Abstract
In bacteria, the sodium ion (Na+) cycle plays a critical role in negotiating the challenges of an extremely alkaline and sodium-rich environment. Alkaliphilic bacteria that grow optimally at high pH values use Na+ for solute uptake and flagellar rotation because the proton (H+) motive force is insufficient for use at extremely alkaline pH. Only three types of electrically driven rotary motors exist in nature: the F-type ATPase, the V-type ATPase, and the bacterial flagellar motor. Until now, only H+ and Na+ have been reported as coupling ions for these motors. Here, we report that the alkaliphilic bacterium Bacillus alcalophilus Vedder 1934 can grow not only under a Na+-rich and potassium ion (K+)-poor condition but also under the opposite condition in an extremely alkaline environment. In this organism, swimming performance depends on concentrations of Na+, K+ or Rb+. In the absence of Na+, swimming behavior is clearly K+- dependent. This pattern was confirmed in swimming assays of stator-less Bacillus subtilis and Escherichia coli mutants expressing MotPS from B. alcalophilus (BA-MotPS). Furthermore, a single mutation in BA-MotS was identified that converted the naturally bi-functional BA-MotPS to stators that cannot use K+ or Rb+. This is the first report that describes a flagellar motor that can use K+ and Rb+ as coupling ions. The finding will affect the understanding of the operating principles of flagellar motors and the molecular mechanisms of ion selectivity, the field of the evolution of environmental changes and stresses, and areas of nanotechnology.
Introduction
Bacterial flagella act as rigid propellers for cell locomotion in liquid and on surfaces, and their rotation is powered by electrochemical gradients of either protons (H+) or sodium ions (Na+) across the bacterial cytoplasmic membrane [1], [2]. The flagellar structure is composed of a helical filament, a basal body that is embedded in the cytoplasmic membrane, and a hook that connects the filament to the basal body. The motor is divided into two parts: the rotor and the stator. The stators are most often called Mot complexes which function as ion channels and act as the energy conversion unit which rotates a flagellar rotor when coupling ions pass through the Mot complexes. Mot complexes have been shown to contain four MotA-like proteins, each of which contains four transmembrane segments (TMS), and two MotB-like proteins, each of which contains a single TMS [1]–[3]. The MotB-like proteins have a major role in determining ion-coupling specificity [4], [5]. In most bacteria H+-coupled motility is widely documented. However, the work of Imae and colleagues in the 1980s revealed that extremely alkaliphilic Bacillus species use only Na+ gradients for motility [6], [7]. Evidence from an increasing number of bacteria indicates that they utilise dual flagellar systems that have different ion specificities that contribute to swimming under particular physical-chemical conditions, such as pH, salinity and viscosity [8]–[12]. Therefore, although extremely alkaliphilic Bacillus species contain only sodium-coupled MotPS and Escherichia coli contains only proton-coupled MotAB, salt-tolerant Vibrio alginolyticus possesses proton-coupled MotAB and sodium-coupled PomAB, and Bacillus subtilis possesses proton-coupled MotAB and sodium-coupled MotPS [13]. In 2008, we reported that alkaliphilic Bacillus clausii KSM-K16 utilises a novel form of dual-ion coupling for motility, in which a single MotAB stator uses both sodium and protons at different pH ranges [14]. Deductions from studies of alignments of the MotB and MotS subunits of sodium- and proton-coupled motility systems facilitated conversion of single-ion coupled stators to dual use and the B. clausii dual-ion stator to single-ion use of either sodium or protons [14]. Recently, Schlegel et al reported that ATP synthesis in Methanosarcina acetivorans can be driven by the H+ or Na+ gradients [15]. No stators that can use potassium ions, which is reported here for the MotPS system of alkaliphilic Bacillus alcalophilus Vedder 1934, have previously been described. This strain, which was isolated from human feces in 1934, is a classic alkaliphilic strain because it was among the very first alkaliphilic “extremophiles” to be described in the literature [16].
The Bacillus alcalophilus Vedder 1934 genome sequence was not yet accessible at the start of this project (BioProject accession number PRJNA13375). Therefore, the uncharacterised flagellar stator genes from this strain were amplified by PCR using primers that were based on the conserved sequences among the motAB and motPS genes of Bacillus species. The sequencing of each PCR product revealed that a single set of genes encoded a MotPS-like pair of proteins (GenBank accession JN561694). Subsequently, one of us (MI) participated in a B. alcalophilus Vedder 1934 genome sequencing project demonstrated that, in fact, only a single mot system is present (This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession ALPT00000000. The version described in this paper is the first version, ALPT01000000). We use the designation “BA-MotPS” for this Mot stator. The BA-MotP and MotS proteins were closely related to the B. subtilis MotP and MotS proteins that constitute the sodium-coupled B. subtilis stator complex (49% and 48% identity and 75% and 68% similarity, respectively). They less closely resembled the B. subtilis MotA and MotB proteins that constitute a proton-coupled Bacillus subtilis stator complex (39% and 29% identity and 61% and 55% similarity, respectively). The swimming behaviour of B. alcalophilus Vedder 1934 was also studied under several conditions. Since this strain is not genetically accessible, we used stator-less mutant strains of B. subtilis (ΔmotAB ΔmotPS) and Escherichia coli RP6894 (ΔmotAB) and a potassium uptake-deficient (Δ(kdpABC) trkΔ1 ΔtrkA) mutant strain of E. coli TK2420 as the host for experiments in which the functional properties of BA-MotPS were studied. We were also able to identify mutational changes in BA-MotS that converted the bi-functional BA-MotPS stator to one that cannot use K+ or Rb+ when the sodium concentration is very low as the native BA-MotPS can.
Materials and Methods
Bacterial strains and growth conditions
The strains and plasmids used in this study are shown in Table 1. Bacillus alcalophilus Vedder 1934 cells and Bacillus pseudofirmus OF4 811MM were grown aerobically at 30°C in malate–yeast extract medium (MYE) at pH 10.5 [17] or KMYE medium. MYE medium contained 50 mM Na2CO3, 1 mM K2HPO4, 0.1 mM MgSO4, 0.01%(w/v) yeast extract, 2.5 mM sodium malate (pH 9.0), and 1%(v/v) trace elements per liter of deionized water (pH 10.5). A similar medium containing 50 mM Na2HPO4 instead of Na2CO3 was occasionally used as MYE medium (pH 7.5), and 50 mM K2CO3 instead of Na2CO3 was occasionally used as KMYE medium (pH 10.5), and KMYE medium containing 50 mM K2HPO4 was occasionally used instead of K2CO3 as KMYE medium (pH 7.5). For assays of swimming speed in liquid, 30 mM Tris-HCl (pH 9.0) containing 5 mM glucose plus the indicated amounts of sodium, potassium or rubidium were used at the indicated values of pH and added EIPA (5-(N-ethyl-N-isopropyl)-amiloride). The actual final Na+ and K+ concentration of each assay medium was determined with a flame photometer (Model AMA-175, Tokyo Koden, Tokyo, Japan) calibrated with standard Na+ solutions of known concentrations. The effects of increasing concentrations of KCl on the growth of E. coli TK2420 were determined as described previously [18] in a defined medium [19]. Bacillus subtilis 168 strain BR151MA (wild type), its derivatives and E. coli strain RP6894 were grown at 37°C in LB medium. E. coli strain TK2420 was grown at 37°C in LBK medium. When necessary, a particular medium was supplemented with erythromycin (0.3 µg/ml), neomycin (7.5 µg/ml), chloramphenicol (5 µg/ml) or ampicillin (100 µg/ml). For assays of swimming speed in liquid, 10 mM phosphate buffer that contained 5 mM glucose and the indicated amounts of sodium and potassium was used at the indicated pH values. For growth assays of E. coli strain TK2420 cells, TK2420 medium (pH 7.0) consisted of 33.6 mM Na2HPO4, 20.3 mM NaH2PO4, 1.1 mM citric acid, 7.6 mM (NH4)2SO4, 6 µM FeSO4, 830 µM MgSO4, 10 mM glucose, 1 µg/ml thiamine, 100 µg/ml ampicillin and 0.06% arabinose was used with the indicated amounts of potassium. The cells were grown at 37°C with shaking, and their growth was monitored by measuring the absorbance at 600 nm.
Table 1. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Description | Source or reference |
| E. coli Strain | ||
| DH5αMCR | F− mcrAΔ1 (mrr-hsd RMS-mcrBC) Φ80dlacZ Δ(lacZYAargF) U169 deoR recA1 endA1 supE44 λthi-1 gyr-496 relA1 | Stratagene |
| TK2420 | F− thi rha lacZ nagA Δ(kdpFAB) Δ(trk-mscL) trkD1 | [49] |
| RP6894 | ΔmotAB | [48] |
| Bacillus subtilis strains | ||
| BR151MA | lys3 trpC2 (wild type) | [50] |
| ΔABΔPS | lys3 trpC2 ΔmotAB::ery ΔmotPS::neo | [13] |
| BA-PS | ΔABΔPS amyE::PmotAB-motAB from Vedder 1934 | This study |
| BA-PS-MotS_M33L | BA-PS introduced a mutation in MotS_M33L | This study |
| Bacillus alcalophilus strain | ||
| Vedder 1934 | wild type, ( = JCM5652) | [16] |
| Bacillus pseudofirmus OF4 strains | ||
| 811M | Met− (wild type) | [51] |
| 811MM | Met− up-motile variant | [45], [52] |
| Plasmids | ||
| pDR67 | amyE integration vector with Cmr gene and Pspac promoter upstream of multiple cloning site | [53] |
| pBAD24 | Expression vector containing PBAD promoter | [54] |
| pDR-PS | pDR67+PmotAB-motPS from Vedder 1934 | This study |
| pDR-PS-MotS_M33L | pDR-PS introduced a mutation in MotS_M33L | This study |
| pBAPS | pBAD24+motPS from Vedder 1934 | This study |
| pBAPS-MotS_M33L | pBAPS introduced a mutation in MotS_M33L | This study |
Construction of plasmid coding B. alcalophilus motPS under B. subtilis PmotAB promoter and its point mutations
For cloning of the B. alcalophilus motPS gene under the B. subtilis PmotAB promoter, we performed gene splicing by overlap extension (gene SOEing) method based on the PCR method. The primers were synthesized based on sequences from B. subtilis and B. alcalophilus and are listed in Table 2. The B. subtilis PmotAB promoter was amplified by PCR using B. subtilis chromosomal DNA as the template with BSAB-BamHI-F primers designed for a BamHI site, and BSpAB-BAPS-R primers introduced a sequence of the N-terminal region of the B. alcalophilus motPS gene. The B. alcalophilus motPS gene was amplified by PCR using B. alcalophilus chromosomal DNA as the template with BSpAB-BAPS-F primers introducing a sequence of the B. subtilis PmotAB promoter and BAPS-SphI-R. These two products share an overlap region. We performed PCR with these two products as the template to join the B. subtilis PmotAB promoter and the B. alcalophilus motPS gene. The amplified fragment was digested by BamHI and SphI and cloned into BamHI and SphI-digested pDR67, yielding pDRPS. To introduce an amino acid substitution into the B. alcalophilus motS gene, we used the gene SOEing method. We synthesized pairs of mutant primers to the sense strand of the B. alcalophilus motS gene, with a mismatch at the mutation site. We amplified the B. alcalophilus motPS under control of a B. subtilis PmotAB promoter by PCR with mutant primers, and cloned it into BamHI and SphI-digested pDR67, yielding pDRPS -MotS_M33L. The presence of the motS mutation was confirmed by DNA sequencing. Each plasmid was used to transform particular mutants to a chloramphenicol-resistance, amylase-negative phenotype for the pDR67 derivative. Recombinant transformants were selected by conventional techniques, and the presence of the insert was confirmed.
Table 2. Oligonucleotides used in this study.
| Primer | Sequence | Accession number and corresponding sequence (nt) |
| BSAB-BamHI-F | 5′-cgcggaTCCGCGACCTCTTGCTTCCGACA-3′ | Z99111 (23924–23902 (minus strand)) |
| BSpAB-BAPS-F | 5′-AAAAAAGGATTTGGTGAAAACTATGAAAAAGCTAGATCTGATGA-3′ | Z99111 (23633–23609 (minus strand)) |
| JN561694 (1458–1480) | ||
| BSpAB-BAPS-R | 5′-TCATCAGATCTAGCTTTTTCATAGTTTTCACCAAATCCTTTTTT-3′ | JN561694 (1480–1458 (minus strand)) |
| Z99111 (23609–23633) | ||
| BAPS-SphI-R | 5′-acAtgcAtgCCCAAGGAATATTGTCTCCTCG-3′ | JN561694 (3020–2999 (minus strand)) |
| PS(BA)-S-M33L-F | 5′-ATGACGTTGATTTTAGTGTTTTTTGTTcTGTTATTTTCGATGTCAGAAAT-3′ | JN561694 (2326–2375) |
| PS(BA)-S-M33L-R | 5′-AAAAAACACTAAAATCAACGTCAT-3′ | JN561694 (2349–2326 (minus strand)) |
| BAPS-NcoI-F | 5′-catGccatggATGAAAAAGCTAGATCTGATGACA-3′ | JN561694 (1458–1481) |
| BAPS-PstI-R | 5′-aaaaCtGcagTTAATAAGTATCATCCTCTTCATA-3′ | JN561694 (2994–2971 (minus strand)) |
Extra nucleotides that were added to introduce restriction sites are underlined.
Substituted nucleotides that were added to introduce point mutation sites are shown by a small letter.
For cloning of the B. alcalophilus motPS genes under an arabinose PBAD promoter of the pBAD24 plasmid, the primers digested by NcoI and PstI were synthesized based on sequences from B. alcalophilus. The B. alcalophilus motPS genes were amplified by PCR using pDRPS and pDRPS-MotS_M33L as the template. These amplified fragments were cloned into NcoI and PstI-digested pBAD24, yielding pBAPS and pBAPS -MotS_M33L.
Measurement of intracellular potassium and sodium concentration
E. coli TK2420 cells were grown at 37°C or 10 hour in TK2420 medium contained 25 mM or 50 mM KCl. The cells were harvested by centrifugation (3,000×g, 10 min, 25°C) and washed by suspension in the same medium. Then, the cells were resuspended in 5 ml of 300 mM sucrose solution, and the cell protein was measured by the Lowry method using 100 µl of the cell suspension. The rest of the suspension was harvested and resuspended in 5 ml of 5% trichloroacetic acid (TCA) solution. After being boiled for 10 min, cell debris was removed by centrifugation and potassium and sodium were measured by atomic absorption spectrometry (Model Z-8000, Hitachi, Japan) that was calibrated with standard potassium and sodium solutions of known concentrations. The intracellular concentration was calculated with an assumed cell volume of 3 µl/mg cell protein [20], [21].
Measurement of swimming speed
For measurement of swimming speed, B. alcalophilus Vedder 1934 cells were grown aerobically on KMYE medium (pH 10.5) overnight at 30°C. The culture was then inoculated into 20 ml of a fresh KMYE medium (pH 10.5) at an A600 of 0.01, and grown aerobically at 30°C. Growth of B. pseudofirmus OF4 811MM cells used MYE medium under the same conditions. Highly motile cells in the late logarithmic phase were harvested by filtration on OMNIPORE membrane filters (0.45 µm) and washed 3-times with 2 ml of the indicated buffer for measurement of swimming speeds. Cells were suspended in 0.5 ml of the same buffer, and incubated at 30°C for 10 min. Cell motility was observed under a dark-field microscope using a Leica DMRE microscope and Progressive 3CCD color video camera system (Model DXC-9000, SONY, Tokyo, Japan) and recorded on videotape. Swimming speed was determined by direct tracing of the moving cells on the video monitor. The ionic strength of the assay buffers increased as the cation concentration was elevated. All results shown are the averages of three independent experiments in which the speed of 20 different cells was measured.
Visualization of flagella
Flagella staining was carried out as described by Aono et al. [22]. B. alcalophilus cells were cultured at 30°C for 7 hours in KMYE medium or for 22 hours in MYE medium, and transferred gently to a microscope slide. The sample was air-dried and treated for 2 minutes with staining solution containing 5% (w/v) tannic acid, 0.75% (w/v) FeCl2, 0.01% NaOH, followed by ammoniac silver nitrate for 30 seconds. Observations of flagella were made using a Leica DMLB100 bright field microscope and Leica DC300F camera, Leica IM50 version 1.20 software (Leica Geosystems, Tokyo, Japan). The number of flagella/cell and flagellar length were measured for 55 cells by using ImageJ version 1.36b software (NIH, Bethesda, Maryland, USA).
Western blot analysis
To investigate the effect of the mutation on the expression level of the stator protein in membrane fractions, three B. subtilis strains, ΔABΔPS, BA-PS and BA-PS-MotS-M33L, and six E. coli strains, RP6894 carrying pBAD24, pBAPS, or pBAPS-MotS-M33L, and TK2420 carrying pBAD24, pBAPS, or pBAPS-MotS-M33L, were grown as described above, harvested and washed in Tris Buffer (50 mM Tris–HCl pH 8.0). Cells were suspended in the same buffer and a protease inhibitor cocktail (SIGMA) was added. Membrane vesicles were prepared by breaking cells with a French Pressure cell [23], [24], except that the buffer used was 50 mM Tris-HCl, pH 8.0. 10 µg of membrane protein/µl from each sample was used for one-dimensional sodium dodecyl sulfate (SDS)-PAGE analyses. The same volume of SDS loading buffer was added to each sample, after which the proteins were separated on 10% polyacrylamide SDS gels (Bio-Rad). The gels were then transferred to nitrocellulose filters (Bio-Rad) electrophoretically by the application of 60 V for 3 h in Tris-glycine-methanol buffer (25 mM Tris, 192 mM glycine, 20% (v/v) methanol [pH 8.3]). The MotP protein from B. alcalophilus was detected by antibodies raised to synthetic peptides that corresponded to a conserved MotP region in B. pseudofirmus OF4 MotP residues 242–253, LEEKLSAFTREK. The corresponding sequence in B. alcalophilus Vedder 1934 is LQEKLNAFNRQK. An additional cysteine was added to the C terminus of the peptide. The peptides were conjugated to keyhole limpet hemocyanin and polyclonal antibodies were raised in rabbits (Operon Biotechnologies, Inc., Tokyo); a purified IgG fraction (Melon Gel IgG Spin Purification Kit, Thermo Fisher Scientific, USA) was used for the analyses. The protein concentration of the broken cell suspension was measured by the Lowry method with BSA as a standard. Goat anti-rabbit horseradish peroxidase (Bio-Rad) was also used as the second antibody for detection of anti-MotP antibody. ECL solution (Amersham Biosciences) was the usual detection reagent. A set of ECL Plus solution (Amersham Biosciences) and Can Get Signal immunoreactions enhancer solution (Toyobo) was used in some experiments, as indicated. A quantitative imaging system, Pluor-S MAX (Bio-Rad), was used for detection and analysis of chemiluminescence images.
Results
Multiple alignments of MotB-type stator proteins from Bacillus and Vibrio species and B. alcalophilus
As illustrated in the alignment in Figure 1, proton-coupled MotB type stator subunits of numerous bacteria contain a conserved valine in the middle of the transmembrane segment. In contrast, the sodium coupled MotS and PomB type stator subunits contain a conserved leucine at the same location. Interestingly, the same site in BA-MotS contains a methionine. This led us to investigate the monovalent cation profiles for support of both growth and swimming of B. alcalophilus.
Figure 1. Stained flagellar of B. alcalophilus and alignment with flagella motor sequences from other bacteria.
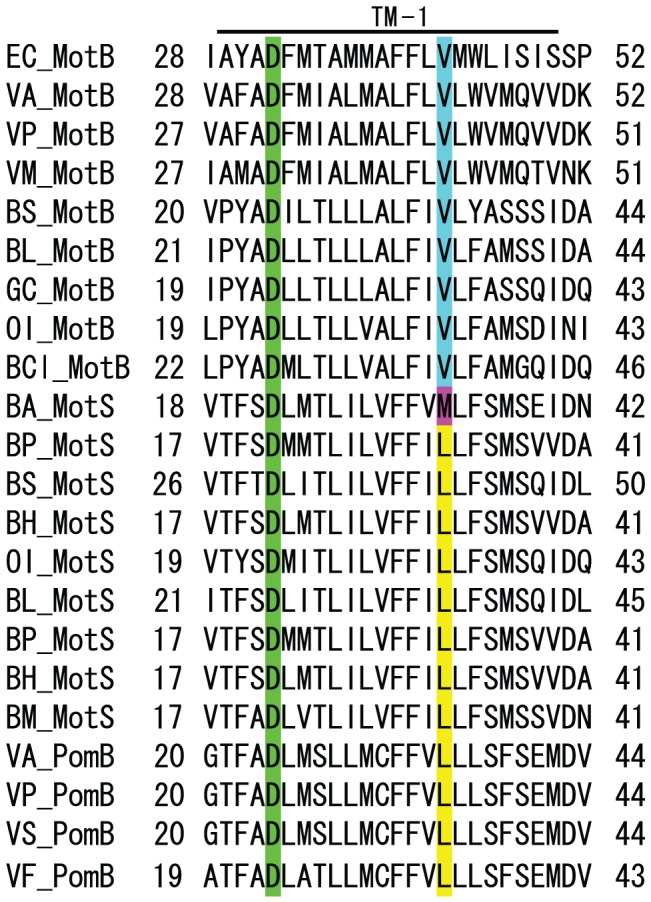
Alignments of the region containing the single transmembrane segment of E. coli MotB (EC_MotB), B. subtilis MotB (BS_MotB) and MotS (BS_MotS), B. licheniformis MotB (BL_MotB) and MotS (BL_MotS), Geobacillus kaustophilus MotB (GK_MotB), Oceanobacillus iheyensis MotB (OI_MotB) and MotS (OI_MotS), B. clausii MotB (BCl_MotB), B. alcalophilus MotS (BA_MotS), B. pseudofirmus MotS (BP_MotB), B. halodurans MotS (BH_MotB), B. megaterium MotB (BM_MotS), V. alginolyticus MotB (VA_MotB) and PomB (VA_PomB), V. parahaemolyticus MotB (VP_MotB) and PomB (VP_PomB), V. mimicus MotB (VM_MotB), V. splendidus PomB (VS_PomB), and V. fisheri PomB (VF_PomB). The position of D32 in EC_MotB is known to be critical for rotation and is highlighted in green. The MotAB of B. clausii can use Na+ instead of H+ to promote flagellar rotation at high pH values. The V37L mutation was critical for sodium selectivity and a combination of the V37L mutation and either the A40S or the G42S mutation was required for production of the BCl-MotB (the ninth line) form that exhibits sodium-coupling at low pH [14]. The position of V43 in EC_MotB (the first line) is conserved among all of the MotB-H+-type proteins and is highlighted in light blue. The position of L32 in BP_MotS (the eleventh line from the top) is conserved among all of the MotS-Na+-type proteins with the exception of BA_MotS and is highlighted in yellow. The same position in B. alcalophilus MotS encodes methionine instead of the conserved leucine residue, and it is highlighted with violet.
Na+- or K+-dependent growth capacities of neutralophilic B. subtilis, alkaliphilic B. pseudofirmus OF4 and alkaliphilic B. alcalophilus at pH 7.5 and 10.5
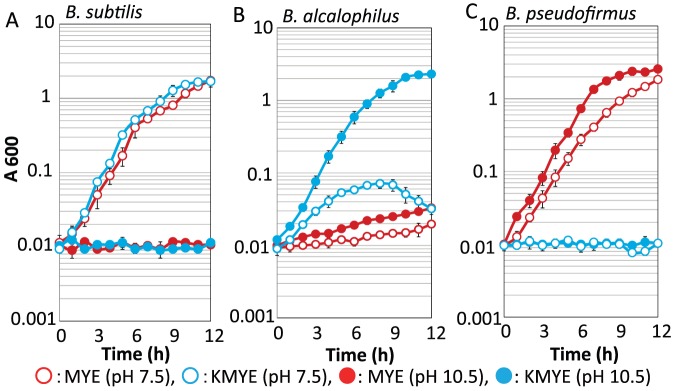
We initially attempted to grow B. alcalophilus in MYE medium, pH 10.5, which is a typical malate-containing growth medium for alkaliphiles; we also used KMYE medium, which contains potassium instead of sodium. As measured using flame photometry, MYE medium is rich in sodium ions (189 mM) and poor in potassium ions (5.5 mM), and KMYE medium is rich in potassium ions (210 mM) and poor in sodium ions (0.7 mM). A similar medium that replaced the Na2CO3 and K2CO3 of the pH 10.5 media with the same molar concentrations of Na2HPO4·7H2O and K2HPO4 was used as the medium for experiments at pH 7.5. The cells of neutralophilic Bacillus subtilis (a control for growth at only neutral pH values) [25], alkaliphilic B. pseudofirmus OF4 (a control for completely Na+-dependent growth) [25] and B. alcalophilus were grown in MYE and KMYE medium at pH 7.5 and 10.5 (Figure 2B). While B. subtilis displays both Na+- and K+-dependent growth at pH 7.5, B. pseudofirmus OF4 displays only Na+-dependent growth at both pH values. Interestingly, B. alcalophilus could grow in all of the media tested, and the best growth was observed in KMYE medium at pH 10.5.
Figure 2. Growth of neutralophilic B. subtilis two alkaliphilic Bacillus species.
(A), (B) and (C) show the growth data for B. subtilis, B. alcalophilus and B. pseudofirmus, respectively. Growth in MYE medium (pH 7.5, red filled circle and pH 10.5, red open circle) and KMYE medium (pH 7.5, light-blue filled circle and pH 10.5, light-blue open circle) at 30°C was monitored at A600. The results are the averages of three independent experiments, with error bars representing the standard deviations.
B. alcalophilus Motility Responds to Added Sodium, Potassium and Rubidium
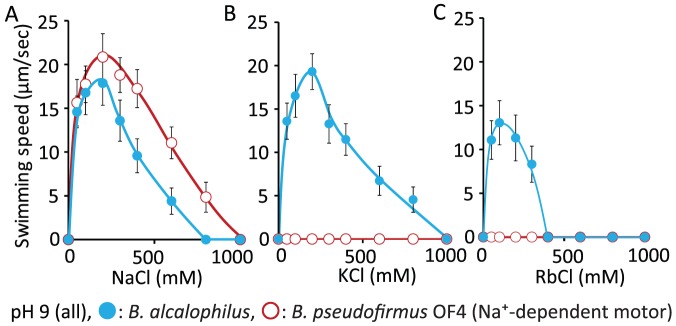
When grown at pH 10.5, cells of B. alcalophilus Vedder 1934 had an average of 3.0±1.2 and 3.0±1.4 flagella per cell, respectively, in MYE and KMYE medium in which Na+ was replaced with K+; the average flagellar length was 5.4±1.9 µm and 6.7±2.1 µm, respectively, in MYE and KMYE. The stained flagella of typical cell in KMYE is shown in Figure S1. To measure swimming speed, B. alcalophilus and B. pseudofirmus OF4 (a control for only Na+-dependent swimming) were grown aerobically on KMYE and MYE medium (pH 10.5) at 30°C, respectively. Swimming speed assays were then conducted in liquid buffer at pH 9.0 (Figure 3). The swimming speeds of B. pseudofirmus OF4 and B. alcalophilus both increased with increasing NaCl up to about 200 mM NaCl. Only swimming of B. alcalophilus speed was also stimulated by KCl or RbCl (Figure 3B, C), exhibiting robust enhancement by up to 200 mM KCl and enhancement of more modest swimming speeds up to 100 mM RbCl. No swimming was observed when B. pseudofirmus OF4 was tested in either KCl or RbCl assay buffer. These data suggest that the flagellar motor of B. alcalophilus can utilise K+, Rb+ and Na+ as coupling ions for the rotation.
Figure 3. Motility of B. pseudofirmus and B. alcalophilus in liquid medium.
B. pseudofirmus OF4 and B. alcalophilus cells in the logarithmic growth phase that were grown at 30°C in MYE medium (pH 10.5) and KMYE medium (pH 10.5), respectively, were harvested and resuspended in 30 mM Tris-HCl (pH 9.0) that contained 5 mM glucose and the indicated sodium (A), potassium (B) or rubidium (C) concentrations as described in the Materials and Methods section. The red line and red open circles show the data for the B. pseudofirmus OF4 strain, and the blue line and blue filled circles show the data for the B. alcalophilus strain. The swimming speed was determined as described in the Materials and Methods section. The results that are shown represent the averages of three independent experiments in each of which the swimming speeds of 20 independent cells as calculated in each experiment. The error bars indicate the standard deviations of the values.
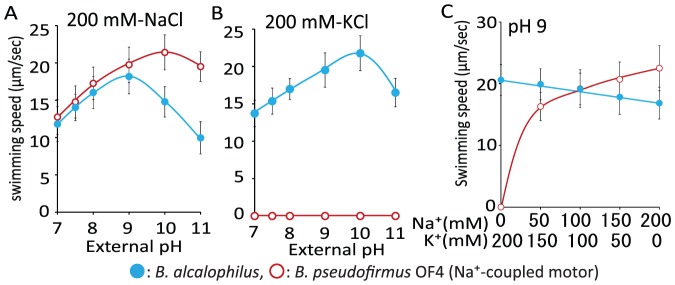
We next performed swimming speed assays in liquid buffer that contained 200 mM Na+ or 200 mM K+ at several different pH values to determine the pH that supported the fastest swimming speed for B. pseudofirmus OF4 and B. alcalophilus (Figure 4A, B). The maximal swimming speed of B. pseudofirmus OF4 in NaCl buffer occurred at pH 10 and the velocity was approximately 22 µm per second. The maximal swimming speed of B. alcalophilus in NaCl buffer was pH 9, and the velocity was approximately 18 µm per second. The maximum velocity of B. alcalophilus in KCl buffer occurred at pH of 10 and the velocity was approximately 22 µm per second. These data suggest that K+-supported motility of B. alcalophilus is similar in speed or slightly faster than Na+-supported motility. We then further probed the cation preference for swimming by B. alcalophilus in comparison with B. pseudofirmus OF4 under conditions in which different Na+/K+ ratios were compared with Na+ and K+ addition alone in the assay media. The swimming speeds of B. alcalophilus and B. pseudofirmus OF4 were measured at pH 9.0 in liquid buffer that contained 200 mM NaCl, 150 mM NaCl plus 50 mM KCl, 100 mM NaCl plus 100 mM KCl, 50 mM NaCl plus 150 mM KCl, or 200 mM KCl (Figure 4C). The velocity of B. pseudofirmus OF4 was clearly dependent upon the Na+ concentration, and no swimming was observed in the absence of Na+. By contrast, the velocity of B. alcalophilus was modestly decreased under elevated Na+ and reduced K+ conditions.
Figure 4. The swimming speed of two alkaliphiles dependent upon pH and concentrations of NaCl and KCl.
The relationship between the swimming speed and several different pH values at 200 mM Na+ (A) or K+ (B) is illustrated. The relationship between swimming speed in 30 mM Tris-HCl containing 5 mM glucose (pH 9.0) and the various indicated concentrations of KCl and NaCl is shown in (C). The red line and red open circles show the data for the B. pseudofirmus OF4 strain, and the blue line and blue filled circles show the data for the B. alcalophilus strain. The swimming speed was determined as described in the Materials and Methods section. The results that are shown represent the averages of three independent experiments in each of which the swimming speeds of 20 independent cells as calculated in each experiment. The error bars indicate the standard deviations of the values.
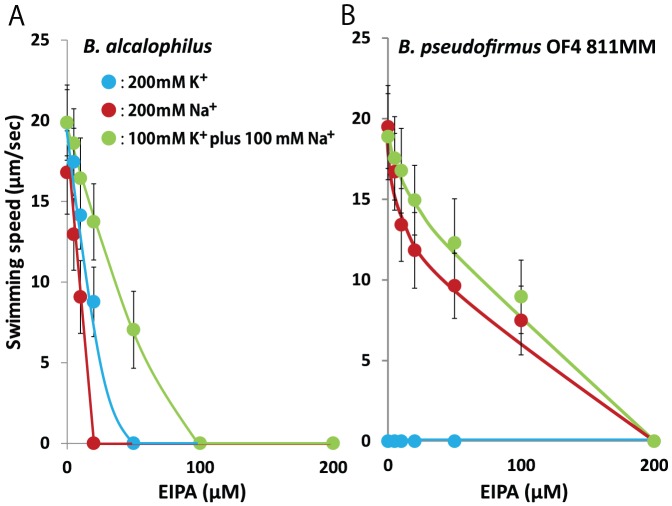
Finally, the effects of the addition of the Na+ channel inhibitor EIPA on the swimming speed of the strains were examined (Figure 5). The swimming speed of B. alcalophilus was sensitive to EIPA in both Na+ and K+ buffer. We previously suggested that EIPA binding sites are present in both the MotP and MotS subunits and that this inhibitor blocks K+ passage [5].
Figure 5. Effect of the Na+ channel inhibitor EIPA on motility.
The red line and red filled circles show the data for the B. pseudofirmus strain, and the blue line and blue filled circles show the data for the B. alcalophilus strain in Tris-HCl buffer that contained 5 mM glucose and 200 mM NaCl (pH 9.0) (A) or 200 mM KCl (pH 9.0) (B). The swimming speed was determined as described in the Materials and Methods. The results that are shown represent the averages of three independent experiments in each of which the swimming speeds of 20 independent cells as calculated in each experiment. The error bars indicate the standard deviations of the values.
A Single Mutation in BA-MotS Change the Profile of the Bi-functional BA-MotPS Stator to a Sodium-only Coupled Stator Profile
The MotB-like proteins are major determinants of the ion-specificity of the B. subtilis MotAB vs. MotPS systems [13], [14]. The next challenge was to change the apparent dual specificity of the B. alcalophilus MotPS system to one that primarily uses sodium or potassium. We hypothesized that replacing the non-consensus BA-MotS-M33 located in the single transmembrane segment with the conserved leucine among sodium type stators might lead to motility dependent only on sodium (Figure 1). To directly compare the swimming properties conferred by BA-MotPS (wild type) with those conferred by BA-MotPS-MotS_M33L, the two flagella-encoding loci, BA-motPS and BA-motPS-MotS_M33L, were introduced into the amyE chromosomal locus of a B. subtilis strain (designated ΔABΔPS) from which both native BS-motAB (motAB of B. subtilis) and BS-motPS (motPS of B. subtilis) were deleted. The resulting B. subtilis mutant strains expressing BA-motPS or BA-motPS-MotS_M33L were named BA-PS and BA-PS-MotS_M33L, respectively. Each gene pair was under control of the B. subtilis motAB promoter. Western blot analyses confirmed the presence of a protein band corresponding to BA-MotP in these strains (Figure. S2). Both stators restored motility to the non-motile ΔABΔPS strain on soft agar plates. We next determined which cations are preferred for flagellar rotation in these strains (Figure 6). The swimming speeds of BA-PS and BA-PS-MotS_M33L were measured at several pH values in phosphate buffer that contained 200 mM NaCl, 150 mM NaCl plus 50 mM KCl, 100 mM NaCl plus 100 mM KCl, 50 mM NaCl plus 150 mM KCl, or 200 mM KCl. We used phosphate buffer instead of Tris buffer for this swimming assay because the motility of B. subtilis is significantly inhibited by Tris buffer. The swimming of BA-PS was observed in the absence of Na+ at all pH values; maximum swimming speed was observed at pH 7.5 and the velocity was approximately 5 µm per second (Figure 6A). The velocity at pH 7 was increased when the Na+ concentration was increased up to 200 mM. The velocity at pH 7.5 and 8 was also stimulated by up to 100 mM and 150 mM Na+ concentrations, respectively. At all pH values, B. subtilis BA-PS, which expressed the native B. alcalophilus stator, exhibited a significant capacity to swim without added sodium but also exhibited stimulation by sodium. The results suggested that the BA-PS rotor retained its original ion-coupling capacity in the heterologous neutralophilic host. No swimming was observed by B. subtilis BA-PS-MotS_M33L in the absence of Na+ at any of the pH values (Figure 6B). At pH 7, swimming of BA-PS-MotS_M33L was clearly dependent upon added sodium ion and required a threshold Na+ concentration of 50 mM. At pH 7.5, stimulation by added sodium was modest. At pH 8, swimming supported by BA-PS-M33L in the neutralophile was stimulated up to 50 mM Na+ and then no swimming was observed above 150 mM Na+ concentrations. The results were consistent with the observations in the native host suggesting that the BA-PS-MotS_M33L stator lost its original potassium-coupling capacity.
Figure 6. The effect of KCl and NaCl on swimming speed of B. subtilis and E.coli strains.
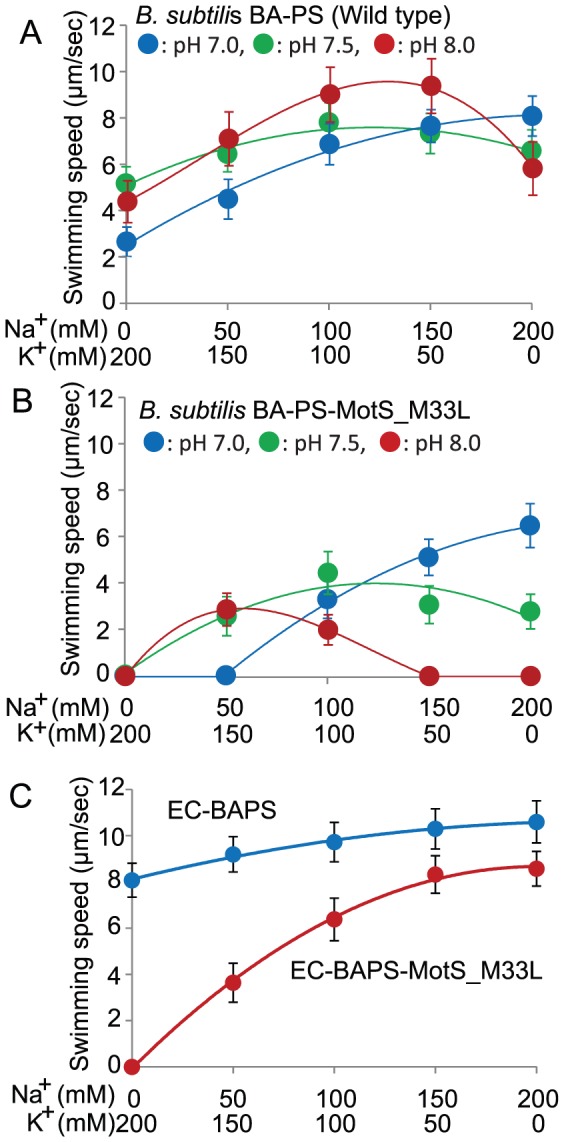
The effect of KCl and NaCl on swimming speed of B. subtilis strains (A) and (B), The velocity was measured at several different pH values in phosphate buffer that contained 200 mM Na+, 150 mM Na+ plus 50 mM K+, 100 mM Na+ plus 100 mM K+, 50 mM Na+ plus 150 mM K+, or 200 mM K+. The blue line and blue filled circles, the green line and green filled circles, and the red line and red filled circles show the data at pH 7.0, 7.5, and 8.0, respectively. The effect of KCl and NaCl on swimming speed of E. coli strain (C) The velocity was measured at pH 7.0 in phosphate buffer that contained 200 mM Na+, 150 mM Na+ plus 50 mM K+, 100 mM Na+ plus 100 mM K+, 50 mM Na+ plus 150 mM K+, or 200 mM K+. The blue line and blue filled circles and the red line and red filled circles show the data for EC-BAPS and EC-BAPS-MotS_M33L, respectively. The swimming speed was determined as described in the Materials and Methods section. The results that are shown represent the averages of three independent experiments in each of which the swimming speeds of 20 independent cells as calculated in each experiment. The error bars indicate the standard deviations of the values.
Both the motPS gene and motPS-MotS_M33L gene of B. alcalophilus complement motility-deficient E. coli RP6894
A stator-defective mutant, E. coli RP6894 was used to expressed the B. alcalophilus motPS genes from a plasmid, pBAPS (generating a transformant designated EC-BAPS) or to express the B. alcalophilus motPS-MotS_M33L genes from plasmid pBAPS-MotS_M33L (generating a strain designated EC-BAPS-MotS_M33L). The E. coli mutant transformants exhibited bands corresponding to BA-MotS in Western analyses (Figure S2). Both of these E. coli transformants exhibited motility, so the swimming velocity of EC-BAPS and EC-BAPS-MotS_M33L was measured at pH 7 in phosphate buffers that contained 200 mM Na+, 150 mM Na+ plus 50 mM K+, 100 mM Na+ plus 100 mM K+, 50 mM Na+ plus 150 mM K+, or 200 mM K+. The velocity of EC-BAPS-MotS_M33L was clearly dependent upon the Na+ concentration, and no swimming was observed in the absence of Na+ (Figure 6C). The velocity of EC-BAPS was not dependent upon added Na+ in the presence of K+ but it was slightly increased with increasing Na+ concentrations in the buffer. The results suggested MotS and MotS_M33L is retaining their distinct ion selectivities when functioning in combination with MotP to power a flagellar motor in E. coli.
Complementation studies in K+-uptake-deficient Escherichia coli mutant strains carrying the motPS gene and the motPS-MotS_M33L gene of B. alcalophilus
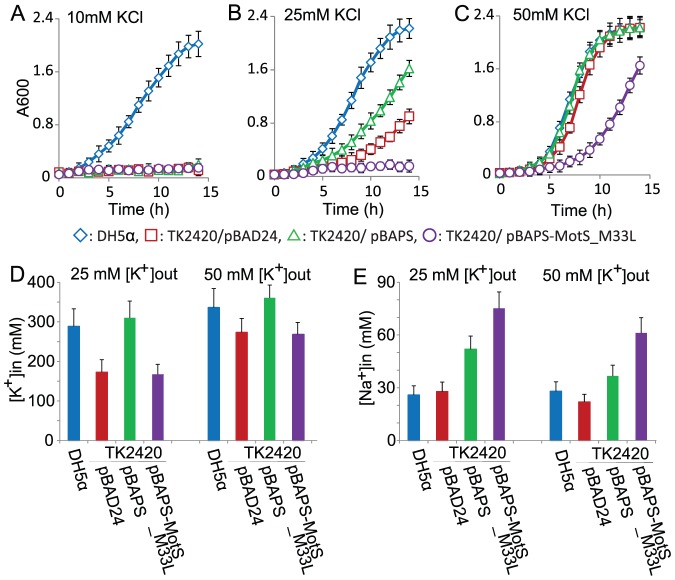
While the profiles of cation dependence of B. alcalophilus MotPS strongly suggested a capacity of the stator to couple motility to K+, the profiles did not provide direct evidence for actual inward translocation of K+ that was dependent upon MotS. We hypothesized that if the stator protein MotPS from B. alcalophilus is incorporated in a functional flagellar motor and takes up K+ during rotation, this K+ uptake would complement the growth defect of K+ uptake-deficient E. coli TK2420. That is, the growth of the cells at growth-limiting K+ concentrations would be better in transformants expressing a functional K+-responsive stator than in the E. coli TK2420 transformed with an empty vector (pBAD24). Further, since the cytotoxicity of Na+ is exacerbated by sub-optimal cytoplasmic concentrations of K+ [25], a transformant expressing the BA-PS-MotS_M33L stator might exhibit even less growth than the empty vector control. As shown in Figure 7, E. coli TK2420 transformed with the empty vector requires >10 mM added K+ for growth whereas an E. coli DH5α (pBAD24) control, with no deficit in K+ uptake, (Figure 7A, B) grows only a little sub-optimally at 10 mM KCl relative to its growth at higher concentrations. By contrast, E. coli TK2420 carrying empty vector (pBAD24), pBAPS and pBAPS-MotS_M33L were all unable to grow in minimal medium with 10 mM K+. In the minimal medium with 25 mM KCl added, the doubling times of E. coli DH5α (pBAD24), and strainTK2420 carrying empty vector (pBAD24) or pBAPS were approximately 1.5, 3.2 or 2.7 h, respectively (Figure 7B). On the other hand, the growth of E. coli TK2420 harboring pBAPS-MotS_M33L was not complemented (Figure 7B). The growth of E. coli TK2420 transformed with either empty vector or pBAPS was enhanced when 50 mM KCl was added to the medium. The two transformants respectively exhibited doubling times of approximately 1.4 or 1.3 h in the presence of added KCl (Figure 7C); these doubling times were comparable to the growth of the DH5α (pBAD24) control. By contrast, the E. coli TK2420 transformant with pBAPS-MotS_M33L exhibited a long lag and a doubling time of 2.1 h.
Figure 7. Effect of KCl on the growth and intracellular ion content of various E. coli TK2420 transformants.
The growth of E. coli strain DH5αMCR transformed with control plasmid pBAD24 (filled blue circles) and E. coli strain TK2420 transformed with pBAD24 (open blue circles), pBAPS (filled red circles) and pBAPS-MotS_M33L (open red circles). Cells were shaken in the TK2420 minimal medium adding 10 mM (A), 25 mM (B) or 50 mM (C) KCl at 37°C under aerobic conditions. Cell growth was monitored at 600 nm. Intracellular [K+] and [Na+] levels in E. coli DH5αMCR transformed with control plasmid pBAD24 (filled light blue bar) and E. coli strain TK2420 transformed with pBAD24 (open light blue filled bar), pBAPS (filled red bar) and pBAPS-MotS_M33L (open red bar). Cells were shaken in the TK2420 minimal medium adding 25 mM (D), or 50 mM (E) KCl at 37°C under aerobic conditions. The results are the averages of three independent duplicate experiments, with error bars representing the standard deviations.
The wild type E. coli (DH5aMCR) clearly grew better than the mutant TK2420 strain carrying the vector alone at sub-optimal [K+]. The osmolality of minimal medium with 25 mM K+ is about 0.2 osmolar. The intracellular K+ content of E. coli cells grown under these conditions had been found to be about 220 mmol K+ per liter of cytoplasmic H2O [21]. The concentration obtained for the wild type in this study (289 mM) is 1.3 times higher than this previously published value but is still in reasonably good agreement with it. The intracellular K+ content of E. coli TK2420 carrying empty vector (pBAD24), pBAPS and pBAPS-MotS_M33L grown in minimal medium with 25 mM K+ was 60%, 107%, and 58% of that of the wild type, respectively (Figure 7D). On the other hand, the intracellular Na+ content of TK2420 carrying empty vector (pBAD24), pBAPS and pBAPS-MotS_M33L grown in minimal medium with 25 mM K+ was 107%, 200%, and 290% of that of the wild type, respectively (Figure 7D). A higher K+ content is presumably why the generation times for E. coli TK2420 carrying pBAPS are better than the TK2420 strain transformed with empty vector. Poor growth of E. coli TK2420 carrying pBAPS-MotS_M33L at 25 mM K+ and growth inhibition at 50 mM K+ presumably result from the combination of reduced K+ uptake capacity at 25 mM K+ and uptake of cytotoxic Na+ at both 25 and 50 mM K+ (Figure 7D, E). The results suggest that BA-MotPS supports actual K+ uptake, with a role as a coupling ion, while the the Na+ uptake catalyzed BA-MotPS-MotS_M33L is consistent with the earlier data that it retains this uptake capacity while having lost a capacity for K+ uptake.
Discussion
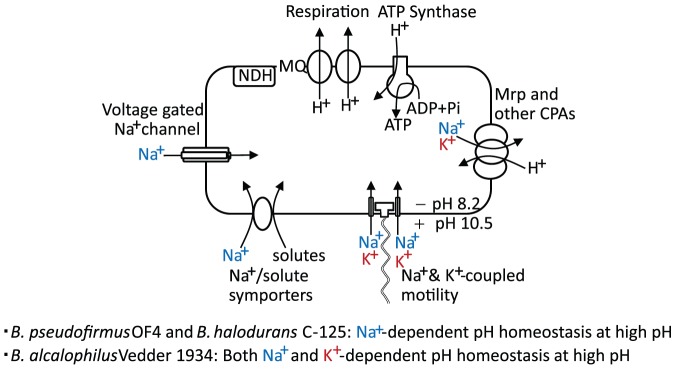
A major finding of this study is that Bacillus alcalophilus Vedder 1934, an alkaliphile strain that was originally isolated from human feces, differs from the most intensively characterized alkaliphilic Bacillus strains, such as B. pseudofirmus OF4 and Bacillus halodurans C-125 in being able to support both cytoplasmic pH homeostasis and flagellar motility at high pH using K+ when the Na+ concentrations are low. In addition, it raises interesting questions about the nature of the potassium cycle that supports pH homeostasis, when Na+ is absent or insufficient. This potassium cycle is expected to differ from the strictly Na+-dependent cycle characterized for both B. pseudofirmus OF4 and B. halodurans C-125. In those two Na+-dependent alkaliphiles, cytoplasmic pH homeostasis at high pH, depends upon use of a hetero-oligomeric Na+/H+ antiporter, the Mrp (or Cation/Proton Antiporter-3 family) antiporter to catalyze net uptake or H+ in exchange cytoplasmic Na+ during aerobic growth on malate (Figure 8) [26], [27]. The antiporters catalyze electrogenic efflux of cytoplasmic Na+ in exchange for a greater number of H+, so that the transmembrane electrical potential (negative inside relative to outside), ΔΨ, can energize the antiport. Coupling of motility, uptake of a large number of solutes and use of a voltage-gated NaChBac type sodium channel all contribute to sufficient return of Na+ to the cytoplasm to ensure ongoing antiport activity in support of pH homeostasis (Figure 8) [26], [28], [29]. Earlier work of Krulwich and colleagues revealed that alkaliphilic B. alcalophilus ATCC27647, which originated from the Vedder 1934 strain, uses electrogenic Na+/H+ antiporter activity and Na+/solute symport to facilitate growth at alkaline pH [30], [31]. However, it is clear from the current findings that a parallel but distinct cycle of potassium is present in B. alcalophilus.
Figure 8. The Na+ and K+ cycles of alkaliphiles.
A diagrammatic illustration of the pH homeostasis capacity of the Na+- and Na+ plus K+-dependent alkaliphiles and elements of their membrane-associated Na+, K+ and H+ translocation pathways.
Although Na+ is usually required for, or greatly enhances growth of alkaliphilic bacteria, some alkaliphiles have long been known to grow in alkaline media that contain K2CO3 but lack Na2CO3 [32], [33]. In addition, in caterpillars, whose gut is known to constitute an alkaline environment, the elevation of gut pH is linked to potassium transport and net accumulation of K2CO3 [34] and termite-derived alkaliphiles show NaCl sensitivity and preference of K+ over Na+ during growth at high pH [35]. While the membranes of some bacteria may be too leaky to K+ to rely on K+-coupled transporters and/or pumps to carry out physiologically important energetic work [36], there are precedents for such usage of potassium in both eukaryotes and in non-alkaliphilic as well as alkaliphilic prokaryotes. K+/H+ antiporters have been reported in both prokaryotes and eukaryotes [37], [38], and potassium has been shown to be a counter-ion for a sodium-proton-glutamate transporter in eukaryotes [39] and for a sodium/proton-potassium antiporter in a non-alkaliphilic Bacillus [40]. Potassium also serves as coupling ion for the KAAT1 potassium/amino acid transporter in the larval insect mid-gut [41] and K+ can also couple to leucine transport in brush border membrane vesicles from Manduca sexta, a well-studied insect model [42]. When B. alcalophilus uses K+ for cytoplasmic pH homeostasis, it is likely that an electrogenic K+/H+ antiporter is employed. It will be of interest to determine whether a K+-translocating multi-subunit Mrp antiporter is of major importance in B. alcalophilus, since Na+-translocating Mrp antiporters have been shown to play critical roles in other alkaliphiles that depend completely upon Na+ for pH homeostasis [26], [27]. K+-translocating Mrp type antiporters have been described in other bacteria [38], [43], [44] but not yet characterized in an alkaliphilic Bacillus. It remains possible that an electrogenic antiporter type other than Mrp has a major role in K+-dependent pH homeostasis in this setting (Figure 8). An electroneutral K+/H+ antiporter was detected in B. alcalophilus [31], but information about electrogenic K+/H+ antiporters is still lacking for this organism. It will be of further interest to ascertain whether there are novel channels or symporters that replenish cytoplasmic potassium and whether B. alcalophilus utilizes, in part, the K+ gradient to augment the ΔΨ as long at the K+ is outwardly directed.
The other major finding in this study, and the most unprecedented one, is the evidence of potassium-coupling to flagellar rotation. The finding that the potassium uptake-defective E. coli strain TK2420 is complemented by the wild-type B. alcalophilus BA-MotPS but not by the mutant BA-MotPS_M33L (Figure 7) supports the conclusion that the K+ is playing the role of coupling ion and being translocated into the cell during flagellar rotation. The consequences of mutating the MotB residue that is uniquely methionine in B. alcalophilus is yet another example verifying the importance of the residue at this particular position for the ion-coupling properties. We note that the B. pseudofirmus OF4 stator tends, in general, to be inhibited more than that of B. alcalophilus at near neutral pH. This inhibition has been attributed to strong competitive inhibition by protons on the Na+-coupled stator of this strain [45], which is also observed with the Na+-coupled symporters of B. pseudofirmus OF4 at pH 7.5 [46]. This putative inhibitory effect, which is less pronounced or absent in the experiments with B. alcalophilus, has been interpreted as one of several indications of the “hard wiring” of B. pseudofirmus OF4 for growth at high pH at the expense of optimized growth at near neutral pH [47]. It is felicitous that the BA-MotPS is functional and retains its native coupling properties in an E. coli strain because this opens up the opportunity for future experiments using single molecular microscopic techniques of analyzing the flagellar motor that are already established in E. coli [48]. Further work may also make it possible to find a mutant form of the stator that lacks or has severely diminished capacity for Na+-coupling as was possible to achieve with the dual function stator of B. clausii [14].
Conclusions
A major finding of the current study is the demonstration that a single bacterial flagellar stator of B. alcalophilus can couple motility to either sodium or potassium ions. This finding may facilitate the identification of additional examples of other cation coupling capacities among the rapidly growing number of bacteria that exhibit natural motility and for which the genomic sequences are available. Further mutagenesis studies and, ultimately, the correlation of these types of data with high-resolution structural data that are related to these and other transporters that catalyse both sodium- and potassium-coupled bioenergetics will facilitate the generation of detailed models of the cation-binding sites, including the basis for competitive proton binding at near neutral pH.
Supporting Information
Stained flagellar of B. alcalophilus . Cells were stained with staining solution that contained 5% (wt/vol) tannic acid as described in Materials and Methods.
(EPS)
Western blot detection of MotP proteins from B. alcalophilus expressed in B. subtilis and E. coli transformants. Three B. subtilis strains, (A) ΔABΔPS, BA-PS and BA-PS-MotS-M33L, and six E. coli strains, (B) RP6894 carrying pBAD24, pBAPS, or pBAPS-MotS-M33L, and (C) TK2420 carrying pBAD24, pBAPS, or pBAPS-MotS-M33L, were grown as described in the Materials and Methods section. Each sample was subjected to SDS-PAGE followed by immunoblotting with anti-MotP antibody. The arrowhead on right-hand side of each panel indicates the MotP protein. The asterisks on right-hand side of the panel A indicate the non-specific bands.
(EPS)
Acknowledgments
We thank Prof. Terry Krulwich and Dr. David Hicks at the Mount Sinai School of Medicine and Dr. Arthur A. Guffanti for their critical discussions and reading of the manuscript. We also thank Prof. Keiichi Namba at Osaka University for using experiment facilities of his laboratory.
Funding Statement
The Strategic Research Foundation Grant-aided for Private Universities and Grant-in-Aid for Scientific Research (B) No. 21370074 of the Ministry of Education, Culture, Sports, Science and Technology of Japan was awarded to MI. NT is a Research Fellow of the Japan Society for the Promotion of Science. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Berg HC (2003) The rotary motor of bacterial flagella. Annu Rev Biochem 72: 19–54. [DOI] [PubMed] [Google Scholar]
- 2. Kojima S, Blair DF (2004) The bacterial flagellar motor: structure and function of a complex molecular machine. Int Rev Cytol 233: 93–134. [DOI] [PubMed] [Google Scholar]
- 3. De Mot R, Vanderleyden J (1994) The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol Microbiol 12: 333–334. [DOI] [PubMed] [Google Scholar]
- 4. Asai Y, Kawagishi I, Sockett RE, Homma M (2000) Coupling ion specificity of chimeras between H+- and Na+-driven motor proteins, MotB and PomB, in Vibrio polar flagella. EMBO J 19: 3639–3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ito M, Terahara N, Fujinami S, Krulwich TA (2005) Properties of motility in Bacillus subtilis powered by the H+-coupled MotAB flagellar stator, Na+-coupled MotPS or hybrid stators MotAS or MotPB. J Mol Biol 352: 396–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hirota N, Imae Y (1983) Na+-driven flagellar motors of an alkalophilic Bacillus strain YN-1. J Biol Chem 258: 10577–10581. [PubMed] [Google Scholar]
- 7. Sugiyama S, Matsukura H, Imae Y (1985) Relationship between Na+-dependent cytoplasmic pH homeostasis and Na+-dependent flagellar rotation and amino acid transport in alkalophilic Bacillus . FEBS Lett 182: 265–268. [DOI] [PubMed] [Google Scholar]
- 8. McCarter LL (2005) Multiple modes of motility: a second flagellar system in Escherichia coli . J Bacteriol 187: 1207–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ito M, Hicks DB, Henkin TM, Guffanti AA, Powers B, et al. (2004) MotPS is the stator-force generator for motility of alkaliphilic Bacillus and its homologue is a second functional Mot in Bacillus subtilis . Mol Microbiol 53: 1035–1049. [DOI] [PubMed] [Google Scholar]
- 10. Atsumi T, Maekawa Y, Yamada T, Kawagishi I, Imae Y, et al. (1996) Effect of viscosity on swimming by the lateral and polar flagella of Vibrio alginolyticus . J Bacteriol 178: 5024–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Paulick A, Koerdt A, Lassak J, Huntley S, Wilms I, et al. (2009) Two different stator systems drive a single polar flagellum in Shewanella oneidensis MR-1. Mol Microbiol 71: 836–850. [DOI] [PubMed] [Google Scholar]
- 12. Doyle TB, Hawkins AC, McCarter LL (2004) The complex flagellar torque generator of Pseudomonas aeruginosa . J Bacteriol 186: 6341–6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ito M, Terahara N, Fujinami S, Krulwich TA (2005) Properties of motility in Bacillus subtilis powered by the H+-coupled MotAB flagellar stator, Na+-coupled MotPS or hybrid stators MotAS or MotPB. J Mol Biol 352: 396–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Terahara N, Krulwich TA, Ito M (2008) Mutations alter the sodium versus proton use of a Bacillus clausii flagellar motor and confer dual ion use on Bacillus subtilis motors. Proc Natl Acad Sci U S A 105: 14359–14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schlegel K, Leone V, Faraldo-Gomez JD, Muller V (2012) Promiscuous archaeal ATP synthase concurrently coupled to Na+ and H+ translocation. Proc Natl Acad Sci U S A 109: 947–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vedder A (1934) Bacillus alcalophilus n. sp.; benevens enkele ervaringen met sterk alcalische voedingsbodems. Antonie Van Leeuwenhoek 1: 143–147. [Google Scholar]
- 17. Sturr MG, Guffanti AA, Krulwich TA (1994) Growth and bioenergetics of alkaliphilic Bacillus firmus OF4 in continuous culture at high pH. J Bacteriol 176: 3111–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wei Y, Southworth TW, Kloster H, Ito M, Guffanti AA, et al. (2003) Mutational loss of a K+ and NH4 + transporter affects the growth and endospore formation of alkaliphilic Bacillus pseudofirmus OF4. J Bacteriol 185: 5133–5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Epstein W, Kim BS (1971) Potassium transport loci in Escherichia coli K-12. J Bacteriol 108: 639–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ito M, Cooperberg B, Krulwich TA (1997) Diverse genes of alkaliphilic Bacillus firmus OF4 that complement K+-uptake-deficient Escherichia coli include an ftsH homologue. Extremophiles 1: 22–28. [DOI] [PubMed] [Google Scholar]
- 21.Bakker E (1993) Cell K+ and K+ transport systems in prokaryotes. In: Bakker E, editor. Alkali cation transport systems in prokaryotes. Boca Raton: CRC Press pp. 205–224.
- 22. Aono R, Ogino H, Horikoshi K (1992) pH-dependent flagella formation by facultative alkaliphilic Bacillus sp. C-125. Biosci Biotechnol Biochem 56: 48–53. [DOI] [PubMed] [Google Scholar]
- 23. Rosen BP (1986) Ion extrusion systems in E. coli . Methods Enzymol 125: 328–386. [DOI] [PubMed] [Google Scholar]
- 24. Swartz TH, Ito M, Ohira T, Natsui S, Hicks DB, et al. (2007) Catalytic properties of Staphylococcus aureus and Bacillus members of the secondary cation/proton antiporter-3 (Mrp) family are revealed by an optimized assay in an Escherichia coli host. J Bacteriol 189: 3081–3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Padan E, Bibi E, Ito M, Krulwich TA (2005) Alkaline pH homeostasis in bacteria: New insights. Biochim Biophys Acta 1717: 67–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krulwich TA, Hicks DB, Swartz TH, Ito M (2007) Bioenergetic adaptations that support alkaliphily. In: Gerday C, Glansdorff N, editors. Physiology and Biochemistry of Extremophiles. Washington, DC: ASM Press. pp. 311–329.
- 27. Kitada M, Kosono S, Kudo T (2000) The Na+/H+ antiporter of alkaliphilic Bacillus sp. Extremophiles 4: 253–258. [DOI] [PubMed] [Google Scholar]
- 28. Fujinami S, Terahara N, Krulwich TA, Ito M (2009) Motility and chemotaxis in alkaliphilic Bacillus species. Future Microbiol 4: 1137–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ito M, Xu H, Guffanti AA, Wei Y, Zvi L, et al. (2004) The voltage-gated Na+ channel NavBP has a role in motility, chemotaxis, and pH homeostasis of an alkaliphilic Bacillus . Proc Natl Acad Sci USA 101: 10566–10571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guffanti AA, Blanco R, Krulwich TA (1979) A requirement for ATP for b-galactoside transport by Bacillus alcalophilus . J Biol Chem 254: 1033–1037. [PubMed] [Google Scholar]
- 31. Guffanti AA, Cohn DE, Kaback HR, Krulwich TA (1981) Relationship between the Na+/H+ antiporter and Na+/substrate symport in Bacillus alcalophilus . Proc Natl Acad Sci USA 78: 1481–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kitada M, Horikoshi K (1987) Bioenergetic properties of alkalophilic Bacillus sp. strain C-59 on an alkaline medium containing K2CO3 . J Bacteriol 169: 5761–5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kitada M, Morotomi S, Horikoshi K, Kudo T (1997) K+/H+ antiporter in alkaliphilic Bacillus sp. no. 66 (JCM 9763). Extremophiles 1: 135–141. [DOI] [PubMed] [Google Scholar]
- 34. Dow JA (1984) Extremely high pH in biological systems: a model for carbonate transport. Am J Physiol Regul Integr Comp Physiol 246: R633–635. [DOI] [PubMed] [Google Scholar]
- 35. Ohkuma M, Shinizu H, Thongaram T, Kosono S, Moriya K, et al. (2003) An alkaliphilic and xylanolytic Paenibacillus species isolated from the gut of a soil-feeding termite. Microbes Environ 18: 145–151. [Google Scholar]
- 36. Buurman ET, McLaggan D, Naprstek J, Epstein W (2004) Multiple paths for nonphysiological transport of K+ in Escherichia coli . J Bacteriol 186: 4238–4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Harvey WR (2009) Voltage coupling of primary H+ V-ATPases to secondary Na+- or K+-dependent transporters. J Exp Biol 212: 1620–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Radchenko MV, Waditee R, Oshimi S, Fukuhara M, Takabe T, et al. (2006) Cloning, functional expression and primary characterization of Vibrio parahaemolyticus K+/H+ antiporter genes in Escherichia coli . Mol Microbiol 59: 651–663. [DOI] [PubMed] [Google Scholar]
- 39. Kanai Y, Hediger MA (2003) The glutamate and neutral amino acid transporter family: physiological and pharmacological implications. Eur J Pharmacol 479: 237–247. [DOI] [PubMed] [Google Scholar]
- 40. Southworth TW, Guffanti AA, Moir A, Krulwich TA (2001) GerN, an endospore germination protein of Bacillus cereus, is an Na+/H+-K+ antiporter. J Bacteriol 183: 5896–5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Castagna M, Shayakul C, Trotti D, Sacchi VF, Harvey WR, et al. (1998) Cloning and characterization of KAAT1, a potassium-coupled amino acid transporter. Proc Natl Acad Sci USA 95: 5395–5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hennigan BB, Wolfersberger MG, Parthasarathy R, Harvey WR (1993) Cation-dependent leucine, alanine, and phenylalanine uptake at pH 10 in brush-border membrane vesicles from larval Manduca sexta midgut. Biochim Biophys Acta 1148: 209–215. [DOI] [PubMed] [Google Scholar]
- 43. Putnoky P, Kereszt A, Nakamura T, Endre G, Grosskopf E, et al. (1998) The pha gene cluster of Rhizobium meliloti involved in pH adaptation and symbiosis encodes a novel type of K+ efflux system. Mol Microbiol 28: 1091–1101. [DOI] [PubMed] [Google Scholar]
- 44. Dzioba-Winogrodzki J, Winogrodzki O, Krulwich TA, Boin MA, Hase CC, et al. (2009) The Vibrio cholerae Mrp system: cation/proton antiport properties and enhancement of bile salt resistance in a heterologous host. J Mol Microbiol Biotechnol 16: 176–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fujinami S, Terahara N, Lee S, Ito M (2007) Na+ and flagella-dependent swimming of alkaliphilic Bacillus pseudofirmus OF4: a basis for poor motility at low pH and enhancement in viscous media in an “up-motile” variant. Arch Microbiol 187: 239–247. [DOI] [PubMed] [Google Scholar]
- 46. Gilmour R, Messner P, Guffanti AA, Kent R, Scheberl A, et al. (2000) Two-dimensional gel electrophoresis analyses of pH-dependent protein expression in facultatively alkaliphilic Bacillus pseudofirmus OF4 lead to characterization of an S-layer protein with a role in alkaliphily. J Bacteriol 182: 5969–5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krulwich T, Liu J, Morino M, Fujisawa M, Ito M, et al.. (2010) Adaptive mechanisms of extreme alkaliphiles; KHorikoshi GA, ABull, F. TRobb and KStetter, editor. Heidelberg: Springer.
- 48. Che YS, Nakamura S, Kojima S, Kami-ike N, Namba K, et al. (2008) Suppressor analysis of the MotB(D33E) mutation to probe bacterial flagellar motor dynamics coupled with proton translocation. J Bacteriol 190: 6660–6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Epstein W, Buurman E, McLaggan D, Naprstek J (1993) Multiple mechanisms, roles and controls of K+ transport in Escherichia coli . Biochem Soc Trans 21: 1006–1010. [DOI] [PubMed] [Google Scholar]
- 50. Grundy FJ, Turinsky AJ, Henkin TM (1994) Catabolite regulation of Bacillus subtilis acetate and acetoin utilization genes by CcpA . J Bacteriol 176: 4527–4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Clejan S, Guffanti AA, Cohen MA, Krulwich TA (1989) Mutation of Bacillus firmus OF4 to duramycin resistance results in substantial replacement of membrane lipid phosphatidylethanolamine by its plasmalogen form. J Bacteriol 171: 1744–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ito M, Hicks DB, Henkin TM, Guffanti AA, Powers BD, et al. (2004) MotPS is the stator-force generator for motility of alkaliphilic Bacillus, and its homologue is a second functional Mot in Bacillus subtilis . Mol Microbiol 53: 1035–1049. [DOI] [PubMed] [Google Scholar]
- 53. Ireton K, Rudner DZ, Siranosian KJ, Grossman AD (1993) Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes Dev 7: 283–294. [DOI] [PubMed] [Google Scholar]
- 54. Guzman CA, Piatti G, Walker MJ, Guardati MC, Pruzzo C (1994) A novel Escherichia coli expression-export vector containing alkaline phosphatase as an insertional inactivation screening system. Gene 148: 171–172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Stained flagellar of B. alcalophilus . Cells were stained with staining solution that contained 5% (wt/vol) tannic acid as described in Materials and Methods.
(EPS)
Western blot detection of MotP proteins from B. alcalophilus expressed in B. subtilis and E. coli transformants. Three B. subtilis strains, (A) ΔABΔPS, BA-PS and BA-PS-MotS-M33L, and six E. coli strains, (B) RP6894 carrying pBAD24, pBAPS, or pBAPS-MotS-M33L, and (C) TK2420 carrying pBAD24, pBAPS, or pBAPS-MotS-M33L, were grown as described in the Materials and Methods section. Each sample was subjected to SDS-PAGE followed by immunoblotting with anti-MotP antibody. The arrowhead on right-hand side of each panel indicates the MotP protein. The asterisks on right-hand side of the panel A indicate the non-specific bands.
(EPS)