Abstract
Intrahepatic cholangiocellular carcinoma (ICC) is the second most common type of primary liver cancer. However, its tumor heterogeneity and molecular characteristics are largely unknown. In this study, we conducted transcriptomic profiling of 23 ICC and combined hepatocellular cholangiocarcinoma tumor specimens from Asian patients using Affymetrix mRNA and Nanostring microRNA microarrays to search for unique gene signatures linked to tumor subtypes and patient prognosis. We validated the signatures in additional 68 ICC cases derived from Caucasian patients. We found that both mRNA and microRNA expression profiles could independently classify Asian ICC cases into two main subgroups, one of which shared gene expression signatures with previously identified hepatocellular carcinoma (HCC) with stem cell gene expression traits. ICC-specific gene signatures could predict survival in Asian HCC cases and independently in Caucasian ICC cases. Integrative analyses of the ICC-specific mRNA and microRNA expression profiles revealed that a common signaling pathway linking miR-200c signaling to epithelial-mesenchymal transition (EMT) was preferentially activated in ICC with stem cell gene expression traits. Inactivation of miR-200c resulted in an induction of EMT while activation of miR-200c led to a reduction of EMT including a reduced cell migration and invasion in ICC cells. We also found that miR-200c and NCAM1 expression were negatively correlated and their expression levels were predictive of survival in ICC samples. NCAM1, a known hepatic stem/progenitor cell marker, was experimentally demonstrated to be a direct target of miR-200c. Conclusion: Our results indicate that ICC and HCC share common stem-like molecular characteristics and poor prognosis. We suggest that the specific components of EMT may be exploited as critical biomarkers and clinically relevant therapeutic targets for an aggressive form of stem cell-like ICC.
Introduction
Primary liver cancer (PLC) is the second most lethal cancer for men in the world.(1) Intrahepatic cholangiocellular carcinoma (ICC) is the second most common type of PLC. While ICC is much less common than hepatocellular carcinoma (HCC), its incidence has increased drastically over the past two decades.(2;3) However, the molecular pathogenesis of ICC is largely unknown. Understanding of the tumor biology of HCC and ICC that contributes to tumor heterogeneity is paramount in developing effective therapies to improve patient outcome.
The cellular origin of HCC and ICC has been subject to intense debate in recent years. It is thought that HCC is derived from hepatocyte while ICC arises from intrahepatic biliary epithelium. However, a mixed form of HCC and ICC, also known as combined hepatocellular cholangiocarcinoma (CHC), has been described to have distinct clinicopathological features but morphological intermediates of HCC and ICC, suggesting that HCC and ICC could share the same cellular origin.(4–6) Recent studies utilizing high resolution genomic approaches have shed light on the revelation of cellular origin of HCC and suggest that a subset of HCC contains stem cell-like features.(7–10) For example, a subset of tumor cells isolated from HCC patients are tumor initiating cells with stem cell traits.(11–14) Moreover, HCC may share an ICC-like gene expression trait.(15) These results are consistent with the cancer stem cell (CSC) hypothesis, which suggests that most tumor cells are derived from undifferentiated cells with stem-like capabilities and that both ICC and HCC may share the same cellular origin of hepatic stem/progenitor cells.
Global mRNA and microRNA profiling approaches have been proven to be effective in identifying genes critical to HCC.(8;9;16–22) In this study, we used both mRNA and microRNA profiling approaches to determine tumor heterogeneity and molecular characteristics of ICC. We found that ICC samples consist of at least two main subtypes that share similar molecular activities with HCC linked to stem cell-like gene expression and patient survival. Integrative genomic analyses revealed that genes and microRNAs involved in epithelial-mesenchymal transition (EMT) are altered in stem-like ICCs. Our results shed light on ICC diagnosis and may open new avenues for therapeutic interventions for targeting poor prognostic ICC patients.
Experimental Procedures
Human Subjects
ICC and CHC tissues were obtained with informed consent from Asian patients who underwent curative resection between 2002 and 2003 at the Liver Cancer Institute and Zhongshan Hospital (Fudan University, Shanghai, China) and between 2008 and 2010 at the Kanazawa University Hospital (Ishikawa, Japan). Sample collection was approved by the Institutional Review Board of the corresponding institutes and recorded by the NIH Office of Human Subjects Research. A total of 23 ICC and CHC cases were used to build mRNA and microRNA signatures. The initial diagnosis was made based on serological test and imaging, and was confirmed histopathologically by pathologists. The characteristics of 68 Caucasian ICC patients from an independent cohort were described recently.(23)
Cell line, culture and transfection
HuCCT1 and HUH28 cell lines were used for miR-200c functional studies. These cell lines were obtained from the Japanese Collection of Research Bioresources Cell Bank and were cultured in RPMI supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 0.1 mg/ml streptomycin, and 2 mmol/L L-glutamine. An immortalized human cholangiocyte-derived cell line, H69, kindly provided by Dr. Gregory Gores (Mayo Clinic), was cultured as previously described.(24) A luciferase reporter containing an upstream 0.9kb fragment of pri-miR-200c was kindly provided by Dr. Li Wang from University of Utah School of Medicine.(25) A detailed description of other transfection reagents, methodologies such as cell culture, cell proliferation and apoptosis assays, luciferase assay, immunohistochemical analysis, and cell migration and invasion assays can be found in the supplemental materials.
Microarray Processing
Total RNA was extracted from frozen tissue using TRIzol (Invitrogen) according to the manufacturer’s protocol. Only RNA samples with good RNA quality as confirmed with the Agilent 2100 Bioanalyzer (Agilent Technologies) were included for array study. Gene expression profiling of 23 tumor samples (16 ICC, 7 CHC), as well as 7 paired non-cancerous liver tissues from ICC patients and 7 benign liver lesions (5 FNH, 2 adenoma) was carried out on Affymetrix GeneChip Human Gene-ST 1.0 arrays according to the manufacturer’s protocol and processed as previously described.(26) Affymetrix gene expression arrays obtained from different platforms were combined with the match probes package in R. Raw gene expression data were normalized using the Robust Multi-array Average (RMA) method and global median centering. For genes with more than one probe set, the mean gene expression was calculated. Total RNA was used for the nCounter microRNA platform. All sample preparation and hybridization was performed according to the manufacturer’s instructions. All hybridization reactions were incubated at 65 °C for a minimum of 12 h. Hybridized probes were purified and counted on the nCounter Prep Station and Digital Analyzer (NanoString) following the manufacturer’s instructions. For each assay, a high-density scan was performed. For platform validation using synthetic oligonucleotides, NanoString nCounter microRNA raw data was normalized for lane-to-lane variation with a dilution series of six spike-in positive controls. The sum of the six positive controls for a given lane was divided by the average sum across lanes to yield a normalization factor, which was then multiplied by the raw counts in each lane to give normalized values. Raw mRNA and microRNA data are accessible through the accession numbers GSE32879 and GSE32957 at the NCBI Gene Expression Omnibus (GEO) database. Other statistical methods can be found in the supplemental materials.
Real-time RT-PCR analysis
Total RNA was subjected to qRT-PCR. Mature microRNAs and other mRNAs were analyzed using the TaqMan microRNA Assays and Gene Expression Assays, respectively, in accordance with manufacturer’s instructions (Applied biosystems, Foster City, CA). All RT reactions were run in a GeneAmp PCR 9700 Thermocycler (Applied biosystems). Probes used for the analyses were as follows: ZEB1, Hs00232783_m1; ZEB2, Hs00207691_m1 ; VIM, Hs00185584_m1; CDH1, Hs01023894_m1; CDH2, Hs00983056_m1; MYC, Hs00905030_m1; Hsa-miR-200c, 002300 ; Hsa-miR-141, 000463 (Applied Biosystems). The experiments were performed in triplicate. The Taqman gene assay for 18s and actin was used to normalize the relative abundance of mRNA. RNU6B RNA was used as a control for miR-200c.
Results
Profiling of mRNA expresssion in ICC and CHC
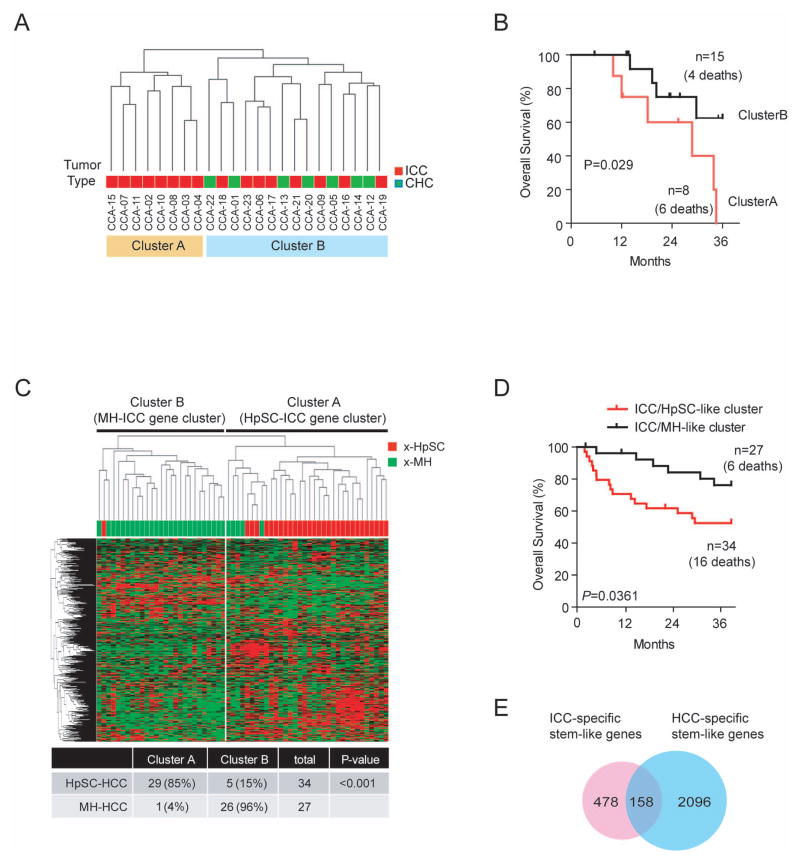
We performed transcriptomic analyses of 30 retrospectively collected ICC and CHC clinical specimens from Chinese (n=13) and Japanese (n=10) patients with seven paired non-tumor liver tissues from ICC patients using Affymetrix GeneChip Human Gene-ST arrays. Five focal nodular hyperplasia (FNH) cases and two adenomas were also included as benign tumors of the liver. Clinical features of these ICC and CHC cases are included in Table S1. Multidimensional scaling analysis revealed that malignant tumor samples were mainly different from benign tumors and non-tumor tissues, suggesting that malignant tumors have a vast different gene expression profile (Figure S1). To determine tumor heterogeneity, unsupervised hierarchical clustering analysis of 23 ICC and CHC samples based on all genes was conducted. The result revealed that tumor samples can be divided into two main groups, i.e., cluster-A and cluster-B (Figure 1A). Kaplan-Meier survival analysis revealed that ICC cases in cluster-A had a shorter survival than those in cluster-B (Figure 1B). These results suggest that gene expression and tumor biology differ significantly among different ICC tumor samples.
Figure 1.
Heterogeneity of Asian ICC and HCC cases based on mRNA expression profiling. (A) Unsupervised hierarchical clustering of 23 ICC and CHC cases based on global mRNA expression using centered correlation and average linkage. The red and orange bars beneath the dendrogram indicate ICC and ICC with CHC features, respectively. ICC cases in cluster-A are referred as HpSC-ICC and ICC cases in Cluster-B are referred as MH-ICC. (B) Kaplan-Meier analysis of ICC cases based on the classification of cluster-A and cluster-B from panel A. (C) Hierarchical clustering of 61 extreme HCC cases (i.e., 34 x-HpSC HCC and 27 x-MH HCC) based on the expression of 636 ICC-specific genes. The red and green bars above heatmap indicate x-HpSC and x-MH, respectively. A summary of HCC cases based on the dendrogram classification with the ICC-specific signature is included. Chi-squared test was used to determine the correct classification. (D) Kaplan-Meir plot of 61 extreme HCC cases based on the classification by the 636-gene signature into HpSC-ICC and MH-ICC clusters. (E) Venn-diagram of stem-like ICC genes and stem-like HCC genes.
We previously identified two HCC subgroups, one resembling gene expression signatures of hepatic stem cells (referring as HpSC-HCC) and the other similar to mature hepatocyte (referring as MH-HCC). To determine if ICC subgroups have different gene expression profiles compared to HCC subgroups, we randomly selected two groups of HCC samples, each consisting of 23 age and gender matched HCC cases from an existing cohort of 246 HCC samples with available Affymetrix data (GEO accession number GSE14520) and compared them to 23 ICC and CHC tumors (Table S1). Unsupervised hierarchical clustering analysis revealed two main branches. All 8 cluster-A ICC samples were grouped with EpCAM+AFP+ HCC cases previously identified having a stem cell-like gene expression trait while all 8 cluster-B ICC samples were grouped with EpCAM− AFP− HCC cases with a mature hepatocyte-like gene expression trait (Figure S2A).(9) Similar results were obtained when ICC/CHC cases were compared to second group of 23 randomly selected HCC cases (Figure S2B). Seven CHC cases were split among two clusters. For the convenience of keeping track these ICC samples, we referred ICC cases in cluster-A as HpSC-ICC (i.e., hepatic stem cell-like ICC) and those in cluster-B as MH-ICC (i.e., mature hepatocyte-like ICC). These results indicate that both ICC and HCC are heterogeneous and their subgroups share similar gene expression profiles.
Next, we performed a class comparison analysis and identified 636 genes that are differentially expressed between 8 HpSC-ICC and 8 MH-ICC cases [univariate p<0.01; false discovery rate (FDR) <0.2] (Table S2). We then tested whether this 636 ICC-specific gene signature could independently classify HCC cases based on HpSC-like or MH-like features. We tested the robustness of the signature to discriminate HpSC-HCC from MH-HCC cases by examining 61 well-defined extreme HCC cases or x-HCC, i.e., those with top quartile EpCAM expression in HCC tissues and with >1000 ng/ml of serum AFP levels vs. those with bottom quartile EpCAM expression and with <20 ng/ml of serum AFP levels (Table S1). Hierarchical clustering analysis revealed that the 636 ICC-specific genes could nicely divide x-HpSC and x-MH HCC cases (Figure 1C) and were associated with HCC survival (Figure 1D). This 636 ICC-specific gene signature was also associated with survival in 139 remaining unstratified HCC cases from the original 246 HCC cases after excluding 46 randomly selected HCC cases and 61 extreme HCC cases used in the initial clustering analysis (Figure S2C). Venn diagram analysis indicated that 158 of 636 ICC-specific genes (25%) overlapped with previously identified stem-like HCC genes (Figure 1E). Consistent with the data in Figure 1C, 158 overlapping genes could significantly discriminate stem-like HCC cases from mature hepatocyte-like HCC cases (p<0.0001) and was associated with HCC survival (P=0.031) (Figure S3). The above data indicate that ICC cases could be classified into two main subtypes that are associated with stem-like or mature hepatocyte like gene expression traits, respectively, and that ICC and HCC may share common gene expression profiles reflecting their cellular origins.
Profiling of microRNA expression in ICC and CHC
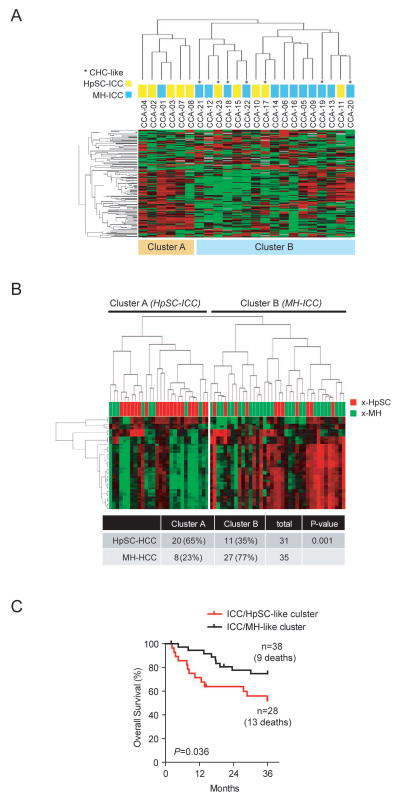
We used the NanoString nCounter microRNA Expression Assay platform to independently examine gene expression profiles of the same 30 ICC and CHC samples used above. Unsupervised clustering analysis based on the expression of all 700 human mature microRNAs revealed that ICCs were again divided into two main clusters, where 5 of 6 ICC cases in cluster A belong to HpSC-ICC and 7 of 10 cases in cluster-B belong to MH-ICC as assigned by mRNA expression (Figure 2A). Class comparison analysis revealed 23 microRNAs to be differentially expressed between HpSC-ICC and MH-ICC (p<0.05) (Table S3). This ICC-specific microRNA signature was further tested for its ability to classify the same HCC cohort described above with available microRNA expression data generated from an independent array platform (GEO accession number: GSE6857). Again, the ICC-specific microRNA signature could significantly discriminate well-defined extreme HCC subgroups and was associated with HCC survival (Figure 2B and 2C).
Figure 2.
Heterogeneity of Asian ICC cases revealed by microRNA expression profiling. (A) Unsupervised hierarchical clustering of 23 Asian ICC cases based on global microRNA expression using centered correlation and average linkage. The yellow and light blue bars beneath the dendrogram indicate HpSC-ICC and MH-ICC subgroups, respectively, as classified by mRNA expression profiling described in Fig 1A. *, CHC-like cases. (B) Hierarchical clustering of 61 extreme HCC cases based on the expression of 23 ICC-specific microRNAs. The red and green bars above heatmap indicate x-HpSC and x-MH, respectively. A summary of HCC cases based on the dendrogram classification with the ICC-specific signature is included. Chi-squared test was used to determine the correct classification. (C) Kaplan Meir plot of 61 extreme HCC cases based on the clustering results of panel B.
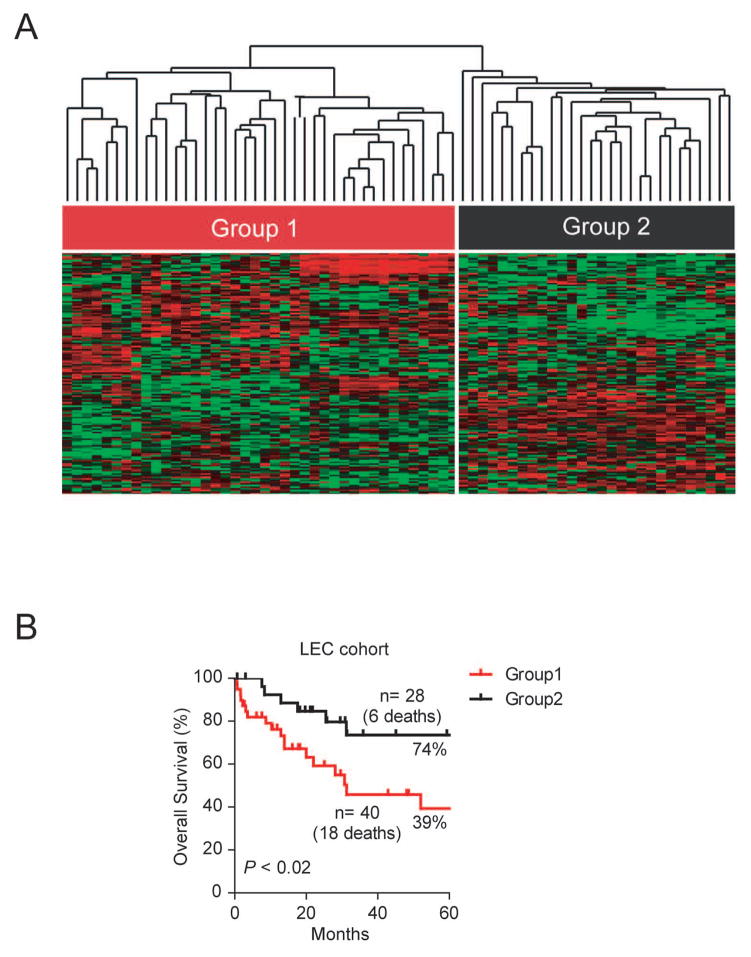
Our results indicate that HpSC-ICC and MH-ICC cases can be independently classified by mRNA and microRNA expression, which suggest that these two subgroups have a clearly measurable difference at the gene expression levels. We hypothesized that those HpSC-ICC tumors share the same stem-like traits with HCC with poor survival, and patients with this type of ICC would have a poor outcome. To determine if ICC-specific gene signature is predictive of ICC patient survival, we performed hierarchical clustering analysis using 158 overlapping genes (described in Figure 1E) in 68 ICC cases from an independent cohort containing Caucasian patients (Figure 3A). Consistently, the 158 overlapping gene signature was significantly associated with patient survival in this cohort (p<0.02) (Figure 3B). Similar results were obtained when all 636 ICC-specific genes were used for this analysis (p<0.04; Figure S4).
Figure 3.
Validation of the ICC-specific gene signature in an independent ICC cohort as a survival predictor. (A) Hierarchical clustering of 68 Caucasian ICC cases based on the expression of 158 overlapping genes between stem-like ICC and stem-like HCC genes using centered correlation and average linkage. The heatmap depicts high (red) and low (green expression of these genes based on a log2 scale. (B) Kaplan-Meier plot of 68 ICC patients based on the dendrogram classification from panel A.
Integrative pathway analysis of ICC-specific mRNA and microRNA
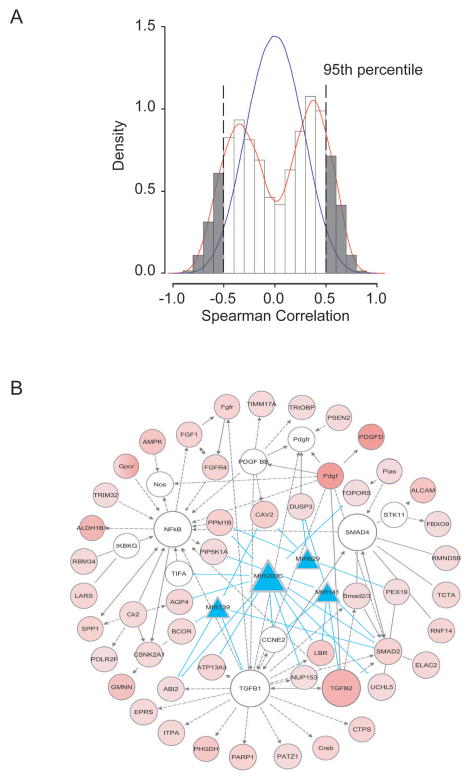
Since microRNA and mRNA are functionally linked, we hypothesized that the expression levels between ICC-specific mRNAs and ICC-specific microRNAs would be highly correlative as they both are associated with the same stem cell-like phenotype. We plotted the density distribution of Spearman correlation coefficients of 636 experimentally-derived genes and 23 experimentally-derived microRNAs (Figure 4A). This analysis revealed that there was a clear enrichment of correlative mRNA-microRNA pairs derived from these signatures since a positive correlative curve shifted to the right and a negative correlative curve shifted to the left when compared to a normal distribution curve derived from a global correlation of all available mRNA and microRNA probes (Figure 4A). A correlation coefficient of 0.5, corresponding to the 95th percentile of the 100-fold random permutations, was used as the cutoff threshold for positive correlation. These results indicated that ICC-specific mRNAs and microRNAs are enriched in the experimentally derived signatures and they are highly correlative.
Figure 4.
Integrative analyses of ICC-specific mRNA and microRNAs based on spearman correlation and Ingenuity Pathway. (A) Correlation between ICC-specific 636 mRNA and 23 microRNA signatures. (B) The top nine gene networks of signaling including TGF-β, Smad4 and NF-κB pathways activated in stem-like ICC tumors. Red shaded ovals represent up-regulated genes in HpSC-ICC tumors, and open ovals represent genes that are not on the list of significant genes but are reported to be associated with the network. Blue shaded triangles represent down-regulated microRNAs specific to HpSC-ICC tumors. The open ovals that are labeled as TGF-β, Smad4 and NF-κB represent molecular nodes related to their respective signaling pathways. Arrows represent positive regulation of gene expression, with solid arrows indicating direct regulation and broken arrows indirect regulation. Blue lines connecting between microRNA and genes represent direct targeting predicted by TargetScan.
To determine if there is any enrichment of affected networks associated with ICC subgroups, we combined significantly correlative mRNA-microRNA pairs and performed pathway analysis using Ingenuity Pathway Analysis (IPA, verson 9.0) that incorporates microRNA-mRNA target relationships from TargetScan. Among 1077 mRNA-microRNA pairs were identified by this analysis, 479 pairs showed negative correlation. Among the top nine networks (Table S4), five microRNAs including miR-200c and miR-141 that are encoded by the same transcript were negatively correlated with genes in the TGF-β, NF-κB and Smad signaling pathways (Figure 4B). A common link between ICC specific mRNA and microRNA seemed to be related to epithelial-mesenchymal transition (EMT) where all three pathways are known regulators. Consistently, known stem cell-related genes such as POU5F1 (Oct4), NANOG, NCAM1 and (PROM1 (CD133) were much more abundantly expressed in HpSC-ICC than MH-ICC cases (Figure S5A). TGFB1 was also significantly elevated in HpSC-ICC compared to MH-ICC. However, no difference in EpCAM expression was observed among these two subgroups. An elevated expression of NCAM1 and TGFB1 in a majority of HpSC-ICC cases was confirmed by immunocytochemistry analysis (IHC) (Figure S5B).
Among the affected networks, it was noticeable that mir-200c appeared a common molecular note linking to EMT as it had a direct interaction with many of the affected genes in this pathway (Figure 4B). Consistently, the expression level of miR-200c was associated with overall survival and disease-free survival in ICC cases (Figure S6). These data suggested that miR-200c may play an important role in maintaining HpSC-like phenotype.
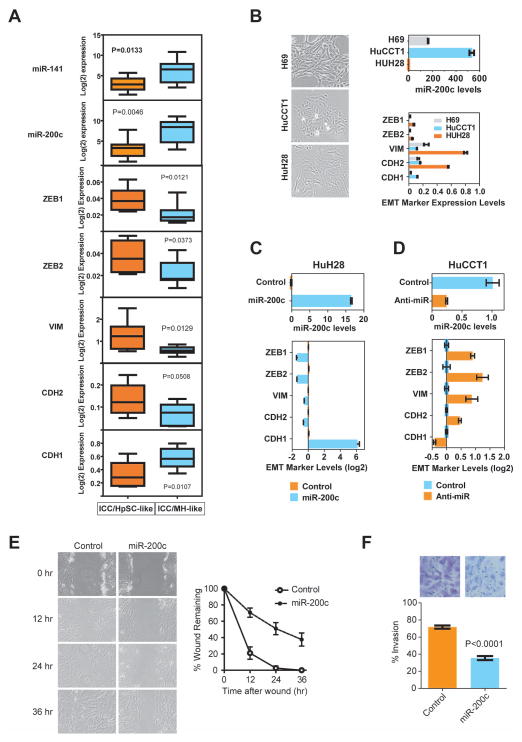
To determine whether EMT was functionally linked to HpSC-ICC cells, we first analyzed representative expression levels of EMT markers in ICC specimens by qRT-PCR. Consistently, mesenchymal markers such as ZEB1, ZEB2, CDH2 and VIM were more abundantly expressed, while an epithelial marker, CDH1, and miR-141/miR-200c were much less abundantly expressed in HpSC-ICC cases as compared to MH-ICC cases (Figure 5A). Next, we determined if an altered miR-200c expression could lead to EMT in ICC cells. We selected two ICC cell lines that represent two opposite ends of the EMT spectrum. A non-malignant H69 cell line derived from normal human intrahepatic cholangiocytes was included as a control.(24) HuH28 cells had fibroblast-like cell morphology with mesenchymal appearances and expressed very low levels of miR-200c but high levels of mesenchymal markers, while HuCCT1 cells had cobblestone-like cell morphology with epithelial appearances and expressed high levels of miR-200c but low levels of mesenchymal markers (Figure 5B). The miR-200c level was also relatively high in H69 cells with epithelial morphology. Transient transfection of miR-200c oligos in HuH28 cells induced a reversed EMT from a cobblestone-like to a mesenchymal-like morphology with a suppression of genes that mediate EMT (Figure 5C). Conversely, transfection of an anti-miR-200c oligo in HuCCT1 resulted in an induction of mesenchymal markers (Figure 5D). In addition, overexpresson of miR-200c suppressed cell migration (Figure 5E) and invasion (Figure 5F) in HuH28 cells. However, miR-200c did not affect cell proliferation and apoptosis in HUH28 cells as measured by MTT and TUNEL assays (Figure S7).
Figure 5.
Inactivation of miR-200c/miR-141 and activation of EMT-related genes are associated with stem-like ICC. (A) Expression analyses of miR-141/miR-200c transcripts and EMT-specific markers based qRT-PCR data in 8 HpSC-ICC and 8 MH-ICC samples classified by gene clustering from Figure 1. The horizontal lines in the box plots represent the median, the boxes represent the interquartile range, and the whiskers represent the 10th and 90th percentiles. A nonparametric test was used to compare the two groups and p values are indicated. (B) Expression of miR-200c and EMT-specific genes in HuH28, HuCCT1 and H69 cells as analyzed by qRT-PCR. (C) Expression of EMT-specific genes in HuH28 cells transduced with miR-200c as analyzed by qRT-PCR. (D) Expression of EMT-specific genes in HuCCT1 cells transduced with an anti-miR-200c oligo as analyzed by qRT-PCR. (E) Cell migration of HuH28 cells transduced with miR-200c as determined by the wound healing assay. (F) Cell invasion of HuH28 cells transduced with miR-200c as determined by the Boyden chamber cell invasion assay. Representative images are shown.
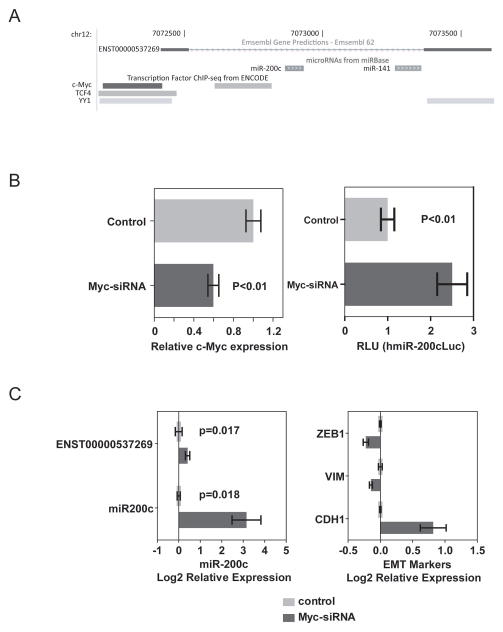
Analyses of the genomic region encoding the human miR-200c/miR-141 locus at the UCSC Genome Browser revealed that miR-200c and miR-141 are derived from a single transcript encoded by a predicted gene (ENST00000537269) (Figure 6A). The available Chip-Seq data revealed several transcriptional factors such as c-Myc and TCF4 to be preferentially bound to the immediate 5′ upstream sequence of the predictive transcription initiation site. To determine whether c-myc directly regulates miR-200c expression, we silenced c-Myc expression with a c-myc specific siRNA in HuH28 cells and examined the activity of a luciferase reporter containing an upstream 0.9 kb fragment of pri-miR-200c (Zhang Y et al, BBRC 2011) (Figure 6B). Consistently, we found that inhibition of c-Myc resulted in an increased hmiR-200cLuc activity. Moreover, c-myc siRNA could effectively induce endogenous miR-200c expression while suppress mesenchymal markers but induce epithelial marker (Figure 6C).
Figure 6.
c-Myc-mediated silencing of miR-200c and induction of EMT. (A) The genome position of ENST00000537269 encoding miR-200c and miR-141, based on Ensembl Gene Predictions using UCSC Genome Browser. (B) Effect of c-Myc siRNA on miR-200cc-Myc expression (left panel) and the miR-200c promoter luciferase activity (right panel) in HuCCT1 cells. (C) Effect of c-Myc siRNA on endogenous levels of miR-200c and induction of EMT-related gene expression in HuCCT1 cells.
NCAM1 as a direct target of miR-200c
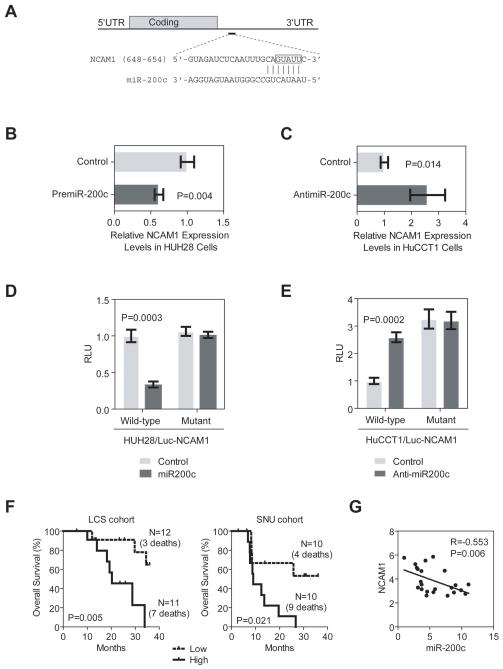
Since several stem/progenitor cell-related genes such as POU5F1, NANOG, MYC, TGFB1, NCAM1 and PROM1 are overexpressed in HpSC-ICC cases (Figure S5), we reasoned that some of these genes may be targets of miR-200c. TargetScan analysis (TargetScanHuman 6.0) revealed that only NCAM1 contained a classical and evolutionarily conserved miR-200c binding site at its 3′UTR (Figure 7A). Ectopic expression of miR-200c in HuH28 cells resulted in a reduction (Figure 7B) while inhibition of miR-200c in HuCCT1 cells lead to an increased expression of NCAM1 (Figure 7C). To further determine whether NCAM1 was a bona fide target of miR-200c-mediated silencing, the miR-200c binding site was cloned into a luciferase reporter. We found that forced expression of miR-200c in HUH28 cells resulted in decreased luciferase activity when a wild-type sequence but not a mutant sequence was present (Figure 7D). Moreover, inhibition of miR-200c in HuCCT1 cells resulted in increased luciferase activity only from a wild-type reporter (Figure 7E). Consistently, ICC cases with high levels of NCAM1 had a worse survival compared to those with low NCAM1 expression (Figure 7F). Moreover, a significant inverse correlation was observed between miR-200c and NCAM1 (Figure 7G).
Figure 7.
Functional interactions between miR-200c and NCAM1. (A) Predicted duplex formation between the 3′UTR sequences of human NCAM1 and miR-200c where vertical bars represent the paired seed sequences. The box highlights the the nucleotides that were changed to CAUAA in a mutant luciferase reporter. (B) Effect of premiR-200c oligo on NCAM1 expression in HuH28 cells as determined by qRT-PCR. (C) Effect of anti-miR-200c oligo on NCAM1 expression in HuCCT1 cells as determined by qRT-PCR. (D) Luciferase activities of wild-type and mutant reporters in HuH28 cells with or without the presence of pre-miR-200c oligo. (E) Luciferase activities of wild-type and mutant reporters in HuCCT1 cells with or without the presence of anti-miR200c oligo. Each experiment was repeated at least three times, and the expression value was shown as the mean ± standard deviation. (F) Kaplan-Meier estimates of overall survival according to expression of NCAM1 in ICC cases from LCS and SNU cohorts. NCAM1 expression values were dichotomized into low and high groups using the within cohort median expression value as a cutoff. (G) Spearman correlation analysis of NCAM1 and miR-200c expression data as determined by mRNA and microRNA arrays.
Discussion
Similar to HCC, ICC is heterogeneous in clinical presentation although our knowledge related to its tumor biology is limited. Several recent studies have begun dissecting the molecular pathogenesis of ICC including functional roles of microRNA in ICC cells.(27;28) Recently, we have utilized global transcriptomic approaches to study HCC heterogeneity and have identified critical genetic loci functionally linked to hepatic CSCs with gene expression profiles resembling normal hepatic stem cells.(7;8) We have also used these approaches to study cholangiocarcinoma.(15;23) In this study, we examined whether ICC and HCC are distinct at the transcriptomic levels. Using two independent transcriptomics approaches, we found that ICC cases from Asian patients can be mainly divided into two subgroups with one resembling of stem-like HCC and other mature hepatocyte-like HCC. Consistently, we found that several known hepatic stem/progenitor cell-specific genes such as POU5F1 (Oct4), NANOG, MYC, TGFB1, NCAM1 and PROM1 are more abundantly expressed in stem-like ICC than mature hepatocyte-like ICC.(29) Moreover, both ICC-specific mRNA and microRNA signatures could independently predict HCC survival as well as ICC prognosis in Caucasian patients. These results are consistent with our recent finding that a subset of HCC may share an ICC-like gene expression trait.(15) Integrative pathway analyses revealed that an altered miR-200c signaling pathway linked to EMT may be responsible for the maintenance of stem-like ICC associated with poor prognosis. For example, we found that two significant microRNAs, i.e., miR-200c and miR-141 encoded by the same transcript, were negatively correlated with genes in the TGF-β, NF-κB and Smad signaling pathways. These two microRNAs share the same seed sequences and are predicted to have similar cellular functions. EMT is an important biological process contributing to embryogenesis and organ development.(30) Recently, components of EMT have been shown to be critical in promoting cancer invasion and metastasis.(31) TGF-β is essential for the induction of EMT during various stages of embryogenesis and plays an important role in carcinoma progression into an invasive state.(32–34) Smad signaling is essential for TGF-β induced EMT.(35) Furthermore, miR-200 family members including miR-141 and miR-200c induce epithelial differentiation, thereby suppressing EMT by inhibiting translation of mRNA for the EMT-activators ZEB1 and ZEB2.(36;37) miR-200 family members are functionally linked to EMT, in part via targeting ZEB1 and ZEB2, as well as cell migration, invasion and tumorigenicity.(36;38) These results suggest that the ZEB1-miR-200 feedback loop is critical for maintaining aggressive tumor features. In addition, we also found that miR-200c directly targets NCAM1. NCAM1 is highly expressed in hepatic stem cells and its function has been tightly linked to EMT.(29;39) Our results are consistent with the hypothesis that the miR-200-EMT gene axle may be functional critical to the development of stem-like ICC. Shared molecular activities including EMT and microRNA among HCC and ICC have been noted in recent publications.(40;41) Interestingly, abnormal regulation of EMT-related genes has been reported to be linked to HCC development.(42–44) However, no evidence has linked miR-200 to HCC development. Consistently, we found no evidence that miR-200c is silenced in stem-like HCC (data not shown). It is plausible that the miR-200c-EMT gene axis is a unique signaling pathway functionally important for stem-like ICC and could be exploited as molecular targets for ICC therapies. In addition, unlike HCC, EpCAM is not a good prognostic biomarker for ICC since its expression is highly elevated in both HpSC-ICC and MH-ICC (data not shown). Further studies involving in-depth analyses of miR-200c and EMT signaling, and utilizing relevant animal models would be needed to test the therapeutic relevance of these targets for ICC.
Human adult livers are believed to be comprised of maturational lineages of cells beginning intrahepatically near the portal triads referred to as canals of Hering.(45) This region is close to intrahepatic bile ducts and is believed to contain liver stem cells. It is suggested that liver stem cells may give rise to bipotent progenitor cells which have the potential to differentiate into both hepatocytic and cholangiocytic lineages. In principle, hepatic stem/progenitor cells could be the common cellular origin for both HCC and ICC. It is hypothesized that cancer progression is driven by the presence of CSC, which is also responsible for treatment resistance and tumor relapse.(46) CSCs have been demonstrated in a growing range of epithelial and other solid organ malignancies, suggesting that the majority of malignancies are dependent on such a compartment.(47) This model is attractive because it may help to address the heterogeneity of HCC and ICC, and could facilitate research strategies to define novel and effective therapies.(48) Consistently, studies on clinicopathological features of ICC suggest that some ICCs could arise from liver stem cells rather than from mature cholangiocytes.(49;50) Moreover, gene expression profiling revealed that some HCC cases contain ICC-like gene expression trait and embryonic stem cell-like traits.(15) These results suggest that certain type of ICC could be derived from the same cell origin that leads to HCC while distinct mechanisms may be evolved in the genesis of ICC.
CHC has been traditionally classified into three subtypes based on the histological description by Allen and Lisa in 1949.(51) These subtypes include type-A (collision or double cancer, which is referred to as separate HCC and ICC arising in the same liver), type-B (contiguous mass, which is referred to as admixed HCC/ICC such as fibrolamellar tumors) and type-C (transitional tumors, which is referred to as a tumor mass with cellular features of both HCC and ICC). In type-A tumors, the HCC and ICC lesions could be interpreted to be originated separately from hepatocyte and bile duct epithelium. Type-B tumors could follow the same mechanism as type-A since it is difficult to distinguish them based on histological data. Since both HCC and ICC cellular features are intimately associated in the type-C tumors, they have been interpreted as arising from the same site and sharing the same cell origin. In our study, two Chinese and five Japanese CHC cases belong to type-B and seven Korean CHC cases(15) belong to type-C. Hierarchical clustering analyses revealed that both type-B and type-C CHC samples could be divided into stem-like ICC and mature hepatocyte-like ICC, which are also associated with survival (Figure S8). While these results are not conclusive due to limited cases, they appear consistent with the hypothesis that both type-B and type-C CHC could originate from the same hepatic progenitor cells shared by HCC and CHC tumors. A new histological subtyping of CHC according to the WHO Classification based on the presence of stem-cell features has been proposed.(52) Long-term follow-up of larger cohorts is needed to define the clinical and biological behavior of all CHC cases.
Our analysis dissecting the heterogeneous ICC based on the expression of stem cell-like signatures could classify ICC cases into subgroups with more uniformed and prognostic phenotypes. In principle, targeting molecular pathways specific to each subpopulation would be more effective for the development of personalized clinical strategies. We suggested that the miR-200c associated EMT pathway and stem-cell activities may contribute to the development of the HpSC-ICC tumors. The association of EMT with poor prognosis is well known in many cancer types. Moreover, recent studies have demonstrated the critical role of miR200c in the control of stem/progenitor cell renewal and differentiation. Our findings are consistent with the hypothesis supporting the pivotal role of miR-200c in the aggressive progression of stem-like ICC.
Supplementary Material
Acknowledgments
This work was supported in part by the Intramural Research Program of the center for Cancer Research, the U.S. National Cancer Institute (Z01 BC 010313 and Z01 BC 010876).
We thank Drs. Gregory J. Gores for H69 cells, Kathleen C. Flanders and Lalage M. Wakefield for anti-TGF-beta1 antibody, and Li Wang for the miR-200c luciferase reporter. We also thank Dr. Xiaolin Wu and members of the microarray core at the NCI-SAIC for help on microarray analysis and Ms. Karen Yarrick for bibliographic assistance.
Abbreviations
- ICC
intrahepatic cholangiocarcinoma
- HCC
hepatocellular carcinoma
- CHC
combined hepatocellular cholangiocarcinoma
- PLC
primary liver cancer
- EMT
epithelial-mesenchymal transition
- CSC
cancer stem cell
- FNH
focal nodular hyperplasia
- GEO
gene expression omnibus
- HpSC-ICC
hepatic stem cell-like ICC
- MH-ICC
mature hepatocyte-like ICC
- x-HCC
extreme HCC
Footnotes
Potential conflict of interest: Nothing to report
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Martin R, Jarnagin W. Intrahepatic cholangiocarcinoma. Current management Minerva Chir. 2003;58:469–478. [PubMed] [Google Scholar]
- 3.Shaib Y, El-Serag HB. The epidemiology of cholangiocarcinoma. Semin Liver Dis. 2004;24:115–125. doi: 10.1055/s-2004-828889. [DOI] [PubMed] [Google Scholar]
- 4.Berthiaume EP, Wands J. The molecular pathogenesis of cholangiocarcinoma. Semin Liver Dis. 2004;24:127–137. doi: 10.1055/s-2004-828890. [DOI] [PubMed] [Google Scholar]
- 5.Komuta M, Spee B, Vander BS, De VR, Verslype C, Aerts R, et al. Clinicopathological study on cholangiolocellular carcinoma suggesting hepatic progenitor cell origin. Hepatology. 2008;47:1544–1556. doi: 10.1002/hep.22238. [DOI] [PubMed] [Google Scholar]
- 6.Zhou H, Wang H, Zhou D, Wang H, Wang Q, Zou S, et al. Hepatitis B virus-associated intrahepatic cholangiocarcinoma and hepatocellular carcinoma may hold common disease process for carcinogenesis. Eur J Cancer. 2010;46:1056–1061. doi: 10.1016/j.ejca.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410–416. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 8.Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY, et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012–1024. doi: 10.1053/j.gastro.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamashita T, Forgues M, Wang W, Kim JW, Ye Q, Jia H, et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68:1451–1461. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
- 10.Cairo S, Wang Y, de RA, Duroure K, Dahan J, Redon MJ, et al. Stem cell-like micro-RNA signature driven by Myc in aggressive liver cancer. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1009009107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma S, Chan KW, Hu L, Lee TK, Wo JY, Ng IO, et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132:2542–2556. doi: 10.1053/j.gastro.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 12.Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, et al. Significance of CD90(+) Cancer Stem Cells in Human Liver Cancer. Cancer Cell. 2008;13:153–166. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 13.Haraguchi N, Ishii H, Mimori K, Tanaka F, Ohkuma M, Kim HM, et al. CD13 is a therapeutic target in human liver cancer stem cells. J Clin Invest. 2010;120:3326–3339. doi: 10.1172/JCI42550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang Y, Kitisin K, Jogunoori W, Li C, Deng CX, Mueller SC, et al. Progenitor/stem cells give rise to liver cancer due to aberrant TGF-beta and IL-6 signaling. Proc Natl Acad Sci U S A. 2008;105:2445–2450. doi: 10.1073/pnas.0705395105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woo HG, Lee JH, Yoon JH, Kim CY, Lee HS, Jang JJ, et al. Identification of a cholangiocarcinoma-like gene expression trait in hepatocellular carcinoma. Cancer Res. 2010;70:3034–3041. doi: 10.1158/0008-5472.CAN-09-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Budhu A, Forgues M, Ye QH, Jia LH, He P, Zanetti KA, et al. Prediction of venous metastases, recurrence and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell. 2006;10:99–111. doi: 10.1016/j.ccr.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 17.Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 18.Ji J, Yamashita T, Budhu A, Forgues M, Jia HL, Li C, et al. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. 2009;50:472–480. doi: 10.1002/hep.22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye QH, Qin LX, Forgues M, He P, Kim JW, Peng AC, et al. Predicting hepatitis B virus-positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning. Nat Med. 2003;9:416–423. doi: 10.1038/nm843. [DOI] [PubMed] [Google Scholar]
- 21.Woo HG, Wang XW, Budhu A, Kim YH, Kwon SM, Tang ZY, et al. Association of TP53 Mutations with Stem Cell-Like Gene Expression and Survival of Patients with Hepatocellular Carcinoma. Gastroenterology. 2011;140:1063–1070.e8. doi: 10.1053/j.gastro.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woo HG, Park ES, Lee JS, Lee YH, Ishikawa T, Kim YJ, et al. Identification of potential driver genes in human liver carcinoma by genomewide screening. Cancer Res. 2009;69:4059–4066. doi: 10.1158/0008-5472.CAN-09-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andersen JB, Spee B, Blechacz BR, Avital I, Komuta M, Barbour A, et al. Genomic and genetic characterization of cholangiocarcinoma identifies therapeutic targets for tyrosine kinase inhibitors. Gastroenterology. 2011 doi: 10.1053/j.gastro.2011.12.005. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park J, Gores GJ, Patel T. Lipopolysaccharide induces cholangiocyte proliferation via an interleukin-6-mediated activation of p44/p42 mitogen-activated protein kinase. Hepatology. 1999;29:1037–1043. doi: 10.1002/hep.510290423. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Yang Z, Whitby R, Wang L. Regulation of miR-200c by nuclear receptors PPARalpha, LRH-1 and SHP. Biochem Biophys Res Commun. 2011;416:135–139. doi: 10.1016/j.bbrc.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Research. 2010;70:10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 28.Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner R, Lozoya O, Wang Y, Cardinale V, Gaudio E, Alpini G, et al. Human hepatic stem cell and maturational liver lineage biology. Hepatology. 2011;53:1035–1045. doi: 10.1002/hep.24157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 32.Oft M, Heider KH, Beug H. TGFbeta signaling is necessary for carcinoma cell invasiveness and metastasis. Curr Biol. 1998;8:1243–1252. doi: 10.1016/s0960-9822(07)00533-7. [DOI] [PubMed] [Google Scholar]
- 33.Heldin CH, Landstrom M, Moustakas A. Mechanism of TGF-beta signaling to growth arrest, apoptosis, and epithelial-mesenchymal transition. Curr Opin Cell Biol. 2009;21:166–176. doi: 10.1016/j.ceb.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 34.Nawshad A, Lagamba D, Polad A, Hay ED. Transforming growth factor-beta signaling during epithelial-mesenchymal transformation: implications for embryogenesis and tumor metastasis. Cells Tissues Organs. 2005;179:11–23. doi: 10.1159/000084505. [DOI] [PubMed] [Google Scholar]
- 35.Pardali K, Moustakas A. Actions of TGF-beta as tumor suppressor and pro-metastatic factor in human cancer. Biochim Biophys Acta. 2007;1775:21–62. doi: 10.1016/j.bbcan.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 36.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 39.Frame MC, Inman GJ. NCAM is at the heart of reciprocal regulation of E-cadherin- and integrin-mediated adhesions via signaling modulation. Dev Cell. 2008;15:494–496. doi: 10.1016/j.devcel.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 40.Seok JY, Na DC, Woo HG, Roncalli M, Kwon SM, Yoo JE, et al. A fibrous stromal component in hepatocellular carcinoma reveals a cholangiocarcinoma-like gene expression trait and EMT. Hepatology. 2012 doi: 10.1002/hep.25570. [DOI] [PubMed] [Google Scholar]
- 41.Meng F, Glaser SS, Francis H, DeMorrow S, Han Y, Passarini JD, et al. Functional analysis of microRNAs in human hepatocellular cancer stem cells. J Cell Mol Med. 2012;16:160–173. doi: 10.1111/j.1582-4934.2011.01282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giannelli G, Bergamini C, Fransvea E, Sgarra C, Antonaci S. Laminin-5 with transforming growth factor-beta1 induces epithelial to mesenchymal transition in hepatocellular carcinoma. Gastroenterology. 2005;129:1375–1383. doi: 10.1053/j.gastro.2005.09.055. [DOI] [PubMed] [Google Scholar]
- 43.Lee TK, Poon RT, Yuen AP, Ling MT, Kwok WK, Wang XH, et al. Twist overexpression correlates with hepatocellular carcinoma metastasis through induction of epithelial-mesenchymal transition. Clin Cancer Res. 2006;12:5369–5376. doi: 10.1158/1078-0432.CCR-05-2722. [DOI] [PubMed] [Google Scholar]
- 44.Jou J, Diehl AM. Epithelial-mesenchymal transitions and hepatocarcinogenesis. J Clin Invest. 2010;120:1031–1034. doi: 10.1172/JCI42615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sigal SH, Brill S, Fiorino AS, Reid LM. The liver as a stem cell and lineage system. Am J Physiol. 1992;263:G139–G148. doi: 10.1152/ajpgi.1992.263.2.G139. [DOI] [PubMed] [Google Scholar]
- 46.Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17:313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 47.Alison MR, Islam S, Wright NA. Stem cells in cancer: instigators and propagators? J Cell Sci. 2010;123:2357–2368. doi: 10.1242/jcs.054296. [DOI] [PubMed] [Google Scholar]
- 48.Oishi N, Wang XW. Novel therapeutic strategies for targeting liver cancer stem cells. Int J Biol Sci. 2011;7:517–535. doi: 10.7150/ijbs.7.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee JH, Rim HJ, Sell S. Heterogeneity of the “oval-cell” response in the hamster liver during cholangiocarcinogenesis following Clonorchis sinensis infection and dimethylnitrosamine treatment. J Hepatol. 1997;26:1313–1323. doi: 10.1016/s0168-8278(97)80467-9. [DOI] [PubMed] [Google Scholar]
- 50.Ishii T, Yasuchika K, Suemori H, Nakatsuji N, Ikai I, Uemoto S. Alpha-fetoprotein producing cells act as cancer progenitor cells in human cholangiocarcinoma. Cancer Lett. 2010;294:25–34. doi: 10.1016/j.canlet.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 51.ALLEN RA, LISA JR. Combined liver cell and bile duct carcinoma. Am J Pathol. 1949;25:647–655. [PMC free article] [PubMed] [Google Scholar]
- 52.Theise ND, Nakashima O, Park YN, Nakanuma Y. Combined hepatocellular-cholangiocarcinoma. In: Bosman FT, Carneiro F, Hruban RH, Theise ND, editors. WHO Classification of Tumours of the Digestive System. Lyon: World Health Organization; 2010. pp. 225–227. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.