Abstract
A family of related proteins in yeast Saccharomyces cerevisiae is known to have in vitro GTPase-activating protein activity on the Rab GTPases. However, their in vivo function remains obscure. One of them, Gyp1p, acts on Sec4p, Ypt1p, Ypt7p, and Ypt51p in vitro. Here, we present data to reveal its in vivo substrate and the role that it plays in the function of the Rab GTPase. Red fluorescent protein-tagged Gyp1p is concentrated on cytoplasmic punctate structures that largely colocalize with a cis-Golgi marker. Subcellular fractionation of a yeast lysate confirmed that Gyp1p is peripherally associated with membranes and that it cofractionates with Golgi markers. This localization suggests that Gyp1p may only act on Rab GTPases on the Golgi. A gyp1Δ strain displays a growth defect on synthetic medium at 37°C. Overexpression of Ypt1p, but not other Rab GTPases, strongly inhibits the growth of gyp1Δ cells. Conversely, a partial loss-of-function allele of YPT1, ypt1-2, can suppress the growth defect of gyp1Δ cells. Furthermore, deletion of GYP1 can partially suppress growth defects associated with mutants in subunits of transport protein particle complex, a complex that catalyzes nucleotide exchange on Ypt1p. These results establish that Gyp1p functions on the Golgi as a negative regulator of Ypt1p.
INTRODUCTION
Rab GTPases form the largest subfamily of small GTPases in the Ras superfamily (Novick and Zerial, 1997). They perform essential functions in different membrane transport pathways in the cell. In the budding yeast Saccharomyces cerevisiae, Ypt1p, Ypt31p/32p, and Sec4p function on the exocytic pathway, whereas Ypt6p, Ypt7p, and Ypt51p/52p/53p function on the endocytic/vacuolar pathway (Lazar et al., 1997). Similar to other small GTPases, Rab GTPases act as molecular switches, cycling between a GTP-bound state and a GDP-bound state. The intrinsic rate of conversion between these two states is low. Guanine nucleotide exchange factors (GEFs) catalyze the exchange of GDP for GTP, whereas GTPase-activating proteins (GAPs) stimulate the hydrolysis of GTP to GDP. The activity of these regulatory proteins can determine where and when the GTPases are active.
Most of the known Rab GAPs share a region of homology, which is likely to represent the catalytic domain (Neuwald, 1997). In S. cerevisiae, there are at least 10 genes encoding proteins containing this RabGAP domain. They are GYP6, GYP7, GYP1, MDR1/MIC1/GYP2, MSB3/GYP3, MSB4/GYP4, YPL249C, YOL112W, YMR192W, and BUB2. The protein products of the first six genes have been shown to have Rab GAP activities in vitro (Strom et al., 1993; Du et al., 1998; Albert and Gallwitz, 1999, 2000; Vollmer et al., 1999). Bub2p, on the other hand, is likely to be one subunit of a two-component GAP for Tem1p, a small GTPase involved in exit from mitosis (reviewed by Hoyt, 2000). One mammalian Rab GAP, GAPCenA, also shares this domain (Cuif et al., 1999). However, the other known mammalian Rab GAPs are not related in their primary sequences to the yeast Rab GAPs (Fukui et al., 1997; Xiao et al., 1997; Liu and Li, 1998).
In vitro mutagenesis studies of Gyp1p and Gyp7p (Albert et al., 1999) and the determination of the structure of the Gyp1p catalytic domain (Rak et al., 2000) have revealed detailed biochemical and structural properties of this Rab GAP family. However, very little is known about their in vivo function, mainly due to the lack of an observable phenotype of their mutants. Deletion of both MSB3 and MSB4 results in slow growth and a partial disorganization of the actin cytoskeleton in a fraction of cells (Bi et al., 2000). The relationship between these phenotypes and the GAP activity of Msb3p and Msb4p is not clear.
The known yeast Rab GAPs have broad and overlapping in vitro substrate specificity. For example, Gyp1p acts almost equally well on Sec4p, Ypt1p, Ypt7p, and Ypt51p (Du et al., 1998; Albert et al., 1999), and Ypt1p is a substrate for both Gyp1p and Msb3p (Albert and Gallwitz, 1999). This overlapping specificity makes it difficult to distinguish the activity of different GAPs in a yeast lysate. Therefore, the previously observed GAP activity for individual Rab proteins in yeast lysates may reflect the combined activity of several GAPs (Walworth et al., 1992; Jones et al., 1998). One question that has not been addressed is whether these GAPs have the same specificity in vivo.
The GTP hydrolysis reactions catalyzed by Rab GAPs could occur at multiple steps of the membrane association/disassociation cycle of Rab GTPases. For example, GTP hydrolysis after vesicle fusion may facilitate the recycling of Rab GTPases by guanine nucleotide disassociation inhibitors; on the other hand, GTP hydrolysis on the vesicle-attached Rab GTPases may prevent fusion. Because hydrolysis at different steps may require distinct Rab GAPs, it is conceivable that Rab GAPs could be either positive or negative regulators of the function of Rab GTPases. We have studied the subcellular localization of Gyp1p and examined its in vivo function by using genetic approaches. In this article, we show that Gyp1p localizes to the yeast Golgi apparatus and functions in vivo as a negative regulator of Ypt1p.
MATERIALS AND METHODS
Media
YPD and synthetic complete (SC) media were as described in Sherman (1991).
Strains and Plasmids
Table 1 lists the genotype of the yeast strains used in this study. The construction of gyp1Δ strains was previously reported (Du et al., 1998). pep4::HIS3 strains were made by polymerase chain reaction (PCR) amplification of the DNA containing the pep4::HIS3 locus from BJ5622 (Jones, 1991) and then introducing the PCR product into our strains by transformation. The vps1Δ::kanMX strain was generated using the same method, transferring the locus from a vps1Δ strain created by the genome deletion project (Winzeler et al., 1999). The vps21Δ::kanMX strain was made by transforming with XhoI/XbaI-digested pSRG97 (Gerrard et al., 2000). The coding region of red fluorescent protein was amplified by PCR from the pDsRed vector (CLONTECH, Palo Alto, CA) and inserted into the BamHI site of a pRS416-based vector containing the TEF promoter and the CYC1 terminator (Mumberg et al., 1995). The resulting plasmid is named pNB1091. The GYP1 open reading frame was cloned in frame between the BamHI and SalI sites in the pNB1091 plasmid to generate pNB1092. To express proteins under the control of the GYP1 promoter and terminator, we cloned 1560 bp of the GYP1 promoter region and 250 bp of the terminator region into pRS315, and created BamHI and SalI sites between the promoter and terminator. The resulting plasmid is designated pNB1093. Fragments encoding Gyp1p(1–637), Gyp1p(212–637), Gyp1p(273–637), Gyp1p(212–630), and Gyp1p(212–620) were cloned into pNB1093 to make constructs expressing Gyp1p of different lengths. R286A and R343K mutations in Gyp1p(212–637) were created using a megaprimer PCR method (Boles and Miosga, 1995). The mutagenic primers were 5′-CAAACAACAGGCGCGTGTATTTTTGGGATA-3′ for the R286A mutation and 5′-GGGGATTTGTCTTCGGTATATCTATCTCTA-3′ for the R343K mutation. All of the above-mentioned constructs were confirmed by sequencing. Plasmids overexpressing Rab GTPases were made by cloning the coding regions from the bacteria expression plasmids (Du et al., 1998) into a pRS413-based vector containing the GPD promoter and the CYC1 terminator (Mumberg et al., 1995). The CEN plasmid containing a genomic clone of YPT1 was made by cloning a 1080-bp BglII/BamHI fragment from pSFNB43 to pRS313.
Table 1.
List of yeast strains used in this study
| Strains | Genotype |
|---|---|
| NY1210 | MATa ura3-52 his3-Δ200 leu2-3,112 |
| NY1211 | MATα ura3-52 his3-Δ200 leu2-3,112 |
| NY2291 | MATa ura3-52 his3-Δ200 leu2-3,112 gyp1Δ∷LEU2 |
| NY2292 | MATa ura3-52 his3-Δ200 leu2-3,112 gyp1Δ∷URA3 |
| NY2393 | MATα ura3-52 his3-Δ200 leu2-3,112 gyp1Δ∷URA3 |
| NY2294 | MATa ura3-52 his3-Δ200 leu2-3,112 bet3∷HIS3+ [LEU2 CEN BET3-GFP; pSFNB516] + [URA3 CEN TEFp-RFP-GYP1; pNB1092] |
| NY2295 | MATa ura3-52 his3-Δ200 leu2-3,112 pep4∷HIS3 |
| NY2296 | MATa ura3-52 his3-Δ200 leu2-3,112 pep4∷HIS3 gyp1Δ∷URA3 |
| NY2297 | MATa/MATα ura3-52/ura3-52 his3-Δ200/his3-Δ200 leu2-3,112/leu2-3,112 gyp1Δ∷URA3/gyp1Δ∷URA3 YPT1/ypt1-2 |
| NY2298 | MATα ura3-52 his3-Δ200 leu2-3,112∷[LEU2 bet5-1] bet5Δ∷HIS3 |
| NY2399 | MATα ura3-52 his3-Δ200 leu2-3,112∷[LEU2 bet5-1] bet5Δ∷HIS3 gyp1Δ∷URA3 |
| NY2300 | MATa ura3-52 his3-Δ200 leu2-3,112 vps1Δ∷kanMX |
| NY2301 | MATa ura3-52 his3-Δ200 leu2-3,112 vps21Δ∷kanMX |
| NY2302 | MATa ura3-52∷[URA3 GALp-YPT1; pSFNB544] his3-Δ200 leu2-3,112 |
| NY2303 | MATa ura3-52∷[URA3 GALp-YPT1; pSFNB544] his3-Δ200 leu2-3,112 gyp1Δ∷LEU2 |
| NY2304 | MATα ura3-52 his3-Δ200 leu2-3,112 ypt1-2 |
| NY2305 | MATα ura3-52 his3-Δ200 leu2-3,112 ypt1-2 gyp1Δ∷URA3 |
Microscopy
Yeast cells were grown in selective medium at 25°C to log phase. Culture (1 ml) was briefly centrifuged in a microfuge tube to pellet the cells. Medium (0.95 ml) was removed and cells were resuspended in the remaining medium. Cell suspension (2 μl) was dropped on a slide and covered with a coverslip. Samples were viewed on a Zeiss Axiophot 2 microscope using a 63× oil-immersion objective (NA 1.4). Images were acquired with a Photometrics Quantix charge coupled device camera by using IPLab for Macintosh software (Scanalytics, Fairfax, VA).
Fractionation of Yeast Lysate
Pep4::HIS3 cells (NY2295 and NY2296) grown in YPD at 25°C (100 A600 units) were harvested at log phase. The cells were washed once with 20 ml of 20 mM HEPES-NaOH, pH 7.5, 20 mM NaN3, 20 mM NaF. The cells were resuspended in 0.95 ml of spheroplasting solution (1.4 M sorbitol, 50 mM KPi, pH 7.5, 10 mM NaN3, 0.4% 2-mercaptoethanol, 0.1 mg/ml zymolyase 100-T [ICN Biomedicals, Irvine, CA]) and incubated at 37°C for 45 min. After cooling it on ice, the suspension was loaded on top of 4 ml of ice-cold 1.7 M sorbitol, 50 mM KPi, pH 7.5, 1× protease inhibitors cocktail (10 μM antipain, 1 μg/ml aprotinin, 30 μM leupeptin, 30 μM chymostatin, 20 μM pepstatin A, 2 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride), and centrifuged at 3000 rpm for 10 min in a GH-3.8 swing-bucket rotor in a Beckman GS-6 centrifuge. Both layers of liquid were removed, and pellet was resuspended in 1 ml of lysis buffer (20 mM tetraethylammonium [TEA]-acetate, pH 7.2, 0.4 M sorbitol, 1 mM EDTA, 1× protease inhibitors cocktail). The suspension was transferred to a 1-ml Dounce grinder cooled on ice. Cells were disrupted with 50 strokes by using the tight pestle. The suspension was centrifuged in a GH-3.8 rotor at 2000 rpm for 3 min to remove unlysed cells. The supernatant was transferred to a fresh tube and used as the total lysate. The protein concentration in the lysate was ∼10 mg/ml.
For the iodixanol floatation experiment, 80 μl of NY2295 lysate was mixed with 320 μl of 50% iodixanol, 20 mM TEA-acetate, pH 7.2, 0.4 M sorbitol, 1 mM EDTA, so that the final concentration of iodixanol was 40%. The mixture (0.1 ml) was loaded to the bottom of an 11 × 34-mm polycarbonate tube (Beckman Instruments, Palo Alto, CA) underneath 0.9 ml of 35% iodixanol, 20 mM TEA-acetate, pH 7.2, 0.4 M sorbitol, 1 mM EDTA. The tubes were centrifuged in a TLA 120.2 rotor at 120,000 rpm for 3 h. Fractions of 130 μl were taken from the top by using a P200 pipette. The protein concentration was determined using the Bio-Rad (Richmond, CA) protein assay. The concentration of iodixanol was determined by absorbance at 244 nm (Schroder et al., 1997).
For the extraction experiment, 100 μl of NY2295 lysate was mixed with 500 μl of lysis buffer, or lysis buffer containing 2.4% Triton X-100, or 4.8 M urea, or 0.12 M Na2CO3, so that the final concentration of the extracting reagents was 2% Triton X-100, 4 M urea, or 0.1 M Na2CO3, respectively. After a 40-min incubation on ice, the mixtures were centrifuged in a TLA 120.2 rotor at 55,000 rpm (100,000 gav) for 30 min.
For the differential centrifugation experiment, 600 μl of NY2295 lysate was centrifuged in an Eppendorf 5402 centrifuge at 11,000 rpm (10,000 × g) for 10 min. The supernatant was transferred to an 11 × 34-mm polycarbonate tube and centrifuged at 55,000 rpm for 20 min.
The sucrose gradient was prepared by layering 1 ml each of 60, 50, 40, 30, 20% sucrose (wt/wt) in 20 mM TEA-acetate, pH 7.2, 1 mM EDTA on top of each other in a 13 × 51-mm ultra-clear tube (Beckman Instruments) and then allowing a gradient to form by diffusion overnight at 4°C. NY2295 lysate (0.3 ml) was loaded on the top of the gradient and centrifuged in a SW 50.1 rotor at 35,000 rpm (120,000 gav) for 20 h. Fractions of 15 drops (∼300 μl) were collected from the bottom of the tube by tube puncturing. The sucrose concentration of each fraction was determined by measuring its refractive index. Densitometric measurement of the Western blot was performed using a GS-700 densitometer (Bio-Rad) and Quantity One software (Bio-Rad).
Antibodies
Purified recombinant Gyp1p (Du et al., 1998) was used to immunize rabbits by Yale Biotechnology Services (New Haven, CT). The anti-Gyp1p serum was affinity purified with Gyp1p coupled to Affi-Gel 10 (Bio-Rad). Antibodies against Ssop, Sncp, and Pep12p have been described (Grote and Novick, 1999). Antibodies against Sed5p, Bet3p, and Trs33p were from Dr. S. Ferro-Novick (Yale University, New Haven, CT). Antibody against Pma1p was from Dr. C.W. Slayman (Yale University). Antibody against yeast alcohol dehydrogenase was purchased from Chemicon International.
CPY Transport Assays
The pulse-chase experiment was performed as described (Govindan et al., 1995). For the overlay assay on YPD plates, freshly saturated cultures were diluted in YPD to A600 = 2. 3 μl of the diluted suspension was spotted on the surface of a YPD plate. After a 3-h incubation at 30°C, a piece of wet nitrocellulose membrane was overlaid on the plate. After 18-h incubation at 30°C, the membrane was lifted and washed with water to remove all the cells. Proteins absorbed on the membrane were detected by immunoblot. For the overlay experiment with YP-raffinose-galactose plates, cells were pregrown and diluted in YP-raffinose medium.
RESULTS
Gyp1p Localizes to Golgi Apparatus
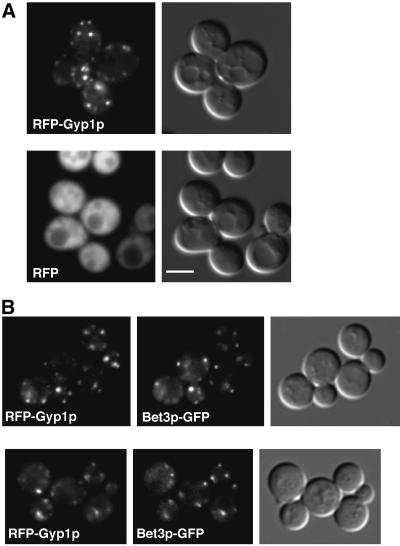
To examine the localization of Gyp1p in yeast, we added a green fluorescent protein (GFP) tag to the N terminus of Gyp1p. The GFP-tagged Gyp1p (GFP-Gyp1p) expressed from the GYP1 promoter showed punctate cytoplasmic localization in live cells (our unpublished observation). This distribution pattern of GFP-Gyp1p was very sensitive to fixation. Once cells were fixed, only evenly diffuse cytoplasmic fluorescence was observed. This sensitivity to fixation was not compatible with colocalization studies that rely on immunofluorescence staining. To perform colocalization experiments, we tagged Gyp1p with red fluorescent protein (RFP, i.e., DsRed; CLONTECH). Because of the low intensity of RFP fluorescence, RFP-tagged Gyp1p (RFP-Gyp1p) had to be expressed at 20 times higher than the endogenous Gyp1p level (our unpublished observation). Nevertheless, RFP-Gyp1p showed the same punctate localization as GFP-Gyp1p, whereas RFP alone gave only a diffuse signal (Figure 1A). Both GFP-Gyp1p and RFP-Gyp1p are fully functional as determined by a plate assay that will be described below.
Figure 1.
Localization of RFP-Gyp1p. (A) RFP-Gyp1p localizes to punctate structures in live cells. RFP-Gyp1p (top) or RFP (bottom) was expressed from a CEN plasmid in gyp1Δ cells (NY2291). The same cells were imaged in fluorescence and differential interference contrast modes. Bar, 4 μm. (B) RFP-Gyp1p partially colocalizes with a cis-Golgi marker, Bet3p-GFP, in live cells. RFP-Gyp1p and Bet3p-GFP were expressed from CEN plasmids in bet3Δ cells (NY2294). The same cells were imaged in fluorescence mode with trimethylrhodamine B isothiocyanate filter and fluorescein isothiocyanate filter and in differential interference contrast mode.
Because the punctate localization of Gyp1p is similar to the localization pattern of yeast Golgi proteins, we performed double labeling experiments by using RFP-tagged Gyp1p and a known resident of the Golgi apparatus, GFP-tagged Bet3p (Bet3p-GFP) (Sacher et al., 1998). Bet3p is a subunit of yeast transport protein particle complex (TRAPP) localized to the cis-Golgi (Sacher et al., 1998; Barrowman et al., 2000). In cells expressing both RFP-Gyp1p and Bet3p-GFP, the distribution of the RFP signal significantly overlapped with that of the GFP signal (Figure 1B). We counted fluorescent spots in 45 cells. There were 228 Gyp1p-positive spots and 202 Bet3p-positive spots. Among them, 140 spots were labeled by both GFP and RFP. In other words, 61% of the RFP-Gyp1 spots were labeled by Bet3-GFP, and 69% of the Bet3-GFP spots were labeled by RFP-Gyp1. The substantial colocalization of Gyp1p and Bet3p indicates that Gyp1p at least partially localizes to Golgi. It is noteworthy that the punctate localization of Bet3-GFP also disappears upon fixation, suggesting that this sensitivity to fixation may be a property of many peripherally bound Golgi proteins.
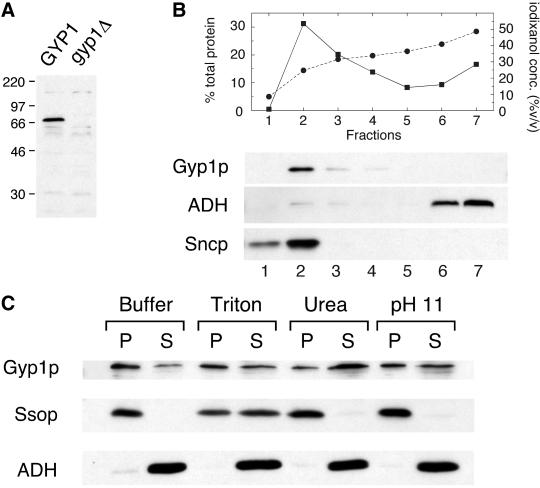
To use an independent approach to assess the localization of Gyp1p, we carried out subcellular fractionation studies. First, we prepared affinity-purified anti-Gyp1p antibody and examined its specificity. When wild-type yeast lysate was separated by SDS-PAGE, a 70-kDa band was detected by this antibody with Western blot, in good agreement with the predicted molecular weight of Gyp1p of 73 kDa (Figure 2A). This band was not detected in the lysate of gyp1Δ cells, confirming that the protein recognized by our antibody is Gyp1p.
Figure 2.
Gyp1p is peripherally associated with membranes. (A) Antibody detection of Gyp1p. Equal amounts of total cell extracts prepared from GYP1 pep4 (NY2295) and gyp1Δ pep4 (NY2296) strains were resolved by SDS-PAGE, blotted, and probed with affinity-purified polyclonal antibody raised against recombinant Gyp1p. The positions of size markers (kDa) are indicated. (B) Lysate of NY2295 was loaded at the bottom of a tube containing 35% iodixanol, and centrifuged in a TLA 120.2 rotor at 120,000 rpm for 3 h. Seven fractions were collected from the top. The percentage of total protein in each fraction (▪) and iodixanol concentration of each fraction (●) was determined. The amount of Gyp1p, ADH, and Sncp in each fraction was determined by Western blot. (C) Lysate of NY2295 was mixed with lysis buffer and lysis buffer containing Triton X-100, urea, and sodium carbonate so that the final concentrations were 2% Triton X-100, 4 M urea, and 0.1 M sodium carbonate (pH 11), respectively. The mixtures were incubated on ice for 40 min and then centrifuged at 100,000 × g for 30 min. Equal amounts of pellet and supernatant fractions were analyzed by SDS-PAGE and Western blot. Urea treatment shifted the distribution of Gyp1p toward the supernatant but did not affect the distribution of the integral membrane protein Ssop.
To determine whether Gyp1p is membrane bound, we used a floatation assay (Figure 2B). Yeast lysate supplemented with iodixanol to 40% (vol/vol) was loaded beneath a layer of 35% iodixanol and centrifuged at 120,000 rpm in a TLA 120.2 rotor for 3 h. Seven fractions were collected from the top. Gyp1p and a membrane marker Sncp floated up to fraction 2, whereas the soluble protein marker alcohol dehydrogenase (ADH) remained at the bottom of the tube. This result indicates that most of the Gyp1p in the lysate was membrane associated. The nature of the interaction between Gyp1p and the membrane was examined by an extraction study (Figure 2C). Most of the Gyp1p in the lysate can be pelleted by centrifugation at 100,000 × g. Some Gyp1p partitioned in the supernatant after 100,000 × g centrifugation. This could be due to the distribution of Gyp1p to very small vesicles that cannot be pelleted at 100,000 × g or perhaps to the disassociation of Gyp1p from the membrane in the absence of iodixanol. Treatment with the nonionic detergent Triton X-100 or a high-pH solution only slightly increased the amount of Gyp1p in the supernatant. However, 4 M urea was found to extract most of the Gyp1p, whereas the integral membrane protein Ssop was still pelleted after this treatment. These results indicate that Gyp1p is peripherally associated with the membrane fraction.
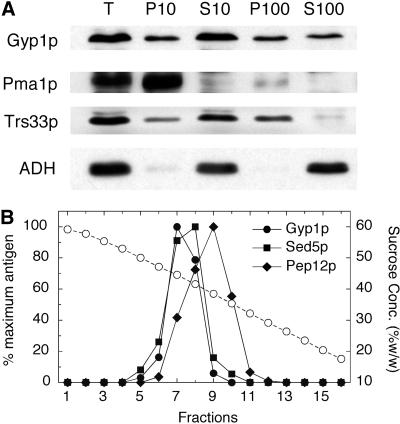
A differential centrifugation experiment was performed to determine the membrane compartment with which Gyp1p is associated (Figure 3A). A yeast lysate was first centrifuged at 10,000 × g, and then the 10,000-g supernatant was further centrifuged at 100,000 × g. It is known that most of the yeast endoplasmic reticulum (ER) and plasma membrane can be pelleted at 10,000 × g. We found that the pelletable Gyp1p was distributed equally between the 10,000- and 100,000-g pellet fractions, whereas the plasma membrane marker Pma1p was almost completely pelleted at 10,000 × g. Therefore, Gyp1p does not behave like a plasma membrane protein. Rather, its sedimentation pattern is similar to that of the Golgi protein Trs33p, which is another subunit of TRAPP (Sacher et al., 1998). Because yeast endosomes also appear as punctate structures by fluorescence microscopy (Gerrard et al., 2000), we compared the fractionation of Gyp1p with Golgi and endosome markers in a sucrose density gradient (Figure 3B). A yeast lysate was loaded on the top of a 20–60% linear sucrose gradient. After centrifugation at 120,000 × g for 20 h, fractions were collected from the bottom of the tube. Gyp1p cofractionated with Sed5p, a Golgi SNARE protein (Banfield et al., 1994). On the other hand, Pep12p, an endosomal SNARE protein (Becherer et al., 1996), peaked at a lower sucrose concentration. As has been previously shown (Sacher et al., 1998), TRAPP subunits Bet3p and Trs33p also cofractionated with Sed5p in this gradient (our unpublished observation). Therefore, our fractionation results are in good agreement with the fluorescence microscopy data, suggesting that Gyp1p localizes to Golgi.
Figure 3.
Subcellular fractionation of Gyp1p. (A) Differential centrifugation of yeast lysate. Total lysate of NY2295 (T) was centrifuged at 10,000 × g to generate supernatant S10 and pellet P10. The S10 supernatant was centrifuged at 100,000 × g to obtain supernatant S100 and pellet P100. Equal amounts of samples were prepared for electrophoresis, separated by SDS-PAGE, and transferred to nitrocellulose. Western blots were probed with polyclonal antibodies against Gyp1p, Pma1p, Trs33p, and ADH. (B) Sucrose gradient fractionation of yeast lysate. Lysate of NY2295 was loaded on the top of a linear 20–60% (wt/wt) sucrose gradient and centrifuged at 120,000 × g for 20 h. Fractions were collected from the bottom. Sucrose concentration (circles) in each fraction was determined using a refractometer. The amounts of Gyp1p, Sed5p, and Pep12p in each fraction were determined by Western blot and densitometry.
GAP Activity of Gyp1p Is Required for Its In Vivo Function
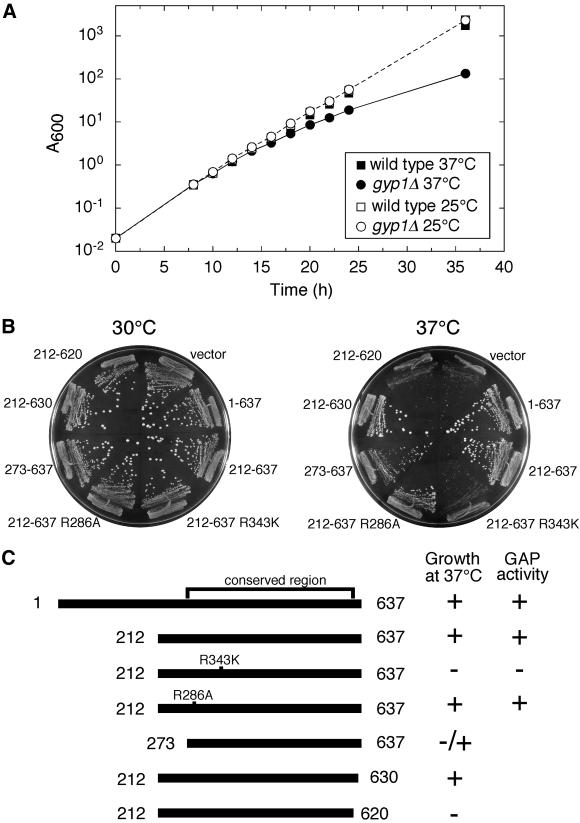
Previously, we found no growth defect when a gyp1Δ strain was grown on rich medium at different temperatures (Du et al., 1998). However, when we examined growth of a gyp1Δ strain on synthetic medium, we noticed a slow growth phenotype at 37°C. A growth curve in liquid SC medium monitored by absorbance at 600 nm is shown in Figure 4A. At 25°C, gyp1Δ cells grow as well as wild-type cells in SC medium. However, the growth rate of gyp1Δ cells started to slow 16 h after a shift from 25 to 37°C. The gyp1Δ cells continued to grow at a slower rate so that the difference in absorbance became more dramatic at later time points. This relatively mild growth defect of gyp1Δ cells at 37°C in synthetic medium can also be observed on solid medium. After 4 d of growth on SC plate at 37°C, the colony size of the gyp1Δ strain was significantly smaller than wild type [Figure 4B, compare the vector transformant with the full-length Gyp1p(1–637) transformant]. gyp1Δ cells also grow slower than wild-type cells on a minimal medium plate at 37°C, but the difference in colony size is not as dramatic as on the SC plate. This synthetic medium growth defect of gyp1Δ cells makes it possible for us to explore the physiological function of different gyp1 mutant alleles.
Figure 4.
GAP activity is essential for the in vivo function of Gyp1p. (A) gyp1Δ has a growth defect in SC medium at 37°C. Wild-type (NY1210) and gyp1Δ (NY2292) cells were grown in SC medium at 25°C to log phase. They were inoculated into 25°C SC medium and 37°C SC medium, so that the final absorbance at 600 nm (A600) was 0.02. The growth was monitored by A600. The cultures were diluted 10 times in fresh medium when the A600 exceeded 1.0. The A600 in the figure reflects the cell density in the original volume, having taken the dilution into consideration. (B) Plate assay to test the ability of different constructs to complement the SC medium growth defect of gyp1Δ. CEN LEU2 plasmids expressing different Gyp1p proteins from the GYP1 promoter were introduced into gyp1Δ cells (NY2292). The transformants were streaked onto SC-LEU plates and incubated at 30 and 37°C. Photos were taken after 4 d. (C) The complementation activity of different Gyp1p proteins correlates with their in vitro GAP activity. The colony size of the full-length Gyp1p construct transformant was scored as +. The colony size of the vector transformant was scored as −. The in vitro GAP activity of Gyp1p(1–637) and Gyp1p(212–637) was determined by measuring the activity of purified recombinant glutathione S-transferase fusion proteins (our unpublished observation). The activity of the R286A and R343K mutants was based on published results (Albert et al., 1999).
We made low copy number CEN plasmid constructs to express Gyp1p proteins under the control of the GYP1 promoter and introduced these constructs by transformation into a gyp1Δ strain to test their in vivo functional activity with the plate assay (Figure 4B). Truncated Gyp1p (residues 212–637) missing the N-terminal third of the open reading frame can complement the growth defect as well as the full-length protein. This truncated Gyp1p contains the catalytic domain (residues 249–630) whose structure has been recently published (Rak et al., 2000). Recombinant Gyp1p(212–637) purified from bacteria has the same GAP activity as full-length Gyp1p (our unpublished data). Further truncation of 61 amino acids from the N terminus resulted in an intermediate colony size. This Gyp1p(273–637) has diminished GAP activity in vitro (our unpublished data). The lower activity is likely to be caused by disturbing the structure of the catalytic domain because the first α-helix of the catalytic domain, α1, and part of α2 are missing from this protein. Truncation from the C terminus of Gyp1p is tolerated less well than truncation from the N terminus. Gyp1p(212–620), lacking half of the final α-helix, α16, completely lost the ability to complement the growth defect. Conserved arginines in the catalytic domain of Gyp1p have been mutagenized and found to have different effects on its catalytic activity (Albert et al., 1999). Arginine 286 is not required for GAP activity, but mutation of this residue lowered the yield of the protein from yeast. Arginine 343 is essential for the GAP activity but not for substrate binding. We introduced the R286A and R343K mutations into GYP1(212–637). Western blot analysis showed that the R343K mutant protein was expressed at the same level as Gyp1p(212–637), whereas the R286A mutant protein was expressed at a lower level (our unpublished observation). However, the R286A mutant can complement the growth defect as well as the wild-type gene, whereas the R343K mutant lost its complementation ability. As summarized in Figure 4C, the ability of different constructs to complement the growth defect correlates directly with their in vitro GAP activity. Therefore, we conclude that the GAP activity is required for the in vivo function of Gyp1p.
Previously, we had observed that overexpression of Gyp1p from a high-copy number plasmid inhibits the growth of certain secretory mutants (Du et al., 1998). The R343K mutation totally abolished this inhibitory effect (our unpublished observation), indicating that this growth inhibition is also dependent on GAP activity.
Gyp1p Is a GAP for Ypt1p In Vivo
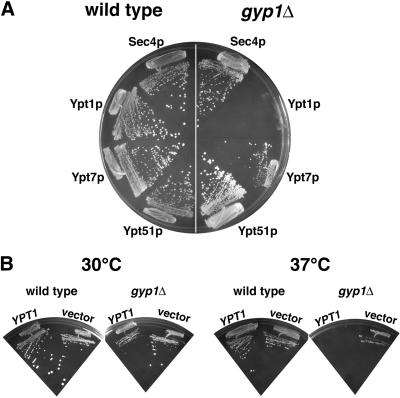
Gyp1p has in vitro GAP activity for Sec4p, Ypt1p, Ypt7p, and Ypt51p (Du et al., 1998). To study the in vivo substrate specificity of Gyp1p, we took a genetic approach. We determined the effect of overexpressing different Rab GTPases on the growth of gyp1Δ cells. We overexpressed the GTPases from the strong glyceraldehyde-3-phosphate dehydrogenase (GPD) promoter on a low-copy number CEN plasmid. Overexpression of Sec4p, Ypt7p, and Ypt51p did not significantly affect the growth of gyp1Δ cells at either 25 or 37°C. However, when a plasmid overexpressing Ypt1p was introduced into gyp1Δ cells, we observed only tiny colonies at 25°C, indicating that overexpression of Ypt1p is toxic to gyp1Δ. To confirm this genetic interaction, we introduced into wild-type and gyp1Δ cells two plasmids at the same time. One plasmid was a 2 μ circle based plasmid containing the URA3 marker and the GYP1 gene, the other plasmid was a CEN plasmid overproducing one Rab GTPase. All of the transformants grew equally well. We streaked the transformants first onto plates containing uracil to allow loss of the 2 μ GYP1 plasmid. Then we streaked the cells onto 5-fluoroorotic acid plates that maintained selection for the CEN plasmid, but selected against Ura+ cells (Figure 5A). Only the cells that can lose the 2 μ GYP1, URA3 plasmid can survive on this plate. The wild-type cells all grew well. The gyp1Δ cells overexpressing Sec4p, or Ypt7p, or Ypt51p also grew well on this plate, whereas gyp1Δ cells overexpressing Ypt1p did not grow. Therefore, the GYP1 gene cannot be lost from a strain overproducing Ypt1p. Although the GTPases were expressed from the same GPD promoter, the protein level may not be the same in the cell. In a previous study (Grote and Novick, 1999), when different hemagglutinin (HA)-tagged Rab GTPases were expressed from the same GAL1 promoter, HA-Ypt1p was expressed at about the same level as HA-Sec4p and HA-Ypt51p. Therefore, the specific inhibitory effect of Ypt1p is not likely to be due to a higher level of expression of Ypt1p compared with the other GTPases.
Figure 5.
Overexpression of Ypt1p specifically inhibits the growth of gyp1Δ. (A) Wild-type (NY1210) and gyp1Δ (NY2291) cells were transformed with a 2 μ URA3 plasmid containing the GYP1 gene, and a CEN HIS3 plasmid overproducing either Sec4p, Ypt1p, Ypt7p, or Ypt51p, from a GPD promoter. Transformants were first streaked on a minimal medium plate containing uracil and then restreaked on minimal medium plate containing 5-fluoroorotic acid. Photos were taken after 4-d incubation at 30°C. (B) Growth of gyp1Δ is inhibited by an extra copy of the YPT1 gene. Wild-type (NY1210) and gyp1Δ (NY2291) cells were transformed with a CEN plasmid containing the YPT1 gene. Transformants were streaked on a minimal medium plate and incubated at 30 and 37°C. Photos were taken after 4 d.
To examine the effect of Ypt1p at a lower expression level, we transformed wild-type and gyp1Δ cells with a low-copy number CEN plasmid expressing Ypt1p from its own promoter. Western blot analysis of lysates showed that transformants of this plasmid express Ypt1p at only 2–3 times the level of the vector-only control (our unpublished observation). At 30°C, this plasmid slightly inhibited growth of gyp1Δ cells on minimal medium (Figure 5B). At 37°C, the inhibitory effect was more dramatic. This result indicates that the growth inhibition is probably not caused by an indirect effect of massive overexpression such as depletion of common protein factors.
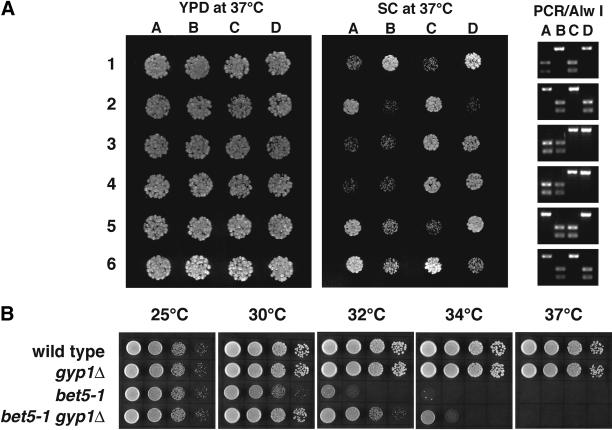
The inhibitory effect of overexpressing Ypt1p on gyp1Δ cells suggests that higher than normal levels of GTP-bound Ypt1p are toxic to the cell. We hypothesize that the growth defect of gyp1Δ at 37°C on synthetic medium may be also caused by a higher than normal level of GTP-bound Ypt1p. A prediction of this hypothesis is that a partial loss-of-function mutation in YPT1 may be able to suppress the growth defect of gyp1Δ. One ypt1 allele suitable to test our hypothesis is ypt1-2 (Bacon et al., 1989). The ypt1-2 mutant does not have a significant growth defect. However, lysates derived from this mutant showed a dramatic defect in a cell-free ER-to-Golgi transport assay. This mutant also showed synthetic negative genetic interactions with other secretory mutants. Therefore, ypt1-2 is a partial loss-of-function allele. We sequenced the ypt1-2 open reading frame and found a single G-to-A point mutation, changing a glycine residue at position 83 to glutamic acid. In most of the small GTPases, the amino acid at this position is a small uncharged residue, either G, V, A, or C. The presence of a negatively charged residue at this position in the Ypt1-2 protein is likely to perturb the protein structure, thereby rendering the protein less active or unstable. The nucleotide change in the ypt1-2 allele also fortuitously abolishes an Alw I restriction site. Therefore, we can distinguish this allele from the wild type by PCR amplification of the YPT1 open reading frame and digestion with Alw I. Wild-type cells give rise to two bands of 380 and 250 bp in agarose gel electrophoresis. The PCR product derived from ypt1-2 cells yielded a single 630-bp band, confirming the loss of the Alw I site. When tetrads from a gyp1Δ/gyp1Δ YPT1/ypt1-2 diploid were dissected, the growth rate at 37°C on synthetic medium showed 2:2 segregation (Figure 6A). All of the slow-growing progeny were gyp1Δ single mutants, whereas all of the fast-growing ones were gyp1Δ ypt1-2 double mutants, indicating that the ypt1-2 mutation does suppress the growth defect of gyp1Δ cells.
Figure 6.
(A) ypt1-2 suppresses the growth defect of gyp1Δ. Tetrads from a gyp1Δ/gyp1Δ YPT1/ypt1-2 diploid (NY2297) were dissected. The same number of cells of each progeny was spotted on YPD and SC plates and incubated at 37°C for 2 d. The 630-bp YPT1 open reading frame from each progeny was amplified by PCR. The PCR products were digested by Alw I. PCR product from YPT1 cells can be digested to produce two bands of 380 and 250 bp. PCR product from ypt1-2 cells cannot be digested. (B) gyp1Δ partially suppresses the growth defect of bet5-1. Tenfold serial dilutions of wild-type (NY1211), gyp1Δ (NY2293), bet5-1 (NY2298), and gyp1Δ bet5-1 (NY2299) were spotted on YPD plates and grown for 36 h at 25, 30, 32, 34, and 37°C.
If Gyp1p acts as a Ypt1p GAP in vivo, we would expect to see genetic interactions between GYP1 and the genes encoding the GEF for Ypt1p. The TRAPP complex was shown recently to have GEF activity on Ypt1p (Wang et al., 2000). Therefore, we crossed a gyp1Δ strain to several temperature-sensitive mutants of TRAPP subunits. We found a mild positive genetic interaction between gyp1Δ and bet5-1 (Figure 6B). The gyp1Δ bet5-1 double mutant has a higher restrictive temperature than the bet5-1 single mutant on YPD medium, suggesting that gyp1Δ can partially suppress the growth defect of bet5-1. We also observed that gyp1Δ can partially suppress a temperature-sensitive bet3 mutant bet3-2 (our unpublished observation).
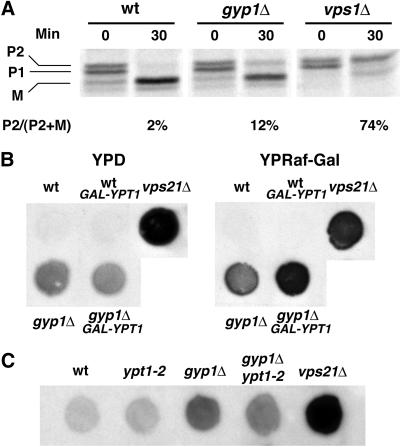
The genetic interaction described above suggests that Gyp1p acts as a negative regulator of Ypt1p function. Ypt1p plays important roles in ER-to-Golgi transport (Segev et al., 1988), intra-Golgi transport (Jedd et al., 1995), and perhaps in post-Golgi transport (Mulholland et al., 1997). The higher level of GTP-bound Ypt1p that we presume to be present in gyp1Δ cells is likely to interfere with some or all of these transport steps. The gyp1Δ strain only exhibits growth defects under particular conditions (synthetic medium at high temperature) and after long incubations, making it difficult to determine the nature of the primary defect. The lack of a growth defect under normal conditions suggests that loss of gyp1 is not likely to severely block the essential exocytic pathway. Indeed, we did not observe any defect in the secretion of invertase from gyp1Δ cells (our unpublished observation). Nor did we see any accumulation of membrane structures when the gyp1Δ cells were grown at 30°C in SC medium and examined by electron microscopy (our unpublished observation). However, when we examined the transport of carboxyl peptidase Y (CPY) by the pulse-chase method, we found that more Golgi form (P2) was present in the gyp1Δ cells at the end of the chase than in the wild-type cells (Figure 7A). This phenotype could be due to the missorting of CPY from the Golgi to the surface, as happens in the vps mutants. To examine whether CPY is secreted from gyp1Δ cells, we performed a CPY overlay assay to measure, in a semiquantitative manner, the amount of CPY secreted. An equal number of cells from different strains was spotted onto a YPD plate and a nitrocellulose membrane was overlayed on top of the plate. The plate was incubated overnight at 30°C, a condition under which gyp1Δ cells grow as well as wild type. The CPY absorbed onto the nitrocellulose membrane was detected by immunoblot with CPY antibody. The gyp1Δ cells secreted more CPY onto the nitrocellulose membrane than did wild-type cells, but not as much as a typical vps mutant, vps21Δ (Figure 7B). When the nitrocellulose membrane was probed with antibody against the cytosolic protein ADH, the same level of minor background signal was detected from each strain, indicating that the higher CPY signal of gyp1Δ cells is not due to cell lysis (our unpublished observation). Because the growth defect of gyp1Δ is probably caused by a higher than normal level of GTP-bound Ypt1p in the cell, we examined whether this mild Vps phenotype is also related to excess Ypt1p function. We introduced into gyp1Δ and wild-type cells a plasmid expressing Ypt1p under the control of the GAL promoter. On YP-raffinose-galactose medium that induces the expression of Ypt1p, more CPY was secreted from the YPT1 plasmid transformant of gyp1Δ cells than from the vector transformant (Figure 7B). Wild-type cells were not affected by the overexpression of Ypt1p. Therefore, analogous to the effect of Ypt1p overexpression on the growth phenotype of gyp1Δ, overexpression of Ypt1p also enhances the Vps phenotype of gyp1Δ cells. Furthermore, we found that the ypt1-2 mutation can partially suppress the Vps phenotype of gyp1Δ cells (Figure 7C), indicating that the missorting of CPY is also caused by the higher level of active Ypt1p in the gyp1Δ cells.
Figure 7.
gyp1Δ missorts CPY to the cell surface. (A) Cells were labeled with 35S for 10 min at 30°C in minimal medium and chased with unlabeled methionine and cysteine for 30 min. CPY was immunoprecipitated from total lysates, separated on SDS-PAGE, and quantified by phosphorimaging. P1, ER form; P2, Golgi form; M, mature form. (B) Overexpression of Ypt1p enhances the CPY missorting phenotype of gyp1Δ. The same number of cells was spotted on YPD and YP-raffinose-galactose plates. Wet nitrocellulose membranes were overlaid on top of the plates. The plates were incubated at 30°C overnight. The amount of CPY absorbed on the membrane was determined by Western blot. (C) ypt1-2 suppresses the CPY missorting phenotype of gyp1Δ. The same number of cells was spotted on YPD plate at 30°C. CPY secretion was determined by the overlay assay.
DISCUSSION
The nucleotide state of small GTPases is determined by the combined action of their GEFs and GAPs. The subcellular localization of these regulatory proteins can play an important role in the function of the GTPases. A classic example is the regulation of nucleocytoplasmic transport by the Ran GTPase cycle (Azuma and Dasso, 2000). The regulatory proteins of Ran have restricted subcellular localizations: RanGAP is localized in the cytosol, whereas RanGEF is a nuclear protein. The localization of these proteins establishes an asymmetric distribution of Ran-GTP and Ran-GDP to the nucleus and cytoplasm, respectively, and thereby determines the directionality of nuclear transport. Another example can be found in the study of yeast spindle position checkpoint (reviewed by Hoyt, 2000). Bub2p and Bfa1p together form a two-component GAP for Tem1p, a small GTPase involved in exit from mitosis. Tem1p, Bub2p, and Bfa1p bind each other and localize on the cytoplasmic face of the spindle pole body (SPB) (Pereira et al., 2000). By contrast, Lte1p, a GEF for Tem1p, is associated with the cortex of the bud. Early in mitosis, Bub2p/Bfa1p GAP keeps SPB-localized Tem1p in a GDP-bound state. Migration of the SPB into the bud puts Tem1p in proximity to its GEF, Lte1p, resulting in the exchange of GDP for GTP and activation of Tem1p function. The fact that Bub2p is related in amino acid sequence to Gyp1p and the other yeast Rab GAPs raises the intriguing possibility that they might also function to spatially regulate the activation of their GTPase substrates.
Our results show that Gyp1p is predominantly membrane bound and is associated with the Golgi complex. Because the Rab GTPases are also attached to membranes and localize to specific membrane compartments, the colocalization of GTPases and their regulatory proteins to the same membrane compartment could significantly increase the efficiency of the nucleotide exchange or hydrolysis reactions. For example, the Sec4p GEF Sec2p colocalizes with Sec4p on post-Golgi secretory vesicles (Walch-Solimena et al., 1997). The C-terminal domain of Sec2p is required for its localization (Elkind et al., 2000). Deletion of the C-terminal domain does not affect the in vitro GEF activity but renders the cell temperature sensitive for growth and secretion.
Localization of Gyp1p to Golgi implies that the yeast Rab GAPs could be more specific in vivo than their in vitro substrate specificity suggests. Among the known in vitro substrates of Gyp1p, Sec4p, Ypt1p, Ypt7p, and Ypt51p, only Ypt1p localizes predominantly to the Golgi (Segev et al., 1988; Wichmann et al., 1992; Brennwald and Novick, 1993; Gerrard et al., 2000). Therefore, it is very likely that in vivo Gyp1p primarily acts on the GTPase that colocalizes with it, Ypt1p. Enhancement of in vivo specificity through compartmentalization may be a common property of this Rab GAP family. Two closely related proteins of this family, Msb3p and Msb4p, localize to the presumptive bud site, the bud tip, and the mother-bud neck (Bi et al., 2000). These are sites of Sec4p localization. Therefore, in spite of the lack of in vitro specificity of Msb3p, it has been proposed that Msb3p could be a Sec4p-specific GAP in vivo (Albert and Gallwitz, 1999).
The gyp1Δ strain does not exhibit a growth defect in rich medium (Du et al., 1998). However, we describe here a slow growth phenotype of gyp1Δ cells in synthetic medium at 37°C. Because we can observe this defect in synthetic complete medium, it is unlikely to be caused by the absence of a specific nutrient. It has been previously noted that synthetic complete medium can enhance phenotypes that are less noticeable in rich medium (Hampsey, 1997). The biological processes affected by the loss of GYP1 are probably more critical or rate limiting when the cells are grown in synthetic medium and at higher than normal temperature. In support of this idea, we have isolated mutants dependent on GYP1 for their growth in rich medium at room temperature (our unpublished data), indicating that Gyp1p also functions under normal growth conditions. The growth phenotype of gyp1Δ cells has also been examined in a genome-scale deletion study (Winzeler et al., 1999) (http://sequence-www.stanford.edu/group/yeast/yeast_deletion_project/Enter_function.html). In that study, the growth rates of homozygous diploid null mutants of 558 nonessential genes were examined in a competitive growth assay. Ten strains were classified as class 0, i.e., they grew at <80% of the average rate in minimal medium at 30°C, but at >95% in rich medium. Eight of them are strains carrying a deletion of a gene involved in amino acid synthesis. The other two strains are gyp1Δ (78% of average rate in minimum medium, 99% in rich) and vps29/pep11Δ (69% in minimum, 100% in rich). Vps29p is a member of the retromer complex essential for the recycling of CPY receptor Vps10p from endosomes to Golgi (Seaman et al., 1998). Strains containing a deletion of several other vps genes also showed significant but weaker minimal medium growth defects in this competitive growth assay; for example, vps5Δ (91% in minimal, 100% in rich), vps8Δ (90% in minimal, 99% in rich), and vps21Δ (91% in minimal, 98% in rich). The gyp1Δ strain has a stronger growth defect but a much weaker CPY missorting defect than these vps mutants, suggesting that the growth phenotype cannot be solely attributed to the missorting of vacuolar proteins.
The identification of a set of conditions that impairs the growth of gyp1Δ cells has allowed us to formally establish that the GAP activity of Gyp1p is required for its in vivo function. The minimal region of Gyp1p that can fully complement the growth defect is approximately the same minimal region sufficient for catalytic activity. Mutation of the catalytically essential arginine residue totally abolished the complementation activity, strongly suggesting that the GAP activity is required for the in vivo function of Gyp1p. This is the first evidence showing that a member of this Rab GAP family actually acts as a GAP in vivo.
We have made three key genetic observations that address the function of Gyp1p. First, we have shown that overexpression of Ypt1p, but not other Rab GTPases, inhibits the growth of gyp1Δ cells. Second, we found that a partial loss-of-function allele of ypt1 suppresses the growth defect of gyp1Δ cells. Finally, we demonstrated that the deletion of gyp1 partially suppresses the growth defect of temperature-sensitive mutants of TRAPP, the GEF for Ypt1p. These genetic data, taken together, suggest that Gyp1p acts on Ypt1p in vivo, a conclusion that is fully consistent with our localization data. Furthermore, the results establish that the in vivo function of Gyp1p is to down-regulate the activity of Ypt1p.
Although it is very likely that Ypt1p is the primary substrate of Gyp1p in vivo, we cannot completely rule out the possibility that Gyp1p may also act on other Rab GTPases such as Ypt51p and Ypt7p. These GTPases are involved in the endosomal/vacuolar system and are not essential for the growth of yeast. Our localization data does not exclude a minor localization of Gyp1p to an intracellular compartment other than Golgi. The genetic test with overexpression constructs based on growth phenotype, therefore, may not be able to detect interactions with the nonessential GTPases.
The genetic interactions that we found between GYP1, YPT1, and TRAPP are analogous to the genetic interactions between Ras and its regulators (Tanaka et al., 1989, 1990). Ira1p and Ira2p are the GAPs for yeast Ras proteins Ras1p and Ras2p. Cdc25p is the Ras GEF. A ras2 null mutation suppresses the sporulation deficiency and heat shock sensitivity phenotypes of both ira1 and ira2 mutations. Disruption of either IRA1 or IRA2 suppresses the lethality of the cdc25 null mutation.
The function of Gyp1p as a negative regulator of Ypt1p implies that the GTP hydrolysis step catalyzed by Gyp1p may not be required for the biological function of Ypt1p, but rather serves to inactivate Ypt1p. Such a role for the GTP hydrolysis mediated by a Rab GTPase has also been proposed based on a study with a xanthosine 5′-triphosphate (XTP) binding mutant of Rab5 (Rybin et al., 1996). That study found that GTP hydrolysis by Rab5 is not required for membrane fusion but for the maintenance of a steady-state level of GTP-bound Rab5.
The synthetic medium growth defect and CPY missorting defect of gyp1Δ cells indicate that maintaining an optimal level of GTP-bound Ypt1p is important for the cell. At present, we do not know the mechanism by which the elevated level of active Ypt1p leads to these defects. Neither of these defects is observed when Ypt1p is overexpressed in wild-type cells, suggesting that the amount of Gyp1p in the cell is not limiting.
The role of GTP hydrolysis by Ypt1p has been previously studied using a GTPase-deficient mutant, ypt1-Q67L (Richardson et al., 1998). A strain with ypt1-Q67L as the only YPT1 gene has almost no growth defect. Overexpressing Ypt1-Q67L protein in wild-type cells has no effect on growth or secretion. Therefore, the authors concluded that GTP hydrolysis is not essential either for Ypt1p-mediated vesicular transport or as a timer to turn off Ypt1p-mediated membrane fusion. It was shown in the same study that Ypt1-Q67L protein has defects in both prenylation and membrane attachment. These defects may offset the effect of reduced GTP hydrolysis. Another possibility is that the Q67L mutation may affect the interaction between Ypt1p and its downstream effectors, rendering the GTP-bound Ypt1-Q67L protein less active and hence less toxic than the wild-type protein. In fact, we found that overexpressing Ypt1-Q67L protein does not affect the growth of even the gyp1Δ cells (our unpublished data).
Proteins sharing the RabGAP domain with Gyp1p are widespread in different organisms. More than 100 proteins containing this domain have been identified (InterPro Entry IPR000195, http://www.ebi.ac.uk/interpro/). Gyp1p itself is highly conserved. Its orthologs exist in Schizosaccharomyces pombe, Caenorhabditis elegans, Drosophila melanogaster, and Arabidopsis thaliana. In fact, the similarity between Gyp1p and its orthologs in other species is much higher than the similarity between Gyp1p and the other yeast Rab GAPs. For example, the RabGAP domain of a human homolog (accession number AL096779) shares 48% identity with Gyp1p (excluding a nonconserved loop region in Gyp1p). In contrast, the closest homolog in yeast, Gyp7p, shares <25% identity with Gyp1p in the same region. It is quite likely that the orthologs of Gyp1p in other species carry out functions similar to those of Gyp1p.
ACKNOWLEDGMENTS
We are grateful to Dr. Susan Ferro-Novick for advice and generously providing plasmids, antibodies, and yeast strains. We thank Jemima Barrowman and Eric Grote for the help with the sucrose gradient experiment. We thank Drs. T.H. Stevens, C.W. Slayman, and R. Piper for plasmids and antibodies. We appreciate the critical reading of the manuscript by Drs. Wei Guo and Eric Grote. This work was supported by grants from the National Institutes of Health to P.N.
Abbreviations used:
- CEN
centromere
- GAP
GTPase-activating protein
- GEF
guanine nucleotide exchange protein
- GFP
green fluorescent protein
- RFP
red fluorescent protein
REFERENCES
- Albert S, Gallwitz D. Two new members of a family of Ypt/Rab GTPase activating proteins. Promiscuity of substrate recognition. J Biol Chem. 1999;274:33186–33189. doi: 10.1074/jbc.274.47.33186. [DOI] [PubMed] [Google Scholar]
- Albert S, Gallwitz D. Msb4p, a protein involved in Cdc42p-dependent organization of the actin cytoskeleton, is a Ypt/Rab-specific GAP. Biol Chem. 2000;381:453–456. doi: 10.1515/BC.2000.059. [DOI] [PubMed] [Google Scholar]
- Albert S, Will E, Gallwitz D. Identification of the catalytic domains and their functionally critical arginine residues of two yeast GTPase-activating proteins specific for Ypt/Rab transport GTPases. EMBO J. 1999;18:5216–5225. doi: 10.1093/emboj/18.19.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y, Dasso M. The role of Ran in nuclear function. Curr Opin Cell Biol. 2000;12:302–307. doi: 10.1016/s0955-0674(00)00093-4. [DOI] [PubMed] [Google Scholar]
- Bacon RA, Salminen A, Ruohola H, Novick P, Ferro-Novick S. The GTP-binding protein Ypt1 is required for transport in vitro: the Golgi apparatus is defective in ypt1 mutants. J Cell Biol. 1989;109:1015–1022. doi: 10.1083/jcb.109.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfield DK, Lewis MJ, Rabouille C, Warren G, Pelham HR. Localization of Sed5, a putative vesicle targeting molecule, to the cis-Golgi network involves both its transmembrane and cytoplasmic domains. J Cell Biol. 1994;127:357–371. doi: 10.1083/jcb.127.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrowman J, Sacher M, Ferro-Novick S. TRAPP stably associates with the Golgi and is required for vesicle docking. EMBO J. 2000;19:862–869. doi: 10.1093/emboj/19.5.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becherer KA, Rieder SE, Emr SD, Jones EW. Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol Biol Cell. 1996;7:579–594. doi: 10.1091/mbc.7.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Chiavetta JB, Chen H, Chen GC, Chan CS, Pringle JR. Identification of novel, evolutionarily conserved Cdc42p-interacting proteins and of redundant pathways linking Cdc24p and Cdc42p to actin polarization in yeast. Mol Biol Cell. 2000;11:773–793. doi: 10.1091/mbc.11.2.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles E, Miosga T. A rapid and highly efficient method for PCR-based site-directed mutagenesis using only one new primer. Curr Genet. 1995;28:197–198. doi: 10.1007/BF00315788. [DOI] [PubMed] [Google Scholar]
- Brennwald P, Novick P. Interactions of three domains distinguishing the Ras-related GTP-binding proteins Ypt1 and Sec4. Nature. 1993;362:560–563. doi: 10.1038/362560a0. [DOI] [PubMed] [Google Scholar]
- Cuif MH, Possmayer F, Zander H, Bordes N, Jollivet F, Couedel-Courteille A, Janoueix-Lerosey I, Langsley G, Bornens M, Goud B. Characterization of GAPCenA, a GTPase activating protein for Rab6, part of which associates with the centrosome. EMBO J. 1999;18:1772–1782. doi: 10.1093/emboj/18.7.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du LL, Collins RN, Novick PJ. Identification of a Sec4p GTPase-activating protein (GAP) as a novel member of a Rab GAP family. J Biol Chem. 1998;273:3253–3256. doi: 10.1074/jbc.273.6.3253. [DOI] [PubMed] [Google Scholar]
- Elkind NB, Walch-Solimena C, Novick PJ. The role of the COOH terminus of Sec2p in the transport of post-Golgi vesicles. J Cell Biol. 2000;149:95–110. doi: 10.1083/jcb.149.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui K, Sasaki T, Imazumi K, Matsuura Y, Nakanishi H, Takai Y. Isolation and characterization of a GTPase activating protein specific for the Rab3 subfamily of small G proteins. J Biol Chem. 1997;272:4655–4658. doi: 10.1074/jbc.272.8.4655. [DOI] [PubMed] [Google Scholar]
- Gerrard SR, Bryant NJ, Stevens TH. VPS21 controls entry of endocytosed and biosynthetic proteins into the yeast prevacuolar compartment. Mol Biol Cell. 2000;11:613–626. doi: 10.1091/mbc.11.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan B, Bowser R, Novick P. The role of Myo2, a yeast class V myosin, in vesicular transport. J Cell Biol. 1995;128:1055–1068. doi: 10.1083/jcb.128.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote E, Novick PJ. Promiscuity in Rab-SNARE interactions. Mol Biol Cell. 1999;10:4149–4161. doi: 10.1091/mbc.10.12.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M. A review of phenotypes in Saccharomyces cerevisiae. Yeast. 1997;13:1099–1133. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1099::AID-YEA177>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Hoyt MA. Exit from mitosis: spindle pole power. Cell. 2000;102:267–270. doi: 10.1016/s0092-8674(00)00031-3. [DOI] [PubMed] [Google Scholar]
- Jedd G, Richardson C, Litt R, Segev N. The Ypt1 GTPase is essential for the first two steps of the yeast secretory pathway. J Cell Biol. 1995;131:583–590. doi: 10.1083/jcb.131.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EW. Tackling the protease problem in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:428–453. doi: 10.1016/0076-6879(91)94034-a. [DOI] [PubMed] [Google Scholar]
- Jones S, Richardson CJ, Litt RJ, Segev N. Identification of regulators for Ypt1 GTPase nucleotide cycling. Mol Biol Cell. 1998;9:2819–2837. doi: 10.1091/mbc.9.10.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar T, Gotte M, Gallwitz D. Vesicular transport: how many Ypt/Rab-GTPases make a eukaryotic cell? Trends Biochem Sci. 1997;22:468–472. doi: 10.1016/s0968-0004(97)01150-x. [DOI] [PubMed] [Google Scholar]
- Liu K, Li G. Catalytic domain of the p120 Ras GAP binds to RAb5 and stimulates its GTPase activity. J Biol Chem. 1998;273:10087–10090. doi: 10.1074/jbc.273.17.10087. [DOI] [PubMed] [Google Scholar]
- Mulholland J, Wesp A, Riezman H, Botstein D. Yeast actin cytoskeleton mutants accumulate a new class of Golgi-derived secretary vesicle. Mol Biol Cell. 1997;8:1481–1499. doi: 10.1091/mbc.8.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Neuwald AF. A shared domain between a spindle assembly checkpoint protein and Ypt/Rab-specific GTPase-activators. Trends Biochem Sci. 1997;22:243–244. doi: 10.1016/s0968-0004(97)01073-6. [DOI] [PubMed] [Google Scholar]
- Novick P, Zerial M. The diversity of Rab proteins in vesicle transport. Curr Opin Cell Biol. 1997;9:496–504. doi: 10.1016/s0955-0674(97)80025-7. [DOI] [PubMed] [Google Scholar]
- Pereira G, Hofken T, Grindlay J, Manson C, Schiebel E. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol Cell. 2000;6:1–10. [PubMed] [Google Scholar]
- Rak A, Fedorov R, Alexandrov K, Albert S, Goody RS, Gallwitz D, Scheidig AJ. Crystal structure of the GAP domain of Gyp1p: first insights into interaction with Ypt/Rab proteins. EMBO J. 2000;19:5105–5113. doi: 10.1093/emboj/19.19.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CJ, Jones S, Litt RJ, Segev N. GTP hydrolysis is not important for Ypt1 GTPase function in vesicular transport. Mol Cell Biol. 1998;18:827–838. doi: 10.1128/mcb.18.2.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybin V, Ullrich O, Rubino M, Alexandrov K, Simon I, Seabra MC, Goody R, Zerial M. GTPase activity of Rab5 acts as a timer for endocytic membrane fusion [see comments] Nature. 1996;383:266–269. doi: 10.1038/383266a0. [DOI] [PubMed] [Google Scholar]
- Sacher M, Jiang Y, Barrowman J, Scarpa A, Burston J, Zhang L, Schieltz D, Yates JR, 3rd, Abeliovich H, Ferro-Novick S. TRAPP, a highly conserved novel complex on the cis-Golgi that mediates vesicle docking and fusion. EMBO J. 1998;17:2494–2503. doi: 10.1093/emboj/17.9.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder M, Schafer R, Friedl P. Spectrophotometric determination of iodixanol in subcellular fractions of mammalian cells. Anal Biochem. 1997;244:174–176. doi: 10.1006/abio.1996.9861. [DOI] [PubMed] [Google Scholar]
- Seaman MN, McCaffery JM, Emr SD. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J Cell Biol. 1998;142:665–681. doi: 10.1083/jcb.142.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev N, Mulholland J, Botstein D. The yeast GTP-binding YPT1 protein and a mammalian counterpart are associated with the secretion machinery. Cell. 1988;52:915–924. doi: 10.1016/0092-8674(88)90433-3. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Strom M, Vollmer P, Tan TJ, Gallwitz D. A yeast GTPase-activating protein that interacts specifically with a member of the Ypt/Rab family. Nature. 1993;361:736–739. doi: 10.1038/361736a0. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Matsumoto K, Toh EA. IRA1, an inhibitory regulator of the RAS-cyclic AMP pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:757–768. doi: 10.1128/mcb.9.2.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Nakafuku M, Tamanoi F, Kaziro Y, Matsumoto K, Toh-e A. IRA2, a second gene of Saccharomyces cerevisiae that encodes a protein with a domain homologous to mammalian ras GTPase-activating protein. Mol Cell Biol. 1990;10:4303–4313. doi: 10.1128/mcb.10.8.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer P, Will E, Scheglmann D, Strom M, Gallwitz D. Primary structure and biochemical characterization of yeast GTPase-activating proteins with substrate preference for the transport GTPase Ypt7p. Eur J Biochem. 1999;260:284–290. doi: 10.1046/j.1432-1327.1999.00192.x. [DOI] [PubMed] [Google Scholar]
- Walch-Solimena C, Collins RN, Novick PJ. Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J Cell Biol. 1997;137:1495–1509. doi: 10.1083/jcb.137.7.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walworth NC, Brennwald P, Kabcenell AK, Garrett M, Novick P. Hydrolysis of GTP by Sec4 protein plays an important role in vesicular transport and is stimulated by a GTPase-activating protein in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2017–2028. doi: 10.1128/mcb.12.5.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Sacher M, Ferro-Novick S. TRAPP stimulates guanine nucleotide exchange on ypt1p. J Cell Biol. 2000;151:289–296. doi: 10.1083/jcb.151.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann H, Hengst L, Gallwitz D. Endocytosis in yeast: evidence for the involvement of a small GTP-binding protein (Ypt7p) Cell. 1992;71:1131–1142. doi: 10.1016/s0092-8674(05)80062-5. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Xiao GH, Shoarinejad F, Jin F, Golemis EA, Yeung RS. The tuberous sclerosis 2 gene product, tuberin, functions as a Rab5 GTPase activating protein (GAP) in modulating endocytosis. J Biol Chem. 1997;272:6097–6100. doi: 10.1074/jbc.272.10.6097. [DOI] [PubMed] [Google Scholar]