Abstract
Recently, the specific hybridization of DNA molecules has been used to construct self-assembled devices, e.g., the mechanical device to mimic cellular protein motors in nature. Here, we present a new light-powered DNA mechanical device based on the photoisomerization of azobenzene moieties and toehold-mediated strand displacement. This autonomous and controllable device is capable of moving towards either end of the track, simply by switching the wavelength of light irradiation, either UV (365nm) or visible (>450nm). This light-controlled strategy can easily solve one main technical challenge for step-wise walking devices: the selection of routes in multipath systems. The principle employed in this study, photoisomerization-induced toehold length switching, could be further useful in the design of other mechanical devices, with the ultimate goal of rivaling molecular motors for cargo transport and macroscopic movement.
Keywords: DNA nanostructure, light control, reversible motion, strand displacement
Nature has evolved various tiny protein machines (e.g., myosin, kinesin and dynein) that travel along a network of tracks within the cell to transport organelles and power cellular motion.1,2 Inspired by these protein-based molecular motors, artificial devices,3-7 especially DNA-based motors, have recently emerged and started to mimic the function of biological motors in cargo transport6 and biosynthesis.7 As biological macromolecules similar in size to molecular motors, and with the added capability of simple programmable assembly, DNAs are suitable for the control of progressive, processive and directional movement at the molecular level.
Energy supply is a major concern for any motor, and various energy sources (e.g., DNA hybridization,8-11 hydrolysis of either DNA/RNA backbones12-15 or ATP molecules16) have been previously explored for DNA walkers. Still, these artificial walkers are in their infancy; they are not as powerful or organized as their natural protein counterparts. In this regard, identification of new types of energy supplies could be meaningful in the development of the next-generation mechanical robots, ideally with more powerful and controllable operation.3
We recently reported a new light energy-powered DNA walker, which is capable of regulated autonomous movement along a nucleic acid track.17 Construction of free-running and autonomous motors, which remain in operation as long as a source of energy is available, is essential to the operation of nanomachines that mimic biological function. However, the capability of autonomous motion sometimes results in decreased controllability. That is, the device cannot be easily stopped at a desired position or time, or once stopped, it is difficult to restart. The availability of an easily-controllable, free-running walking device would also be significant for the future design of nanorobots to perform multiple and complex functions. We have demonstrated that photochemical energy sources can serve as inputs in the operation of nanosized DNA walkers, where light-initiated chemical cleavage allows precise control of the speed and motion of autonomous nanorobots.17,18
The directional movement of the above-mentioned light-driven walker stems from the “burnt-bridge” mechanism, whereby the anchorage sites on the track are irreversibly consumed during the motion. However, as a result, the reversible, cyclic operation of the walking device has been eliminated, preventing the walker from mimicking natural molecular motors (e.g., kinesin) that change the directionality of movement towards both ends of the tracks,2 and seriously restricting future usage of the device for multiple trips. Advancing the functions of the walking system to facilitate reversible and multi-directional locomotion is one goal of DNA engineering. In the present work, we describe a new light-driven walking system, which enables the DNA walker to travel in either direction on the track, with precise controllability, by using different wavelengths of light irradiation.
RESULTS AND DISCUSSION
The movement of the DNA walker is guided by the prescriptive landscape. Through precise specification of walker/ environment interactions, directional locomotion can be realized without a cleavage process. In DNA strand exchange, one prehybridized strand in a DNA duplex is displaced by an invading strand. The rate of strand exchange can be quantitatively controlled by varying the length of toeholds (short single-stranded domains to initiate displacement).19,20 Based on toehold-mediated strand displacement, we demonstrate in this study the reversible, autonomous and controllable movement of DNA walkers along an azobenzene-incorporated prescriptive track.
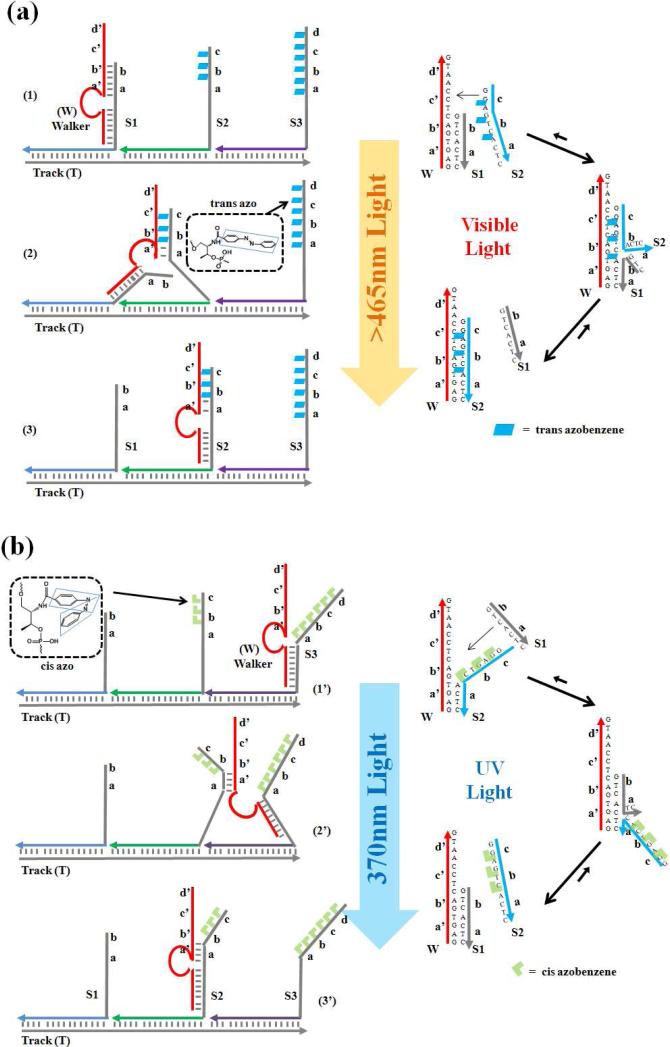
The walking system consists of three parts: a single-stranded DNA track (T), several photosensitive anchorage sites (e.g., S1 and S2), and a DNA walker (W) (Figure 1). The track contains three 21-nucleotide (nt) long binding regions that are complementary to the recognition tags of the respective anchorage sites. Through specific DNA interaction, the track organizes the anchorage sites into one self-assembled construction, with the walker (W) binding sites extending from the track. After assembly, the distance between two adjacent anchorage sites corresponds to two helical pitches, roughly 7nm. The construction of the overall walking system was verified by native polyacrylamide gel electrophoresis (Figure S1 in the Supporting Information).
Figure 1.
Principle of the DNA-based walking device. Walker, anchorage sites, and track form the walking system through self-assembly; the locomotion of the walker is realized through toehold-mediated strand displacement. (a) Visible light irradiation (azobenzene, trans) triggers walker motion in the direction of S1→S3; (b) UV light (azobenzene, cis) induces the reverse movement of S3→S1. (Domains a, b, c and d are complementary to a’, b’, c’ and d’, respectively).
The DNA walker contains two legs, a searching leg and a holding leg, and they are linked by a DNA oligomer as the body. The two motion legs are designed to respectively bind the two extender segments of each anchorage site. The holding leg binds the 16-nt extender segment, which is identical in all anchorage sites to prevent the walker from leaving the track. The other extender segment, which serves as the toehold to mediate the locomotion (strand displacement) of the walker, is designed to be complementary with the searching leg of the DNA walker.
Various numbers of azobenzene moieties were incorporated into different extender segments. The photoinduced isomerization of azobenzene molecules has been broadly used to induce significant conformational and biochemical changes in nucleic acids,21-25 peptides and proteins.26 The light-driven cis/trans isomerization of the azobenzene moiety is wavelength-dependent: UV light at 365nm drives the trans-to-cis conversion, while visible light at around 465nm corresponds to the cis-to-trans isomerization. When incorporated into the DNA structure, the UV light-induced cis-form lowers the binding affinity of the DNA duplex based on the spatial hindrance to DNA hybridization. In contrast, visible light irradiation reverses the isomerization and enables DNA duplex binding. By introducing azobenzene moieties into the toehold domain of extender segments, the moving direction of the walker (i.e., the strand-displacement pathway towards the longer toehold binding site) is controllable by irradiation with different wavelengths of light (Figure 1).
The directional locomotion of the DNA walker was investigated using a fluorescence resonance energy transfer (FRET) assay. As the FRET acceptor, Dabcyl quencher was labeled on the walker's holding leg, and fluorescence donors, 6-carboxyfluorescein (6-FAM) and tetramethylrhodamine (TAMRA), were attached to anchorage sites S1 and S2, respectively (Figure S2 in the Supporting Information). Because FRET efficiency is proportional to the inverse 6th power of the donor-acceptor distance, FRET is expected to occur most efficiently when the walker steps onto the respective anchorage site, thereby bringing FAM (or TAMRA) and Dabcyl together.
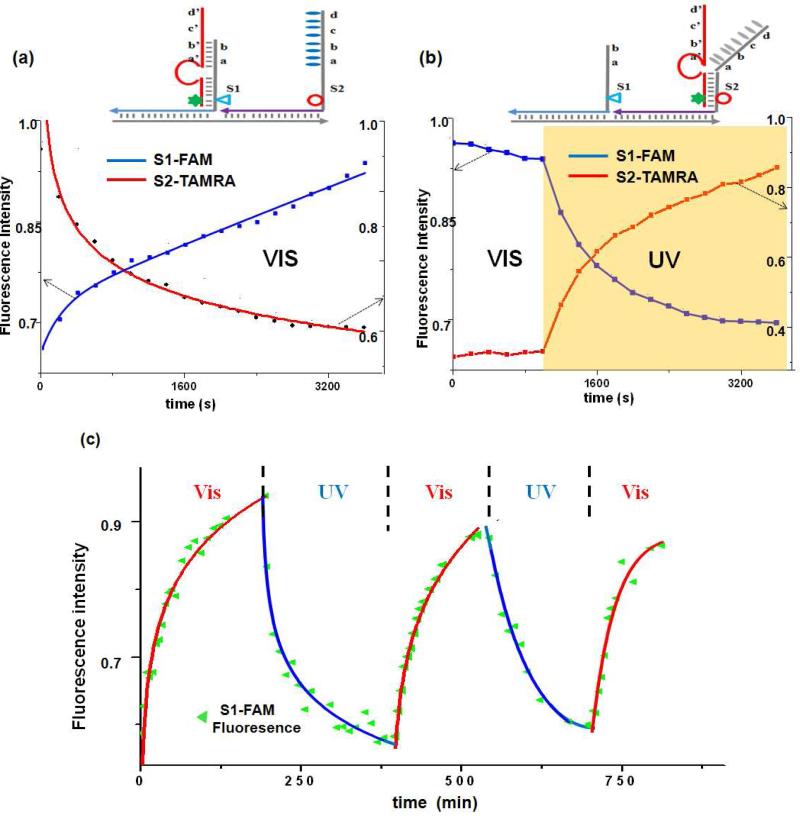
We first tested the feasibility of our method by using the simplest two-step photocontrollable locomotion (Figure 2a). The FAM-labeled S1 anchorage site containing 7 nucleotides (7-nt) served as the toehold domain for binding the searching leg of the DNA walker. In comparison, the TAMRA-labeled S2 site containing a longer toehold region (14-nt) also included nine azobenzene moieties (one after every two bases, see Experimental Section). Such azobenzene/nucleic acid ratio has previously been proven to be highly efficient in regulating DNA hybridization efficiency.21-25 Based on the “nearest-neighbor model” of azobenzene-incorporated DNA duplex formation,25 a full thermodynamic description has been presented and successfully used to predict the melting temperature of azobenzene-incorporated DNA duplexes. Under visible light, the trans-form of azobenzene facilitates DNA duplex binding, and the longer toehold within the S2 anchorage mediates the directional motion of walker from S1 to S2 site. In contrast, UV light irradiation induces the trans-to-cis isomerization of azobenzene and reduces the length of the available toehold within the S2 anchorage site to reverse the direction of motion (S2 to S1).
Figure 2.
Progressive and reversible walking demonstrated by FRET assay. Dabcyl (quencher, star)-labeled walker moves on the anchorage site labeled with fluorophore TAMRA (circle) and FAM (triangle). Fluorescence intensity (515nm for FAM, λex= 488nm; 580nm for TAMRA, λex= 550nm) was monitored during (a) S1→S2 locomotion (continuous visible irradiation), (b) S2→S1 locomotion (visible irradiation for 1000s followed by UV irradiation), and (c) the reversible movement of the walker during the alternate period of UV and visible irradiation.
Using locomotion from S2 to S1 as an example, the assumption was experimentally tested. The walker was specifically positioned on the S2 site through separately prepared T(track)-S1 and S2-W(walker) conjugates (Figure S1 in the Supporting Information). Mixing the conjugates immediately prior to light irradiation ensured that the S2 anchorage would be the initiating site (the largest distribution binding position) for the walker. Indeed, consistent with our expectation, directional walking pathways, i.e., S1→S2 under visible light (azobenzene trans) and S2→S1 under UV light (azobenzene cis), were observed (Figure 2). When initiated at the S2 site, W was expected to stay under the visible light, instead of stepping onto the S1 site. Following the expected motional mode, irradiation for the first 1000s with visible light (450nm, Figure 2b) did not have a great effect on the fluorescence from either FAM or TAMRA; however, UV light irradiation (365nm) immediately initiated the quenching of the FAM signal and the recovery of TAMRA fluorescence.
As control, it was demonstrated that the same UV light does not affect the fluorescence signal of either dye (data not shown); the fluorescence change was indeed associated with locomotion of the walker from S2 to S1 site. In another control experiment, we demonstrated that the free anchorage site (with the same legs as S2) under the same conditions would not disturb migration of the walker between S1 and S2 along the track (Figure S3 in the Supporting Information). This observation is important in proving that the DNA walker remains attached to the same track during the entire operation. Such “processive” locomotion3 prevents formation of a “bridge” between two tracks and forces the walker to move a longer distance along the track.
The reversible movement of the walker along this photocontrollable track was also verified. As shown in Figure 2c, by alternating the wavelength of light (450nm or 365nm), the direction of locomotion was switched accordingly. At the end of each cycle of light irradiation, the device can be reset, and the walker is able to repeatedly perform similar mechanical motion. It is noteworthy that such reversible locomotion cannot be achieved by movements based on the above-mentioned “burnt-bridge” mechanism whereby the track is irreversibly damaged during passage. Furthermore, no waste molecules (neither DNA duplex27,28 nor chemical triggers29,30) accumulate during the reversible operation, an important consideration for the continuous long-term usage of the walking device.
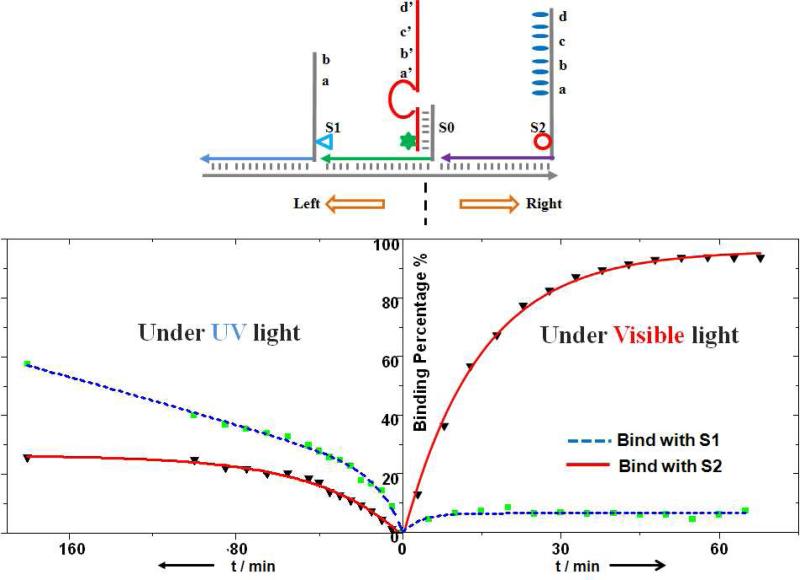
When given a selection of routes in multipath systems, DNA walkers based on the “burnt bridge” mechanism will lose control of direction, with equal chance of moving on either path. This difficulty is viewed as one of main challenges for active DNA nanostructures.10,31,32 However, we have demonstrated that route selection can be easily achieved in this azobenzene-modified walking device via irradiation with different wavelengths of light. As shown in Figure 3, a S0 anchorage site was introduced between the above-mentioned S1 and S2 sites, as the initiating binding site (crossing point) for the walker. When irradiated with visible light, after 50 minutes incubation, c.a. 94% of the walker strands move to the S2 site (longer toehold and faster, more stable binding), and only 6% of the strands bind to S1. In another study, under UV light, as expected, more walker strands turned to the S1 site (69% at equilibrium) compared with S2 (31%, see Figure S4 in the Supporting Information). With UV light irradiation, it was also proven that 1) it takes longer for walkers to choose the route and 2) the final walker strand distribution is less controllable. This result is associated with the fact that a smaller toehold length difference (comparing that between S1 and S0 sites with that of between S2 and S0) can result in a longer time to reach equilibrium and a smaller equilibrium constant.19,20 Potentially, this process could be further optimized by varying the numbers of toehold nucleotides and azobenzenes in the anchorage sites.
Figure 3.
Programmed control of route selection at a junction. Left turn (S0→S1) was achieved under UV light irradiation, and right turn (S0→S2) was induced by visible light. The fractions of the walker (labeled with Dabcyl, star) moving towards S2 (labeled with TAMRA, circle) or S1 (labeled with FAM, triangle) were monitored based on the fluorescence intensity change and further calculated using a standard calibration curve.
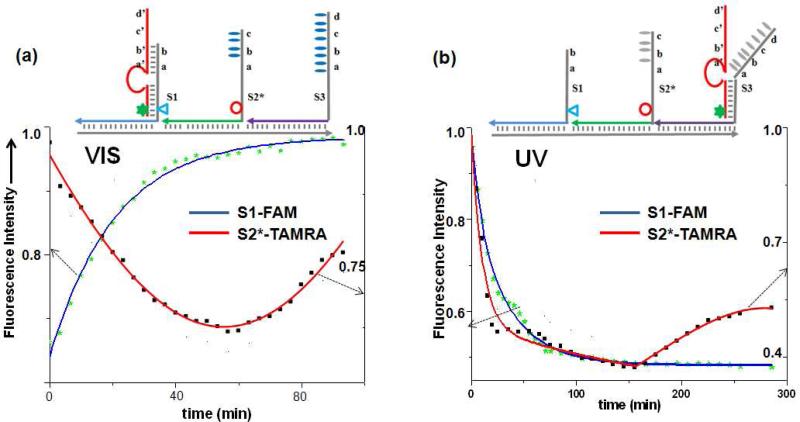
Finally, we wanted to prove that movement over a longer distance can be achieved by logical programming of the anchorage sites. As an example, an S2* anchorage site was introduced between S1 and S3 sites (with the same extender segments as S2, Experimental Section), with the distance from each site as 7nm (Figure 4a). With visible irradiation, S2* has a 10-nt-long segment in between S1 (7-nt) and S3 (14-nt) to bind with the walker's searching leg. The toehold length difference mediates the movement in the direction of S1→S2*→S3. Moreover, these three anchorage sites were designed with various numbers of azobenzene moieties: 0, 3 and 9 for S1, S2* and S3, respectively. These numbers were selected so that the binding strength with the DNA walker followed the order of S1>S2*>S3 under UV light,25 because trans-to-cis isomerization of the azobenzene moieties results in a different degree of change to the three sites. Based on the switched toehold length and dynamic binding affinity, the walker should preferentially move in the direction of S3→S2*→S1, reversing the previous movement.
Figure 4.
Progressive operation of the walker on a 3-step track. Motion was demonstrated in the direction of S1→S2*→S3 under visible light, and it was reversed to follow the S3→S2*→S1 direction with UV irradiation. Fluorescence intensity was monitored at 515nm for FAM (triangle)-labeled S1 site (λex= 488nm) and at 580nm for TAMRA (circle)-labeled S1* site (λex= 550nm).
As demonstrated from the fluorescence measurements (Figure 4), when walking was initiated at S1 site (visible light irradiation), about an hour on average was required to pass through TAMRA-modified S2* (minimum fluorescence) and further move on to the S3 site (recovered TAMRA fluorescence). Under UV light irradiation, the reverse motion was demonstrated, starting from S3 site, passing through S2* (minimum TAMRA fluorescence), and finally reaching the S1 destination (recovered TAMRA fluorescence), proving that the walker can move in either direction along a prescriptive photo-switchable track.
CONCLUSIONS
Based on the isomerization of azobenzene moieties and toehold-mediated strand displacement, we have designed a light-controlled DNA walking device that can move to either end of the oligonucleotide track, and the direction of motion can be switched using different wavelengths of light. Compared with other reported DNA walkers,3-17 this new strategy not only preserves the autonomous and controllable movement, but also provides a reusable track, making it feasible to reset the device after the complete trip, as observed in nature for kinesin and myosin.1,2 So far, the rate and step number of this device are still limited, which might be due to the imperfect photoisomerization efficiency of the azobenzene moieties. However, the principle of photoisomerization-induced toehold length switching could be further used in designing other autonomous DNA-based walking devices, where sequential and controllable release of the toehold is required.8-11 Moreover, as the first study (based on our knowledge) of using photoswitchable molecules for alternating the DNA toehold binding regions, this system can be employed for external control of the route selection in multi-pathway systems, e.g., “crossing” or “T junction”, for more advanced walking devices that can rival molecular motors in nature.
MATERIALS AND METHODS
Oligonucleotides
All oligonucleotides were synthesized using an ABI 3400 DNA synthesizer (Applied Biosystems, Inc., Foster City, CA). The DNA sequences employed in this study: Track T1-2*-3: 5’-ACC ATC TGT GGC ATA GCA GCG AGT ATC TAA CGC ATG GAA GCG TCG ATC TTG AGC ATT GGA GCG-3’; T1-0-2: 5’-AGT ATC TAA CGC ATG GAA GCG ACC ATC TGT GGC ATA GCA GCG TCG ATC TTG AGC ATT GGA GCG-3’; T1-2: 5’-AGT ATC TAA CGC ATG GAA TCG ATC TTG AGC ATT GGA GCG-3’; Anchorage site S0: 5’-GTC CGA ATC AGC ACT TTC GCT GCT ATG CCA CAG ATG GT-3’; S1: 5’-GTC ACT CTTGTC CGA ATC AGC ACT –FAM-TTC GCT CCA ATG CTC AAG ATC GA-3’; S2: 5’-CA Z-TT-Z-GG-Z-AG-Z-TC-Z-AC-Z-TC-Z-TT-Z-GT-Z-C CGA ATC AGC ACT –TAMRA-TTC GCT TCC ATG CGT TAG ATA CT-3’; S2*: 5’-GGZ-AG-Z-TC-Z-ACT CTTGTC CGA ATC AGC ACT –TAMRA-TTC GCT TCC ATG CGT TAG ATA CT-3’; S3: 5’-CA-Z-TT-Z-GG-Z-AG-Z-TC-Z-AC-Z-TC-Z-TT-Z-GT-Z-C CGA ATC AGC ACT TTC GCT GCT ATG CCA CAG ATG GT-3’; Walker W: 5’-Dabcyl-AGT GCT GAT TCG GAC AGG CTA GCT ACA ACG AGA GTG ACT CCA ATG-3’ (T1-2 = track to arrange anchorage site in the order of S1-S2; Z = azobenzene-modified position in the anchorage site).
Manipulation of the Walking System
Using S3→S2*→ S1 as an example, the formation of the walking system was as follows. After separate annealing (95°C-10°C, over 20 min) in 25mM Tris buffer (pH=7.5, containing 50mM NaCl and 5mM MgCl2), T-S1-S2* and S3-W conjugates were mixed to give a final concentration for each sequence of 0.1μM, and the sample was irradiated with either a 6W portable UV lamp (60 Hz with center wavelength at 365 nm and measured light source power around 0.2mW) or a table lamp (60Hz, 120V with 60W bulb and measured light source power 30mW). The irradiation power employed was measured using a power meter (Newport Corp., Irvine, CA). During irradiation, the temperature of the sample was maintained at 23.0°C using a water bath (ThermoNESLAB Inc.). After a series of irradiation times, the samples were removed and subjected to FRET experiments.
FRET Measurements
A FluoroMax-4 Spectrofluorometer with a temperature controller (Jobin Yvon) was used for all steady-state fluorescence measurements. After a series of irradiation time periods, fluorescence intensities at 515nm (FAM, λex=488nm; slit number=5nm, 0.2s integration time) and 580nm (TAMRA, λex=550nm; slit number=5nm, 0.2s integration time) were recorded, with temperature controlled at 23°C.
Native PAGE Analysis
The binding and three-dimensional structures of the DNA walking system were observed using native PAGE gel. Normally the gel was run in 8-10% acrylamide (containing 19/1 acrylamide/bisacrylamide) solution with 1×TBE/15mM Mg2+ buffer, at 100V constant voltage for 1 hour (4°C). After that, the gel was stained 30 min using Stains-All (Sigma-Aldrich) to image the position of DNA. Photographic images were obtained under visible light with a digital camera.
Supplementary Material
Acknowledgements
We acknowledge Dr. K. R. Williams for manuscript review. This work is supported by grants awarded by the National Institutes of Health (GM066137, GM079359 and CA133086) and by NSF. M.Y. would like to thank the Eastman Co. Fellowship (Kingsport, TN) for support.
Footnotes
Supporting Information Available: Schematic represent of the sequence of DNA for constructing walking system, native gel electrophoresis to demonstrate walking system construction, FRET between Dabcyl/FAM and between Dabcyl/TAMRA, processive motion of the walker along the track, and fluorescence measurement and calibration curve for right/left turning choice. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Schliwa M, Woehlke G. Molecular Motors. Nature. 2003;422:759–765. doi: 10.1038/nature01601. [DOI] [PubMed] [Google Scholar]
- 2.Roostalu J, Hentrich C, Bieling P, Telley I-A, Schiebel E, Surrey T. Directional Switching of the Kinesin Cin8 Through Motor Coupling. Science. 2011;332:94–99. doi: 10.1126/science.1199945. [DOI] [PubMed] [Google Scholar]
- 3.Von Delius M, Leigh D-A. Walking Molecules. Chem. Soc. Rev. 2011;40:3656–3676. doi: 10.1039/c1cs15005g. [DOI] [PubMed] [Google Scholar]
- 4.Bath J, Turberfield A-J. DNA Nanomachines. Nat. Nanotechnol. 2007;2:275–284. doi: 10.1038/nnano.2007.104. [DOI] [PubMed] [Google Scholar]
- 5.Barrell M-J, Compana A-G, von Delius M, Geertsema E-M, Leigh D-A. Light-Driven Transport of a Molecular Walker in Either Direction Along a Molecular Track. Angew. Chem., Int. Ed. 2011;50:285–290. doi: 10.1002/anie.201004779. [DOI] [PubMed] [Google Scholar]
- 6.Gu H, Chao J, Xiao S, Seeman N-C. A Proximity-based Programmable DNA Nanoscale Assembly Line. Nature. 2010;465:202–205. doi: 10.1038/nature09026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He Y, Liu D-R. Autonomous Multistep Organic Synthesis in a Single Isothermal Solution Mediated by a DNA Walker. Nat. Nanotechnol. 2010;5:778–782. doi: 10.1038/nnano.2010.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin P, Choi H-M-T, Calvert C-R, Pierce N-A. Programming Biomolecular Self-Assembly Pathways. Nature. 2008;451:318–322. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- 9.Omabegho T, Sha R, Seeman N-C. A Bipedal DNA Brownian Motor with Coordinated Legs. Science. 2009;324:67–71. doi: 10.1126/science.1170336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muscat R-A, Bath J, Turberfield A-J. A Programmable Molecular Robot. Nano Lett. 2011;11:982–987. doi: 10.1021/nl1037165. [DOI] [PubMed] [Google Scholar]
- 11.Green S-J, Bath J, Turberfield A-J. Coordinated Chemomechanical Cycles: a Mechanism for Autonomous Molecular Motion. Phys. Rev. Lett. 2008;101:238101. doi: 10.1103/PhysRevLett.101.238101. [DOI] [PubMed] [Google Scholar]
- 12.Lund K, Manzo A-J, Dabby N, Michelotti N, Johnson-Buck A, Nangreave J, Taylor S, Pei R, Stojanovic MN, Walter N-G, et al. Molecular Robots Guided by Prescriptive Landscapes. Nature. 2010;465:206–210. doi: 10.1038/nature09012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian Y, He Y, Chen Y, Yin P, Mao C. A DNAzyme that Walks Processively and Autonomously Along a One-Dimensional Track. Angew. Chem., Int. Ed. 2005;44:4355–4358. doi: 10.1002/anie.200500703. [DOI] [PubMed] [Google Scholar]
- 14.Bath J, Green S-J, Tuberfield A-J. A Free–Running DNA Motor Powered by a Nicking Enzyme. Angew. Chem., Int. Ed. 2005;44:4358–4361. doi: 10.1002/anie.200501262. [DOI] [PubMed] [Google Scholar]
- 15.Wickham S-F-J, Endo M, Katsuda Y, Hidaka K, Bath J, Sugiyama H, Turberfield A-J. Direct Observation of Stepwise Movement of a Synthetic Molecular Transporter. Nat. Nanotechnol. 2011;6:166–169. doi: 10.1038/nnano.2010.284. [DOI] [PubMed] [Google Scholar]
- 16.Yin P, Yan H, Daniell X-G, Turberfield A-J, Reif J-H. A Unidirectional DNA Walker that Moves Autonomously Along a DNA Track. Angew. Chem., Int. Ed. 2004;43:4906–4911. doi: 10.1002/anie.200460522. [DOI] [PubMed] [Google Scholar]
- 17.You M, Chen Y, Zhang X, Liu H, Wang K, Wang R, Williams KW, Tan W. An Autonomous and Controllable Light-Driven DNA Walking Device. Angew. Chem., Int. Ed. 2012;51:2457–2460. doi: 10.1002/anie.201107733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.You M, Zhu Z, Liu H, Gulbakan B, Han D, Wang R, Williams KR, Tan W. Pyrene-Assisted Efficient Photolysis of Disulfide Bonds in DNA-based Molecular Engineering. ACS Appl. Mater. Interfaces. 2010;2:3601–3605. doi: 10.1021/am1007886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang D-Y, Seelig G. Dynamic DNA Nanotechnology Using Strand-Displacement Reactions. Nat. Chem. 2011;3:103–113. doi: 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- 20.Genot A-J, Zhang D-Y, Bath J, Turberfield A-J. Remote Toehold: a Mechanism for Flexible Control of DNA Hybridization Kinetics. J. Am. Chem. Soc. 2011;133:2177–2182. doi: 10.1021/ja1073239. [DOI] [PubMed] [Google Scholar]
- 21.Liang X, Mochizuki T, Asanuma H. A Supra-Photo Switch Involving Sandwiched DNA Base Pairs and Azobenzenes for Light-Driven Nanostructures and Nanodevices. Small. 2009;5:1761–1768. doi: 10.1002/smll.200900223. [DOI] [PubMed] [Google Scholar]
- 22.Zhou M, Liang X, Mochizuki T, Asanuma H. A Light-Driven DNA Nanomachine for Efficiently Photoswitching RNA Digestion. Angew. Chem., Int. Ed. 2010;49:2167–2170. doi: 10.1002/anie.200907082. [DOI] [PubMed] [Google Scholar]
- 23.Kang H, Liu H, Phillips J-A, Cao Z, Kim Y, Chen Y, Yang Z, Li J, Tan W. Single-DNA Molecular Nanomotor Regulated by Photons. Nano Lett. 2009;9:2690–2696. doi: 10.1021/nl9011694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.You M, Wang R, Zhang X, Chen Y, Wang K, Peng L, Tan W. Photo-Regulated DNA-Enzymatic Nanostructures by Molecular Assembly. ACS Nano. 2011;5:10090–10095. doi: 10.1021/nn204007y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Asanuma H, Matsunaga D, Komiyama M. Clear-Cut Photo-Regulation of the Formation and Dissociation of the DNA Duplex by Modified Oligonucleotide Involving Multiple Azobenzenes. Nucleic. Acids. Symp. Ser. 2005;49:35–36. doi: 10.1093/nass/49.1.35. [DOI] [PubMed] [Google Scholar]
- 26.Schierling B, Noel A-J, Wende W, Hien Le T, Volkov E, Kubareva E, Oretskaya T, Kokkinidis M, Rompp A, Spengler B, et al. Controlling the Enzymatic Activity of a Restriction Enzyme by Light. Proc. Natl. Acad. Sci. USA. 2010;107:1361–1366. doi: 10.1073/pnas.0909444107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shin J, Pierce N-A. A Synthetic DNA Walker for Molecular Transport. J. Am. Chem. Soc. 2004;126:10834–10835. doi: 10.1021/ja047543j. [DOI] [PubMed] [Google Scholar]
- 28.Sherman W-B, Seeman N-C. A Precisely Controlled DNA Biped Walking Device. Nano Lett. 2004;4:1203–1207. [Google Scholar]
- 29.Wang Z, Elbaz J, Willner I. DNA Machines: Bipedal Walker and Stepper. Nano Lett. 2011;11:304–309. doi: 10.1021/nl104088s. [DOI] [PubMed] [Google Scholar]
- 30.Wang C, Ren J, Qu X. A Stimuli Responsive DNA Walking Device. Chem. Comm. 2011;47:1428–1430. doi: 10.1039/c0cc04234j. [DOI] [PubMed] [Google Scholar]
- 31.Pinheiro A-V, Han D, Shih W-M, Yan H. Challenges and Opportunities for Structural DNA Nanotechnology. Nat. Nanotechnol. 2011;6:763–772. doi: 10.1038/nnano.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wickham S-F-J, Bath J, Katsuda Y, Endo M, Hidaka K, Sugiyama H, Turberfield A-J. A DNA-based Molecular Motor that Can Navigate a Network of Tracks. Nat. Nanotechnol. 2012;7:169–173. doi: 10.1038/nnano.2011.253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.