Abstract
Visceral adipose tissue (VAT) is a source of inflammatory cytokines that in obese subjects may contribute to low-level systemic inflammation and development of metabolic syndrome. Expansion of VAT involves adipocyte hyperplasia and hypertrophy and requires breakdown of the extracellular matrix (ECM) and increased vascular outgrowth. To investigate changes of gene expression associated with VAT expansion and the role of combined genetics and diet we implemented gene microarray analyses of VAT in NONcNZO10 (NZ10) and control SWR/J mice subjected to control chow (CD) or a diet of high protein and fish oil (HPO). NZ10 mice on CD showed increased body weight, hyperglycemia and hyperinsulinemia at 25 weeks whereas those on HPO diet retained normal insulin levels and were normoglycemic. Two-way ANOVA revealed a significant interaction between diet and strain on blood glucose, serum insulin and percent fat but not for body weight. Microarray heat-maps revealed a remarkable combined effect of genetics and diet on genes that regulate ECM as well as angiogenic genes. RT-PCR confirmed markedly increased expression of Matrix Metalloproteinases (MMPs) -2, -3, -11, and -12, VEGF-A and C, Von Willebrand Factor (VWF) and PPARγ selectively in the NZ10/CD group. MMP-7 was significantly decreased. Protein levels of MMP-2, 3 and 9 were significantly increased in the VA of NZ10 mice fed CD while those of MMP-7 were down-regulated. Microarrays also revealed diet-dependent 2–4-fold increased expression of all 4 Tissue inhibitor of Metalloproteinases (TIMP) isoforms in NZ10 mice. Two-way ANOVA confirmed strongly interactive roles of diet and genetics on fat deposition and progression of T2D in this polygenic mouse model.
Keywords: Adipose, Gene Expression, high protein fish oil diet, Matrix Metalloproteinase, angiogenesis
Introduction
NONcNZO10/LtJ (NZ10) is a new polygenic mouse model of obesity-induced type 2 diabetes (T2D) that develops maturity-onset insulin resistance and hyperglycemia (1). The obesity/T2D phenotype of NZ10 mice is relatively diet-independent, the phenotype is supported even on moderately low fat chow and the mice do not have overtly dysregulated leptin gene expression (2, 3). The NZ10 mice differ from the parent NZO/HILt strain in acquiring adiposity primarily in VAT and this may contribute to the T2D susceptibility phenotype (4, 5).
During the development of obesity, adipose tissue is remodeled, a process that requires changes in the expression of genes that regulate adipogenesis, adipocyte hypertrophy, and angiogenesis. Proteolysis of the extracellular matrix (ECM) plays a key role in adipose expansion and includes changes in the activities of matrix metalloproteinases (MMPs) and tissue inhibitors of MMPs (TIMPs) (6–10). Overexpression of TIMPs inhibits adipogenesis but may promote lipid accumulation and hypertrophy of mature adipocytes and although controversial, some studies suggest an essential role for enhanced TIMP activity (9, 10). There is discrepancy over the contributions of individual MMPs and TIMPs to adipose expansion. Increased mRNA levels of MMP-3, -11, -12, -13, -14 and TIMP-1 were reported in gonadal fat depots of nutritionally-induced obese wild type mice whereas MMP-7, -9, -16, and -24 were decreased and there was no change in the levels of MMP-2, -10, -15, -17, -19, -23, TIMP-2 and -3 (11). Increased mRNA levels of MMP-2, MMP-3, MMP-12, MMP-14, and MMP-19 were reported in white adipose tissue of obese ob/ob and db/db mice; MMP-7 mRNA again was decreased (12). In db/db mice it was found that omega-3 fatty acid supplements significantly prevented obesity and blocked the expression of MMP-12 and -14 mRNAs (7). Several studies have described increased levels of MMP-2 and -9 in adipose and serum of obese human subjects (13, 14). Adipose expansion in animal models and humans is associated with active angiogenesis and elevated expression of associated genes (15, 16).
In the present study we used microarray, RT-PCR and western blotting to quantify MMPs, TIMPs and angiogenic genes in VAT of NZ10 mice. The results indicate strong interaction between diet and genetics on VA deposition and T2D phenotypes with selective diet/genetics-dependent induction of MMPs 2, 3, 11, 12, multiple angiogenic genes and all 4 TIMPs. Consistent with 2 other murine models MMP-7 was downregulated in the VAT of NZ10 mice fed HD.
Materials and Methods
Animals and diets
Four-week old male NZ10 mice and their non-diabetic SWR/J counterparts were purchased from The Jackson Laboratory. After 2 weeks of acclimatization mice were randomized into 2 groups (at least 10 mice per group) to receive CD or HPO diets for a period of 19 weeks. CD is a semi-defined open formula custom diet with macronutrients based on that of Purina 5K20 that provides 27% calories from fat, 19% from protein and 54% from carbohydrate. This diet contains lower fat than the standard western style diet for rodents such as Purina 5TJN (40% fat). The HPO diet is Purina LabDiet 5TWH, based on the South Beach diet and optimized for high protein and “good” fat (omega-3) with casein, canola and menhaden fish oils. Food was provided ad-lib and weekly food intake was measured by providing pre-weighed chow; there was no significant difference in food consumption (data not shown). For tissue harvest, mice were fasted for 12-h, sacrificed by cervical dislocation and the VA depots including epididymal, perirenal, and retroperitoneal rapidly dissected and processed for RNA analysis. Percent body fat was estimated using Lunar PIXImus II densitometer before sacrifice. Blood was collected from 12 hr-fasted mice at age 25 weeks for measurement of glucose and plasma insulin.
RNA preparation
Total RNA was isolated from VAT using TRIzol Reagent (Invitrogen, Carlsbad, CA). The samples were processed using an RNeasy Mini Kit (Qiagen, Valencia, CA) and digested with RNase free DNase (Qiagen, Valencia, CA). The integrity and size distribution of total RNA was monitored using an Agilent 2100 bioanalyzer (Santa Clara, CA). The concentration of RNA was determined by spectrophotometry, using Nanodrop-1000 (Nanodrop Technologies, Wilmington, DE). Reverse transcription was performed on 200ng/μl of RNA with High-Capacity cDNA Reverse Transcription kit using random primers (Applied Biosystems, Foster City, CA).
Microarray
Details of our cRNA and micro-array procedures are described elsewhere (17). Equal amounts of RNA were pooled from 4 animals per group for each experimental group. Hybridizations were implemented by Ocean Ridge Biosciences (Palm Beach, Florida) using a CodeLink Bioarray (Applied Microarrays, Inc. Tempe, Arizona) using standard procedures. For the microarray data statistical analysis, samples were binned into two treatment groups (NZ10, SWR). The log2-transformed and normalized spot intensities for the 25,173 detectable probes were examined for differences between the treatment groups by 2-way ANOVA using National Institute of Ageing (NIA) Array Analysis software. The ANOVA was conducted using the Bayesian Error Model and 10 degrees of freedom. Statistical significance was determined using the False Discovery Rate (FDR) method. A total of 181 probes showed significant differences with P<0.05 (see online Supplemental Table). Principal Component Analysis was performed on the 25,173 detectable probes using NIA software.
Real Time (RT) PCR
Gene expression levels were assayed by RT-PCR using the ABI 7900HT thermal cycler with custom assays (8 pooled samples per group). The following Assay IDs Mm00439506_m1, Mm01168406_g1, Mm00485048_m1, Mm00500554_m1, Mm01168420_m1, Mm00437306_m1, Mm01202432_m1, Mm00550376_m1, Mm01184322_m1 were used for MMP-2, 3, 11, 12, VegfA, C, VWF and PPARγ genes respectively. Eukaryotic 18S rRNA (Assay ID Hs99999901_s1) was used for normalization.
Western Blotting
Samples (3 mice per group) containing equal amounts of protein were subjected to SDS-PAGE and western blotting as described previously (18). Antibodies were MMP-2 (Santa Cruz Biotechnology, CA), MMP-3 (Abcam, MA), MMP-7 (Cell Signaling Technology, MA), MMP-9 (Abcam, MA) Beta-Actin (Chemicon, CA) antibodies.
Statistical analysis
Data are expressed as means ± SEM. Differences between two groups were analyzed by Student’s t test and Mann-Whitney U test. A two-way between groups ANOVA was used to evaluate diet and genetic strain interaction effects for dependent variables; a significant interaction was interpreted by a subsequent simple-effects analysis with Bonferroni correction.
Results and Discussion
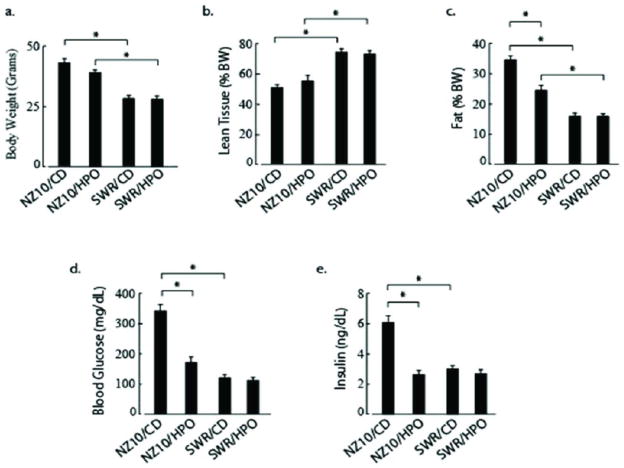
NZ10 mice fed CD for 19 weeks displayed hyperglycemia and hyperinsulinemia with significantly increased percent fat compared to their counterparts fed HPO (Figure 1). Two-way ANOVA revealed very significant interaction between diet and genetic strain for blood glucose, serum insulin and percent fat (all p<0.01). The interaction effect between strain and diet did not reach significance for body weight (p=0.10) or lean tissue (p=0.32) but there was a significant main effect of strain on both of the latter parameters (p<0.001). For SWR mice there was no significant diet-related differences in blood glucose, insulin level, fat deposition or body weight. Elevated blood glucose and plasma insulin in the NZ10-CD group were almost normalized by HPO diet. There were no apparent adverse effects (behavior, grooming, feeding) of long-term HPO feeding in either group.
Figure 1.
Physiological and Biochemical Parameters of NZ10 and SWR mice after 19 weeks on diet. 1a. Body weight; 1b Percent lean tissue by PIXimus densitometry; 1c Percent fat by Luna PIXImus densitometry; 1d Fasting blood glucose; 1e Serum Insulin. Data represent mean ± SEM and were analyzed by 2-way ANOVA as described in Methods (* p<0.01; n=10 or larger).
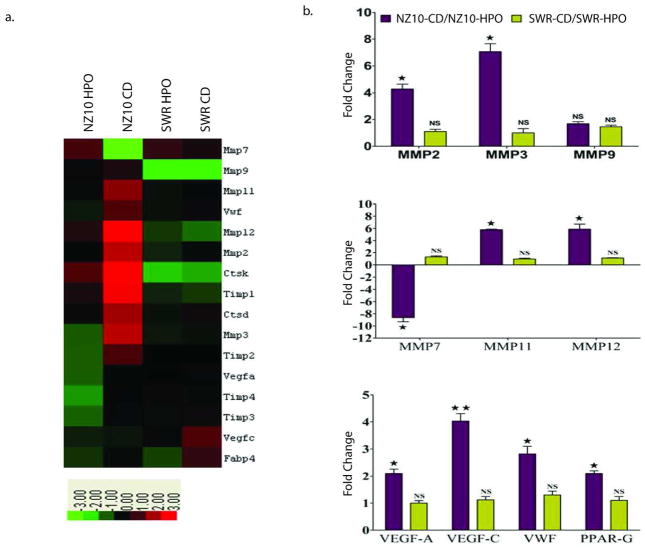
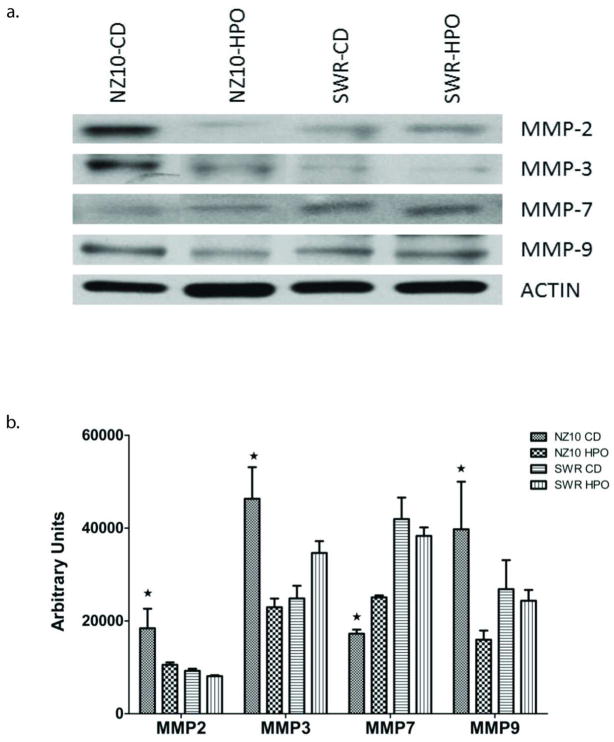
Microarray heat maps (Figure 2a) suggested diet and strain-dependent increase in the levels of MMPs and angiogenic genes as well as TIMPs and cathepsins D and K. Responsive MMP and angiogenic genes were confirmed and quantified using RT-PCR (Fig. 2b). The following transcripts were increased by 3–7-fold in VAT of NZ10 mice fed CD relative to their counterparts fed HPO: MMP-2, 3, 11, 12, VEGF-A, VEGF-C and VWF. Protein expression of pro-MMP-2, 3 and 9 was also significantly increased in the VAT of NZ10 mice fed CD while, pro-MMP-7 protein expression was reduced in the VAT of both groups of NZ10 mice (Fig 3). These results are consistent with previous reports of increased levels of MMPs -3, -11, and -12 in the VA of obese wild type mice (11) and elevated MMP2 in obese db/db mice and humans (12). It was recently reported that MMPs 3 and 12 are part of a panel of MMPs that are induced by inflammatory cytokines (19). None of these transcripts were diet-regulated in the SWR group. MMP9 expression was not regulated but MMP-7 transcripts were markedly decreased in NZ10-CD relative to NZ10-HPO or SWR groups. Two other studies have reported similar findings for MMP-7 in different models suggesting that decreased MMP-7 may be an obesity marker (11, 12). Microarrays indicated increased levels of TIMPs-1–4 transcripts in the NZ10-CD group relative to HPO or SWR groups (see Table 1) suggesting components of diet and genetics in the regulation of these transcripts. A complete list of differentially expressed genes is presented in the Online Supplement (Fig. S1).
Figure 2.
(a) Microarray Analysis. Heat maps show differences in transcript levels of MMPs, TIMPs and angiogenic factors in the VAT of NZ10 and SWR mice fed either diet for 19 weeks. Pooled visceral adipose (n=4) was used for performing the microarrays. MMP: matrix metalloproteinase; VWF: Von Willebrand Factor; CtsK: Cathepsin K; CtsD: Cathepsin D; TIMP: Tissue Inhibitor of Metalloproteinase; FABP4: Fatty acid binding protein 4. Color scale represents fold increase (red) or decrease (green) from baseline. (b) RT-PCR Analysis of MMPs and angiogenic transcripts comparing effect of diet on transcripts levels in VAT after 19 weeks. Top panel, ratios of MMP 2, 3 and 9 transcript levels; middle panel, ratios of MMP 7, 11 and 12 transcript levels; bottom panel, ratios of VEGF, VWF and PPAR-γ transcript levels. Dark bars: NZ10-CD/NZ10-HPO; Light bars: SWR-CD/SWR-HPO. Data represent mean ± SEM (* p<0.05; ** p< 0.01; *** p<0.001; n=8).
Figure 3.
Effect of CD and HPO diet on the protein expression of MMP-2, 3, 7 and 9 in the visceral adipose of NZ10 and SWR mice at 25 weeks. (a) Representative western blots (b) quantification of blots by densitometry showing mean ± SEM; *p<0.05 comparing HPO and SWR (n=3).
Table 1. Diet-regulated genes from microarray.
Comparison of selected diet-regulated genes in NZ10 and SWR mouse strains by Microarray. The Table shows the ratio of transcript levels for the CD vs. HPO diet after 19 weeks on diet for each mouse strain.
| NZ10 CD/NZ10 HFO | SWR CD/SWR HFO | |
|---|---|---|
| MMP2 | 4.41 | 1.35 |
| MMP3 | 9.13 | 1.09 |
| MMP7 | −44.32 | −1.35 |
| MMP8 | 3.43 | 1.60 |
| MMP9 | 1.37 | 1.00 |
| MMP11 | 3.53 | 1.20 |
| MMP12 | 5.86 | 0.74 |
| MMP19 | 2.91 | 0.47 |
| MMP25 | 2.04 | 1.44 |
| TIMP1 | 4.63 | 0.87 |
| TIMP2 | 4.38 | 0.98 |
| TIMP3 | 2.23 | 1.27 |
| TIMP4 | 2.83 | 0.82 |
| CTSD | 3.41 | 1.39 |
| CTSK | 8.17 | 1.14 |
| CTSZ | 3.36 | 0.77 |
| CTSB | 2.71 | 0.73 |
| FABP4 | 3.01 | 0.88 |
| PPARG | 4.32 | 0.96 |
Previous work has shown positive correlations between PPARγ gene expression and the obese phenotype of mice fed high fat (20). Our studies also showed a marked increase in VA expression of PPAR-γ only in the NZ10-CD group confirming PPAR-γ as a genetics- and diet-responsive transcript also in this model.
Conclusion
Our results confirm that a diet of high protein and fish/vegetable oil decreases the risk of obesity and T2D in NONcNZ10 mice. The studies highlight roles for MMPs 2, 3, 11, and 12, provide further evidence that depressed expression of MMP-7 is a marker of obesity and implicate all 4 TIMPs as well as cathepsins in VA expansion in this model. Because some humans are genetically predisposed to obesity and T2D, NZ10 mice may provide an appropriate model for dissecting the underlying genes that confer the susceptibility phenotype. This is the first study to describe diet-regulation of obesity-related genes in any murine model of polygenic susceptibility for obesity and T2D. Further studies with larger sample sizes are warranted to accurately define the molecular basis of diet-regulated obesity in these mice.
Supplementary Material
Supplement Table (S1). List of differentially expressed transcripts by Microarray. The Table includes all mouse probes with signals above threshold in at least one sample and significant with False Discovery rate < 5% (181 probes). Values indicate probe intensities (background subtracted, Log2-transformed and normalized) for each group.
Acknowledgments
Supported by grant # 1RO1HL092499 from the NIH and by a grant from the Florida Heart Research Institute.
References
- 1.Reifsnyder PC, Leiter EH. Deconstructing and reconstructing obesity-induced diabetes (diabesity) in mice. Diabetes. 2002;51:825–832. doi: 10.2337/diabetes.51.3.825. [DOI] [PubMed] [Google Scholar]
- 2.Cho RY, Kim JH, Park YS, et al. Hyperglycemia, maturity-onset obesity, and insulin resistance in NONcNZ010/LtJ males, a new mouse model of type 2 diabetes. Am J Physiol Endocrinol Metab. 2007;293:E327–E336. doi: 10.1152/ajpendo.00376.2006. [DOI] [PubMed] [Google Scholar]
- 3.Leiter EH, Reifsnyder PC, Xiao Q, Mistry J. Adipokine and insulin profiles distinguish diabetogenic and non-diabetogenic obesities in mice. Obesity. 2007;15:1961–8. doi: 10.1038/oby.2007.234. [DOI] [PubMed] [Google Scholar]
- 4.Herberg L. Insulin resistance in abdominal and subcutaneous obesity: comparison of C57BL/6J-ob/ob with New Zealand obese mice. In: Shafrir E, Renold A, editors. Frontiers in Diabetes Research: Lessons from Animal Diabetes II. London, UK: John Libbey & Company Ltd; 1988. pp. 367–373. [Google Scholar]
- 5.Zhou J, Martin R, Tulley R, et al. Failure to ferment dietary resistant starch in specific mouse models of obesity results in no body fat loss. J Agric Food Chem. 2009;57:8844– 8851. doi: 10.1021/jf901548e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiu J, Cheng R, Zhou XY, Zhu JG, Qin DN, Kou CZ, et al. Gene expression profiles of adipose tissue of high-fat diet-induced obese rats by cDNA microarrays. Mol Biol Rep. 2010;37:3691–5. doi: 10.1007/s11033-010-0021-6. [DOI] [PubMed] [Google Scholar]
- 7.Huber J, Loffler M, Bilban M, Reimers M, Kadi A, Todoric J, et al. Prevention of highfat diet-induced adipose tissue remodeling in obese diabetic mice by n-3 polyunsaturated fatty acids. Int J Obesity. 2007;31:1004–13. doi: 10.1038/sj.ijo.0803511. [DOI] [PubMed] [Google Scholar]
- 8.Meissburger B, Stachorski L, Roder E, Rudofsky G, Wolfrum C. Tissue inhibitor of matrix metalloproteinase 1 (TIMP1) controls adipogenesis in obesity in mice and in humans. Diabetologia. 2011;54:1468–79. doi: 10.1007/s00125-011-2093-9. [DOI] [PubMed] [Google Scholar]
- 9.Minematsu T, Huang L, Ibuki A, Nakagami G, Akase T, Sugama J, Nagase T, Yoshimura K, Sanada H. Altered Expression of Matrix Metalloproteinases and Their Tissue Inhibitors in Matured Rat Adipocytes in Vitro. Biol Res Nurs. 2011 Jun 16; doi: 10.1177/1099800411410870. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 10.Meissburger B, Stachorski L, Roder E, Rudofsky G, Wolfrum C. Tissue inhibitor of matrix metalloproteinase 1 (TIMP1) controls adipogenesis in obesity in mice and in humans. Diabetologia. 2011;54:1468–79. doi: 10.1007/s00125-011-2093-9. [DOI] [PubMed] [Google Scholar]
- 11.Maquoi E, Munaut C, Colige A, Collen D, Lijnen HR. Modulation of adipose tissue expression of murine matrix metalloproteinases and their tissue inhibitors with obesity. Diabetes. 2002;51:1093–101. doi: 10.2337/diabetes.51.4.1093. [DOI] [PubMed] [Google Scholar]
- 12.Chavey C, Mari B, Monthouel MN, Bonnafous S, Anglard P, Van Obberghen E, Tartare-Deckert S. Matrix metalloproteinases are differentially expressed in adipose tissue during obesity and modulate adipocyte differentiation. J Biol Chem. 2003;278:11888–96. doi: 10.1074/jbc.M209196200. [DOI] [PubMed] [Google Scholar]
- 13.Van Hul M, Lijnen HR. A functional role of gelatinase A in the development of nutritionally induced obesity in mice. J Thromb Haemost. 2008;6:1198–206. doi: 10.1111/j.1538-7836.2008.02988.x. [DOI] [PubMed] [Google Scholar]
- 14.Derosa G, Ferrari I, D’Angelo A, Tinelli C, Salvadeo SA, Ciccarelli L, Piccinni MN, Gravina A, Ramondetti F, Maffioli P, Cicero AF. Matrix metalloproteinase-2 and -9 levels in obese patients. Endothelium. 2008;15:219–24. doi: 10.1080/10623320802228815. [DOI] [PubMed] [Google Scholar]
- 15.Daquinag AC, Zhang Y, Kolonin MG. Vascular targeting of adipose tissue as an antiobesity approach. Trends Pharmacol Sci. 2011;32:300–7. doi: 10.1016/j.tips.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 16.Lee MJ, Wu Y, Fried SK. Adipose tissue remodeling in pathophysiology of obesity. Curr Opin Clin Nutr Metab Care. 2010;13:371–6. doi: 10.1097/MCO.0b013e32833aabef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson A, Shehadeh L, Yu H, Webster KA. Age-Related Changes in the Molecular Genetic Profiles of Murine Bone Marrow Mesenchymal Stem Cells. BMC Genomics. 2010;11:229. doi: 10.1186/1471-2164-11-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chopra I, Li HF, Wang H, Webster KA. Phosphorylation of the insulin receptor by AMP-activated protein kinase (AMPK) promotes ligand-independent activation of the insulin signalling pathway in rodent muscle. Diabetologia. 2012;55:783–94. doi: 10.1007/s00125-011-2407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Hara A, Lim FL, Mazzatti DJ, Trayhurn P. Microarray analysis identifies matrix metalloproteinases (MMPs) as key genes whose expression is up-regulated in human adipocytes by macrophage-conditioned medium. Pflugers Arch. 2009;458(6):1103–14. doi: 10.1007/s00424-009-0693-8. [DOI] [PubMed] [Google Scholar]
- 20.Kubota Y, Terauchi H, Miki K, et al. PPARγ mediates high-fat diet-induced adipocyte hypertrophy and insulin resistance. Mol Cell. 1999;4:597–609. doi: 10.1016/s1097-2765(00)80210-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement Table (S1). List of differentially expressed transcripts by Microarray. The Table includes all mouse probes with signals above threshold in at least one sample and significant with False Discovery rate < 5% (181 probes). Values indicate probe intensities (background subtracted, Log2-transformed and normalized) for each group.