Abstract
Background
Adverse neurodevelopmental sequelae are reported among children who undergo early cardiac surgery to repair congenital heart defects (CHD). APOE genotype has previously been determined to contribute to the prediction of these outcomes. Understanding further genetic causes for the development of poor neurobehavioral outcomes should enhance patient risk stratification and improve both prevention and treatment strategies.
Methods
We performed a prospective observational study of children who underwent cardiac surgery before six months of age; this included a neurodevelopmental evaluation between their fourth and fifth birthdays. Attention and behavioral skills were assessed through parental report utilizing the Attention Deficit-Hyperactivity Disorder-IV scale preschool edition (ADHD-IV), and Child Behavior Checklist (CBCL/1.5-5), respectively. Of the seven investigated, three neurodevelopmental phenotypes met genomic quality control criteria. Linear regression was performed to determine the effect of genome-wide genetic variation on these three neurodevelopmental measures in 316 subjects.
Results
This genome-wide association study identified single nucleotide polymorphisms (SNPs) associated with three neurobehavioral phenotypes in the postoperative children ADHD-IV Impulsivity/Hyperactivity, CBCL/1.5-5 PDPs, and CBCL/1.5-5 Total Problems. The most predictive SNPs for each phenotype were: a LGALS8 intronic SNP, rs4659682, associated with ADHD-IV Impulsivity (P = 1.03×10−6); a PCSK5 intronic SNP, rs2261722, associated with CBCL/1.5-5 PDPs (P = 1.11×10−6); and an intergenic SNP, rs11617488, 50 kb from FGF9, associated with CBCL/1.5-5 Total Problems (P = 3.47×10−7). 10 SNPs (3 for ADHD-IV Impulsivity, 5 for CBCL/1.5-5 PDPs, and 2 for CBCL/1.5-5 Total Problems) had p<10−5.
Conclusions
No SNPs met genome-wide significance for our three neurobehavioral phenotypes; however, 10 SNPs reached a threshold for suggestive significance (p<10−5). Given the unique nature of this cohort, larger studies and/or replication are not possible. Studies to further investigate the mechanisms through which these newly identified genes may influence neurodevelopment dysfunction are warranted.
Introduction
Congenital heart defects (CHDs) are the most common human birth defect, with an incidence of 8 per 1000 live births (30,000–40,000 cases annually in the United States). Approximately one-third of children with CHD require early surgical intervention. Improved surgical outcomes and intensive care have resulted in longer life expectancies for many subjects with CHD. However, studies have identified adverse neurological and functional outcomes in many survivors of neonatal and infant cardiac surgery [1], [2], [3], [4]. Of these, neurodevelopmental dysfunction is the most common outcome in survivors of surgical palliation of CHD [5]. Within our cohort, 30% and 22% of 381 subjects who underwent cardiac surgery in infancy scored in the clinically significant range for inattention and hyperactivity/impulsivity, respectively, for the Attention-Deficit/Hyperactivity Disorder-IV scale, preschool edition (ADHD-IV), [6] at five years of age [7]. In addition, 15% of the cohort was in the clinically significant range for pervasive developmental problems (PDPs) [7] as determined by the Child Behavior Checklist (CBCL) [8] at five years of age. Follow-up on this group of children between ages five and 10 years of age further identified 30% and 29% of 109 children who underwent cardiac surgery in infancy as having clinically significant scores for the Attention-Deficit Hyperactivity Disorder IV Scale (ADHD-IV) of inattention and hyperactivity/impulsivity, respectively, as ascertained by both reports from parents and/or teachers [9]. Moreover, 49% of the 109 subjects were receiving some sort remedial academic services and 15% had been assigned to a special education classroom.
Apolipoprotein E, coded by the APOE gene, is an important regulator of cholesterol metabolism and also a neuroresiliency gene. Prior work in this cohort of 550 neonates with CHD treated surgically in the first 6 months of life identified an association between the APOE ε2 allele and poorer neurodevelopmental outcomes of 244 children evaluated at 1 year of age survivors [10]. Of the original cohort, 381 underwent a detailed neurodevelopmental evaluation at 4 to 5 years of age. The APOE ε2 allele was associated with increased behavioral problems, impaired social interactions, and restricted behavior patterns at age 4–5 years [7], in addition to predicting poorer outcomes at 1 year of age [10].
With regard to genetics, ADHD is a well-studied neurodevelopmental outcome. A variable number tandem repeat (VNTR) of 7 alleles in the dopamine D4 receptor gene (DRD4) and a 148-bp microsatellite repeat in the dopamine D5 receptor gene (DRD5) have been consistently associated with ADHD [11], [12], [13], [14], [15], [16]. A VNTR polymorphism in the 3′-untranslated region (UTR) of DAT1 (also known as SLC6A3), a carrier of dopamine that removes it from the synaptic cleft, has also been repeatedly associated with ADHD [11], [12], [15], [17], [18]. More recently, genome-wide association studies (GWAS) have identified SNPs in CDH13, GFOD1, and TLL that may be associated with ADHD, although replication was not present or not attempted in these studies [19], [20], [21].
A genome-wide association study of neurodevelopmental outcomes in a cohort of children who underwent major cardiac surgery has not been previously reported. Thus, our goal with this study was to conduct a GWAS to provide an unbiased assessment for common genetic variants that contribute strongly to common neurodevelopmental phenotypes in our unique cohort of children.
Methods
Ethics Statement
Subjects were collected at the Children’s Hospital of Philadelphia (CHOP) on a protocol approved by the Institutional Review Boards of CHOP and the University of Washington from 10/1998–04/2003. Informed, written consent was obtained from parents or guardians of all the subjects.
Study Design
This was genome-wide analysis of a previously described prospective cohort [7], [10] to identify gene regions potentially affecting neurobehavioral outcomes for preschool-aged subjects (4–5 years of age) after cardiac surgery in infancy. Subjects who were ≤6 months of age and undergoing major surgical treatment of CHDs with cardiopulmonary bypass with or without deep hypothermic circulatory arrest (DHCA) were eligible for enrollment. Exclusion criteria for the original cohort included: (1) multiple congenital anomalies, (2) recognizable genetic or phenotypic syndrome, and (3) language other than English spoken in the home.
Of the 550 subjects initially enrolled in the study, 381 returned for 4-year neurodevelopmental follow-up. Of those not returning, 64 subjects were deceased and 105 individuals were lost to follow-up. Of these 381 eligible subjects, an additional 51 individuals were excluded due to the possible presence of a genetic or chromosomal syndrome as evaluated by a senior board-certified medical geneticist. As well, 14 subjects lacked APOE genotypes, other covariate data, or neurodevelopmental outcome phenotype data and were therefore removed from consideration in the analyses, leaving a total of 316 subjects for genome-wide analysis.
Genetic Evaluation
Subjects were evaluated by a genetic dysmorphologist at the 1-year and/or 4-year evaluations, with additional clinical genetic evaluation or testing performed as indicated. Due to the difficulty in recognizing neonatal dysmorphic features, some subjects were enrolled for whom a diagnosis of a genetic syndrome was made at a later date. Subjects were classified as either: having no definite genetic syndrome or chromosomal abnormality (normal), suspected genetic syndrome (suspect), or a definite genetic syndrome or chromosomal abnormality (genetic). Following this classification, each patient’s genetics records were individually reviewed by a second senior board-certified medical geneticist, also blinded to the GWAS data, to determine whether subjects were to be included or excluded from the current analysis, which focuses on nonsyndromic subjects. Due to this review, 51 subjects with known or suspected genetic abnormalities were excluded from analysis due to the potential for genetic confounding effects.
Operative Management
Details about the operative management of patients in this cohort have been previously reported [7], [10], [22], [23].
Data Collection
Data on preoperative factors that might affect neurobehavioral outcomes independently, including gestational age, birth head circumference, and birth weight, were obtained from hospital records. Weight and age at surgery were recorded for the initial operation and for subsequent procedures with cardiopulmonary bypass. Operative variables were recorded, including the durations of cardiopulmonary bypass and DHCA, lowest nasopharyngeal temperature, and hematocrit level after hemodilution [7], [10], [22], [23]. Demographic and clinical characteristics of the cohort are presented in Table 1 .
Table 1. Baseline and operative characteristics of the cohort.
| Baseline Characteristics | Cohort Subset (n = 316) |
| Gender, n (%) | |
| Female | 136 (43.0%) |
| Male | 180 (57.0%) |
| Ethnicity, n (%) | |
| Asian/Pacific Islander, Hispanic, or other ancestry | 37 (11.7%) |
| African ancestry, not Hispanic | 70 (22.2%) |
| European ancestry, not Hispanic | 209 (66.1%) |
| Gestational Age, mean ± SD, weeks | 38.6±1.93 |
| Birth weight, mean ± SD, kg | 3.18±0.59 |
| Birth head circumference, mean ± SD, cm | 33.7±1.98 |
| APOE Genotype, n (%) a | |
| ε2 | 43 (13.6%) |
| ε3 | 187 (59.2%) |
| ε4 | 86 (27.2%) |
| Diagnostic Class, n (%) | |
| I: 2 ventricles, no arch obstruction | 160 (50.6%) |
| II: 2 ventricles, arch obstruction | 36 (11.4%) |
| III: 1 ventricle, no arch obstruction | 31 (9.8%) |
| IV: 1 ventricle, arch obstruction | 89 (28.2%) |
| Specific CHD Diagnoses, n (%) | |
| Hypoplastic Left Heart Syndrome | 87 (27.5%) |
| Tetralogy of Fallot | 52 (16.5%) |
| Transposition of the Great Arteries | 30 (9.5%) |
| Ventricle Septal Defect | 28 (8.9%) |
| Ventricle Septal Defect, Coarctation of Aorta | 15 (4.7%) |
| Single Ventricle | 23 (7.3%) |
| Other Distinct CHD Diagnoses | 81 (25.6%) |
| Preoperative intubation, n (%) | 91 (28.8%) |
| Preoperative length of stay, mean ± SD, days | 2.16±2.74 |
| Age at first operation, mean ± SD, days | 41.3±53.9 |
| Weight at first operation, mean ± SD, kg | 3.91±1.26 |
| Total cardiopulmonary bypass time in first operation, mean ± SD, min | 66.2±40.4 |
| Use of DHCA, n (%) | 180 (56.9%) |
| Total DHCA time in first operation, mean ± SD, min | 39.8±16.7 |
| Hematocrit level after hemodilution in first operation, mean ± SD, % | 28.0±3.99 |
| Postoperative length of stay after first operation, mean ± SD, days | 10.70±9.91 |
Abbreviations: SD = standard deviation. DHCA = deep hypothermic circulatory arrest.
APOE genotypes were categorized into three groups: ε2 (ε2ε2, ε2ε3, or ε2ε4), ε3 (ε3ε3), and ε4 (ε3ε4 or ε4ε4).
Four-year Neurodevelopmental Examinations
Neurodevelopmental evaluations were performed between the fourth and fifth birthdays. Growth measurements (weight, length, and head circumference) were recorded. A health history was obtained, focusing on the incidence of interim illnesses, hospitalizations, neurologic events or interim evaluations, current medication use, and parental concerns about health. Parents were asked specifically whether they had ever been told that their child had autism, Asperger syndrome, pervasive developmental disorder not otherwise specified, or ADHD. Attention and other behavioral skills were assessed through parental report by using the Child Behavior Checklist for ages 1.5 to 5 years (CBCL/1.5-5) and the ADHD Rating Scale-IV, Preschool Version (ADHD-IV).
The CBCL/1.5-5 is a questionnaire used to obtain parental reports of behavior problems and prosocial adaptive skills demonstrated within the previous 6 months [8]. Responses are grouped to produce 7 narrow-band problem scores and 5 Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV) oriented scores. The pervasive developmental problems (PDPs) score is one of the DSM-IV oriented scores resulting from the CBCL/1.5-5 and has been previously utilized to identify preschoolers at risk for autism [24]. Other scores from the CBCL/1.5-5 include a total problem score and 2 broad-band indices (internalizing and externalizing problems). The CBCL/1.5-5 total problems score consists of the sum of the scores for the 99 specific problem items on the form plus the highest scores for any written-in responses to item 100. All raw scores were transformed to T-scores with mean = 50 and these T-scores were used as the outcomes of analyses. Of these CBCL/1.5-5 scores, five previously analyzed outcome scores (attention problems, internalizing problems, pervasive developmental problems (PDPs), withdrawn behavior, and total problems) [7] were tested for genome-wide association (see Table 2 ).
Table 2. Improved estimates for four-year neuropsychological outcomes in children with isolated congenital heart defects.
| Domain | Test | Score, Mean ± SD | Not excluding all geneticsyndromes (n = 381) | Excluding all (likely)genetic syndromes(n = 316) |
| Social Skills | CBCL/1.5-5 PDPs | 54.6±7.21 | 15% | 9.5% |
| CBCL/1.5-5 withdrawn | 54.5±6.76 | 11% | 8.5% | |
| Attention | ADHD Rating Scale-IV Inattention | 5.83±5.00 | 30% | 24.7% |
| CBCL/1.5-5 attention problems | 53.8±6.10 | 12% | 7.9% | |
| ADHD Rating Scale-IV Hyperactivity | 6.84±5.20 | 22% | 21.5% | |
| Behavior | CBCL/1.5-5 total problems | 47.7±11.15 | 18% | 15.2% |
| CBCL/1.5-5 internalizing problems | 48.7±11.27 | 22% | 19.3% |
Abbreviations. SD = standard error. ADHD = attention deficit/hyperactivity disorder. CBCL = Child Behavior Checklist for ages 1.5 to 5 years. PDPs = pervasive developmental problems.
The ADHD Rating Scale-IV, Preschool Version, is an 18-item questionnaire utilizing a Likert scale (0– not at all, to 3– very often) that requires parents to rate the frequency of ADHD symptom occurrences, as defined by the DSM-IV [6]. This scale was specifically developed for children 3 to 6 years of age. Normative data was collected from a sample of 907 children. Mean scores (not transformed) are provided for inattention and hyperactivity/impulsivity. Both the raw ADHD-IV scores for inattention and hyperactivity/impulsivity were analyzed for genome-wide association (see Table 2 ).
Subjects were classified as having normal, at-risk, or clinically significant scores for both the CBCL/1.5-5 and ADHD Rating Scale-IV scales in accordance with prior published guidelines and reports [6], [7], [8], [9].
Genotyping
Whole blood or buccal swab samples were obtained before the operation and were stored at 4°C. 550,000 SNPs were genotyped utilizing the Illumina HumanHap 550 k BeadChip performed at the University of Pennsylvania Center for Applied Genomics. Quality control was performed at the University of Washington. Copy number variation (CNV) data was not included in these analyses. The SNP data was filtered out for patient call rate <97% and genotype call rate <99%, minor allele frequency (MAF) cutoff of <0.5% (for power considerations), and Hardy-Weinberg equilibrium (HWE) p<10−6. After these filters, 514,139 SNPs remained with a genotyping rate of 99.772% for 478 subjects, of which 316 are included in these analyses, as detailed above.
APOE Genotyping
Genomic DNA was prepared and was used for determination of APOE genotypes using a previously published method [10], [25]. APOE genotypes were classified into three groups as follows: ε2 (ε2ε2, ε2ε3, or ε2ε4) ε3 (ε3ε3), or ε4 (ε4ε3 or ε4ε4), and the APOE ε2 genotype was included as a covariate in the linear regression model for all phenotypes based on considerable prior published evidence within this cohort that the APOE ε2 genotype is associated with markedly detrimental neurodevelopmental outcomes [7], [10], [22], [23], [26], [27]. Nine subjects had the ε2ε4 genotype and were included in analyses grouped in the ε2 group, rather than excluded, due to the overall small sample size, prior work in this cohort demonstrating significant ε2 effects on outcomes but modest or null ε4 effects [7], [10], [22], [23], [26], [27], and because APOE was not a study endpoint.
Analysis
All analyses were performed in PLINK [28], with graphics produced by R (http://r-project.org). Genotypes were coded using an additive model. Due to the mixed genetic descent of the cohort (see Table 1 for demographic information, including genetic ancestry), the first 3 principal component eigenvectors from principal components analysis (PCA) were utilized to adjust for potential population stratification. Of the 7 neurodevelopmental outcomes tested (see Table 2 ), only three (ADHD-IV Hyperactivity/Impulsivity, CBCL/1.5-5 PDPs, and CBCL/1.5-5 Total Problems) had a genomic inflation factor lambda <1.03 and are presented, as the high genomic inflation factor in other phenotypes could reflect false positive results. The outcome of all linear regression analyses in the GWAS were the continuous score values from the CBCL/1.5-5 and ADHD-IV scales.
Linear regression was performed on each outcome to obtain a residual phenotype value, adjusted for the following potentially confounding variables: the first 3 principal component eigenvectors for race, gender, gestational age, birth weight, birth head circumference, APOE ε2 genotype, diagnostic class, preoperative intubation, preoperative length of stay, age at first operation, weight at first operation, total cardiopulmonary bypass time, use of DHCA, total DHCA time, hematocrit at first operation, and postoperative length of stay. These surgical covariates had previously been determined to influence outcomes [22], [23], [29]. The residual phenotype then underwent further linear regression for evaluation of genotype effects utilizing an additive model.
Maternal education and socioeconomic status are important modifiers of neurodevelopmental outcomes [30], [31]. However, they were not included as covariates in analyses due to a poor correlation with the specific outcomes of ADHD-IV Impulsivity, CBCL/1.5-5 PDPs, and CBCL/1.5-5 Total Problems.
Results
Demographic and clinical characteristics of the study cohort of 316 subjects are shown in Table 1 . Table 2 displays the mean, standard deviation, and percentage of our 316 subjects who were either at risk or clinically significant for the selected five CBCL/1.5-5 and two ADHD-IV neurodevelopmental phenotypes that had been previously studied in this cohort [7]. In comparison to a prior publication from this cohort [7], this subset has a lower percentage of subjects at risk or clinically significant for all phenotypes, likely due to the exclusion of 51 subjects with likely or confirmed genetic syndrome.
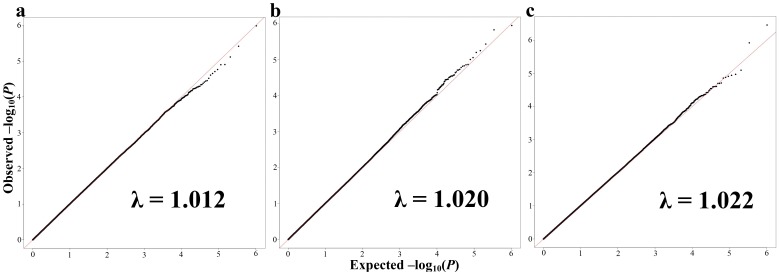
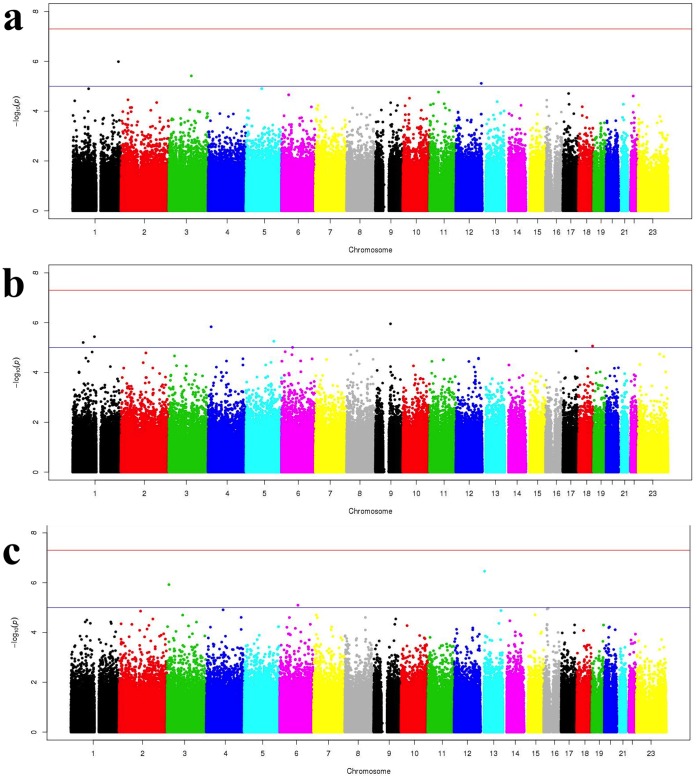
Linear regression analyses of 514,139 SNPs in 316 subjects resulted in three adjusted phenotypes (ADHD-IV Hyperactivity/Impulsivity, CBCL/1.5-5 PDPs, and CBCL/1.5-5 Total Problems) passing our quality control for genomic inflation factor lambda <1.03 when adjusting for the first three principal components and the covariates described in the Methods section. Figure 1A–C displays the Q-Q plot of expected and observed log -10 p-values for the ADHD-IV Impulsivity (1A), CBCL/1.5-5 Scale for PDPs (1B), and CBCL/1.5-5 Total Problems (1C). For these three residual phenotypes, no SNPs had genome-wide significance of p<5×10−8. 10 suggestive SNPs had a p<10−5 and are presented in Table 3 . Three SNPs with p<10−5 were found for ADHD-IV Impulsivity Scale, five SNPs for CBCL/1.5-5 PDPs, and two SNPs for CBCL/1.5-5 Total Problems Score. Manhattan plots of the –log P value for all 514,139 SNP, for ADHD-IV Impulsivity, CBCL/1.5-5 PDPs, and Total Problems, respectively, are presented in Figure 2A–C .
Figure 1. Genome-wide QQ plots of expected and observed –log(P) values for all SNPs analyzed in linear regression covariate-adjusted models for the selected four-year neuropsychological domains.
Panel A presents the QQ plot for the phenotype, ADHD-IV Impulsivity Scale. Panel B presents the QQ plot for the phenotype, CBCL/1.5-5 PDPs. Panel C presents the QQ plot for the phenotype, CBCL/1.5-5 Total Problems. λ = genomic inflation factor.
Table 3. Top SNP Results for Selected Four-Year Outcomes.
| SNP | Chr | Positiona | ClosestReferenceGeneb | Distance to Closest Reference Gene | Minor/Major Alleles | MAF | EA only Betac,d | Betac | EA only P d | P |
| Top GWAS SNPs associated with ADHD-IV Hyperactivity/Impulsivity Scale c | ||||||||||
| rs4659682 | 1 | 233,008,289 | LGALS8 | – | G/A | 0.042 | 2.146 | 2.313 | 2.33×10−4 | 1.03×10−6 |
| rs7625411 | 3 | 114,294,118 | (GTPBP8) | 72,873 bp | C/T | 0.177 | 2.956 | 2.779 | 4.98×10−4 | 3.84×10−6 |
| rs543533 | 12 | 128,025,510 | NLRP9P | – | C/T | 0.377 | −2.157 | −2.07 | 9.67×10−5 | 7.67×10−6 |
| Top GWAS SNPs associated with CBCL/1.5-5 PDPs c | ||||||||||
| rs2261722 | 9 | 76,099,135 | PCSK5 | – | C/T | 0.317 | 3.037 | 3.137 | 2.82×10−5 | 1.11×10−6 |
| rs10516292 | 4 | 15,425,972 | (BST1) | 15,907 bp | C/T | 0.219 | 3.688 | 3.92 | 2.63×10−4 | 1.47×10−6 |
| rs11206315 | 1 | 54,455,429 | SSBP3 | – | G/A | 0.157 | 4.264 | 3.982 | 1.01×10−5 | 6.25×10−6 |
| rs12965975 | 18 | 71,438,271 | (C18orf62) | 169,694 bp | C/T | 0.393 | 1.723 | 3.086 | 0.0277 | 8.68×10−6 |
| rs228144 | 6 | 55,739,721 | BMP5 | – | C/T | 0.112 | 3.142 | 4.407 | 3.23×10−3 | 9.83×10−6 |
| Top GWAS SNPs associated with CBCL/1.5-5 Total Problems c | ||||||||||
| rs11617488 | 13 | 21,091,962 | (FGF9) | 51,253 bp | C/T | 0.275 | −4.453 | −5.245 | 5.13×10−4 | 3.47×10−7 |
| rs9879307 | 3 | 9,845,065 | TTLL3 | – | C/T | 0.317 | −3.592 | −0.2014 | 9.94×10−3 | 1.2×10−6 |
Chr = chromosome. EA = European Ancestry. GWAS = genome-wide association study. MAF = minor allele frequency. SNP = single nucleotide polymorphism.
Position information and annotation from reference assembly 36.3.
SNPs not found within a gene region are represented in parentheses, e.g. (GTPBP8).
All beta-coefficients and p-values are the result of linear regression, where the outcome was the residual phenotype, adjusted for by the covariates listed in the methods.
Analysis performed on the majority genetic descent group (EA) of the cohort (n = 209), adjusting for all covariates in the methods.
Figure 2. Manhattan plots of –log(P) for association of SNPs and chromosomal position for all SNPs analyzed in linear regression covariate-adjusted models.
Panel A presents the Manhattan plot for the phenotype, ADHD-IV Impulsivity Scale. Panel B presents the Manhattan plot for the phenotype, CBCL/1.5-5 PDPs. Panel C presents the Manhattan plot for the phenotype, CBCL/1.5-5 Total Problems. Red horizontal line represents 5×10−8 threshold for genome-wide significance. Blue horizontal line indicates the 10−6 threshold for suggestive genome-wide significance.
As this cohort utilized subjects of diverse genetic ancestry due to small sample size while adjusting for the first 3 principle components to correct for the inherent population stratification of this analysis, we sought to determine the effects of the 10 suggestive SNPs in the majority European ancestry (EA) subset (n = 209) of the cohort (see Table 3 ). All SNP p-values remained suggestive of significance. Sensitivity analyses between the EA subset and all other subjects of mixed genetic ancestry showed no significant differences in direction of SNP effect.
As previous studies into the genetics of ADHD have implicated DRD4, DRD5, and DAT1 we explored their effects in our cohort. Rs3758653 and rs11246226 from our dataset were located near DRD4 (upstream and downstream, respectively) and rs27072 was found within DAT1. No SNP tagged DRD5. None of these SNPs were associated with ADHD-IV Impulsivity in this cohort (p>0.10 for all three SNPs). While rs27072 is likely strong proxy marker for the VNTR associated with ADHD in DAT1 as it is located 480 bases upstream of the VNTR, the cited VNTR in DRD4 is located in the third (of 4) exon in DRD4, with neither of these two SNPs in strong LD with it. Thus, the 550 K data does not comprehensibly test the effects of DRD4 or DRD5 in this cohort.
Discussion
Neurodevelopmental disorders are the most common sequelae following surgical palliation of CHD during infancy, with >30% of survivors scoring within the clinically significant range for attention problems in a recent study [32]. With improved surgical outcomes and techniques leading to vastly improved lifespan, a growing goal of independence has arisen in the families of nonsyndromic CHD subjects. However, while intellectual outcome measures vary from poor to excellent, CHD subjects have more problems with social involvement, school performance, and total competence in comparison to control subjects [33]. The Boston Circulatory Arrest Study reported that older post-surgical CHD subjects had difficulty integrating or coordinating the skills to accomplish higher-order goals, such as producing connected discourse or applying math concepts to solve problems [1], [2], [3], [29], [34], [35]. As a result, neurodevelopmental issues, with numerous effects across multiple spectrums of day-to-day life, can be a significantly limiting consequence in subjects who undergo surgical treatment of CHD.
We have completed the first GWAS for neurodevelopmental phenotypes in a unique cohort of 316 prospectively collected subjects, without known or likely genetic syndromes, who underwent cardiac surgery at less than six months of age. Although no SNP reached genome-wide significance (p<5×10−8); in total, 10 SNPs reached a threshold for suggestive significance (p<10−5) for one of three behavioral phenotypes: three for ADHD-IV Impulsivity, five for CBCL/1.5-5 PDPs, and two for CBCL/1.5-5 Total Problems.
This cohort is both the largest of its kind and also unique in its inclusion criteria, as it was designed to identify markers in genes that may affect the recovery of the brain after surgical palliation of a wide spectrum of CHDs and therefore adjusted for numerous potentially confounding inpatient and surgical variables, making it not possible to replicate these results in a similar, independent cohort. Of note, the Boston Circulatory Arrest Study had 155 eligible subjects and examined cognitive and behavioral outcomes after repair of transposition of the great arteries [1], [2], [3], [29], [34], [35]. As a result, we present our results as potential candidate genes involved in the pathogenesis of neurobehavioral problems in children who underwent cardiac surgery as infants, with the caveat that further investigation is required. However, interesting candidates did emerge.
The top SNPs for each outcome studied highlighted interesting candidate genes. Two SNPs, rs4659682 and rs2261722, associated with ADHD-IV Hyperactivity/Impulsivity and CBCL/1.5-5 PDP scores, respectively, were located in gene regions. Rs4659682 (p = 1.03×10−6 with ADHD-IV Hyperactivity/Impulsivity score) is a LGALS8 intronic SNP, located in the 2nd of 12 total introns. LGALS8 encodes Galectin-8, a beta-galactoside-binding lectin with a conserved carbohydrate recognition domain [36]. The galectin family of proteins have been implicated in development [37], [38], growth regulation and apoptosis [37], [39], and other functions [36]. Rs2261722 (p = 1.11×10−6 with CBCL/1.5-5 PDP score) is a PCSK5 intronic SNP, located in the 17th of 35 total introns. PCSK5 encodes a member of the subtilisin-like proprotein convertase family that processes proteins into their active forms, such as nerve growth factor [40]. As such, PCSK5 is involved in neurodevelopment [40] and has been associated with a decrease in ventricular volume in a case-control cohort collected for Alzheimer’s disease and Mild Cognitive Impairment [41]. Finally, rs11617488 is an intergenic SNP (3.47×10−7 with CBCL/1.5-5 Total Problems), approximately 50 kb from FGF9. FGF9 is a member of the fibroblast growth factor family of proteins and has broad range of activities, including CNS development [42], embryogenesis [43], cell repair [44], and cell growth [45]. Of the other SNPs that predicted outcomes at p<10−5, rs10516292, an intergenic SNP approximately 16 kb from BST1 stands out for its potential relevance to neurodevelopment and neuroresiliency, due to numerous independent studies and meta-analyses associating BST1 variants and Parkinson’s disease [46], [47], [48].
The three SNPs identified for association with ADHD-IV Impulsivity score have not previously been identified in prior GWAS for ADHD [19], [20], [49]. The lack of replication of our results in other published GWAS for ADHD likely reflects several differences between our study and the previously published ones. Foremost, the cohorts are fundamentally different, as our study was designed to identify genes that may affect neuroresiliency in children after surgery to repair CHDs, and therefore adjusted for numerous inpatient and surgical variables that could act as confounders. As an example, APOE effects on neurodevelopmental phenotypes previously identified in this cohort are not found in healthy children [7], [10], [50]. APOE effects on neurodevelopmental outcomes have been found in children with other stresses, including lead exposure, oxygen deprivation, malnutrition, and cerebral palsy [51], [52], [53], [54], [55]. Additionally, the analysis strategy is divergent, as Hinney et al., Neale et al., and Lasky-Su et al. all utilized case-control GWAS approaches, though Lasky-Su et al. did analyze quantitative measures of ADHD. Third, the phenotype definition varied, as Hinney et al. analyzed all 3 subtypes of ADHD (primarily inattentive, primarily impulsive/hyperactive, and combined types), while Neale et al. and Lasky-Su et al. utilized only combined type ADHD. Lasky-Su et al. utilized the Long Version of Conner’s Parent and Teacher Rating scales and results from Parental Accounts of Childhood Symptoms interviews as their quantitative outcomes. Fourth, the neurobehavioral phenotypic diversity is far greater in our cohort, with subjects that not only score in the clinically significant range for ADHD-IV Impulsivity score, but also for CBCL/1.5-5 PDPs and total problems, and other four-year neurodevelopmental phenotypes previously reported [7]. Thus, the independent results presented in this article could potentially indicate that unique genetic resiliency pathways impacting ADHD that are important when a similar environmental insult has occurred.
This study represents the first time within this cohort that we have re-estimated the prevalence of clinically significance for 4-year neurodevelopmental behavioral phenotypes without 51 subjects with possible or definite genetic syndromes. Excluding these children, we have noted a lower proportion of subjects that are clinically significant or at risk than reported previously [7]. Thus, the current data presented provides a better risk assessment for nonsyndromic CHD patients.
The CBCL has been the most commonly used instrument in studying neurodevelopmental outcomes in CHD subjects after surgery [8]. With regard to the phenotypes studied in this cohort, the CBCL/1.5-5 PDP score has been reported as having good predictive validity in screening preschoolers at risk for autism spectrum disorder [24]. In contrast, results using the CBCL/1.5-5 Total Problems score have not been widely reported. This is likely because it is a broad-band index representing a wide spectrum, thus having high sensitivity for neurodevelopmental problems but low specificity for any particular disorder.
This study has focused on exploring the genetic basis for the development of neurobehavioral disability in children following congenital heart surgery, while accounting for the known effects of APOE. The degree of disability is expected to be multifactorial, involving genetic and environmental factors, and particularly, their interaction. While we removed subjects with likely genetic syndromes, some infants may have had unrecognized genetic disorders that influenced both structural heart and neurodevelopmental phenotypes. Moreover, there is increasing evidence that brain development of some children with CHD is abnormal in utero, resulting in microcephaly [56] and a structurally immature brain at birth [57]. In addition to these genetic factors, the repair of CHD here included surgical intervention, which necessitated the use of cardiopulmonary bypass or deep hypothermic circulatory arrest. We have shown that many operative and pre- and post-operative factors predict variation in neurodevelopmental outcomes. [58], [59], [60]. The sum of these environmental factors is observed in neuroimaging and neuropathological studies that demonstrate ischemic periventricular white matter injury as a result of surgical intervention [61], [62]. By removing subjects with likely genetic syndromes and adjusting for the effects of APOE and surgical covariates, we focused our efforts not on identifying genetic variation that causes CHD, but on genetic variation that predicts neuroresiliency or lack thereof, in the face of enormous physiological stress. The genetic factors identified in these circumstances may not predict similar outcomes in cases where this physiological challenge was not present. This is the case for APOE genotype, which does not predict outcomes in children who are not exposed to such a challenge [50].
Some limitations of this study must be considered. First, the cohort is 66.1% of European ancestry. Although we utilized all subjects who met our criteria, including subjects of African and Asian ancestry while adjusting for the first 3 principal components, variants that do not occur in European ancestry subjects are underpowered here. As well, while the analysis strategy adjusted for population stratification within the cohort by adjusting for the first three principal component eigenvectors, there remains the possibility of false positive results. However, when we removed all genetic ancestries except for the majority European ancestry, we observed p-values that were suggestive of significance. Ideally, these GWAS results would be replicated in a separate and independent cohort to decrease false positive results. However, this was not possible in our unique cohort, and as a result we are presenting our results as candidate genes for the development of neurobehavioral phenotypes after neonatal cardiac surgery. It is possible that any true associations here, like the APOE association we previously reported, will be replicated in other cohorts with neurological insults [51], [52], [53]. Strengths of this unique cohort include the robust neurodevelopmental phenotyping, which allows for investigation of phenotypes not readily available from the electronic medical records or in other retrospective studies. As well, in comparison with the Boston Circulatory Study, which specifically focused on patients with transposition of the great arteries [1], [2], [3], [29], [34], [35], our cohort includes data across a spectrum of nonsyndromic CHD cases.
In conclusion, the suggestive results presented in this unique study offer candidate genes that may be implicated in the pathogenesis of neurobehavioral problems in a child as sequelae of neonatal cardiac surgery or other neurological insult. Given the potential long-term morbidity of these neurodevelopmental problems, further study of these candidate genes is warranted. Identification of genetic variation underlying differential susceptibility provides a window into novel pathways that may provide guidance in developing therapies and preventative strategies in children with CHD. As was the case for APOE, identified as a pediatric neuroresiliency gene in this cohort, these genes may affect outcomes of a broader range of neurological insults, such as lead exposure and hypoxia.
Acknowledgments
We would like to thank all subjects and families for their participation. Genotyping was performed at and supported by the Center for Applied Genomics at the Children’s Hospital of Philadelphia. DSK was supported by the Sarnoff Cardiovascular Research Fellowship for Medical Students.
Funding Statement
This work was supported by a grant from the Fannie E. Rippel Foundation, an American Heart Association National Grant-in-Aid (9950480N), HL071834 from the National Institutes of Health, and a Washington State Life Sciences Discovery Award to the Northwest Institute for Genetic Medicine. DSK was supported in part by the Sarnoff Cardiovascular Research Fellowship for Medical Students. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bellinger DC, Jonas RA, Rappaport LA, Wypij D, Wernovsky G, et al. (1995) Developmental and Neurologic Status of Children after Heart Surgery with Hypothermic Circulatory Arrest or Low-Flow Cardiopulmonary Bypass. New England Journal of Medicine 332: 549–555. [DOI] [PubMed] [Google Scholar]
- 2. Bellinger DC, Wypij D, Kuban KCK, Rappaport LA, Hickey PR, et al. (1999) Developmental and Neurological Status of Children at 4 Years of Age After Heart Surgery With Hypothermic Circulatory Arrest or Low-Flow Cardiopulmonary Bypass. Circulation 100: 526–532. [DOI] [PubMed] [Google Scholar]
- 3. Bellinger DC, Wypij D, duPlessis AJ, Rappaport LA, Jonas RA, et al. (2003) Neurodevelopmental status at eight years in children with dextro-transposition of the great arteries: The Boston Circulatory Arrest Trial. The Journal of Thoracic and Cardiovascular Surgery 126: 1385–1396. [DOI] [PubMed] [Google Scholar]
- 4. Limperopoulos C, Majnemer A, Shevell MI, Rosenblatt B, Rohlicek C, et al. (2001) Functional Limitations in Young Children With Congenital Heart Defects After Cardiac Surgery. Pediatrics 108: 1325–1331. [DOI] [PubMed] [Google Scholar]
- 5. Wernovsky G, Shillingford AJ, Gaynor JW (2005) Central nervous system outcomes in children with complex congenital heart disease. Curr Opin Cardiol 20: 94–99. [DOI] [PubMed] [Google Scholar]
- 6.Merrell K (2003) Preschool and Kindergarten Behavior Scales. Austin, TX: Psychological Corp.
- 7. Gaynor JW, Nord AS, Wernovsky G, Bernbaum J, Solot CB, et al. (2009) Apolipoprotein E Genotype Modifies the Risk of Behavior Problems After Infant Cardiac Surgery. Pediatrics 124: 241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Achenbach TM, Ruffle TM (2000) The Child Behavior Checklist and related forms for assessing behavioral/emotional problems and competencies. Pediatr Rev 21: 265–271. [DOI] [PubMed] [Google Scholar]
- 9. Shillingford AJ, Glanzman MM, Ittenbach RF, Clancy RR, Gaynor JW, et al. (2008) Inattention, Hyperactivity, and School Performance in a Population of School-Age Children With Complex Congenital Heart Disease. Pediatrics 121: e759–e767. [DOI] [PubMed] [Google Scholar]
- 10. Gaynor JW, Gerdes M, Zackai EH, Bernbaum J, Wernovsky G, et al. (2003) Apolipoprotein E genotype and neurodevelopmental sequelae of infant cardiac surgery. The Journal of Thoracic and Cardiovascular Surgery 126: 1736–1745. [DOI] [PubMed] [Google Scholar]
- 11. Faraone SV, Perlis RH, Doyle AE, Smoller JW, Goralnick JJ, et al. (2005) Molecular Genetics of Attention-Deficit/Hyperactivity Disorder. Biological Psychiatry 57: 1313–1323. [DOI] [PubMed] [Google Scholar]
- 12. Gizer IR, Ficks C, Waldman ID (2009) Candidate gene studies of ADHD: a meta-analytic review. Hum Genet 126: 51–90. [DOI] [PubMed] [Google Scholar]
- 13. Faraone SV, Doyle AE, Mick E, Biederman J (2001) Meta-analysis of the association between the 7-repeat allele of the dopamine D(4) receptor gene and attention deficit hyperactivity disorder. Am J Psychiatry 158: 1052–1057. [DOI] [PubMed] [Google Scholar]
- 14. Li D, Sham PC, Owen MJ, He L (2006) Meta-analysis shows significant association between dopamine system genes and attention deficit hyperactivity disorder (ADHD). Hum Mol Genet 15: 2276–2284. [DOI] [PubMed] [Google Scholar]
- 15. Maher BS, Marazita ML, Ferrell RE, Vanyukov MM (2002) Dopamine system genes and attention deficit hyperactivity disorder: a meta-analysis. Psychiatr Genet 12: 207–215. [DOI] [PubMed] [Google Scholar]
- 16. Lowe N, Kirley A, Hawi Z, Sham P, Wickham H, et al. (2004) Joint Analysis of the DRD5 Marker Concludes Association with Attention-Deficit/Hyperactivity Disorder Confined to the Predominantly Inattentive and Combined Subtypes. The American Journal of Human Genetics 74: 348–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Purper-Ouakil D, Wohl M, Mouren MC, Verpillat P, Ades J, et al. (2005) Meta-analysis of family-based association studies between the dopamine transporter gene and attention deficit hyperactivity disorder. Psychiatr Genet 15: 53–59. [DOI] [PubMed] [Google Scholar]
- 18. Yang B, Chan RCK, Jing J, Li T, Sham P, et al. (2007) A meta-analysis of association studies between the 10-repeat allele of a VNTR polymorphism in the 3′-UTR of dopamine transporter gene and attention deficit hyperactivity disorder. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics 144B: 541–550. [DOI] [PubMed] [Google Scholar]
- 19. Neale BM, Lasky-Su J, Anney R, Franke B, Zhou K, et al. (2008) Genome-wide association scan of attention deficit hyperactivity disorder. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics 147B: 1337–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lasky-Su J, Neale BM, Franke B, Anney RJL, Zhou K, et al. (2008) Genome-wide association scan of quantitative traits for attention deficit hyperactivity disorder identifies novel associations and confirms candidate gene associations. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics 147B: 1345–1354. [DOI] [PubMed] [Google Scholar]
- 21. Lesch KP, Timmesfeld N, Renner TJ, Halperin R, Roser C, et al. (2008) Molecular genetics of adult ADHD: converging evidence from genome-wide association and extended pedigree linkage studies. J Neural Transm 115: 1573–1585. [DOI] [PubMed] [Google Scholar]
- 22.Gaynor JW, Wernovsky G, Jarvik GP, Bernbaum J, Gerdes M, et al.. (2007) Patient characteristics are important determinants of neurodevelopmental outcome at one year of age after neonatal and infant cardiac surgery. J Thorac Cardiovasc Surg 133: 1344–1353, 1353 e1341–1343. [DOI] [PMC free article] [PubMed]
- 23. Fuller S, Nord AS, Gerdes M, Wernovsky G, Jarvik GP, et al. (2009) Predictors of impaired neurodevelopmental outcomes at one year of age after infant cardiac surgery. Eur J Cardiothorac Surg 36: 40–47. [DOI] [PubMed] [Google Scholar]
- 24. Muratori F, Narzisi A, Tancredi R, Cosenza A, Calugi S, et al. (2011) The CBCL 1.5-5 and the identification of preschoolers with autism in Italy. Epidemiol Psychiatr Sci 20: 329–338. [DOI] [PubMed] [Google Scholar]
- 25. Tardiff BE, Newman MF, Saunders AM, Strittmatter WJ, Blumenthal JA, et al. (1997) Preliminary Report of a Genetic Basis for Cognitive Decline After Cardiac Operations. The Annals of Thoracic Surgery 64: 715–720. [DOI] [PubMed] [Google Scholar]
- 26. Gaynor JW, Gerdes M, Nord AS, Bernbaum J, Zackai E, et al. (2010) Is cardiac diagnosis a predictor of neurodevelopmental outcome after cardiac surgery in infancy? J Thorac Cardiovasc Surg 140: 1230–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zeltser I, Jarvik GP, Bernbaum J, Wernovsky G, Nord AS, et al. (2008) Genetic factors are important determinants of neurodevelopmental outcome after repair of tetralogy of Fallot. J Thorac Cardiovasc Surg 135: 91–97. [DOI] [PubMed] [Google Scholar]
- 28. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, et al. (2007) PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. The American Journal of Human Genetics 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wypij D, Newburger JW, Rappaport LA, duPlessis AJ, Jonas RA, et al. (2003) The effect of duration of deep hypothermic circulatory arrest in infant heart surgery on late neurodevelopment: The Boston Circulatory Arrest Trial. J Thorac Cardiovasc Surg 126: 1397–1403. [DOI] [PubMed] [Google Scholar]
- 30. Msall ME, Bier JA, LaGasse L, Tremont M, Lester B (1998) The vulnerable preschool child: the impact of biomedical and social risks on neurodevelopmental function. Semin Pediatr Neurol 5: 52–61. [DOI] [PubMed] [Google Scholar]
- 31.Wickremasinghe AC, Hartman TK, Voigt RG, Katusic SK, Weaver AL, et al.. (2011) Evaluation of the ability of neurobiological, neurodevelopmental and socio-economic variables to predict cognitive outcome in premature infants. Child Care Health Dev. [DOI] [PubMed]
- 32.Miatton M, De Wolf D, Francois K, Thiery E, Vingerhoets G (2007) Neuropsychological performance in school-aged children with surgically corrected congenital heart disease. J Pediatr 151: 73–78, 78 e71. [DOI] [PubMed]
- 33. Hovels-Gurich H, Konrad K, Wiesner M, Minkenberg R, Herpertz-Dahlmann B, et al. (2002) Long term behavioural outcome after neonatal arterial switch operation for transposition of the great arteries. Archives of Disease in Childhood 87: 506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Visconti KJ, Bichell DP, Jonas RA, Newburger JW, Bellinger DC (1999) Developmental Outcome After Surgical Versus Interventional Closure of Secundum Atrial Septal Defect in Children. Circulation 100: II–145-II-150. [DOI] [PubMed] [Google Scholar]
- 35. Newburger JW, Jonas RA, Wernovsky G, Wypij D, Hickey PR, et al. (1993) A Comparison of the Perioperative Neurologic Effects of Hypothermic Circulatory Arrest versus Low-Flow Cardiopulmonary Bypass in Infant Heart Surgery. New England Journal of Medicine 329: 1057–1064. [DOI] [PubMed] [Google Scholar]
- 36. Bidon N, Brichory F, Bourguet P, Le Pennec JP, Dazord L (2001) Galectin-8: a complex sub-family of galectins (Review). Int J Mol Med 8: 245–250. [DOI] [PubMed] [Google Scholar]
- 37. Hadari YR, Arbel-Goren R, Levy Y, Amsterdam A, Alon R, et al. (2000) Galectin-8 binding to integrins inhibits cell adhesion and induces apoptosis. J Cell Sci 113 (Pt 13): 2385–2397. [DOI] [PubMed] [Google Scholar]
- 38. Kolundzic N, Bojic-Trbojevic Z, Radojcic L, Petronijevic M, Vicovac L (2011) Galectin-8 is expressed by villous and extravillous trophoblast of the human placenta. Placenta 32: 909–911. [DOI] [PubMed] [Google Scholar]
- 39. Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F (2012) Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 482: 414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Seidah NG, Benjannet S, Pareek S, Savaria D, Hamelin J, et al. (1996) Cellular processing of the nerve growth factor precursor by the mammalian pro-protein convertases. Biochem J 314 (Pt 3): 951–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Furney SJ, Simmons A, Breen G, Pedroso I, Lunnon K, et al. (2011) Genome-wide association with MRI atrophy measures as a quantitative trait locus for Alzheimer's disease. Mol Psychiatry 16: 1130–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Puk O, Moller G, Geerlof A, Krowiorz K, Ahmad N, et al. (2011) The pathologic effect of a novel neomorphic Fgf9(Y162C) allele is restricted to decreased vision and retarded lens growth. PLoS ONE 6: e23678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hellingman CA, Koevoet W, Kops N, Farrell E, Jahr H, et al. (2010) Fibroblast growth factor receptors in in vitro and in vivo chondrogenesis: relating tissue engineering using adult mesenchymal stem cells to embryonic development. Tissue Eng Part A 16: 545–556. [DOI] [PubMed] [Google Scholar]
- 44. Warburton D, Perin L, Defilippo R, Bellusci S, Shi W, et al. (2008) Stem/progenitor cells in lung development, injury repair, and regeneration. Proc Am Thorac Soc 5: 703–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yin Y, Wang F, Ornitz DM (2011) Mesothelial- and epithelial-derived FGF9 have distinct functions in the regulation of lung development. Development 138: 3169–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Saad M, Lesage S, Saint-Pierre A, Corvol JC, Zelenika D, et al. (2011) Genome-wide association study confirms BST1 and suggests a locus on 12q24 as the risk loci for Parkinson's disease in the European population. Hum Mol Genet 20: 615–627. [DOI] [PubMed] [Google Scholar]
- 47. Nalls MA, Plagnol V, Hernandez DG, Sharma M, Sheerin UM, et al. (2011) Imputation of sequence variants for identification of genetic risks for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet 377: 641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Simon-Sanchez J, van Hilten JJ, van de Warrenburg B, Post B, Berendse HW, et al. (2011) Genome-wide association study confirms extant PD risk loci among the Dutch. Eur J Hum Genet 19: 655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hinney A, Scherag A, Jarick I, Albayrak Ö, Pütter C, et al. (2011) Genome-wide association study in German patients with attention deficit/hyperactivity disorder. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics 156: 888–897. [DOI] [PubMed] [Google Scholar]
- 50. Rask-Nissila L, Jokinen E, Terho P, Tammi A, Hakanen M, et al. (2002) Effects of diet on the neurologic development of children at 5 years of age: The STRIP project. The Journal of Pediatrics 140: 328–333. [DOI] [PubMed] [Google Scholar]
- 51. Wright RO, Hu H, Silverman EK, Tsaih SW, Schwartz J, et al. (2003) Apolipoprotein E Genotype Predicts 24-Month Bayley Scales Infant Development Score. Pediatr Res 54: 819–825. [DOI] [PubMed] [Google Scholar]
- 52. Arai Y, Mizuguchi M, Ikeda K, Takashima S (1996) Transient expression of apolipoprotein-E in neonates with pontosubicular neuron necrosis. Acta Neuropathologica 91: 396–399. [DOI] [PubMed] [Google Scholar]
- 53. McAdoo JD, Warner DS, Goldberg RN, Vitek MP, Pearlstein R, et al. (2005) Intrathecal administration of a novel apoE-derived therapeutic peptide improves outcome following perinatal hypoxic‚Äìischemic injury. Neuroscience Letters 381: 305–308. [DOI] [PubMed] [Google Scholar]
- 54. Oria RB, Patrick PD, Blackman JA, Lima AAM, Guerrant RL (2007) Role of apolipoprotein E4 in protecting children against early childhood diarrhea outcomes and implications for later development. Medical Hypotheses 68: 1099–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kuroda MM, Weck ME, Sarwark JF, Hamidullah A, Wainwright MS (2007) Association of Apolipoprotein E Genotype and Cerebral Palsy in Children. Pediatrics 119: 306–313. [DOI] [PubMed] [Google Scholar]
- 56.Licht DJ, Shera DM, Clancy RR, Wernovsky G, Montenegro LM, et al.. (2009) Brain maturation is delayed in infants with complex congenital heart defects. J Thorac Cardiovasc Surg 137: 529–536; discussion 536–527. [DOI] [PMC free article] [PubMed]
- 57. Shillingford AJ, Ittenbach RF, Marino BS, Rychik J, Clancy RR, et al. (2007) Aortic morphometry and microcephaly in hypoplastic left heart syndrome. Cardiology in the Young 17: 189–195. [DOI] [PubMed] [Google Scholar]
- 58. Markowitz SD, Ichord RN, Wernovsky G, Gaynor JW, Nicolson SC (2007) Surrogate markers for neurological outcome in children after deep hypothermic circulatory arrest. Semin Cardiothorac Vasc Anesth 11: 59–65. [DOI] [PubMed] [Google Scholar]
- 59. Gaynor JW, Nicolson SC, Jarvik GP, Wernovsky G, Montenegro LM, et al. (2005) Increasing duration of deep hypothermic circulatory arrest is associated with an increased incidence of postoperative electroencephalographic seizures. J Thorac Cardiovasc Surg 130: 1278–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gaynor JW, Gerdes M, Zackai EH, Bernbaum J, Wernovsky G, et al. (2003) Apolipoprotein E genotype and neurodevelopmental sequelae of infant cardiac surgery. J Thorac Cardiovasc Surg 126: 1736–1745. [DOI] [PubMed] [Google Scholar]
- 61. Andropoulos DB, Hunter JV, Nelson DP, Stayer SA, Stark AR, et al. (2010) Brain immaturity is associated with brain injury before and after neonatal cardiac surgery with high-flow bypass and cerebral oxygenation monitoring. J Thorac Cardiovasc Surg 139: 543–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mahle WT, Tavani F, Zimmerman RA, Nicolson SC, Galli KK, et al. (2002) An MRI study of neurological injury before and after congenital heart surgery. Circulation 106: I109–114. [PubMed] [Google Scholar]