Abstract
Lafora disease (LD) is a fatal, autosomal recessive neurodegenerative disorder that results in progressive myoclonus epilepsy. A hallmark of LD is the accumulation of insoluble, aberrant glycogen-like structures called Lafora bodies. LD is caused by mutations in the gene encoding the E3 ubiquitin ligase malin or the glucan phosphatase laforin. Although LD was first described in 1911, its symptoms are still lacking a consistent molecular explanation and consequently a cure is far from being achieved. Some data suggest that malin forms a functional complex with laforin. This complex promotes the ubiquitination of proteins involved in glycogen metabolism and misregulation of pathways involved in this process results in Lafora body formation. In addition, recent results obtained from both cell culture and LD mouse models have highlighted a role of the laforin-malin complex in the regulation of ER-stress and protein clearance pathways. These results suggest that LD should be considered as a novel member of the group of protein clearance diseases such as Parkinson’s, Huntington’s, or Alzheimer’s, in addition to being a glycogen metabolism disease. Herein, we review the latest results concerning the role of malin in LD and attempt to decipher its function.
Keywords: Laforin, malin, glucan phosphatase, Lafora disease, Lafora bodies, glycogen, autophagy, ER stress, Ubiquitination, E3-ubiquitin ligase
INTRODUCTION
Lafora progressive myoclonus epilepsy (Lafora disease, LD; OMIM 254780) is a rare, fatal neurodegenerative disorder characterized by epilepsy, neurodegeneration and accumulation of polyglucosan inclusions in brain and other peripheral tissues [1]. It was described in 1911 by the Spanish neurologist Gonzalo R. Lafora, who described the presence of dark and intense inclusions in post-mortem preparations of patients that he called “amyloid bodies” [2]. While amyloid was later shown to be proteinaceous, the term originally referred to material that stained similar to plant starch [3]. The “amyloid bodies” of LD were shown to be water-insoluble glucans that are in fact very similar to plant starch and were named Lafora bodies (LBs) ([4], [5]). LD manifests during adolescence with generalized tonic-clonic seizures, myoclonus, absences, drop attacks and visual hallucinations. A progressive dementia with apraxia, aphasia and visual loss follows, leading patients to a vegetative state and death, usually within the first decade from the onset of the disease ([6], [7], [8]).
LD-causing mutations have been identified in two genes, EPM2A, encoding the glucan phosphatase laforin, and EPM2B/NHLRC1, encoding the E3 ubiquitin ligase malin ([9], [10], [11]). The identification of the malin and laforin genes and determining their biochemical activities were key steps in unraveling the cellular mechanisms that cause LD; however, there was not an obvious link between the phosphatase laforin and the E3 ligase malin. An emerging theme came from multiple labs showing that malin ubiquitinates substrates in a laforin-dependent manner, suggesting that malin and laforin form a functional complex ([12], [13], [14], [15]). These biochemical results are consistent with LD clinical data: patients carrying mutations in either EPM2A or EPM2B are phenotypically indistinguishable. Therefore, laforin and malin likely function in the same physiological pathway.
In the last decade, several laboratories have described possible functions of laforin and malin in cell physiology. While incomplete, these data are beginning to elucidate the molecular bases of the pathophysiology of LD. These results suggest that LD has strong similarities with more frequent neurological disorders like Parkinson’s, Huntington’s, and Alzheimer’s. In this review we discuss the current knowledge of the E3 ubiquitin ligase malin in Lafora disease.
1.- E3-ubiquitin ligase activity of malin
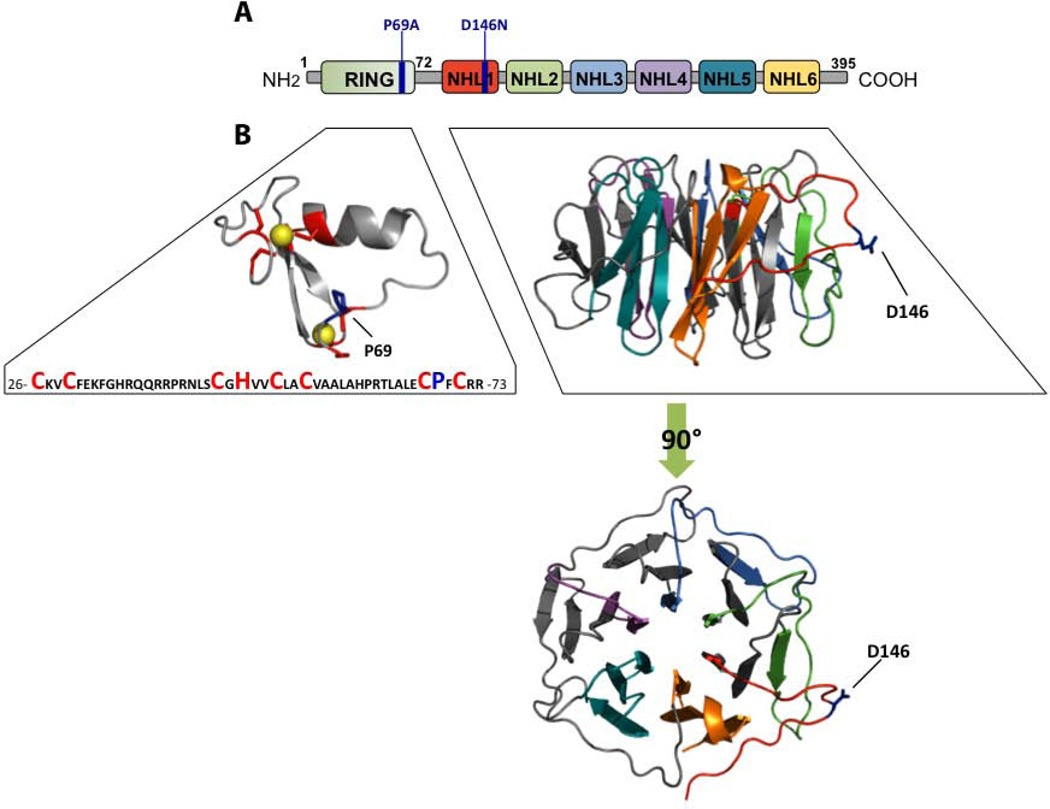
In 2003, Minassian and colleagues [11] described that mutations in EPM2B result in LD. EPM2B is located in chromosome 6q22.3 and contains a single exon encoding a 395 amino acid protein named malin. Malin is an E3-ubiquitin ligase with a zinc finger RING (Really Interesting New Gene) domain of the C3HC4 type at the N-terminus and six NHL domains (also present in NCL1, HT2A and LIN-41 proteins) at the C-terminus. The NHL domains are predicted to fold into a β-propeller structure that mediates protein-protein interactions (Fig. 1).
Fig. 1.
A) Schematic depicting of the domains present in malin, numbering refers to human malin. The most frequent Lafora disease mutation observed in EPM2B-LD patients encodes malin-P69A. The malin-D146N mutation is involved in binding to laforin. B) RING and NHL-containing domains (amino acids 26–72 and 113–393, respectively) were submitted to the ESyPred3D server and modeled using the structure of human TRIM32 RING motif (PDB:2ct2) (RING model) and M. tuberculosis PknD (PDB:1rwl) (NHL model) as templates. Structural models were displayed using PyMOL. The position of critical residues of the zinc finger C3HC4 type in the RING domain are indicated in red; spheres represent zinc atoms; the position of the P69A mutation is also depicted. The six NHL domains are predicted to fold into a β-propeller structure; the position of the D146N mutation, which affects the malin-laforin interaction, is also indicated.
Ubiquitination is one of the most common post-translational modifications of proteins. It occurs by the addition of ubiquitin monomers to a lysine of a target protein by a process involving three different steps: 1) activation of ubiquitin by the E1-ubiquitin activating enzyme; 2) transfer of the activated ubiquitin to the E2-ubiquitin conjugating enzyme; and 3) attachment of ubiquitin to a lysine of the substrate either directly via a HECT-type E3 ligase or indirectly in the case of RING-type E3 ligases ([16], [17]). The reaction results in the attachment of a single ubiquitin moiety to one lysine (monoubiquitination), a single ubiquitin to several lysines (multiubiquitination), or several ubiquitin molecules to one lysine (polyubiquitination). In the latter case, new ubiquitin molecules are linked to previous moieties using any of the seven internal lysine residues present in ubiquitin (K6, K11, K27, K29, K33, K48, and K63). Depending on the type of linkage present in the polyubiquitin chain, the substrate is targeted to the proteasome for destruction (K48-linked chains) or the modification causes changes in cell signalling, trafficking, and/or interactions with other proteins ([16], [17], [18]).
As indicated above, malin is a RING-type E3-ubiquitin ligase. Among the 38 E2-conjugating enzymes encoded in the human genome, malin is able to interact in vitro with UbcH2, UbcH5a, UbcH5c and UbcH6 and not with UbcH1, UbcH3, UbcH5b, UbcH7, UbcH10 and UbcH13 [12]. However, the E2s that participate in the in vivo ubiquitination process mediated by malin are still unknown. Identification of the endogenous E2(s) is an important point to be resolved since the topology of the polyubiquitin chains present in a substrate depends both on the type of the E2s and the E3s ([16], [17]). We and others have reported that malin mediates the incorporation of both K48- and K63-linked ubiquitin chains in different substrates, but it is unknown which E2(s) participates in these events ([12], [19], [20], see below).
2.- Mutations in EPM2B
Approximately 60 different mutations in the EPM2B have been associated with LD to date [21]. Some of these mutations affect the enzymatic activity of malin, whereas others affect the interaction between malin and another protein. The most prevalent mutation is P69A, in the RING motif of malin. The malin-D146N mutation disrupts the interaction with laforin without altering the ubiquitinating activity of malin [15] (Fig. 1). This mutation impairs the formation of a functional laforin-malin complex ([14], [15]) and leads to aberrant glycogen accumulation [22]. Although patients with mutations in laforin or malin show similar clinical manifestations, those carrying mutations in the EPM2B have a less severe progression of the disease and live longer. In fact, the mutation D146N has been associated with a slower progression of the disease ([23], [24], [25]).
3.- Malin phylogeny
From an evolutionary point of view, the fact that malin forms a functional complex with laforin raised the question of whether these two proteins share a common phylogenetic lineage or whether they evolved independently. Laforin is conserved in all vertebrates, a select group of protozoans, and in two invertebrate genomes; however, the gene encoding laforin is absent from most protozoan and invertebrate genomes such as yeast, fly and worm ([26], [27]). A recent study investigated malin phylogeny and found that the malin gene is exclusively found in vertebrate genomes and the cephalochordate Branchiostoma floridae [28]. Thus, the distribution of malin does not correlate with that of laforin, suggesting that laforin has a malin-independent function(s) [28]. This study also reported an evolutionary relationship between malin and TRIM32, an E3-ubiquitin ligase that belongs to the TRIM family (TRIpartite Motif-containing). Both malin and TRIM32 exhibit a similar modular structure, containing a functional RING motif and six NHL-protein interaction motifs. Species distribution of malin and TRIM32 and the exon-intron composition of both proteins further corroborated the possibility of a common phylogenetic origin. Moreover, some functional redundancies were also discovered: TRIM32 is capable of ubiquitinating some malin substrates in overexpression cell culture systems, but with different polyubiquitin chain topology. However, this redundancy was not reciprocal since specific TRIM32 substrates were not ubiquitinated by the laforin-malin complex. Cumulatively, the phylogenetic studies suggest that malin initially evolved from ancestral TRIM genes and developed a particular E3 ubiquitin ligase activity, possibly co-evolving with laforin as a binding partner in vertebrate species [28].
4.- Subcellular localization of malin
Attempts to define the subcellular localization of endogenous malin have been unsuccessful due to the lack of a reliable α-malin antibody. Ganesh and co-workers, using overexpression studies of malin fused to GFP indicated that malin is located at the endoplasmic reticulum and that upon treatment of the cells with the proteasome inhibitor MG132, malin forms perinuclear aggregates that are also immunoreactive against ubiquitin, ubiquitin-conjugating enzymes, chaperones and proteasome subunits [29]. The same group showed later that malin predominantly localizes to the nucleus and this localization does not change upon subjecting the cells to heat shock or glucose starvation ([30], [31]). Cheng et al. reported that low-level expression of malin-myc localizes to the nucleus and gross overexpression results in malin-myc perinuclear ER-like localization [32]. Although these studies offer insights into malin function, there have been no reports to date that demonstrate endogenous malin localization.
5.- Malin interacting partners and physiological pathways involved
5.1.- Laforin
As mentioned above, laforin and malin form a functional complex and are likely involved in the regulation of multiple pathways. The first indication that malin interacted with laforin came from yeast two-hybrid experiments ([12], [13], [32], [33]). The physiological relevance of this interaction is highlighted by the fact that the Lafora disease mutation malin-D146N abolishes the malin-laforin interaction without affecting the E3-ubiquitin ligase activity of malin [15]. The malin-laforin interaction was confirmed by co-immunoprecipitation of the two proteins, and the interaction was shown to be direct using purified recombinant proteins [12]. Moreover, work from Guinovart and colleagues elegantly reported a mechanism whereby the malin-laforin complex inhibits neuronal glycogen synthesis [14]. Despite these convincing studies, the lack of an α-malin antibody has impeded the confirmation of the malin-laforin complex in a truly physiological context.
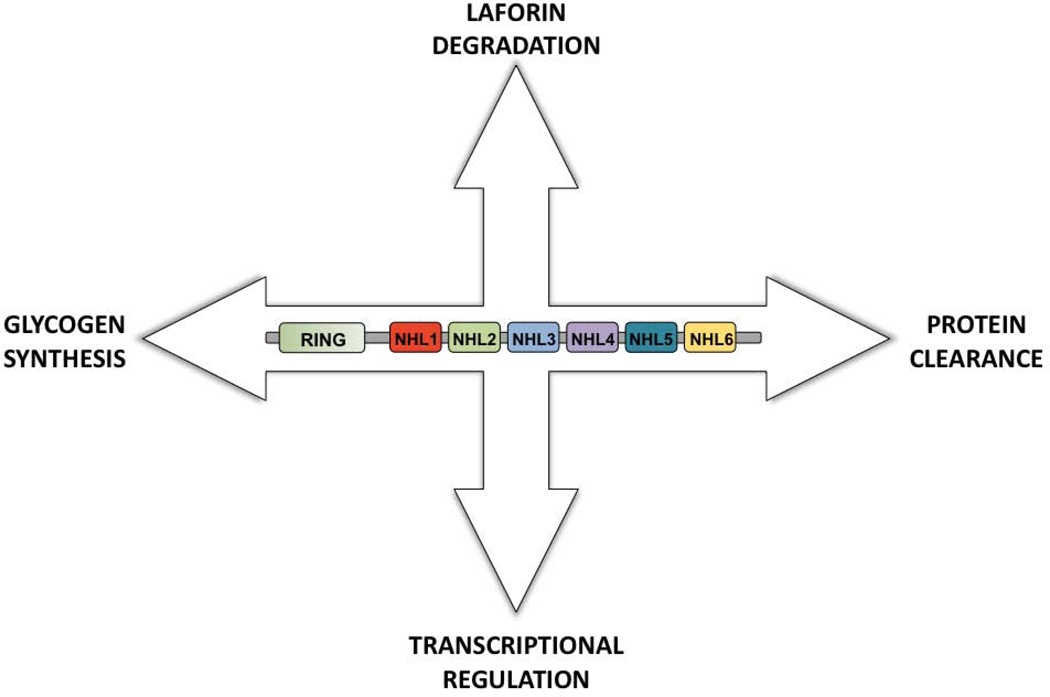
Laforin and malin exhibit an intriguing relationship because while they form a complex, malin also ubiquitinates laforin and targets it for degradation [12]. Malin-directed degradation of laforin is counter-intuitive since loss of either gene results in LD. However, patient mutations in malin result in increased levels of laforin [11], and similar results are observed in malin-deficient mouse models ([34], [35], [36], [37]). While we and others interpret these results to mean that malin promotes the degradation of laforin, others in the field disagree (see below). An additional layer of complexity arises from the fact that one group reported that malin is more abundant in the presence of laforin [14]. These results suggest that laforin increases the stability of malin in spite of laforin being degraded by malin (Fig. 2).
Fig. 2.
Schematic summarizing the action of malin on different proteins and in different pathways. See text for details.
We reported that the interaction between laforin and malin is enhanced by the AMP-activated protein kinase (AMPK) [15]. AMPK phosphorylates laforin at residue Ser25 and this modification increases the interaction between laforin and malin [38]. Conditions that trigger the activation of AMPK such as glucose starvation, improve the interaction between laforin and malin ([15], [31]). These results predict that AMPK activation would lead to lower levels of R5/PTG and glycogen synthase, known substrates of the laforin-malin complex. However, DePaoli-Roach et al found no change in R5/PTG levels in mice under conditions that activate AMPK [34]. Thus, more work is required to define more precisely the role of AMPK in laforin-malin regulation.
5.2.- Enzymes involved in glycogen synthesis
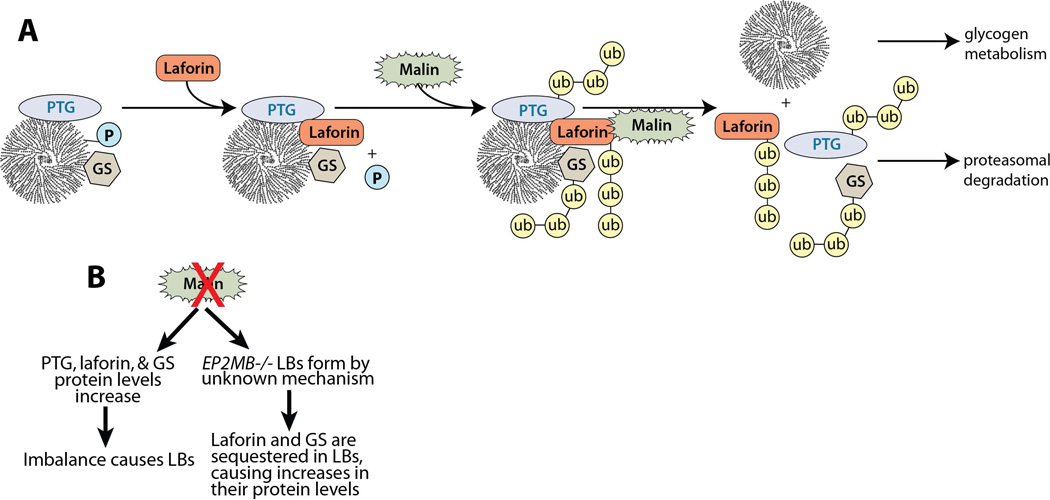
One of the first identified substrates of the laforin-malin complex was R5/PTG ([14], [15], [39]). R5/PTG, encoded by PPP1R3C gene, is a targeting subunit of protein phosphatase 1 (PP1), directing PP1 to glycogen. R5/PTG-targeted PP1 dephosphorylates glycogen synthase, activating it, and thus is an activator of glycogen synthesis. R5/PTG-directed PP1 also dephosphorylates and inhibits glycogen phosphorylase. Therefore, misregulation of R5/PTG affects both synthesis and breakdown of glycogen ([40], [41], [42]). Results from three labs demonstrated that the laforin-malin complex ubiquitinates R5/PTG, decreases R5/PTG protein levels, and downregulates glycogen levels. The laforin-malin complex also interacts with the PP1 binding partners GL (PPP1R3B) and R6 (PPP1R3D), but not GM (PPP1R3A) ([39], [43]). In addition to affecting PP1 activity, the laforin-malin complex ubiquitinates and downregulates the activity of glycogen synthase (GS) [14] and glycogen debranching enzyme (GDE/AGL) [32]. Jana and colleagues recently reported that the laforin-malin complex controls glycogen synthesis by ubiquitinating and promoting the proteasomal degradation of neuronatin, an 81 amino acid protein that stimulates glycogenesis [44]. These reports strongly suggest a role for the laforin-malin complex in the regulation of glycogen synthesis. For this reason, it was proposed that in the absence of a functional laforin-malin complex glycogen synthesis would proceed without proper coordination and would result in the accumulation of poorly branched polyglucosan species, i.e. Lafora bodies (Fig. 2), (Fig. 3).
Fig. 3.
A) Proposed mechanism of action for malin in glycogen metabolism. The protein targeting to glycogen (PTG) subunit of PP1 and glycogen synthase (GS) both bind glycogen particles during normal glycogen metabolism. During glycogen synthesis, GS incorporates a phosphate (P) in glycogen on approximately 1/10,000 glucose monomers. Laforin is targeted to the glycogen particle via its CBM and liberates phosphate from glycogen. The malin-laforin interaction is enhanced by laforin-Ser25 phosphorylation via AMP-activated protein kinase. Once bound to laforin, malin ubiquitinates laforin, PTG, and GS. This ubiquitination triggers the release of all three enzymes from the glycogen particle, targets them for proteasome-dependent degradation, and allows glycogen metabolism to proceed normally. GS, glycogen synthase; P, phosphate group; PTG/R5, protein targeting to glycogen subunit of protein phosphatase 1; ub, ubiquitin. B) Predicted mechanisms that result in increased levels of glycogen synthase, PTG, and/or laforin in Epm2b-deficient mice.
Although the above work is from multiple labs using different systems, these results are largely based on overexpression of laforin and malin in cell cultures and on in vitro results utilizing recombinant proteins. In contrast to these results, mice lacking either malin or laforin that are 3 to 6 months of age do not show increased levels of glycogen synthase or R5/PTG ([34], [35], [45]). However, a more recent report on this matter indicates that in the brain of 11 month old mice lacking malin, there is an increase in the levels of glycogen synthase [36], suggesting that the laforin-malin complex does downregulate the levels of proteins involved in glycogen synthesis. Obviously, more work is needed to reconcile these results (Fig. 3).
5.3.- Malin in ER-stress and protein clearance
In addition to the role that the laforin-malin complex has in glycogen homeostasis, the complex has additional roles in several other pathways. Multiple labs have reported that the laforin-malin complex plays a role in protecting cells from ER-stress conditions ([46], [47]). In cell culture models depleted of malin or laforin there is increased ER-stress response that eventually leads to decreased proteasome function and increased apoptosis, which could be important factors in the development of LD ([46], [47]) (Fig. 2).
It has also been reported that laforin and malin form a functional complex with Hsp70 and that this macro-complex suppresses the cytotoxicity produced by the accumulation of misfolded proteins (i.e., expanded polyglutamine proteins, and α-synuclein) [48]. It was proposed that laforin interacts with both Hsp70 and misfolded proteins while recruiting malin to trigger the ubiquitination of these proteins and targeting them for degradation. These results suggest that the laforin-malin complex could be considered as a new component of the neuronal response to misfolded proteins. If correct, the laforin-malin complex could have similar functions in protein clearance as those reported for other E3-ubiquitin ligases such as parkin, CHIP, dorfin and E6-AP [48]. Given these results, it has been proposed that one of the primary causes of Lafora disease may be the inability to eliminate misfolded proteins, and for this reason, the disease should be considered as a novel member of the group of protein clearance diseases ([8], [29]) (Fig. 2).
Additionally, it was recently described that the laforin-malin complex is a positive regulator of autophagy. Cellular and mouse models lacking either laforin or malin show a decrease in autophagy, likely due to an impairment in autophagosome formation ([37], [49]). In both cases, there are decreased content of autophagic vesicles and lower levels of the LC3-II autophagic marker. As a result of autophagic dysfunction, there are increased levels of the p62 autophagic marker in both cases. The autophagic dysfunction observed in models lacking either laforin or malin may lead to the accumulation of diverse autophagic substrates that would contribute to cell stress and cell death. Rodriguez de Cordoba and colleagues recently reported autophagy defects in 16-day-old mice lacking malin [37]. Therefore, these defects may occur at a very early stage of the disease and these results further highlight the similarities to other more common neurological disorders that present similar autophagic impairment like Parkinson’s, Alzheimer’s or Huntington’s [50] (Fig. 2). Thus, a common reoccurring theme is the similarities between Lafora disease and these disorders.
5.4.- Malin and transcriptional regulation
Two reports recently implicate malin in transcriptional regulation. One group demonstrated that laforin and malin form a ternary complex with the co-chaperone CHIP (C-terminus Hsp70 Interacting Protein). This interaction improved the stability of malin [51] and was necessary for the heat shock response mediated by the transcription factor HSF1 (Heat Shock Factor 1) [30]. These authors reported that laforin translocates to the nucleus after heat shock, requiring both CHIP and HSF1 for this nuclear translocation. Once inside the nucleus, the laforin-malin complex is required for the function of HSF1 as a transcriptional regulator. These results indicate that the laforin-malin complex is required for full protection against heat-shock-induced cell death and provide another link between laforin-malin and cellular responses to stress [30] (Fig. 2).
In the second report, malin was shown to interact with dishevelled2 (Dvl2), a key component of the Wnt signalling pathway. Dvl2 is a cytosolic protein that regulates β-catenin shuttling to the nucleus where β-catenin mediates transcription of Wnt target genes. The authors reported that malin enhances K48- and K63-linked ubiquitination of Dvl2, promotes its degradation via the proteasome and autophagy, and inhibits Wnt signalling. These results suggest a possible dysregulation of Wnt signalling in Lafora disease [20]. Loss of function of malin may increase Wnt signalling in developing or adult brain leading to abnormal synaptic differentiation, synaptic plasticity, or other neurogenic defects (Fig. 2).
6.- Animal models lacking malin
Multiple animal models lacking malin have been utilized to study LD. The first animal model of Lafora disease due to mutations in EPM2B was that of miniature wirehaired dachshunds. LD is caused in these dogs by the expansion of a 12-nucleotide sequence in the region between the RING finger and the NHL domains. This expansion, containing from 14 to 26 repetitions, severely decreases malin mRNA levels (900 times lower than wild type) [52]. The expansion was found in many Canidae species, but not in closely related Arctoidae or Felidae and was only commonly found in miniature wirehaired dachshunds. Although these dogs provided valuable insights into LD, the difficulty in their handling and breeding does not make them an appropriate model to study the mechanistic cause of human LD.
In addition to the canine model, multiple mouse models lacking Epm2b have been generated. DePaoli-Roach et al., analyzed 3-month-old mice lacking Epm2b and reported that these mice develop Lafora bodies in brain, heart, and skeletal muscle [34]. Additionally, they observed no increase in glycogen synthase, R5/PTG, or glycogen debranching enzyme in these animals and they did not observe any changes in enzymatic activities of glycogen metabolism enzymes; specifically, glycogen synthase and glycogen phosphorylase enzymatic activities were unchanged. However, they did observe increases in laforin protein levels. They reported that laforin from wild-type animals was found in the soluble fraction after a low-speed spin, but the increased laforin protein in Epm2b−/− mice was observed in a low speed insoluble pellet. They proposed that the absence of malin promotes the accumulation of LBs by an unknown mechanism and that LBs sequester laforin and protect it from degradation by another source than malin-directed degradation. Thus, they argued that the increase in laforin occurs indirectly in malin deficient mice and not because malin is the E3 ligase for laforin. However, as described above, we previously reported that malin ubiquitinates and promotes the degradation of laforin in cell culture and recapitulated these results using purified recombinant proteins in vitro [12].
Shortly after the first report, Minassian and colleagues published a second mouse model lacking Epm2b [35]. They analyzed 6-month-old animals and they also reported that the mice develop LBs in brain, skeletal muscle, and liver, indicating that the mouse model recapitulates the disease. They too reported that glycogen synthase levels and activity are unchanged in mice lacking Epm2b. Similar to DePaoli-Roach et al., they reported higher levels of laforin in their mice and an increase of laforin in the insoluble fraction, but they did not observe a decrease of laforin in the soluble fraction. Thus, they did not observe a redistribution of laforin, but did observe increased laforin in the insoluble pellet. They too argue that the increase in laforin in malin-deficient mice is a result of laforin being “trapped” in LBs. Thus, they argue that malin does not regulate the protein levels of laforin, but that laforin levels are increased in malin-deficient mice by the indirect accumulation of laforin in LBs. Additionally, they reported that malin-deficient mice have increased levels of glycogen phosphate as compared to wild-type animals, but not as high as in laforin-deficient mice. The authors hypothesize that lack of malin results in LB formation only in part due to increased glycogen phosphorylation, and that malin has an additional function in regulating glycogen metabolism that contributes to LD.
At a later date, Guinovart and colleagues reported their results from the analysis of 11-month-old Epm2b−/− mice [36]. They confirmed the presence of LBs reported by the earlier groups, but extended these findings with elegant microscopy focused on the hippocampus. They found LBs in both neurons and astrocytes. Importantly, they reported that LBs in soma and processes of parvalbumin-positive interneurons were accompanied by progressive loss of these neurons and neurophysiological alterations, providing a direct link between LBs and impairment of hippocampal function. Additionally, they found increased levels of glycogen synthase in 11-month-old malin-deficient mice, but did not observe increases in glycogen synthase activity. [36]. This report suggests that the age of the mice studied may explain the differences in reported results.
Recently, a fourth mouse model lacking Epm2b was reported by Rodriguez de Cordoba and colleagues [37] Phenotypically, at six months of age, these mice were similar to those reported above: they accumulate LBs in different areas of the brain and they exhibit higher levels of laforin. This group extended our understanding of LD by analyzing young, 16-day-old Epm2b−/− mice. They found that in 16-day-old malin-deficient mice there are no visible LBs present in brain, yet laforin levels are increased and the increased laforin is found in the soluble fraction. As the mice aged, LBs appeared and laforin became enriched in the insoluble fraction, possibly forming part of these LBs. The authors then investigated autophagy in this model and reported that Epm2b−/− mice exhibit a dysfunction in autophagy in animals as young as 16 days old. Therefore they concluded that autophagy is one of the first determinants that is impaired in Lafora disease. In addition, these authors reported neurological and behavioural abnormalities in Epm2b−/− mice, such as reduced spontaneous motor activity and coordination, abnormal clasping upon tail suspension and reduced object recognition task [37].
In conclusion, several animal models of LD caused by the lack of malin are now available. These animals recapitulate most of the features present in LD patients, thus they are good models to study the pathophysiology of the disease. The initial reports utilizing these models have observed some differences, but future studies will likely result in a unifying theme describing how the loss of malin results in LBs and Lafora disease.
7.- PERSPECTIVES
Greater than 100 papers have been published on Lafora disease in the last five years. In spite of the advances during this time, there are still holes in our understanding of the cellular basis of the disease and controversies that need to be resolved. For example, what is the role of malin in the regulation of glycogen synthesis? Pioneering work by the Dixon and Roach labs established that laforin is a glucan phosphatase, that acts as a control mechanism to eliminate the phosphates that glycogen synthase introduces erroneously into the glycogen molecule ([26], [53], [54], [55]). Thus, there is a direct path where loss of laforin leads to hyperphosphorylation of glycogen that eventually turns into a Lafora body causing LD.
Similar hyperphosphorylated glycogen is observed in mouse models lacking malin; however, these animals also contain higher levels of laforin. Therefore, these results are difficult to reconcile. As discussed above, there is no consensus as to whether or not malin regulates protein levels of glycogen metabolism enzymes. Thus, the mechanism for LB formation in the absence of malin is still unresolved.
Another unresolved point is the consequence of the laforin-malin mediated ubiquitination of substrates. Some reports indicate that ubiquitinated substrates are targeted for proteasomal degradation (e.g. laforin, R5/PTG, and glycogen synthase). However, ubiquitination of other substrates does not target them for proteasomal degradation (e.g. β-subunit of the AMPK complex). How can one reconcile these results? One possibility is that the laforin-malin complex introduces different ubiquitin chains depending on the E2 that assists in substrate ubiquitination. This mechanism would explain why in some cases the result of the action of the laforin-malin complex is the degradation of the substrate whereas in other cases the substrate is not degraded. Recent reports indicate that the laforin-malin complex has a positive role in autophagy. As it has been reported that autophagy mediates the degradation of proteins that are labelled with K63-linked ubiquitin chains, it could be possible that the modification mediated by the laforin-malin complex would target substrates to be degraded by the lysosome.
Over the past decade, our understanding of Lafora disease has proceeded very rapidly. The latest results suggest that protein clearance may be a key pathway defective in LD. For this reason, LD may be considered as a novel member of protein clearance diseases (i.e., Parkinson’s, Alzheimer’s, and Huntington’s.) in addition to being a glycogen metabolism disease. If results concerning the mechanism of LD continue at their current pace, then our understanding of the molecular basis of this pathology will soon allow a rational therapeutic approach for this devastating disease.
ACKNOWLEDGMENTS
C.R-M. is supported by a contract from la Fundació La Marató de TV3. This work was supported by a grant from the Spanish Ministry of Education and Science (SAF2011-27442), a grant from la Fundació La Marato de TV3 (ref. 100130) and a grant from Generalitat Valenciana (Prometeo 2009/051) to P.S. and the National Institutes of Health grants R00NS061803, P20RR020171, R01NS070899 and University of Kentucky College of Medicine startup funds to M.S.G.
REFERENCES
- 1.Gentry MS, Dixon JE, Worby CA. Lafora disease: insights into neurodegeneration from plant metabolism. Trends Biochem Sci. 2009;34:628–639. doi: 10.1016/j.tibs.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lafora GR, Glueck B. Beitrag zur histogpathologie der myoklonischen epilepsie. Gesamte Neurol Psychiatr. 1911;6:1–14. [Google Scholar]
- 3.Virchow R. Die cellularpathologie inihrer Begründung auf physsiologische and pathologische Gewebelehre. Hirschwald. 1858 [Google Scholar]
- 4.Harriman DG, Millar JH, Stevenson AC. Progressive familial myoclonic epilepsy in three families: its clinical features and pathological basis. Brain. 1955;78:325–349. doi: 10.1093/brain/78.3.325. [DOI] [PubMed] [Google Scholar]
- 5.Yokoi S, Austin J, Witmer F, Sakai M. Studies in myoclonus epilepsy (Lafora body form). I. Isolation and preliminary characterization of Lafora bodies in two cases. Arch Neurol. 1968;19:15–33. doi: 10.1001/archneur.1968.00480010033002. [DOI] [PubMed] [Google Scholar]
- 6.Minassian BA. Lafora's disease: towards a clinical, pathologic, and molecular synthesis. Pediatr Neurol. 2001;25:21–29. doi: 10.1016/s0887-8994(00)00276-9. [DOI] [PubMed] [Google Scholar]
- 7.Ganesh S, Puri R, Singh S, Mittal S, Dubey D. Recent advances in the molecular basis of Lafora's progressive myoclonus epilepsy. J Hum Genet. 2006;51:1–8. doi: 10.1007/s10038-005-0321-1. [DOI] [PubMed] [Google Scholar]
- 8.Delgado-Escueta AV. Advances in lafora progressive myoclonus epilepsy. Current neurology and neuroscience reports. 2007;7:428–433. doi: 10.1007/s11910-007-0066-7. [DOI] [PubMed] [Google Scholar]
- 9.Minassian BA, Lee JR, Herbrick JA, Huizenga J, Soder S, Mungall AJ, Dunham I, Gardner R, Fong CY, Carpenter S, et al. Mutations in a gene encoding a novel protein tyrosine phosphatase cause progressive myoclonus epilepsy. Nat Genet. 1998;20:171–174. doi: 10.1038/2470. [DOI] [PubMed] [Google Scholar]
- 10.Serratosa JM, Gomez-Garre P, Gallardo ME, Anta B, de Bernabe DB, Lindhout D, Augustijn PB, Tassinari CA, Malafosse RM, Topcu M, et al. A novel protein tyrosine phosphatase gene is mutated in progressive myoclonus epilepsy of the Lafora type (EPM2) Hum Mol Genet. 1999;8:345–352. doi: 10.1093/hmg/8.2.345. [DOI] [PubMed] [Google Scholar]
- 11.Chan EM, Young EJ, Ianzano L, Munteanu I, Zhao X, Christopoulos CC, Avanzini G, Elia M, Ackerley CA, Jovic NJ, et al. Mutations in NHLRC1 cause progressive myoclonus epilepsy. Nat Genet. 2003;35:125–127. doi: 10.1038/ng1238. [DOI] [PubMed] [Google Scholar]
- 12.Gentry MS, Worby CA, Dixon JE. Insights into Lafora disease: malin is an E3 ubiquitin ligase that ubiquitinates and promotes the degradation of laforin. Proc Natl Acad Sci U S A. 2005;102:8501–8506. doi: 10.1073/pnas.0503285102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lohi H, Ianzano L, Zhao XC, Chan EM, Turnbull J, Scherer SW, Ackerley CA, Minassian BA. Novel glycogen synthase kinase 3 and ubiquitination pathways in progressive myoclonus epilepsy. Hum Mol Genet. 2005;14:2727–2736. doi: 10.1093/hmg/ddi306. [DOI] [PubMed] [Google Scholar]
- 14.Vilchez D, Ros S, Cifuentes D, Pujadas L, Valles J, Garcia-Fojeda B, Criado-Garcia O, Fernandez-Sanchez E, Medrano-Fernandez I, Dominguez J, et al. Mechanism suppressing glycogen synthesis in neurons and its demise in progressive myoclonus epilepsy. Nature neuroscience. 2007;10:1407–1413. doi: 10.1038/nn1998. [DOI] [PubMed] [Google Scholar]
- 15.Solaz-Fuster MC, Gimeno-Alcaniz JV, Ros S, Fernandez-Sanchez ME, Garcia-Fojeda B, Criado Garcia O, Vilchez D, Dominguez J, Garcia-Rocha M, Sanchez-Piris M, et al. Regulation of glycogen synthesis by the laforin-malin complex is modulated by the AMP-activated protein kinase pathway. Hum Mol Genet. 2008;17:667–678. doi: 10.1093/hmg/ddm339. [DOI] [PubMed] [Google Scholar]
- 16.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nature reviews. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 18.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. 'Protein Modifications: Beyond the Usual Suspects' review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moreno D, Towler MC, Hardie DG, Knecht E, Sanz P. The laforin-malin complex, involved in Lafora disease, promotes the incorporation of K63-linked ubiquitin chains into AMP-activated protein kinase beta subunits. Molecular biology of the cell. 2010;21:2578–2588. doi: 10.1091/mbc.E10-03-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma J, Mulherkar S, Mukherjee D, Jana NR. Malin Regulates Wnt Signaling Pathway through Degradation of Dishevelled2. J Biol Chem. 2012;287:6830–6839. doi: 10.1074/jbc.M111.315135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh S, Ganesh S. Lafora progressive myoclonus epilepsy: a meta-analysis of reported mutations in the first decade following the discovery of the EPM2A and NHLRC1 genes. Human mutation. 2009;30:715–723. doi: 10.1002/humu.20954. [DOI] [PubMed] [Google Scholar]
- 22.Couarch P, Vernia S, Gourfinkel-An I, Lesca G, Gataullina S, Fedirko E, Trouillard O, Depienne C, Dulac O, Steschenko D, et al. Lafora progressive myoclonus epilepsy: NHLRC1 mutations affect glycogen metabolism. J Mol Med (Berl) 2011;89:915–925. doi: 10.1007/s00109-011-0758-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gómez-Abad C, Gómez-Garre P, Gutiérrez-Delicado E, Saygi S, Michelucci R, Tassinari CA, Rodriguez de Cordoba S, Serratosa JM. Lafora disease due to EPM2B mutations. A clinical and genetic study. Neurology. 2005;64:982–986. doi: 10.1212/01.WNL.0000154519.10805.F7. [DOI] [PubMed] [Google Scholar]
- 24.Baykan B, Striano P, Gianotti S, Bebek N, Gennaro E, Gurses C, Zara F. Late-onset and slow-progressing Lafora disease in four siblings with EPM2B mutation. Epilepsia. 2005;46:1695–1697. doi: 10.1111/j.1528-1167.2005.00272.x. [DOI] [PubMed] [Google Scholar]
- 25.Franceschetti S, Gambardella A, Canafoglia L, Striano P, Lohi H, Gennaro E, Ianzano L, Veggiotti P, Sofia V, Biondi R, et al. Clinical and genetic findings in 26 Italian patients with Lafora disease. Epilepsia. 2006;47:640–643. doi: 10.1111/j.1528-1167.2006.00479.x. [DOI] [PubMed] [Google Scholar]
- 26.Gentry MS, Dowen RH, 3rd, Worby CA, Mattoo S, Ecker JR, Dixon JE. The phosphatase laforin crosses evolutionary boundaries and links carbohydrate metabolism to neuronal disease. The Journal of cell biology. 2007;178:477–488. doi: 10.1083/jcb.200704094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gentry MS, Pace RM. Conservation of the glucan phosphatase laforin is linked to rates of molecular evolution and the glucan metabolism of the organism. BMC evolutionary biology. 2009;9:138. doi: 10.1186/1471-2148-9-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roma-Mateo C, Moreno D, Vernia S, Rubio T, Bridges TM, Gentry MS, Sanz P. Lafora disease E3-ubiquitin ligase malin is related to TRIM32 at both the phylogenetic and functional level. BMC evolutionary biology. 2011;11:225. doi: 10.1186/1471-2148-11-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mittal S, Dubey D, Yamakawa K, Ganesh S. Lafora disease proteins malin and laforin are recruited to aggresomes in response to proteasomal impairment. Hum Mol Genet. 2007;16:753–762. doi: 10.1093/hmg/ddm006. [DOI] [PubMed] [Google Scholar]
- 30.Sengupta S, Badhwar I, Upadhyay M, Singh S, Ganesh S. Malin and laforin are essential components of a protein complex that protects cells from thermal stress. J Cell Sci. 2011;124:2277–2286. doi: 10.1242/jcs.082800. [DOI] [PubMed] [Google Scholar]
- 31.Singh PK, Singh S, Ganesh S. The Laforin-Malin Complex Negatively Regulates Glycogen Synthesis by Modulating Cellular Glucose Uptake via Glucose Transporters. Mol Cell Biol. 2012;32:652–663. doi: 10.1128/MCB.06353-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng A, Zhang M, Gentry MS, Worby CA, Dixon JE, Saltiel AR. A role for AGL ubiquitination in the glycogen storage disorders of Lafora and Cori's disease. Genes Dev. 2007;21:2399–2409. doi: 10.1101/gad.1553207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandez-Sanchez ME, Criado-Garcia O, Heath KE, Garcia-Fojeda B, Medrano-Fernandez I, Gomez-Garre P, Sanz P, Serratosa JM, Rodriguez de Cordoba S. Laforin, the dual-phosphatase responsible for Lafora disease, interacts with R5 (PTG), a regulatory subunit of protein phosphatase-1 that enhances glycogen accumulation. Hum Mol Genet. 2003;12:3161–3171. doi: 10.1093/hmg/ddg340. [DOI] [PubMed] [Google Scholar]
- 34.DePaoli-Roach AA, Tagliabracci VS, Segvich DM, Meyer CM, Irimia JM, Roach PJ. Genetic depletion of the malin E3 ubiquitin ligase in mice leads to lafora bodies and the accumulation of insoluble laforin. J Biol Chem. 2010;285:25372–25381. doi: 10.1074/jbc.M110.148668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turnbull J, Wang P, Girard JM, Ruggieri A, Wang TJ, Draginov AG, Kameka AP, Pencea N, Zhao X, Ackerley CA, et al. Glycogen hyperphosphorylation underlies lafora body formation. Annals of neurology. 2010;68:925–933. doi: 10.1002/ana.22156. [DOI] [PubMed] [Google Scholar]
- 36.Valles-Ortega J, Duran J, Garcia-Rocha M, Bosch C, Saez I, Pujadas L, Serafin A, Canas X, Soriano E, Delgado-Garcia JM, et al. Neurodegeneration and functional impairments associated with glycogen synthase accumulation in a mouse model of Lafora disease. EMBO molecular medicine. 2011;3:667–681. doi: 10.1002/emmm.201100174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Criado O, Aguado C, Gayarre J, Duran-Trio L, Garcia-Cabrero AM, Vernia S, San Millan B, Heredia M, Roma-Mateo C, Mouron S, et al. Lafora bodies and neurological defects in malin-deficient mice correlate with impaired autophagy. Hum Mol Genet. 2012;21:1521–1533. doi: 10.1093/hmg/ddr590. [DOI] [PubMed] [Google Scholar]
- 38.Roma-Mateo C, Solaz-Fuster MC, Gimeno-Alcaniz JV, Dukhande V, Donderis J, Worby CA, Marina A, Criado O, Koller A, Rodriguez de Cordoba S, et al. Laforin, a dual specificity protein phosphatase involved in Lafora disease, is phosphorylated at Ser25 by AMP-activated protein kinase. Biochem J. 2011;439:265–275. doi: 10.1042/BJ20110150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Worby CA, Gentry MS, Dixon JE. Malin decreases glycogen accumulation by promoting the degradation of protein targeting to glycogen (PTG) J Biol Chem. 2008;283:4069–4076. doi: 10.1074/jbc.M708712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Printen JA, Brady MJ, Saltiel AR. PTG, a protein phosphatase 1-binding protein with a role in glycogen metabolism. Science. 1997;275:1475–1478. doi: 10.1126/science.275.5305.1475. [DOI] [PubMed] [Google Scholar]
- 41.Brady MJ, Printen JA, Mastick CC, Saltiel AR. Role of protein targeting to glycogen (PTG) in the regulation of protein phosphatase-1 activity. J Biol Chem. 1997;272:20198–20204. doi: 10.1074/jbc.272.32.20198. [DOI] [PubMed] [Google Scholar]
- 42.Roach PJ, Skurat AV, Harris RA. Regulation of glycogen metabolism. Comprehensive Physiology. 2011:609–647. [Google Scholar]
- 43.Vernia S, Solaz-Fuster MC, Gimeno-Alcaniz JV, Rubio T, Garcia-Haro L, Foretz M, de Cordoba SR, Sanz P. AMP-activated protein kinase phosphorylates R5/PTG, the glycogen targeting subunit of the R5/PTG-protein phosphatase 1 holoenzyme, and accelerates its down-regulation by the laforin-malin complex. J Biol Chem. 2009;284:8247–8255. doi: 10.1074/jbc.M808492200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma J, Rao SN, Shankar SK, Satishchandra P, Jana NR. Lafora disease ubiquitin ligase malin promotes proteasomal degradation of neuronatin and regulates glycogen synthesis. Neurobiology of disease. 2011;44:133–141. doi: 10.1016/j.nbd.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 45.Tagliabracci VS, Girard JM, Segvich D, Meyer C, Turnbull J, Zhao X, Minassian BA, Depaoli-Roach AA, Roach PJ. Abnormal metabolism of glycogen phosphate as a cause for Lafora disease. J Biol Chem. 2008;283:33816–33825. doi: 10.1074/jbc.M807428200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vernia S, Rubio T, Heredia M, Rodriguez de Cordoba S, Sanz P. Increased endoplasmic reticulum stress and decreased proteasomal function in lafora disease models lacking the phosphatase laforin. PloS one. 2009;4:e5907. doi: 10.1371/journal.pone.0005907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rao SN, Maity R, Sharma J, Dey P, Shankar SK, Satishchandra P, Jana NR. Sequestration of chaperones and proteasome into Lafora bodies and proteasomal dysfunction induced by Lafora disease-associated mutations of malin. Hum Mol Genet. 2010;19:4726–4734. doi: 10.1093/hmg/ddq407. [DOI] [PubMed] [Google Scholar]
- 48.Garyali P, Siwach P, Singh PK, Puri R, Mittal S, Sengupta S, Parihar R, Ganesh S. The malin-laforin complex suppresses the cellular toxicity of misfolded proteins by promoting their degradation through the ubiquitin-proteasome system. Hum Mol Genet. 2009;18:688–700. doi: 10.1093/hmg/ddn398. [DOI] [PubMed] [Google Scholar]
- 49.Aguado C, Sarkar S, Korolchuk VI, Criado O, Vernia S, Boya P, Sanz P, de Cordoba SR, Knecht E, Rubinsztein DC. Laforin, the most common protein mutated in Lafora disease, regulates autophagy. Hum Mol Genet. 2010;19:2867–2876. doi: 10.1093/hmg/ddq190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Polajnar M, Zerovnik E. Impaired autophagy: a link between neurodegenerative diseases and progressive myoclonus epilepsies. Trends Mol Med. 2011;17:293–300. doi: 10.1016/j.molmed.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 51.Rao SN, Sharma J, Maity R, Jana NR. Co-chaperone CHIP stabilizes aggregate-prone malin, a ubiquitin ligase mutated in Lafora disease. J Biol Chem. 2010;285:1404–1413. doi: 10.1074/jbc.M109.006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lohi H, Young EJ, Fitzmaurice SN, Rusbridge C, Chan EM, Vervoort M, Turnbull J, Zhao XC, Ianzano L, Paterson AD, et al. Expanded repeat in canine epilepsy. Science. 2005;307:81. doi: 10.1126/science.1102832. [DOI] [PubMed] [Google Scholar]
- 53.Worby CA, Gentry MS, Dixon JE. Laforin, a dual specificity phosphatase that dephosphorylates complex carbohydrates. J Biol Chem. 2006;281:30412–30418. doi: 10.1074/jbc.M606117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tagliabracci VS, Turnbull J, Wang W, Girard JM, Zhao X, Skurat AV, Delgado-Escueta AV, Minassian BA, Depaoli-Roach AA, Roach PJ. Laforin is a glycogen phosphatase, deficiency of which leads to elevated phosphorylation of glycogen in vivo. Proc Natl Acad Sci U S A. 2007;104:19262–19266. doi: 10.1073/pnas.0707952104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tagliabracci VS, Heiss C, Karthik C, Contreras CJ, Glushka J, Ishihara M, Azadi P, Hurley TD, DePaoli-Roach AA, Roach PJ. Phosphate incorporation during glycogen synthesis and Lafora disease. Cell Metab. 2011;13:274–282. doi: 10.1016/j.cmet.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]