After almost 2 decades of relatively unsuccessful attempts to identify genes for complex genetic diseases by means of linkage mapping, the application of genome-wide association studies (GWAs) over the past 5 years has identified a wealth of novel disease associations. This success was made possible by a paradigm shift and by a change in technology. GWAs investigate common diseases using case–control or cohort studies rather than extended or nuclear families for rarer diseases. Microarray technologies enable fast and accurate genotyping of millions of single nucleotide polymorphisms (SNPs) in a short time. In contrast, genome-wide family-based linkage studies had much lower resolution, with standard panels including only hundreds of microsatellite markers.
The success of GWAs has been overwhelming. As of May 2012, the catalog of published GWAs (Hindorff et al. 2012) included over 1,250 papers and more than 6,400 single nucleotide polymorphisms (SNPs). Although the era of GWAs started only 5 years ago, by the end of 2007, five published GWAs already had more than 1,000 citations (Web of Knowledge), the most significant, with the highest number of citations, being the Wellcome Trust Case Control Consortium (2007) study.
Unexpectedly, many of the identified associations did not map to genes but to gene deserts, and the biology underlying these discoveries is rarely immediately apparent. The most prominent finding among these is the association of coronary artery disease (CAD) and myocardial infarction (MI) to 9p21.3 (Samani et al. 2007; Schunkert et al. 2008, 2011), which is the best replicated locus for CAD and MI. Although a large antisense non-coding RNA in the INK4 locus (named ANRIL) was identified as early as 2007 (Pasmant et al. 2007), the function of the gene desert locus for CAD/MI was unknown until quite recently. Specifically, Harismendy et al. (2011) identified enhancers in 9p21, and found that the CAD risk alleles of two SNPs were located in one of these enhancers and that the variants disrupt a binding site for STAT1. Binding of STAT1 inhibits CDKN2BAS expression, which is reversed by short interfering RNA knockdown of STAT1. Harismendy et al. (2011) also demonstrated interactions with other loci.
The CAD/MI GWA findings directly informed studies that identified biological processes important to disease etiology, and it took 4 years to understand the function of the 9p21.3 CAD locus. Although just one of many loci that have been identified using the single marker analysis in a simple case–control study design, the story of the chromosome 9p21.3 locus for CAD/MI points to the importance and value of functional studies following the detection of associations in GWAs.
The success of GWAs using a large number of unrelated individuals has only been made possible by the great advancements in microarray technology. However, it has also required new developments in statistical methodology. As pointed out by Cardon and Palmer (2003) and others, the importance of population stratification as a cause of non-replicated association outcomes led to a great shift in association study design in the 1990s, away from the traditional case–control approach towards the more costly and less efficient family-based designs. However, family-based association designs are often neither practical nor plausible, especially for pharmacogenetic studies, including personalized medicine. It is therefore only natural that approaches that allow adjustments for population stratification in case control or cohort studies are highly cited (Price et al. 2006; Pritchard and Rosenberg 1999; Pritchard et al. 2000).
Considering the critical contribution of the technological and methodological developments in the story of the chromosome 9p21.3 CAD locus, we can anticipate that the post-GWAs era requires both novel study designs and new methods for analysis to complement the established study designs and methods because of the complexity in genetics. This special issue therefore addresses both of these important aspects.
The first three articles by Almasy (2012), Altmann et al. (2012), and Wijsman (2012) deal with next generation sequencing (NGS), i.e., high-throughput sequencing studies, the technology succeeding GWAs. Within a very few years, single-strand whole-genome sequences will be available at reasonable cost, and will become the final and standard DNA typing technology used in both research and Medical Genetics. As soon as this technology becomes the standard measurement of genetic information, genetics will move from a technology-driven to a phenotype-driven scientific discipline. As pointed out by Almasy (2012), “the hopes for advancement of the field were invested in maximizing the informativeness of the genotype data available for study with phenotype often simplified to a case–control [scenario], … [attention in] the post-GWAS era … will now turn to the phenotype, half of the phenotype-genotype connection.” Although the concept of a “phenome project” was proposed some time ago (Freimer and Sabatti 2003), we are now able to start such an initiative, the “natural successor to the human genome project.” A wealth of data is available from high-throughput methodologies, such as transcriptomics, proteomics, and metabolomics. At the same time, it is clear that the future success of genetic studies will require well-defined, interpretable, and commonly accepted phenotype definitions because “the complete catalog of the human genome sequence does not tell us what that sequence does.” In her presentation, Almasy (2012) discusses both well-known and novel approaches to reducing heterogeneity to investigate groups of related phenotypes in order to optimize the use of phenotypes in the various types of genetic studies.
Although whole-genome sequencing will most likely be the standard approach for genetic studies in the near future, sequencing technologies are still improving rapidly. Data processing and management, quality control of sequencing data as well as data sharing through the Internet are not simple, and a single standard has not yet emerged. This is in contrast to GWAs, for which these processes have become standardized over the past years. One of the major aims in sequencing studies is to identify genetic variations, such as SNPs and insertions/deletions, for further genetic analyses. Müller-Myhsok and colleagues provide a beginner’s guide to SNP calling from high-throughput sequencing studies (Altmann et al. 2012). To this end, they reviewed the essential modules for SNP calling. The process begins with base calling, followed by quality control, mapping of the reads to the reference genome, and post alignment and post mapping (which include visualization and post-processing of the alignments to allow for additional base quality recalibration), and finally, the variant calling procedure itself. Of course, this bioinformatics pipeline just represents the starting point for subsequent work. For example, the investigator might wish to filter identified variants by a set of customized criteria. One important observation by Altmann et al. (2012) is that the final SNP calls are substantially affected by the choice of the specific software package used for SNP variant calling. These findings indicate that more research in bioinformatics will be required to establish a reliable easy-to-use pipeline for variant calling. In 2007, when the first major GWA studies were published, Clerget-Darpoux and Elston (2007) in an editorial provocatively asked, “should we stop collecting family data, forget that we inherit our genes from our parents and ignore the fundamental laws of genetic transmission as being unnecessary for gene discovery? Can we hope to anticipate, understand and treat the many diseases to which humans are prone, simply by finding genomic variants that differ between those who have and those who do not have disease?” Clerget-Darpoux and Elston (2007) argued that the common disease-common variant GWAs approach fails to identify rare variants, although we know that “multiple rare variants in a disease gene can play an important role in disease susceptibility.” One such example is the work by Fitze et al. (2002, 2003), who studied mutations in the RET proto-oncogene in patients with Hirschsprung’s disease. They found that a mutation of the RET gene, which is in cis with the −5A allele in the promoter, leads to a “milder” Hirschsprung phenotype, while the −5G allele together with a mutation in cis results in a “more severe” form of Hirschsprung’s disease.
Even more importantly, family information is relevant to refine the genetic model and for estimation of disease risk. In the third article of this issue, on high-throughput sequencing studies, Wijsman (2012) discusses the importance of extended pedigrees for the analysis of rare variants. The large pedigree design is practical, efficient, and well suited for investigating rare variations; also see Bailey-Wilson and Wilson (2011). More specifically, unless the sample sizes are very large, rare functionally relevant variants with large effect size will be difficult to detect in population-based studies (Wilson and Ziegler 2011). However, causal variants will co-segregate with a trait in family-based studies, and because of the increased presence of a specific causal variant in relatives, the effect size will no longer be overwhelmed by its low population frequency. As a result, the ability to detect a rare variant will be increased. As Wijsman observes, in comparison with population-based designs, the extended pedigree design “requires only a small fraction of the sample size needed to identify rare variants of interest, and many highly suitable, well-understood, and available statistical and computational tools already exist.” A further advantage of this approach is the ready availability of samples of large pedigrees with large amounts of phenotypic data for high-throughput sequencing.
The second set of articles focuses on study designs. Cortessis et al. (2012) summarizes the current state of epigenetics. In their work, the authors do not follow the broad definition of epigenetics that has recently been used “to describe any non-genetic mechanism influencing phenotype” (Cortessis et al. 2012). Instead, they adopt more specific definitions, differentiating epigenetic processes that stably affect gene expression through mechanisms not involving the primary nucleotide sequence from epigenetic states, the configuration of chromatin and DNA marks utilized by these processes.
In the first part of their work, Cortessis et al. (2012) review determinants of epigenetic states. They emphasize that even within an individual there are many epigenomes across tissues and cell types, and they change over time. Nevertheless, there are also remarkable consistencies in epigenetic patterns, and DNA methylation patterns are highly conserved, making the empirical study of epigenetic phenomena reasonable and possible. With appropriate limitations to the scope of research questions and interpretation of molecular mechanisms, use of available tissue samples (e.g., peripheral blood) from population studies and biobanks may lead to an initial understanding of the epigenetic contribution of common diseases. Thomas et al. subsequently provide a comprehensive review of human studies for epigenetic phenomena and discuss the many methodological challenges. Measuring the exposure is one of these because of the long time period between exposure and disease onset. The biospecimen itself presents another challenge. Currently, cultured cell lines are used in most epigenetic studies, except for DNA methylation studies, which can use a small amount of fresh tissue. Improvements in epigenome measurement technologies would be necessary for population studies, because it is still not feasible in practice to obtain large amounts of tissues. In the study of environmental influences on epigenetics, the challenge of measurement technology must be addressed for both the epigenetic profiles, as well as the environmental exposures in large population samples. Even the microarray approaches available present such a great heterogeneity, in that each platform has its own statistical analysis methods; for a review of the statistical methods used for methylation analysis with microarrays, the reader may refer to Siegmund (2011).
In the final section of their article, Cortessis et al. (2012) discuss the pros and cons of various designs for studying epigenetic phenomena. They highlight the strengths of twin studies for epigenetics, argue that case–control studies “are fundamentally flawed,” and point to settings in which epigenetic phenomena should be studied in extended pedigrees.
In the subsequent article, Aschard et al. (2012), discuss genome-wide environmental interaction (GWEI) studies. Although some of the challenges in GWEI studies are similar to those of epigenetics, such as exposure measurement or change of exposure dose over time, others differ. For example, population-based designs, such as prospective cohorts or prospective case–control studies, are of great importance for investigating GWEI. Another important difference is that the primary aim of GWEI studies is to study the statistical effect of interactions between genetic markers and environmental factors on disease. In contrast, epigenetic studies primarily deal with the identification of differences in epigenetic profiles, which can be the molecular mediator of the gene-environment interaction, between two groups of individuals.
Pharmacogenetic studies constitute a special case of gene-environment interaction studies, where interactions between genes and drugs are studied to improve treatment efficacy and/or to reduce adverse effects. Ritchie (2012) takes an epidemiological approach to pharmacogenetics. She compares three study designs: the standard randomized parallel group design with two groups, the prospective measurement of treatment response with case–control studies (in each of which treatment responders are compared with non-responders), and the type of cross-sectional study in which treatment responders and non-responders are identified by records in biobanks. One important advantage of pharmacogenetic studies over general GWEI studies is that the genotyping can be very targeted if the mechanism of action of the drug is known, and small-scale chips specific to these known mechanisms have been developed by several companies and academic groups. DNA markers are applied to the selection of targeted therapies in pharmacogenetic studies, as well as to other areas of personalized medicine such as diagnosis and prognosis. In their review of personalized medicine using DNA biomarkers, Ziegler et. al. delineate and compare three broad categories of biomarkers, DNA markers (SNPs, microsatellites, etc.), DNA tumor biomarkers (e.g., presence or absence of a specific mutation in the tumor), and general biomarkers (gene expression, protein expression, etc.). The authors stress the importance of distinguishing between prognostic biomarkers, which help in predicting the progress of a disease, and predictive biomarkers (more generally termed companion diagnostic tests), which are connected with treatment response, as in pharmacogenetic studies. They subsequently discuss phases of diagnostic and prognostic biomarker studies, offering a slightly extended phase approach compared to standard phase models (see, e.g., Pepe et al. 2001). In the extended model, three distinct initial sub-phases allow for the separation of studies on analytical validity from retrospective validation using an independent new set of samples. Finally, Ziegler and colleagues discuss the suitability of clinical trial designs for predictive biomarkers. One of the scenarios, termed the “individual profile design,” may specifically arise for DNA biomarkers. In the extreme, the number of SNP profiles might be identical to the number of subjects randomized in the study. At the same time, the number of available therapies might be identical to the number of different profiles. As the authors emphasize, in this case, it is not a single therapy but a therapy strategy that is evaluated. Although this study design perfectly reflects the paradigm of individualized treatment, it poses new challenges to the regulators about the precise application of the many different therapies.
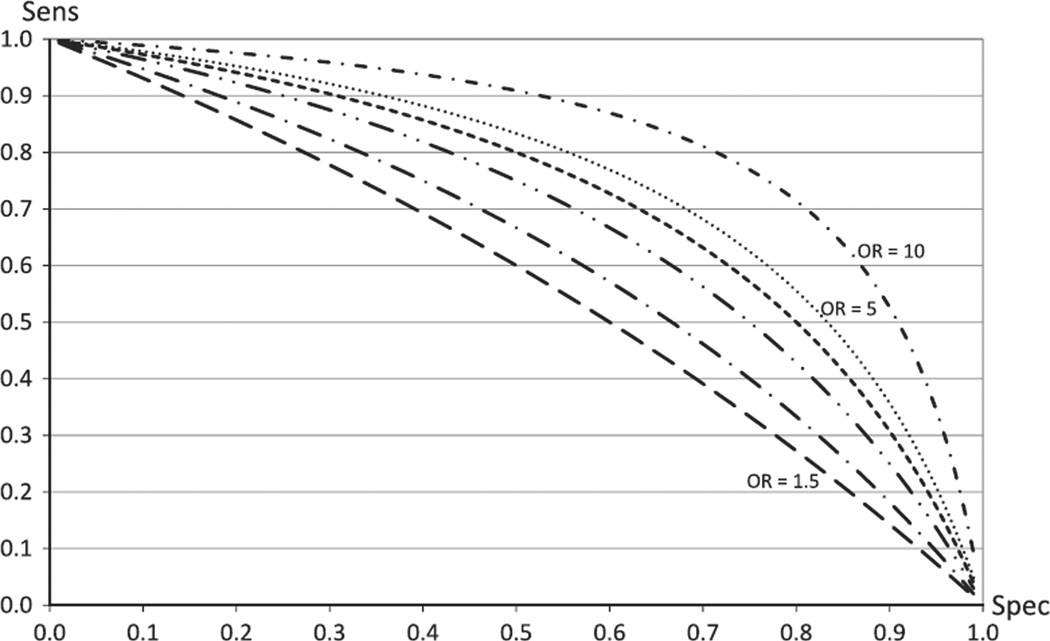
The final group of articles focuses on analytical methods. Kruppa et al. (2012) provide an overview on classification and risk estimation using machine learning methods (sometimes also termed data mining). Two of the major applications of the work presented in this article are prognosis and diagnosis using biomarkers, such as data from GWAs. The authors emphasize that SNPs of genome-wide significance, which may play important roles in disease etiology, can be poor classifiers, a phenomenon that can be observed from a relationship between the odds ratio (OR) and sensitivity (sens) and specificity (spec) (Pepe et al. 2004):
The relationship between the OR, a measure of association, and sens and spec, two measures of diagnostic accuracy, is depicted in Fig. 1. Kruppa et al. (2012) illustrate that “if a SNP has a high sensitivity of 0.9 and a strong association of OR = 3.0, the specificity is only 0.25” (also see Cook 2007; Wald et al. 1999).
Fig. 1.
Relationship between strength of association and classification accuracy. Sensitivity (sens) is plotted as a function of specificity (spec) for odds ratios (OR) varying from 1.5, 2, 3, 4, 5, to 10
Kruppa et al. (2012) discuss in detail the construction and evaluation of multimarker rules, i.e., how to obtain classifiers or probability estimates by using many SNPs. To this end, the authors consider the ACCE model (Gudgeon et al. 2007; Haddow and Palomaki 2004) developed by the Centers for Disease Control (CDC) and argue that no single measure or classification threshold is sufficient to judge the clinical validity of a “multimarker rule,” or using the terminology of Ziegler et al. (2012), the clinical validity of a diagnostic or prognostic test. They thus implicitly argue against the use of a single threshold for the area under the curve of 0.8 or 0.95 for a test to be clinically valid. A final important aspect of their article is their consideration of nonparametric approaches for probability estimation using machine learning methods. In fact, the model-free, nonparametric probability estimation problem has long been considered difficult (Devroye et al. 1996), and it has only recently been solved for standard machine learning methods, such as versions of random forests (Biau 2012).
Zaitlen and Kraft (2012) consider an entirely different application of GWA data, estimating the proportion of phenotypic variance explained by genetic factors, i.e., the heritability. Heritability estimation with the aid of twin studies and family studies was popular until genetic markers became abundant at high numbers. Until very recently, heritability estimates were solely based on phenotypic information, and no marker information was utilized. It therefore comes at no surprise that the assumptions underlying the classical estimation approaches are rather strict (as discussed in detail, e.g., by Ziegler and König 2010), and it is likely that heritability estimates using the traditional approach were biased and sometimes even overly optimistic. In fact, the wide range of estimates, e.g., for the quantitative trait body mass index ranging from around 80 % for twin studies to as low as 5 % for family studies (Bouchard and Perusse 1993), clearly illustrates the limitations of this approach.
An alternative means to estimate heritability using genotype data has recently been offered by Yang and Visscher in the context of GWAs (Yang et al. 2010), which Zaitlen and Kraft observe has “broad implications for the genetic architecture of phenotypes as well as the future success of GWAs” (Zaitlen and Kraft 2012). In their review, Zaitlen and Kraft examine this novel approach, in which observed DNA variation from genotyping or sequencing is used for heritability estimation. They also discuss the assumptions and the impacts on estimates if assumptions are not met.
The review of Vansteelandt and Lange (2012) focuses on the developments in the causal inference literature relevant to the analysis of genetic association studies. Of course, causality is directly related to Hill’s (1965) criteria for causation [also see its revision by Howick et al. (2009)], and one of the ultimate proofs of causality in population genetics relies on the use of families. More specifically, the aim is to demonstrate a deviation from Mendel’s second law, the law of independent assortment, which assumes the independence of one trait from another. Family designs provide a clear proof of this deviation, thus of causality, because families allow direct observation of the inheritance of phenotypes and genotypes from parents to their offspring. However, most GWAs and most genetic epidemiological studies lack the familial component; therefore, causality needs to be considered by employing a different approach. Although the concepts of causal inference are widely discussed in the epidemiological and biostatistical literature, they seem to be insufficiently discussed in genetic epidemiology. Vansteelandt and Lange (2012) nicely introduce the basic concepts and discuss the assumptions that must be satisfied for a causal interpretation to hold. To illustrate the complexity of causality, the authors provide interesting examples showing that commonly used statistical methods can be misleading for inferring causal genetic effects.
Sun (2012) focuses on the recent development of pathway- and network-based methods, which hold the promise of integrating multiple biologically related genetic variants into a single analytical model. Complex traits involve multiple genes from different biological processes. These inter-connected genes often function synergistically within a biological system to contribute to a trait. Most GWAs only examine the association of a single SNP at a time and ignore the connectivity between the genes. Biological networks and pathways have been constructed to represent the functional or physical connectivity among genes, based on accumulated biological knowledge. In this review, Sun summarizes approaches to incorporating the pathway and network into the GWAs analysis. On the one hand, pathway and network information may assist in identifying a set of functionally related genetic associations with a trait, frame the hypothesis of a genetic association, and provide the putative functional roles of the findings. On the other hand, the statistical inference from the GWA data can also infer novel biological relationship among genes. Although still in their infancy, these pathway- and network-based methods have demonstrated their utility in analyzing GWA data, and can complement the limitations of the single SNP association approach.
High-throughput technologies such as next generation sequencing (NGS) are generating more data in a more cost-effective way than ever before. However, the deluge of data will not necessarily improve our knowledge of the genetics of human diseases, unless the corresponding issues in study design and methodology are well considered and properly developed. The success of GWAs demonstrates the tremendous value of efforts in design and method development. We anticipate that further investments in these fields, as discussed in detail in this special issue, will lead to continuous progress in understanding human diseases in the post-GWAs era.
Acknowledgments
The authors gratefully acknowledge France Gagnon and Sarah Namer for their invaluable organizational support in the preparation of this Special Issue. AZ acknowledges funding by the European Union (BiomarCare, grant no.: HEALTH-2011-278913), the German Ministry of Education and Research (CARDomics, grant nos.: 01KU0908A and 01KU0908B; Phenomics, grant no.: 0315536F), and the DFG excellence cluster “Inflammation at Interfaces.” YVS was partly supported by National Institutes of Health grant HL100245 and MD005964.
Footnotes
Conflicts of interest None.
Contributor Information
Andreas Ziegler, Email: ziegler@imbs.uni-luebeck.de, Institut für Medizinische Biometrie und Statistik, Universität zu Lübeck, Universitätsklinikum Schleswig–Holstein, Campus Lübeck, Maria-Goeppert-Str. 1, 23562 Lübeck, Germany.
Yan V. Sun, Email: yvsun@emory.edu, Department of Epidemiology, Department of Biomedical Informatics, Emory University, Atlanta, GA, USA.
References
- Almasy L. The role of phenotype in gene discovery in the whole genome sequencing era. Hum Genet. 2012 doi: 10.1007/s00439-012-1191-1. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann A, Weber P, Bader D, Preuß M, Binder EB, Müller-Myhsok B. A beginners guide to SNP calling from high-throughput DNA-sequencing data. Hum Genet. 2012 doi: 10.1007/s00439-012-1213-z. [DOI] [PubMed] [Google Scholar]
- Aschard H, Lutz S, Maus B, Duell EJ, Fingerlin T, Chatterjee N, Kraft P, Van Steen K. Challenges and opportunities in genome-wide environmental interaction (GWEI) studies. Hum Genet. 2012 doi: 10.1007/s00439-012-1192-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Wilson JE, Wilson AF. Linkage analysis in the next-generation sequencing era. Hum Hered. 2011;72:228–236. doi: 10.1159/000334381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biau G. Analysis of a random forests model. J Mach Learn Res. 2012;13:1063–1095. [Google Scholar]
- Bouchard C, Perusse L. Genetics of obesity. Annu Rev Nutr. 1993;13:337–354. doi: 10.1146/annurev.nu.13.070193.002005. [DOI] [PubMed] [Google Scholar]
- Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet. 2003;361:598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- Clerget-Darpoux F, Elston RC. Are linkage analysis and the collection of family data dead? Prospects for family studies in the age of genome-wide association. Hum Hered. 2007;64:91–96. doi: 10.1159/000101960. [DOI] [PubMed] [Google Scholar]
- Cook NR. Use and misuse of the receiver operating characteristic curve in risk prediction. Circulation. 2007;115:928–935. doi: 10.1161/CIRCULATIONAHA.106.672402. [DOI] [PubMed] [Google Scholar]
- Cortessis VK, Thomas DC, Levine AJ, Breton CV, Mack TM, Siegmund KD, Haile RW, Laird PW. Environmental epigenetics: prospects for studying epigenetic mediation of exposure-response relationships. Hum Genet. 2012 doi: 10.1007/s00439-012-1189-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devroye L, Györfi L, Lugosi G. A probabilistic theory of pattern recognition. Berlin: Springer; 1996. [Google Scholar]
- Fitze G, Cramer J, Ziegler A, Schierz M, Schreiber M, Kuhlisch E, Roesner D, Schackert HK. Association between c135G/A genotype and RET proto-oncogene germline mutations and phenotype of Hirschsprung’s disease. Lancet. 2002;359:1200–1205. doi: 10.1016/S0140-6736(02)08218-1. [DOI] [PubMed] [Google Scholar]
- Fitze G, Appelt H, König IR, Görgens H, Stein U, Walther W, Gossen M, Schreiber M, Ziegler A, Roesner D, Schackert HK. Functional haplotypes of the RET proto-oncogene promoter are associated with Hirschsprung disease (HSCR) Hum Mol Genet. 2003;12:3207–3214. doi: 10.1093/hmg/ddg354. [DOI] [PubMed] [Google Scholar]
- Freimer N, Sabatti C. The human phenome project. Nat Genet. 2003;34:15–21. doi: 10.1038/ng0503-15. [DOI] [PubMed] [Google Scholar]
- Gudgeon JM, McClain MR, Palomaki GE, Williams MS. Rapid ACCE: experience with a rapid and structured approach for evaluating gene-based testing. Genet Med. 2007;9:473–478. doi: 10.1097/gim.0b013e3180a6e9ef. [DOI] [PubMed] [Google Scholar]
- Haddow JE, Palomaki GE. A model process for evaluating data on emerging genetic tests. In: Khoury MJ, Little J, Burke W, editors. Human genome epidemiology: scope and strategies. New York: Oxford University Press; 2004. pp. 217–233. [Google Scholar]
- Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, Heintzman N, Ren B, Fu XD, Topol EJ, Rosenfeld MG, Frazer KA. 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature. 2011;470:264–268. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill AB. The environment and disease: association or causation? Proc R Soc Med. 1965;58:295–300. doi: 10.1177/003591576505800503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff JA, Junkins HA, Mehta JP, Manolio TA. [Accessed May 16, 2012];A catalog of published genome-wide association studies. 2012 http://www.genome.gov/26525384.
- Howick J, Glasziou P, Aronson JK. The evolution of evidence hierarchies: what can Bradford Hill’s ‘guidelines for causation’ contribute? J R Soc Med. 2009;102:186–194. doi: 10.1258/jrsm.2009.090020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruppa J, Ziegler A, König IR. Risk estimation and risk prediction using machine learning methods. Hum Genet. 2012 doi: 10.1007/s00439-012-1194-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, Bieche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- Pepe MS, Etzioni R, Feng Z, Potter JD, Thompson ML, Thornquist M, Winget M, Yasui Y. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001;93:1054–1061. doi: 10.1093/jnci/93.14.1054. [DOI] [PubMed] [Google Scholar]
- Pepe MS, Janes H, Longton G, Leisenring W, Newcomb P. Limitations of the odds ratio in gauging the performance of a diagnostic, prognostic, or screening marker. Am J Epidemiol. 2004;159:882–890. doi: 10.1093/aje/kwh101. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Rosenberg NA. Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association mapping in structured populations. Am J Hum Genet. 2000;67:170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MD. The success of pharmacogenomics in moving genetic association studies from bench to bedside: implementation of personalized medicine in the post-GWAS era. Hum Genet. 2012 doi: 10.1007/s00439-012-1221-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann H-E, Barrett JH, König IR, Stevens S, Szymczak S, Trégouët D-A, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Balmforth AJ, Baessler A, Ball SG, Strom TM, Brænne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H Wellcome Trust Case Control Consortium, Cardiogenics Consortium. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schunkert H, Götz A, Braund P, Tregouet D-A, McGinnis R, Mangino M, Linsel-Nitschke P, Cambien F, Hengstenberg C, Stark K, Blankenberg S, Tiret L, Ducimetiere P, Schreiber S, El Mokhtari NE, Hall AS, Dixon RJ, Goodall AH, Liptau H, Pollard H, Schwarz DF, Hothorn LA, Wichmann H-E, König IR, Fischer M, Meisinger C, Ouwehand W, Cardiogenics Consortium. Deloukas P, Thompson JR, Erdmann J, Ziegler A, Samani NJ. Repeated replication and a prospective meta-analysis of the association between chromosome 9p21.3 and coronary artery disease. Circulation. 2008;117:1675–1684. doi: 10.1161/CIRCULATIONAHA.107.730614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schunkert H, König IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AFR, Barbalic M, Gieger C, Absher D, Aherrahrou Z, Allayee H, Altshuler D, Anand SS, Andersen K, Anderson JL, Ardissino D, Ball SG, Balmforth AJ, Barnes TA, Becker DM, Becker LC, Berger K, Bis JC, Boekholdt SM, Boerwinkle E, Braund PS, Brown MJ, Burnett MS, Buysschaert I, Carlquist JF, Chen L, Cichon S, Codd V, Davies RW, Dedoussis G, Dehghan A, Demissie S, Devaney JM, Diemert P, Do R, Doering A, Eifert S, El Mokhtari NE, Ellis SG, Elosua R, Engert JC, Epstein SE, de Faire U, Fischer M, Folsom AR, Freyer J, Gigante B, Girelli D, Gretarsdottir S, Gudnason V, Gulcher JR, Halperin E, Hammond N, Hazen SL, Hofman A, Horne BD, Illig T, Iribarren C, Jones GT, Jukema JW, Kaiser MA, Kaplan LM, Kastelein JJP, Khaw K-T, Knowles JW, Kolovou G, Kong A, Laaksonen R, Lambrechts D, Leander K, Lettre G, Li M, Lieb W, Loley C, Lotery AJ, Mannucci PM, Maouche S, Martinelli N, McKeown PP, Meisinger C, Meitinger T, Melander O, Merlini PA, Mooser V, Morgan TM, Mühleisen TW, Muhlestein JB, Münzel T, Musunuru K, Nahrstaedt J, Nelson CP, Nöthen MM, Olivieri O, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund KD. Statistical approaches for the analysis of DNA methylation microarray data. Hum Genet. 2011;129:585–595. doi: 10.1007/s00439-011-0993-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YV. Integration of biological networks and pathways with genetic association studies. Hum Genet. 2012 doi: 10.1007/s00439-012-1198-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vansteelandt S, Lange C. Causation and causal inference in genetic epidemiology. Hum Genet. 2012 doi: 10.1007/s00439-012-1208-9. [DOI] [PubMed] [Google Scholar]
- Wald NJ, Hackshaw AK, Frost CD. When can a risk factor be used as a worthwhile screening test? BMJ. 1999;319:1562–1565. doi: 10.1136/bmj.319.7224.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijsman EM. The role of large pedigrees in an era of high-throughput sequencing. Hum Genet. 2012 doi: 10.1007/s00439-012-1190-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AF, Ziegler A. Lessons learned from Genetic Analysis Workshop 17: transitioning from genome-wide association studies to whole-genome statistical genetic analysis. Genet Epidemiol. 2011;35:S107–S114. doi: 10.1002/gepi.20659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME, Visscher PM. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitlen N, Kraft P. Heritability in the GWAS era. Hum Genet. 2012 doi: 10.1007/s00439-012-1199-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler A, König IR. A statistical approach to genetic epidemiology: concepts and applications: with an e-learning course by Friedrich Pahlke. Second edn. Weinheim: Wiley-VCH; 2010. [Google Scholar]
- Ziegler A, Koch A, Krockenberger K, Großhennig A. Personalized medicine using DNA biomarkers: A review. Hum Genet. 2012 doi: 10.1007/s00439-012-1188-9. [DOI] [PMC free article] [PubMed] [Google Scholar]