Abstract
The intrauterine environment can impact the developing infant by altering the function of the placenta through changes to the epigenetic regulatory features of this tissue. Genetic variation, too, may impact infant development or may modify the relationship between epigenetic alterations and infant outcomes. To examine the association of this variation with early life infant neurodevelopment, we examined the extent of DNA methylation of the glucocorticoid receptor gene (NR3C1) promoter and a common SNP in the promoter region in a series of 186 placentas from healthy newborn infants. We associated these molecular features with specific summary measures from the NICU Network Neurobehavioral Scales. After controlling for genotype and confounders, we identified significant associations of NR3C1 methylation with infant quality of movement (P=0.05) and with infant attention (P=0.05), and a potential interaction between methylation and genotype on infant attention score. These results suggest that epigenetic alteration of the NR3C1 gene in the placentas of genetically susceptible infants can have impacts on neurodevelopment which may have lifelong impact on neurobehavioral and mental health outcomes. Further research is needed to more precisely define these relationships and the interaction between epigenetic alterations and genetic variations on infant health.
Keywords: Epigenetic, glucocorticoid receptor, placenta, neurodevelopment, human, attention, quality of movement
It is now well established that the environment experienced during in-utero development is linked to health and disease throughout life (McMillen & Robinson, 2005). Although initially focused on cardiovascular and metabolic diseases, these relationships have been expanded to include numerous health outcomes, including neurodevelopment and mental health (Champagne, 2010; Roth & Sweatt, 2011; Tremblay, 2010; Van den Bergh, 2011). The mechanisms through which the intrauterine environment elicits these effects are the subject of active investigation by ourselves and others (Robins, Marsit, Padbury, & Sharma, 2011).
Epigenetic regulation constitutes the control of gene expression that does not involve changes in DNA sequence. There are a number of known modes of epigenetic regulation, the most prominent of which are DNA methylation, imprinting, histone modification and miRNA. These epigenetic processes due to their intimate link to development, their relative stability, and their function in gene control have been recognized as candidate mechanisms for the molecular basis of the developmental origins of health and disease (Dolinoy, Das, Weidman, & Jirtle, 2007; Mathers & McKay, 2009).
Neurodevelopment is tightly linked to the intrauterine environment. The placenta serves as the source of fetal nutrients, water, gas exchange, excretion, and immune regulation. These effects are modulated by simultaneous production of many pregnancy related hormones, proteins and growth factors, as well as an array of neuropeptide hormones that are analogous to those produced by the hypothalamus and pituitary of the brain, including GnRH, TRH, GHRH, CRH, and oxytocin (J. H. Liu, 2009). Through action on the HPA axis, stress hormones can affect the regulation of the neuroendocrine environment leading to physiologic and behavioral outcomes that can persist into adulthood (Slone-Wilcoxon & Redei, 2004; Welberg & Seckl, 2001). Thus, the placenta, as a key regulator of the intrauterine environment, may play a critical role in mediating appropriate neurodevelopment. Recent work from our group suggests that the intrauterine environment, represented by infant growth, may elicit these effects through epigenetic modulation of the placenta (Banister et al., 2011; Filiberto et al., 2011). This study aimed to expand these observations to better characterize how epigenetic alteration of the glucocorticoid receptor gene in human placenta, as well as polymorphic variation of this gene, are associated with infant neurodevelopmental outcome. We utilized the sensitive and prospectively validated NICU Network Neurobehavioral Scales (NNNS) to assess newborn neurobehavioral performance including neurological integrity, behavioral functioning, and signs of stress/abstinence (J. Liu et al., 2010).
METHODS
Study Population and Sample Collection
Subjects involved in this study are part of the ongoing Rhode Island Child Health Study (Marsit et al., 2011), which enrolls mother-infant pairs following delivery at Women and Infants Hospital of Rhode Island. Term infants born small for gestational age (SGA, lowest 10th percentile), or large for gestational age (LGA, highest 10th percentile), based on birthweight and gestational age and calculated from the Fenton growth chart (Fenton, 2003) are selected and enrolled along with an appropriate for gestational age (AGA) infant matched on infant gender, gestational age (±3 days), and maternal age (±2 years). Only singleton, viable infants are included in the study. Other exclusion criteria included maternal age of less than 18 years, a life-threatening medical complication in the mother, and congenital or chromosomal abnormality of the infant. A structured chart review was used to collect information from the maternal inpatient medical record from delivery. The mothers then underwent an interviewer-administered structured questionnaire to obtain demographic, lifestyle and exposure histories.
Within 2 hours of delivery, 12 fragments of placenta tissue, 3 from each of 4 quadrants (totaling approximately 1 g of tissue) were excised from each subject. Samples were taken from the maternal side of the placenta 2 cm from the umbilical cord insertion site and were free of maternal decidua. The samples were placed immediately in RNAlater solution (Life Technologies, Grand Island, NY) and stored at 4°C. At least 72 hours later, placenta samples were removed from RNAlater, blotted dry, snap-frozen in liquid nitrogen, and samples from all regions were homogenized into a single sample using a mortar and pestle. The samples are stored in at −80°C until needed for examination. DNA was extracted from homogenized placenta samples using the QIAmp DNA Mini Kit (Qiagen, Inc., Valencia, CA) following manufacturer’s protocols and purified DNA was quantified using a ND-1000 spectrophotometer (Nanodrop, Wilmington, DE).
NICU Network Neurobehavioral Scales Assessment
Items for the NNNS were scored using previously established summary scores (B. Lester & Tronick, 2004). For this examination, we focused on the 4 summary scores (attention, habituation, stress abstinence and quality of movement) shown to be most sensitive to the intrauterine environment in previous studies (J. Liu, et al., 2010). For this analysis, 186 subjects with NNNS data, enrolled between September 2009 and December 2010, were examined. All subjects provided written informed consent for participation under appropriate protocols approved by the Institutional Review Boards for Women and Infants’ Hospital and Brown University.
SNP Genotyping
Allelic discrimination was performed using Taqman® allelic discrimination on a 7300 Real Time PCR System for the single nucleotide polymorphism, rs41423247 (BclI) in the NR3C1 gene (which encodes the glucocorticoid receptor), located upstream of the transcriptional start site.
DNA Bisulfite Modification and Pyrosequencing
We sought to interrogate the 13 CpG sites in the NR3C1 exon 1F promoter region, previously shown to demonstrate DNA methylation variability associated with maternal depression and cortisol response in infant cord blood (Oberlander et al., 2008) and with infant growth status in placenta(Filiberto, et al., 2011). DNA samples (1μg) were bisulfite-modified using the EZ DNA methylation Kit (Zymo Research, CA, USA) following the manufacturer’s protocol. Pyrosequencing was performed on PCR product amplified from bisulfite-modified DNA as described previously (Filiberto, et al., 2011). The primers for amplification were Forward-5′-TTT TTT TTT TGA AGT TTT TTT A-3′ and Reverse 5′-Biotin-CCC CCA ACT CCC CAA AAA-3′. The first sequencing primer was designed to sequence the first five CpG sites (5′-GAG TGG GTT TGG AGT-3′), and the second sequencing primer was designed to sequence the following eight CpG sites (5′-AGA AAA GAA TTG GAG AAA TT-3′)for a total of thirteen sites sequenced. Percent DNA methylation at each CpG site was quantified using the Pyro Q-CpG software, version 1.0.9. (Qiagen). Bisulfite conversion controls were included on each sequencing read. In order for the sample’s methylation extent to be called, the bisulfite conversion rate must be >93%, and for all samples examined the conversion rate was >95%. Approximately 10% of samples were randomly repeated and the correlation coefficient for these repeats was greater than 0.9.
Gene Expression
Total RNA was extracted from an aliquot of the homogenized placenta sample using the RNeasy mini kit (Qiagen) following manufacturer’s protocols and was quantified using a ND-1000 spectrophotometer (Nanodrop, Wilmington, DE). RNA samples were aliquoted and stored at −80°C and samples were thawed only once for expression analysis. Expression of the NR3C1 mRNA was measured using a commercially available Taqman Gene Expression Assay (Applied Biosystems, Valencia, CA, part number: Hs00353740_m1) with GAPDH (part number: Hs00266705_g1) serving as a referent on BioRad CFX Connect Real-Time PCR Detection System (BioRad, Hercules, CA). All reactions were run in triplicate on the same plate, and the sample with the lowest expression served as a reference sample to allow normalization using the ΔΔCt method.
Statistical methods
A generalized linear model (GLM) was used to compare the mean DNA methylation at each of the 13 CpG sites, as well as overall DNA methylation, in the NR3C1 gene to both growth status independently and to four behavioral outcomes on the NNNS. From the NNNS subscales we examined habituation, stress, quality of movement and attention. Three models were used for testing: 1) for an association between methylation at each site and each of the four outcomes; 2) for association between methylation at each site and each of the four outcomes while controlling for variations in genotype (SNP); 3) for association between methylation and NNNS outcomes while controlling for variations in genotype as well as covariates previously identified as relevant, including infant growth status (SGA, AGA, LGA), infant sex, delivery method, maternal age, tobacco use during pregnancy and maternal insurance as a marker of socioeconomic status. Associations between DNA methylation and NNNS scores were considered significant at the p <0.05 level. To test for multiplicative interaction, a cross-term was included in the model and compared to a model without the interaction term using a likelihood ratio test. The correlation between mean NR3C1 methylation and expression was assessed using a Spearman’s rank correlation.
RESULTS
The demographics of the 186 infants that were studied are shown in Table 1. Based on the design of the cohort and our enrollment strategy, the population is over-represented by small and large for gestational age infants (SGA and LGA, respectively). All infants enrolled were at or near term gestation, with a mean gestational age of 39.1 weeks. There was a greater proportion of female infants, and a greater proportion with mothers on public insurance programs; the majority of infants were white. Only 5% of mothers reported tobacco use during pregnancy. Also included in Table 1 are the descriptive statistics of the summary scores for the 4 NNNS measures examined in this study.
Table 1.
Population Demographic and Clinical Characteristics and NNNS Summary Scores
| Variable | N | % |
|---|---|---|
| Growth Status | ||
| SGA | 53 | 28 |
| AGA | 100 | 54 |
| LGA | 33 | 18 |
| Infant Gender | ||
| Female | 101 | 55 |
| Male | 85 | 45 |
| Delivery Method | ||
| Vaginal | 105 | 56 |
| Caesarean Section | 35 | 19 |
| Unknown | 46 | 25 |
| Maternal Ethnicity | ||
| White | 124 | 67 |
| African American | 19 | 10 |
| Other | 39 | 21 |
| Unknown | 4 | 2 |
| Maternal Insurance | ||
| Public | 101 | 54 |
| Private | 85 | 46 |
| Maternal Tobacco Use During Pregnancy | ||
| No | 176 | 95 |
| Yes | 8 | 4 |
| Unknown | 2 | 1 |
| Variable | N | Mean | Std. Dev. | Median | Min | Max |
|---|---|---|---|---|---|---|
| Birth weight (g) | 186 | 3240.8 | 685.1 | 3132.5 | 1705 | 4730.0 |
| Gestational Age (weeks) | 186 | 39.1 | 1.1 | 39.2 | 37.0 | 41.2 |
| Maternal Age (yrs) | 186 | 28.3 | 5.8 | 29.0 | 18.0 | 40.0 |
| APGAR Score 1 min | 183 | 7.7 | 1.2 | 8.0 | 1 | 9 |
| APGAR Score 5 min | 183 | 8.9 | 0.8 | 9.0 | 0 | 10 |
| Habituation | 97 | 7.0 | 1.5 | 7.3 | 1.0 | 9.0 |
| Attention | 160 | 4.1 | 1.2 | 4.1 | 1.6 | 6.2 |
| Stress Abstinence | 186 | 0.21 | 0.07 | 0.20 | 0.06 | 0.41 |
| Quality of Movement | 186 | 4.0 | 0.69 | 4.1 | 2.3 | 5.5 |
Methylation of NR3C1 exon 1F was assessed through quantitative bisulfite pyrosequencing of DNA from placenta samples from each of the infants involved in this study. Descriptive statistics of the distribution of methylation at each of the 13 CpG sites examined and the mean of the region are provided in Supplementary Table 1.
Associations between methylation at each of the individual CpGs and the mean overall with the 4 selected NNNS scores were assessed with linear regression, and Table 2 provides the result of those analyses. A significant positive association between increased methylation at the CpG site 10 and increased infant habituation was identified. In contrast, greater levels of methylation at CpG site 7 are significantly associated with lower stress abstinence scores. Significant positive associations were also observed between infant quality of movement scores and methylation at CpG sites 6, 7, and 8, with increases of nearly a standard deviation in quality of movement associated with a 10% increase in methylation extent at these CpG sites. Although there were no statistically significant associations between methylation at any NR3C1 sites and infant attention scores, all sites showed a negative trend and there were borderline significant associations for CpG site 5 and overall for the region (Table 2). This indicates that increased methylation of the G NR3C1 promoter was associated with reduced infant attention.
Table 2.
Linear regression analysis examining the association of selected NNNS Summary Scores as the dependent variable and NR3C1 methylation at individual CpG positions as the independent variable.
| Habituation (n=97) | Stress (n=185) | Quality of Movement (n=185) | Attention (n=159) | |||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| NR3C1 CpG Position | Estimate (per 10%) | P | Estimate (per 10%) | P | Estimate (per 10%) | P | Estimate (per 10%) | P |
|
| ||||||||
| Position 1 | 0.01 | 1.0 | 0.02 | 0.4 | 0.1 | 0.6 | −0.4 | 0.2 |
| Position 2 | 0.1 | 0.8 | −0.01 | 0.5 | 0.3 | 0.1 | −0.3 | 0.5 |
| Position 3 | 1.2 | 0.1 | −0.03 | 0.1 | 0.1 | 0.3 | −0.06 | 0.8 |
| Position 4 | −0.4 | 0.6 | 0.01 | 0.6 | 0.1 | 0.7 | −0.07 | 0.9 |
| Position 5 | 0.4 | 0.4 | 0.02 | 0.2 | −0.01 | 1.0 | −0.6 | 0.06 |
| Position 6 | 0.3 | 0.7 | −0.01 | 0.5 | 0.6 | 0.01 | −0.1 | 0.7 |
| Position 7 | 0.9 | 0.1 | −0.04 | 0.04 | 0.6 | 0.01 | −0.4 | 0.3 |
| Position 8 | 0.7 | 0.2 | 0.01 | 0.3 | 0.4 | 0.02 | −0.3 | 0.4 |
| Position 9 | 0.2 | 0.6 | −0.002 | 0.9 | 0.1 | 0.4 | −0.2 | 0.3 |
| Position 10 | 0.9 | 0.03 | 0.002 | 0.9 | −0.01 | 0.9 | −0.3 | 0.2 |
| Position 11 | 0.4 | 0.1 | −0.003 | 0.8 | −0.06 | 0.6 | −0.2 | 0.3 |
| Position 12 | 0.7 | 0.08 | −0.005 | 0.6 | −0.02 | 0.9 | −0.2 | 0.3 |
| Position 13 | 0.5 | 0.08 | −0.02 | 0.1 | 0.002 | 1.0 | −0.2 | 0.4 |
| Mean across all positions | 1.4 | 0.07 | −0.02 | 0.5 | 0.4 | 0.2 | −0.9 | 0.1 |
Note: Significant Correlations are indicated using italics.
To examine if infant genotype at a common polymorphic region in the NR3C1 promoter (rs41423247) might mediate the association between methylation and NNNS outcome, we performed allelic discrimination assays to determine the infant genotype at this SNP. Overall, 44% of infants were homozygous for the predominant allele, GG, at this SNP, 45% were heterozygous, and 11% were homozygous for the minor variant, CC. Genotype frequencies were in Hardy-Weinberg equilibrium. No significant association was detected between the extent of methylation at any of the exon 1F CpGs examined and genotype.
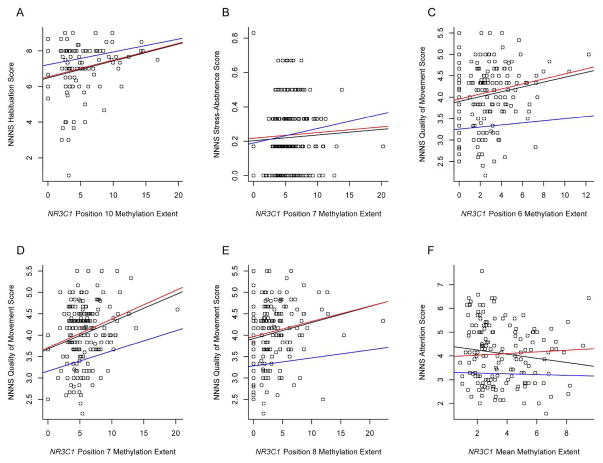
The independent associations between NR3C1 methylation and genotype were examined utilizing generalized linear models for each of the individual CpG methylation marks and NNNS scores controlled for genotype (Table 3, Genotype Adjusted Model). We further controlled for additional confounders in the fully adjusted models (Table 3, Fully Adjusted Model). The positive association between CpG site 10, methylation and habituation remained significant when genotype was controlled and there was little change to estimate of the effect between the uncontrolled and genotype-controlled models, suggesting no confounding of this relationship by genotype (Figure 1). When covariates were included, the significance of this association was lost and the estimate of the slope attenuated (Table 3 and Figure 1). Similarly the significant negative association between stress abstinence and methylation at CpG site 7 was retained in the genotype-controlled model and was no longer significant when additional confounders were included.
Table 3.
Multivariable linear regression models examining the association between NR3C1 exon 1F methylation at individual sites as independent variables and selected NNNS summary scores as the dependent variables controlled for NR3C1 genotype (Genotype-adjusted model) and for NR3C1 genotype and additional covariates (Fully Adjusted Model.
| Genotype Adjusted | Fully Adjusted | |||
|---|---|---|---|---|
|
| ||||
| Estimate (slope) | P | Estimate (slope) | P | |
|
| ||||
| Habituation | ||||
| NR3C1 Position 10, per 10% | 0.92 | 0.05 | 0.69 | 0.3 |
| Genotype | ||||
| Wt | Referent | Referent | ||
| Heterozygote | −0.01 | 1.0 | 0.17 | 0.7 |
| Variant | −0.65 | 0.2 | −0.45 | 0.5 |
| Stress | ||||
| NR3C1 Position 7, per 10% | −0.04 | 0.03 | −0.02 | 0.4 |
| Genotype | ||||
| Wt | Referent | Referent | ||
| Heterozygote | 0.02 | 0.1 | 0.03 | 0.05 |
| Variant | −0.002 | 0.9 | 0.015 | 0.5 |
| Quality of Movement | ||||
| NR3C1 Position 6, per 10% | 0.59 | 0.01 | 0.25 | 0.4 |
| Genotype | ||||
| Wt | Referent | Referent | ||
| Heterozygote | −0.16 | 0.1 | −0.20 | 0.1 |
| Variant | 0.05 | 0.8 | 0.002 | 1.0 |
| NR3C1 Position 7, per 10% | 0.67 | 0.0005 | 0.47 | 0.05 |
| Genotype | ||||
| Wt | Referent | Referent | ||
| Heterozygote | −0.16 | 0.1 | −0.21 | 0.1 |
| Variant | 0.11 | 0.5 | 0.03 | 0.9 |
| NR3C1 Position 8, per 10% | 0.36 | 0.02 | 0.21 | 0.3 |
| Genotype | ||||
| Wt | Referent | Referent | ||
| Heterozygote | −0.13 | 0.2 | −0.18 | 0.2 |
| Variant | 0.09 | 0.6 | 0.02 | 0.9 |
| Attention | ||||
| NR3C1 Mean, per 10% | −0.7 | 0.2 | −1.2 | 0.05 |
| Genotype | ||||
| Wt | Referent | Referent | ||
| Heterozygote | −0.22 | 0.3 | −0.01 | 1.0 |
| Variant | 0.52 | 0.1 | 0.60 | 0.09 |
Note: Significant Correlations are indicated using italics.
The genotype adjusted model includes only methylation extent at the individual CpG site and genotype. The fully adjusted model includes m methylation extent at the individual CpG site, genotype, infant growth status (small for gestational age, appropriate for gestional age, large for gestational age), infant sex, delivery method, maternal age, tobacco use during pregnancy and maternal insurance as a markers of socioeconomic status.
Figure 1.
Scatterplots of the correlations between the extent of methylation of specific CpG positions or the mean (x-axis) and their association with NNNS summary scores. Lines on each plot represent the fits of the linear models used to examine the association where black line=bivariate association between methylation and NNNS score, red line=association between methylation and NNNS score adjusted for NR3C1 genotype, and blue line= association between methylation and NNNS score adjusted for NR3C1 genotype, infant growth status (small for gestational age, appropriate for gestional age, large for gestational age), infant sex, delivery method, maternal age, tobacco use during pregnancy and maternal insurance as a markers of socioeconomic status. (A) NR3C1 position 6 methylation and NNNS habituation summary score, (B) NR3C1 position 7 methylation and NNNS stress-abstinence summary score, (C) NR3C1 position 6 methylation and NNNS quality of movement summary score, (D) NR3C1 position 7 methylation and NNNS quality of movement summary score, (E) NR3C1 position 8 methylation and NNNS quality of movement summary score, (F) NR3C1 mean methylation across all sites and NNNS quality of movement summary score.
The positive association between quality of movement scores and methylation at CpG sites 6, 7, and 8 each also remained significant in the models controlling for genotype. When controlling for additional confounders, only the association between quality of movement and methylation at CpG site 7, was significant, suggesting both independent and correlated effects of methylation at this site and genotype (Table 3 and Figure 1). Higher overall methylation across NR3C1 exon 1F was significantly associated with lower attention scores, in the fully adjusted model, while there was also a borderline association between variant genotype and increased attention scores.
To determine if there was a modification of the association between NR3C1 methylation and attention scores by genotype, we modeled the multiplicative interaction of these independent effects. Homozygous wildtype and heterozygous individuals demonstrated a reduction in attention score of 0.7 for each 10% increase in mean methylation of the NR3C1 exon 1F, while homozygous variant individuals demonstrated a decrease in attention of only 0.1 for every 10% increase in GR mean methylation (Table 4). The likelihood ratio test for interaction suggested a trend for a multiplicative interaction (p=0.06).
Table 4.
Effect modification of association between GR methylation and NNNS Attention Summary Score by GR genotype.
| Genotype Category | NR3C1 Mean Methylation Estimate (Slope) | P |
|---|---|---|
| WT/WT & WT/Var | −0.74 | 0.11 |
| Var/Var | −0.1 | 0.11 |
Note: Likelihood ratio test for interaction, Pint=0.06
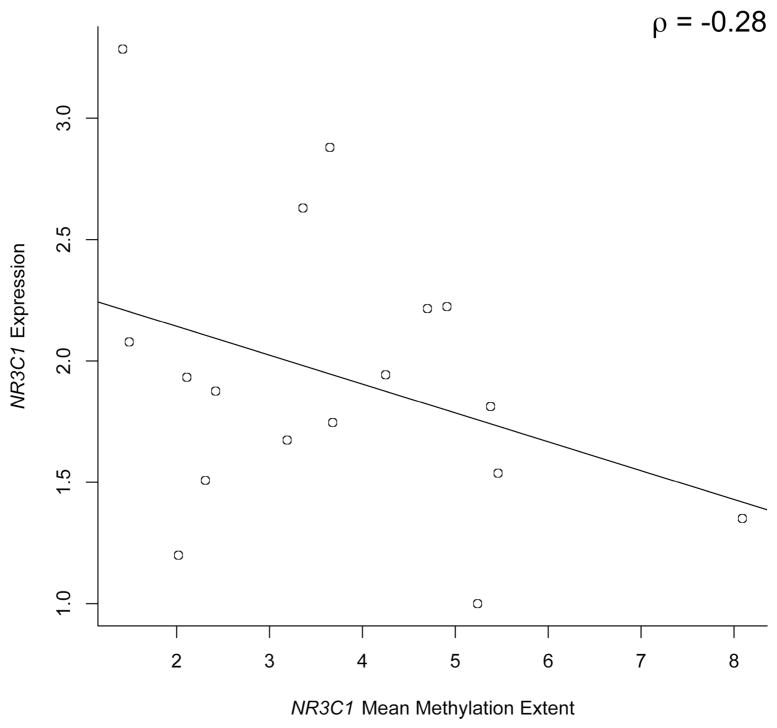
We have previously demonstrated, in vitro, that methylation of the exon 1F region was associated with expression in a choricarcinoma cell line (Filiberto, et al., 2011), but to examine if methylation of this region was correlated to gene expression in primary placenta samples, we examined the mRNA expression of NR3C1 in a subset of 17 placenta tissue samples with high quality available RNA and whose methylation spanned a range of values. We observed a negative correlation (Spearman’s ρ=−0.28), with samples demonstrating a greater mean methylation extent exhibiting reduced expression of NR3C1 mRNA (Figure 2).
Figure 2.
Relationship between NR3C1 gene expression and NR3C1 mean methylation across all sites in human placenta samples (n=17). The result of a Spearman correlation analysis (ρ) is depicted on the figure.
DISCUSSION
There is increasing evidence that epigenetic alterations and changes in gene expression in the placenta are associated with changes in later development, including infant neurodevelopment (B. M. Lester & Padbury, 2009; Marsit, et al., 2011). Stress factors during pregnancy, resulting from various adverse intrauterine environments, are associated with reduced placental 11beta-hydroxysteroid dehydrogenase type 2 (11β-HSD2) and norepinephrine transporter (NET) gene expression potentially through epigenetic alterations, and in turn, result in increased circulating catecholamine levels in the fetus and placental microenvironment, increased fetal exposure to cortisol, and dysregulation of the infant’s HPA axis and neurobehavior (B. M. Lester & Padbury, 2009; Sarkar et al., 2001). Factors as subtle as maternal mood have been shown to impact the placental expression of key neuroactive genes such as the serotonin transporter (Ponder et al., 2011). Research in animal models and humans has suggested that behavior and the environment can impact the epigenetic control of another critical gene in the HPA axis, the glucocorticoid receptor, at exon 1F, and that variation in the pattern of DNA methylation in this region is associated with functional alteration of NR3C1 expression resulting in later life behavior (Oberlander, et al., 2008; Seckl & Meaney, 2006; Weaver et al., 2004). Our previous work suggested that the environment, captured by measures of in-utero growth, is associated with alteration of methylation of this gene region in the placenta and that altered methylation can affect expression of the glucocorticoid receptor in placental cells (Filiberto, et al., 2011). This study specifically examined if epigenetic variation in the placenta at the NR3C1 exon 1F as well as genetic variation in the promoter region of NR3C1 was associated with neurobehavioral measures assessed at birth, using the NICU Network Neurobehavioral Scales. As this study was modest in sample size, we focused specifically on 4 measures known to be particularly predictive of later neurobehavioral outcome (J. Liu, et al., 2010) and which we have previously seen to be associated with variation in other epigenetic regulatory mechanisms in the placenta (Marsit, et al., 2011). We demonstrated that there may be a relationship between epigenetic variation in the placental glucocorticoid receptor promoter and infant neurodevelopmental outcome. Our results further suggest that a polymorphism in the NR3C1 gene may modify these associations.
We observed a significant positive association between methylation at CpG site 10 and infant habituation scores, which remained significant when genotype was included in the model. As habituation scores reflect the infant’s ability to adapt to their environment such as blocking out disruptive noises or lights during sleep, this could indicate a relationship between methylation and these capacities. Controlling for additional confounders, including birth weight, gender, ethnicity, and maternal factors decreased the effect estimate, and reduced significance, suggesting that some of these factors may be confounders of this association. As the stress/abstinence scores examine coping mechanisms critical in habituation, it is not surprising that these findings are consistent with the association observed between methylation at CpG 7 and lower stress abstinence scores. Again, this association between site 7 methylation and stress/abstinence was no longer evident when covariates were added to the model, suggesting that demographic characteristics or environmental factors may confound this association.
Infants with higher quality of movement scores on the NNNS were more likely to have higher methylation at CpG sites, 6, 7 and 8, indicating that methylation at these sites is protective (i.e. improves infant’s movement and reflexes). The fact that all three sites demonstrated a significant association of similar magnitude suggests that they may act in concert to effect glucocorticoid receptor expression and function. A TFBLAST search of the Transfac transcription factor binding site database indicates an SP1 binding site starting between position 7 and 8, and suggests a potential mechanism through which methylation may be effecting transcription factor binding at this region, similar to the effect that has been demonstrated with the NGF1-A binding site encompassing positions 3 and 4 (McGowan et al., 2009). Additional transcription factors with binding sites in this region include SRY, E2F, ISRE, MZF1, and p300, suggesting that the effects of specific site methylation may be by altering the binding of these transcription factors.
This association, specifically with methylation at CpG 7 and quality of movement scores remains significant in the fully adjusted model, and further indicates an important role for methylation at the regions and NR3C1 control leading to downstream motor control. Some studies have suggested that increased cortisol exposure during particular periods of intrauterine development are correlated with reduced neuromuscular maturity (Ellman et al., 2008). Our observed increased glucocorticoid receptor methylation may be a compensatory mechanism in order to protect the fetus from potential glucorticoid overload and thereby allow appropriate neuromuscular and/or neurobehavioral development, as we observe associated with increased methylation. Our previous work examining promoter methylation of placental HSD11B2, the enzyme responsible for converting active cortisol to cortisone, demonstrated an inverse association between methylation and infant quality of movement (Marsit, Maccani, Padbury, & Lester, 2012), further suggestive that the NR3C1 methylation we are observing may be compensating for increased cortisol load by reducing binding and thus activation opportunities.
Contrary to the associations with habituation, stress/abstinence, and quality of movement, the association between methylation of the NR3C1 and attention scores on the NNNS is inverse; higher methylation was associated with poorer outcomes on the NNNS attention scores. In fact, adjustment for confounders improved the significance of the association both overall and for specific sites, suggesting that there may be a complicated interplay between genetics, environment, and epigenetic regulation on this neurobehavioral outcome. This negative association may also be consistent with the other observed associations in this study. For example, an infant who is better able to block out or ignore disruptions (e.g exhibits greater habituation) may be less likely to pay attention to environmental cues. Thus, protective methylation marks at individual sites may accumulate, resulting in poorer performance in other areas of the exam. Conversely, an infant who scores well on attention may lack some of the protective methylation marks associated with better habituation scores. Genotype also appeared to modify the association between methylation of NR3C1 exon 1F and attention scores, with homozygous variant infants at the SNP in the NR3C1 promoter less susceptible to the effect of methylation on attention than the homozygous wildtype and heterozygous infants. This study was not powered for the examination of interaction, yet we still observed a borderline statistically significant interaction between methylation and NR3C1 genotype. It is unlikely that this genotype affects the levels of methylation at this region, as we did not observe any association between genotype and extent of methylation (data not shown). Instead, it may affect the impact of methylation on GR expression. Genetic variation at this SNP has been proposed as a modifier of HPA activity (Derijk, 2009), although has not been directly associated with cortisol response (Bouma et al., 2011). Our results suggest that DNA methylation status combined with genotype may be more predictive of phenotype at this gene. Future studies with greater sample sizes are needed to more precisely quantify this interaction.
Using a subset of the sample population, we also examined the relationship between methylation of this region and NR3C1 expression in primary placenta samples, observing an inverse association. We note expected greater variability in expression amongst placentas with the lowest levels of methylation, and the absence of methylation alone does not necessarily signify expression, but signifies an opportunity for expression, assuming the presence of the appropriate signals to activate the gene. As such, expression is much more indicative of the environment experienced immediately prior to collection, in this case, delivery of the infant. Methylation, on the other hand, as a more stable epigenetic mark is likely to be less influenced by near-term events, and likely represents a more long term indicator of the intrauterine environment (Hanson & Gluckman, 2008; Maccani & Marsit, 2009).
This study is one of the first to link epigenetic alteration of the glucocorticoid receptor in the placenta and genetic variation of the NR3C1 promoter to infant neurobehavioral development. These results highlight the critical importance of placental function on infant development and underscore the hypothesis that the maternal environment can influence lifetime health of the infant through epigenetic modification of the placenta. It is also important to highlight that epigenetic processes are part of normal development and that many of the associations noted here might shed light onto normal as well as aberrant developmental pathways. There are limitations to this study, as, like all population-based studies, we cannot definitively define the mechanisms which link the intrauterine environment to epigenetic alterations and then specifically to these outcomes. The inclusion of additional intermediate biomarkers reflective of function, such as cortisol measures, could lend support to the associations observed. This study has also focused on only one gene involved in the complex development of the HPA axis. More research using larger populations and with more comprehensive characterizations of the placental epigenome and infant genome should be conducted to confirm and further explore the findings presented here, as well as to better identify what, if any, particular environmental factors during pregnancy may be influencing the molecular patterns examined.
Acknowledgments
Thanks to Gilda Ferro and Joyce Lee for their hard work in recruitment of subjects into this study and the support of the staff of the Brown Center for the Study of Children at Risk for their efforts. This work was supported by NIH grants from the NCRR (P20 RR018728) and the NIMH (R01 MH094609).
Footnotes
The authors have no conflicts of interest to declare.
References
- Banister CE, Koestler DC, Maccani MA, Padbury JF, Houseman EA, Marsit CJ. Infant growth restriction is associated with distinct patterns of DNA methylation in human placentas. [Research Support, N.I.H., Extramural] Epigenetics : official journal of the DNA Methylation Society. 2011;6(7):920–927. doi: 10.4161/epi.6.7.16079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouma EM, Riese H, Nolte IM, Oosterom E, Verhulst FC, Ormel J, Oldehinkel AJ. No associations between single nucleotide polymorphisms in corticoid receptor genes and heart rate and cortisol responses to a standardized social stress test in adolescents: the TRAILS study. [Research Support, Non-U.S. Gov’t] Behavior genetics. 2011;41(2):253–261. doi: 10.1007/s10519-010-9385-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne FA. Epigenetic perspectives on development: Evolving insights on the origins of variation. Developmental psychobiology. 2010;52(4):e1–e3. doi: 10.1002/dev.20443. [DOI] [PubMed] [Google Scholar]
- Derijk RH. Single nucleotide polymorphisms related to HPA axis reactivity. [Research Support, Non-U.S. Gov’t Review] Neuroimmunomodulation. 2009;16(5):340–352. doi: 10.1159/000216192. [DOI] [PubMed] [Google Scholar]
- Dolinoy DC, Das R, Weidman JR, Jirtle RL. Metastable epialleles, imprinting, and the fetal origins of adult diseases. [Research Support, N.I.H., Extramural Research Support, U.S. Gov’t, Non-P.H.S. Review] Pediatric research. 2007;61(5 Pt 2):30R–37R. doi: 10.1203/pdr.0b013e31804575f7. [DOI] [PubMed] [Google Scholar]
- Ellman LM, Schetter CD, Hobel CJ, Chicz-Demet A, Glynn LM, Sandman CA. Timing of fetal exposure to stress hormones: effects on newborn physical and neuromuscular maturation. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] Developmental psychobiology. 2008;50(3):232–241. doi: 10.1002/dev.20293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton TR. A new growth chart for preterm babies: Babson and Benda’s chart updated with recent data and a new format. BMC pediatrics. 2003;3:13. doi: 10.1186/1471-2431-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiberto AC, Maccani MA, Koestler D, Wilhelm-Benartzi C, Avissar-Whiting M, Banister CE, Marsit CJ. Birthweight is associated with DNA promoter methylation of the glucocorticoid receptor in human placenta. [Research Support, N.I.H., Extramural] Epigenetics : official journal of the DNA Methylation Society. 2011;6(5):566–572. doi: 10.4161/epi.6.5.15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson MA, Gluckman PD. Developmental origins of health and disease: new insights. [Research Support, Non-U.S. Gov’t Review] Basic & clinical pharmacology & toxicology. 2008;102(2):90–93. doi: 10.1111/j.1742-7843.2007.00186.x. [DOI] [PubMed] [Google Scholar]
- Lester B, Tronick E. The Neonatal Intensive Care Unit Network Neurobehavioral Scale. Pediatrics. 2004;113(Suppl 3 Pt 2):631–695. [PubMed] [Google Scholar]
- Lester BM, Padbury JF. Third pathophysiology of prenatal cocaine exposure. Dev Neurosci. 2009;31(1–2):23–35. doi: 10.1159/000207491. 000207491 [pii] [DOI] [PubMed] [Google Scholar]
- Liu J, Bann C, Lester B, Tronick E, Das A, Lagasse L, Bada H. Neonatal neurobehavior predicts medical and behavioral outcome. Pediatrics. 2010;125(1):e90–98. doi: 10.1542/peds.2009-0204. peds.2009–0204 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JH. Endocrinology of Pregnancy. In: Resnik Ca., editor. Maternal-Fetal Medicine, Principles and Practice. 6. Philadelphia: Saunders Elsevier; 2009. pp. 111–124. [Google Scholar]
- Maccani MA, Marsit CJ. Epigenetics in the placenta. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Review] American journal of reproductive immunology. 2009;62(2):78–89. doi: 10.1111/j.1600-0897.2009.00716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsit CJ, Lambertini L, Maccani MA, Koestler DC, Houseman EA, Padbury JF, Chen J. Placenta-Imprinted Gene Expression Association of Infant Neurobehavior. The Journal of pediatrics. 2011 doi: 10.1016/j.jpeds.2011.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsit CJ, Maccani MA, Padbury JF, Lester BM. Placental 11-Beta hydroxysteroid dehydrogenase methylation is associated with newborn growth and a measure of neurobehavioral outcome. PLoS One. 2012;7(3):e33794. doi: 10.1371/journal.pone.0033794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathers JC, McKay JA. Epigenetics - potential contribution to fetal programming. [Research Support, Non-U.S. Gov’t Review] Advances in experimental medicine and biology. 2009;646:119–123. doi: 10.1007/978-1-4020-9173-5_13. [DOI] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonte B, Szyf M, Meaney MJ. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. [Comparative Study Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] Nature neuroscience. 2009;12(3):342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillen IC, Robinson JS. Developmental origins of the metabolic syndrome: prediction, plasticity, and programming. Physiol Rev. 2005;85(2):571–633. doi: 10.1152/physrev.00053.2003. 85/2/571 [pii] [DOI] [PubMed] [Google Scholar]
- Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3(2):97–106. doi: 10.4161/epi.3.2.6034. 6034 [pii] [DOI] [PubMed] [Google Scholar]
- Ponder KL, Salisbury A, McGonnigal B, Laliberte A, Lester B, Padbury JF. Maternal depression and anxiety are associated with altered gene expression in the human placenta without modification by antidepressant use: Implications for fetal programming. Developmental psychobiology. 2011;53(7):711–723. doi: 10.1002/dev.20549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins JC, Marsit CJ, Padbury JF, Sharma SS. Endocrine disruptors, environmental oxygen, epigenetics and pregnancy. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Review] Frontiers in bioscience. 2011;3:690–700. doi: 10.2741/e279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Sweatt JD. Annual Research Review: Epigenetic mechanisms and environmental shaping of the brain during sensitive periods of development. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Review] Journal of child psychology and psychiatry, and allied disciplines. 2011;52(4):398–408. doi: 10.1111/j.1469-7610.2010.02282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Tsai SW, Nguyen TT, Plevyak M, Padbury JF, Rubin LP. Inhibition of placental 11beta-hydroxysteroid dehydrogenase type 2 by catecholamines via alpha-adrenergic signaling. Am J Physiol Regul Integr Comp Physiol. 2001;281(6):R1966–1974. doi: 10.1152/ajpregu.2001.281.6.R1966. [DOI] [PubMed] [Google Scholar]
- Seckl JR, Meaney MJ. Glucocorticoid “programming” and PTSD risk. Ann N Y Acad Sci. 2006;1071:351–378. doi: 10.1196/annals.1364.027. [DOI] [PubMed] [Google Scholar]
- Slone-Wilcoxon J, Redei EE. Maternal-fetal glucocorticoid milieu programs hypothalamic-pituitary-thyroid function of adult offspring. Endocrinology. 2004;145(9):4068–4072. doi: 10.1210/en.2004-0473. en.2004-0473 [pii] [DOI] [PubMed] [Google Scholar]
- Tremblay RE. Developmental origins of disruptive behaviour problems: the ‘original sin’ hypothesis, epigenetics and their consequences for prevention. Journal of child psychology and psychiatry, and allied disciplines. 2010;51(4):341–367. doi: 10.1111/j.1469-7610.2010.02211.x. [DOI] [PubMed] [Google Scholar]
- Van den Bergh BR. Developmental programming of early brain and behaviour development and mental health: a conceptual framework. Developmental medicine and child neurology. 2011;53(Suppl 4):19–23. doi: 10.1111/j.1469-8749.2011.04057.x. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7(8):847–854. doi: 10.1038/nn1276. nn1276 [pii] [DOI] [PubMed] [Google Scholar]
- Welberg LA, Seckl JR. Prenatal stress, glucocorticoids and the programming of the brain. J Neuroendocrinol. 2001;13(2):113–128. doi: 10.1046/j.1365-2826.2001.00601.x. jne601 [pii] [DOI] [PubMed] [Google Scholar]