, fli1a, scl, and gata1 function independent of etsrp. 7 Fli1a, scl, and gata1 expression are all reduced in foxc1a/b morphants (Figure 7E-J). To test whether other mesoderm or non-mesodermal tissues were disrupted, expression of pax2a, a gene expressed in the intermediate mesoderm and central nervous system, was examined. As previously reported, pax2a expression is largely normal in the nervous system and intermediate mesoderm in foxc1a/b morphants, with the exception of the pronephric primordium (Figure 7K-L).30 Additional paraxial mesoderm genes mespa, mespb, deltaC, and par1 were examined in foxc1a/b morphants and behaved as previously reported (Online Figure VIII).30 Quantitative RT-PCR results support the in situ hybridization data, demonstrating significant decreases in etsrp, fli1a, scl, and gata1 expression in foxc1a/b morphants (Figure 7M). These results demonstrate a significant decrease in the early expression of primitive erythroid and angioblast genes when foxc1a/b are knocked down.
Figure 7. Morpholino knockdown of foxc1a/b results in decreased angioblasts and primitive erythrocytes.
(A-L) Dorsal view flatmounts, anterior up, of 6-8 somite stage embryos injected with control morpholino, C-MO, or morpholinos to knockdown expression of both foxc1a and foxc1b, dMO. In situ hybridization for tg(−2.3etsrp:gfp) or the genes noted at the center of each pair of panels was performed to assay the affects of foxc1a/b knockdown. Representative images of more than 10 embryos per group. Note the decreased expression of tg(−2.3etsrp:gfp), etsrp, fli1a, scl, and gata1 in dMO treated embryos especially at the posterior lateral plate mesoderm (bracketed). Expression of pax2a in the intermediate mesoderm is not affected by foxc1a/b knockdown except at the pronephric primordia, arrow in L, and nervous system expression is unaffected. (M) Quantitative RT-PCR demonstrates reduced expression of etsrp, fli1a, scl, and gata1 in dMO treated embryos as compared to C-MO treated embryos. All groups were normalized to β-actin and expression in uninjected wildtype embryos was set at 100%. Asterisks (*) denote statistically significant changes as determines with Students t-test (p<0.05).
Discussion
Etsrp is the most upstream transcription factor in the regulatory hierarchy of angioblasts. To identify the signals and factors that induce angioblasts from mesoderm we have analyzed regulatory regions in the etsrp promoter. Bioinformatic analysis identified two evolutionarily conserved, noncoding sequences near the etsrp locus. One region, up1, is located 2.3 kb upstream of the transcription start site of etsrp. The second, int2, is within the second intron of etsrp. We tested and confirmed that both of these sequences function as enhancers for angioblast gene expression. Surprisingly deletion of these two conserved enhancers did not abolish the expression of the etsrp:gfp transgene. We then mapped a very short, 35 base-pair, proximal enhancer that was responsible for the remaining etsrp promoter expression.
Multiple enhancers for a single gene are common. Sometimes the enhancers drive partial spatial or temporal expression that when summed with other enhancers give the full gene expression. In the case of etsrp, we have identified three enhancers that drive expression in angioblasts and endothelial cells. There is some bias towards elevated artery or vein expression from each enhancer suggesting different signaling pathways may converge on each enhancer to drive robust etsrp expression during development. When each enhancer is isolated, expression is much weaker than in the context of the tg(−2.3etsrp:gfp) with all three enhancers present. Although we have found that tg(−2.3etsrp:gfp) is sufficient for strong expression in angioblasts, a recently published BAC transgenic line appears to be even more robust.32 It may be possible that other distant “shadow” enhancers are able to drive etsrp expression; however our bioinformatic analysis suggests that they are not evolutionarily conserved if they do exist.
To identify the transcription factors regulating etsrp expression, we focused on the up1 enhancer. By breaking down the up1 sequence into multiple overlapping EMSA probes we demonstrated that several protein complexes from endothelial cells bind to up1. One protein binding site was identified as a FOX consensus site and zebrafish Foxc1a and Foxc1b were found to bind by both EMSA and ChIP assays. In zebrafish, foxc1a/b has established functions in mesodermal, vascular, and mesenchymal development.12, 28, 30 Although vascular anomalies have been previously reported in foxc1a/b morphant embryos, the effects of foxc1a/b knockdown on early angioblast gene expression has not been described. Here, we have demonstrated that the early expression of etsrp, fli1a, and scl are disrupted when foxc1a/b are knocked down. Reduced expression of primitive erythrocyte gene gata1 suggests that both blood and vascular lineages are affected by loss of foxc1a/b. In addition, a previously reported disturbance in pax2a expression in the anterior pronephric region was confirmed.
It was surprising that knockdown of foxc1a/b had a large effect on etsrp expression, while deletion of up1 had relatively little effect on transgene expression. One possible explanation for this finding is that foxc1a/b binds at non-conserved sites or is recruited to the promoter through non-direct DNA binding interactions. Another possibility is that foxc1a/b has an indirect function in inducing angioblasts, possibly by generating signals in the paraxial mesoderm which is also defective in foxc1a/b morphants. In either case, our results highlight the importance of foxc1a/b in multiple mesodermal cell lineages and suggest that Foxc1a/b functions directly at the etsrp promoter.
In mammals, FoxC1 and FoxC2 are the homologs of zebrafish foxc1a/b. Null mice generated for each gene display significantly overlapping phenotypes. Both null mice have skeletal, eye, kidney, and cardiovascular problems.33, 34 The cardiovascular defects of FoxC1/C2 double null mice are reminiscent of the Etv2 null phenotype; although FoxC1/FoxC2 double null mice have visible blood while Etv2 null mice are completely anemic. The genetic relationship between these genes has yet to be studied in mouse models. However, it has been reported that FoxC2 and Etv2 have common downstream targets at a conserved FOX:ETS enhancer binding site.12 In fact, a mammalian Mef2c enhancer containing this double binding site is capable of driving expression in zebrafish vasculature.12 Our results suggest that FoxC1/2 may function upstream of Etv2 in addition to the established shared downstream function.
The reduced expression of primitive erythrocyte marker gata1 in foxc1a/b morphant embryos was somewhat surprising given that blood cells have been reported to be present in both null mice and morphant zebrafish. However, a FOX:ETS binding site has been reported at the SCL/TAL locus and Scl is directly upstream of Gata1.12, 35 Although this site likely drives expression in angioblasts, it is possible the site also functions in primitive erythrocytes in conjunction with FoxC2 and non-Etv2 ETS proteins. Another possibility is that non-ETS dependent FOX binding sites are present in the multiple enhancers driving Scl expression.36 A third possibility is an indirect effect of foxc1a/b knockdown on scl expression. The paraxial mesoderm is defective in foxc1a/b morphants and this tissue directly regulates the specification of primitive erythrocytes in zebrafish.37 Thus, the defect in scl and gata1 expression may be due to missing signals from the developing somites. This idea awaits further study.
In conclusion, foxc1a/b functions directly upstream of etsrp and upstream of scl in zebrafish mesoderm to specify angioblasts and primitive erythrocytes. This finding bridges the knowledge gap in molecular events underlying the mesoderm to angioblast transition. It may also have clinical implications as FOXC1 is linked to Axenfeld-Rieger anomaly and glaucoma38 while FOXC2 is linked to lymphedema-distichiasis syndrome39, 40 both diseases are associated with circulatory defects. Recently FOXC2 has been suggested to be an important mediator of tumor angiogenesis.41, 42 The link between FOXC1, FOXC2, and ETV2 in these diseases may be a clinically important avenue of study in the future.
Supplementary Material
Novelty and Significance.
What is known?
Etsrp/etv2 is an ETS family transcription factor that is a key regulator of endothelial cell development in embryos.
Loss of Etsrp/etv2 function results in embryonic lethality due to cardiovascular defects.
Ectopic expression of Etsrp/etv2 results in the induction of endothelial genes in non-endothelial cells.
What new information does this article contribute?
A new transgenic zebrafish line was developed using the etsrp/etv2 promoter to express GFP in the early endothelial cells.
Three regulatory regions were identified within the promoter to contribute to proper etsrp/etv2 expression.
The Foxc1a/b transcription factors were found to regulate etsrp/etv2 espression in the early endothelial cells through one of the regulatory regions.
Summary.
Etsrp/etv2 is a critical regulator of blood vessel development, however, little is known about how this gene is itself regulated. To identifiy upstream regulators we have generated transgenic zebrafish using the etsrp/etv2 promoter to drive GFP expression in the developing vasculature. Within the promoter sequence we have identified three regions necessary for robust expression. One regulatory region contains a forkhead transcription factor consensus binding site. We demonstrate that Foxc1a/b can bind to this site in vitro and in vivo and knockdown of these proteins results in decreased etsrp/etv2 expression as well as other blood and blood vessel genes. Previously, Foxc1a/b was found to cooperate with Etsrp/etv2 to regulate many important vascular genes. Here we find that foxc1a/b is also upstream of etsrp/etv2 in the genetic hierarchy of endothelial cell development. This provides a direct genetic link from mesoderm to endothelial cell. Previous work has demonstrated that the BMP, Notch, and Wnt signaling pathways regulate etsrp/etv2 in embryonic stem cells. It will be interesting to determine in future studies if the identified regulatory regions respond to these pathways through Foxc1a/b or other transcription factors in vivo.
Acknowledgements
The authors thank Anqi Liu for maintenance of zebrafish lines, the UCLA Orthopedic Hospital Research Center for the use of their real-time qPCR machine, Andrew Harmon for helping generate the tg(int2-gata2:gfp) lines and screening for founder fish, and Yesenia Rios, Zahra Tehrani, and Michaela Patterson for feedback on the manuscript.
Sources of Funding
M.B.V. was supported by NIH-T32 HL08634 and NIH-T32 HL69766. S.L. was supported by NIH-5R01DK054508-13.
Non-standard Abbreviations and Acronyms
- ALPM
Anterior lateral plate mesoderm
- ChIP
Chromatin immunoprecipitation
- DA
Dorsal aorta
- EMSA
Electrophoretic mobility shift assay
- FOX
Forkhead box
- GFP
Green fluorescent protein
- hpf
Hours post fertilization
- HUVEC
Human umbilical vein endothelial cell
- PAE
Porcine aortic endothelial cell
- PCV
Posterior cardinal vein
- PLPM
Posterior lateral plate mesoderm
- RT-PCR
Reverse transcriptase polymerase chain reaction
Footnotes
Disclosures
None.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sumanas S, Jorniak T, Lin S. Identification of novel vascular endothelial-specific genes by the microarray analysis of the zebrafish cloche mutants. Blood. 2005;106:534–541. doi: 10.1182/blood-2004-12-4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stainier DY, Weinstein BM, Detrich HW, 3rd, Zon LI, Fishman MC. Cloche, an early acting zebrafish gene, is required by both the endothelial and hematopoietic lineages. Development. 1995;121:3141–3150. doi: 10.1242/dev.121.10.3141. [DOI] [PubMed] [Google Scholar]
- 3.Sumanas S, Lin S. Ets1-related protein is a key regulator of vasculogenesis in zebrafish. PLoS Biol. 2006;4:e10. doi: 10.1371/journal.pbio.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong KS, Proulx K, Rost MS, Sumanas S. Identification of vasculature-specific genes by microarray analysis of etsrp/etv2 overexpressing zebrafish embryos. Dev Dyn. 2009;238:1836–1850. doi: 10.1002/dvdy.21990. [DOI] [PubMed] [Google Scholar]
- 5.Gomez GA, Veldman MB, Zhao Y, Burgess S, Lin S. Discovery and characterization of novel vascular and hematopoietic genes downstream of etsrp in zebrafish. PLoS One. 2009;4:e4994. doi: 10.1371/journal.pone.0004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pham VN, Lawson ND, Mugford JW, Dye L, Castranova D, Lo B, Weinstein BM. Combinatorial function of ets transcription factors in the developing vasculature. Dev Biol. 2007;303:772–783. doi: 10.1016/j.ydbio.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren X, Gomez GA, Zhang B, Lin S. Scl isoforms act downstream of etsrp to specify angioblasts and definitive hematopoietic stem cells. Blood. 2010;115:5338–5346. doi: 10.1182/blood-2009-09-244640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sumanas S, Gomez G, Zhao Y, Park C, Choi K, Lin S. Interplay among etsrp/er71, scl, and alk8 signaling controls endothelial and myeloid cell formation. Blood. 2008;111:4500–4510. doi: 10.1182/blood-2007-09-110569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Haro L, Janknecht R. Functional analysis of the transcription factor er71 and its activation of the matrix metalloproteinase-1 promoter. Nucleic Acids Res. 2002;30:2972–2979. doi: 10.1093/nar/gkf390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee D, Park C, Lee H, Lugus JJ, Kim SH, Arentson E, Chung YS, Gomez G, Kyba M, Lin S, Janknecht R, Lim DS, Choi K. Er71 acts downstream of bmp, notch, and wnt signaling in blood and vessel progenitor specification. Cell Stem Cell. 2008;2:497–507. doi: 10.1016/j.stem.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferdous A, Caprioli A, Iacovino M, Martin CM, Morris J, Richardson JA, Latif S, Hammer RE, Harvey RP, Olson EN, Kyba M, Garry DJ. Nkx2-5 transactivates the ets-related protein 71 gene and specifies an endothelial/endocardial fate in the developing embryo. Proc Natl Acad Sci U S A. 2009;106:814–819. doi: 10.1073/pnas.0807583106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Val S, Chi NC, Meadows SM, Minovitsky S, Anderson JP, Harris IS, Ehlers ML, Agarwal P, Visel A, Xu SM, Pennacchio LA, Dubchak I, Krieg PA, Stainier DY, Black BL. Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell. 2008;135:1053–1064. doi: 10.1016/j.cell.2008.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komuro I, Izumo S. Csx: A murine homeobox-containing gene specifically expressed in the developing heart. Proc Natl Acad Sci U S A. 1993;90:8145–8149. doi: 10.1073/pnas.90.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene nkx2-5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 15.Chen JN, Fishman MC. Zebrafish tinman homolog demarcates the heart field and initiates myocardial differentiation. Development. 1996;122:3809–3816. doi: 10.1242/dev.122.12.3809. [DOI] [PubMed] [Google Scholar]
- 16.Schoenebeck JJ, Keegan BR, Yelon D. Vessel and blood specification override cardiac potential in anterior mesoderm. Dev Cell. 2007;13:254–267. doi: 10.1016/j.devcel.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peterkin T, Gibson A, Patient R. Common genetic control of haemangioblast and cardiac development in zebrafish. Development. 2009;136:1465–1474. doi: 10.1242/dev.032748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiong JW, Yu Q, Zhang J, Mably JD. An acyltransferase controls the generation of hematopoietic and endothelial lineages in zebrafish. Circ Res. 2008;102:1057–1064. doi: 10.1161/CIRCRESAHA.107.163907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Westerfield M. The zebrafish book: A guide for the laboratory use of zebrafish (brachydanio rerio) M. Westerfield; Eugene, OR: 1993. [Google Scholar]
- 20.Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The tol2kit: A multisite gateway-based construction kit for tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- 21.Meng A, Jessen JR, Lin S. Transgenesis. Methods Cell Biol. 1999;60:133–148. [PubMed] [Google Scholar]
- 22.Jowett T. Analysis of protein and gene expression. Methods Cell Biol. 1999;59:63–85. doi: 10.1016/s0091-679x(08)61821-x. [DOI] [PubMed] [Google Scholar]
- 23.Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. Pipmaker--a web server for aligning two genomic dna sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. Ecr browser: A tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;32:W280–286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi J, Dong L, Ahn J, Dao D, Hammerschmidt M, Chen JN. Foxh1 negatively modulates flk1 gene expression and vascular formation in zebrafish. Dev Biol. 2007;304:735–744. doi: 10.1016/j.ydbio.2007.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- 27.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, Voss N, Stegmaier P, Lewicki-Potapov B, Saxel H, Kel AE, Wingender E. Transfac and its module transcompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skarie JM, Link BA. Foxc1 is essential for vascular basement membrane integrity and hyaloid vessel morphogenesis. Invest Ophthalmol Vis Sci. 2009;50:5026–5034. doi: 10.1167/iovs.09-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Topczewska JM, Topczewski J, Solnica-Krezel L, Hogan BL. Sequence and expression of zebrafish foxc1a and foxc1b, encoding conserved forkhead/winged helix transcription factors. Mech Dev. 2001;100:343–347. doi: 10.1016/s0925-4773(00)00534-7. [DOI] [PubMed] [Google Scholar]
- 30.Topczewska JM, Topczewski J, Shostak A, Kume T, Solnica-Krezel L, Hogan BL. The winged helix transcription factor foxc1a is essential for somitogenesis in zebrafish. Genes Dev. 2001;15:2483–2493. doi: 10.1101/gad.907401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Detrich HW, 3rd, Kieran MW, Chan FY, Barone LM, Yee K, Rundstadler JA, Pratt S, Ransom D, Zon LI. Intraembryonic hematopoietic cell migration during vertebrate development. Proc Natl Acad Sci U S A. 1995;92:10713–10717. doi: 10.1073/pnas.92.23.10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Proulx K, Lu A, Sumanas S. Cranial vasculature in zebrafish forms by angioblast cluster-derived angiogenesis. Dev Biol. 2010 doi: 10.1016/j.ydbio.2010.08.036. [DOI] [PubMed] [Google Scholar]
- 33.Kume T, Deng K, Hogan BL. Murine forkhead/winged helix genes foxc1 (mf1) and foxc2 (mfh1) are required for the early organogenesis of the kidney and urinary tract. Development. 2000;127:1387–1395. doi: 10.1242/dev.127.7.1387. [DOI] [PubMed] [Google Scholar]
- 34.Kume T, Jiang H, Topczewska JM, Hogan BL. The murine winged helix transcription factors, foxc1 and foxc2, are both required for cardiovascular development and somitogenesis. Genes Dev. 2001;15:2470–2482. doi: 10.1101/gad.907301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vyas P, McDevitt MA, Cantor AB, Katz SG, Fujiwara Y, Orkin SH. Different sequence requirements for expression in erythroid and megakaryocytic cells within a regulatory element upstream of the gata-1 gene. Development. 1999;126:2799–2811. doi: 10.1242/dev.126.12.2799. [DOI] [PubMed] [Google Scholar]
- 36.Dhami P, Bruce AW, Jim JH, Dillon SC, Hall A, Cooper JL, Bonhoure N, Chiang K, Ellis PD, Langford C, Andrews RM, Vetrie D. Genomic approaches uncover increasing complexities in the regulatory landscape at the human scl (tal1) locus. PLoS One. 2010;5:e9059. doi: 10.1371/journal.pone.0009059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rohde LA, Oates AC, Ho RK. A crucial interaction between embryonic red blood cell progenitors and paraxial mesoderm revealed in spadetail embryos. Dev Cell. 2004;7:251–262. doi: 10.1016/j.devcel.2004.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tumer Z, Bach-Holm D. Axenfeld-rieger syndrome and spectrum of pitx2 and foxc1 mutations. Eur J Hum Genet. 2009;17:1527–1539. doi: 10.1038/ejhg.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishimura DY, Swiderski RE, Alward WL, Searby CC, Patil SR, Bennet SR, Kanis AB, Gastier JM, Stone EM, Sheffield VC. The forkhead transcription factor gene fkhl7 is responsible for glaucoma phenotypes which map to 6p25. Nat Genet. 1998;19:140–147. doi: 10.1038/493. [DOI] [PubMed] [Google Scholar]
- 40.Mears AJ, Jordan T, Mirzayans F, Dubois S, Kume T, Parlee M, Ritch R, Koop B, Kuo WL, Collins C, Marshall J, Gould DB, Pearce W, Carlsson P, Enerback S, Morissette J, Bhattacharya S, Hogan B, Raymond V, Walter MA. Mutations of the forkhead/winged-helix gene, fkhl7, in patients with axenfeld-rieger anomaly. Am J Hum Genet. 1998;63:1316–1328. doi: 10.1086/302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hayashi H, Sano H, Seo S, Kume T. The foxc2 transcription factor regulates angiogenesis via induction of integrin beta3 expression. J Biol Chem. 2008;283:23791–23800. doi: 10.1074/jbc.M800190200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sano H, Leboeuf JP, Novitskiy SV, Seo S, Zaja-Milatovic S, Dikov MM, Kume T. The foxc2 transcription factor regulates tumor angiogenesis. Biochem Biophys Res Commun. 2010;392:201–206. doi: 10.1016/j.bbrc.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.