Abstract
Next-generation sequencing technologies have expanded the experimental possibilities for studying the genome-wide regulation of transcription by nuclear receptors, their collaborating transcription factors, and coregulators. These technologies allow investigators to obtain abundance and DNA sequence information in a single experiment. In this review, we highlight proven and potential uses of next-generation sequencing in the study of gene regulation by nuclear receptors. We also provide suggestions on how to effectively leverage this technology in a collaborative environment.
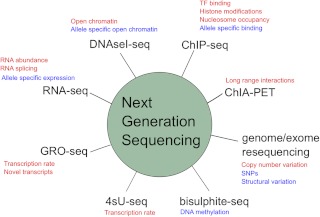
Next-generation sequencing (NGS) technologies, which yield large numbers (20–200 million) of short (25–100 bp) DNA sequences, create a broad variety of research possibilities for experimental and computational biologists. This flexibility arises through a combination of the two complementary aspects of NGS data: sequence characterization and sequence read counting. Figure 1 summarizes the NGS-based experiments that capitalize on these two aspects. Here we review the applications of NGS technologies to the study of transcriptional and epigenetic gene regulation by nuclear receptors. This review does not cover every application of NGS, but focuses on those with demonstrated applications in this field or, in our view, those that hold the greatest potential.
Fig. 1.
Next-generation sequencing applications in studies of transcriptional regulation. Applications in red make use mainly of the quantification of abundance, whereas applications in blue make use of sequence-based observations.
Gene Expression Profiling
The flexibility derived from mRNA sequencing (RNA-seq) leads to experimental considerations previously not encountered in microarray studies. RNA-seq allows an investigator to obtain higher accuracy through deeper sequencing, although at a higher cost. A tradeoff therefore arises: whether to sequence many samples at a low coverage or to sequence fewer samples at a greater depth. Interestingly, this choice has a strong influence on the results, because longer or more abundant transcripts are more likely to be detected as differentially expressed, especially at low sequencing depth (1, 2). The representation of short genes expressed at lower levels will increase with sequencing depth. Although this bias is evident in RNA-seq data, systematic biases also occur in DNA-microarray technologies. Discrepancies in differentially expressed gene sets found in RNA-seq and microarray experiments (3) are therefore common. Differences in systematic biases or batch effects in microarray and NGS place microarray studies at an advantage in terms of leveraging the abundance of microarray gene expression profiles and data analysis methods that have became available over the last decade.
NGS is superior (4, 5) for experiments examining the sequence aspect of transcription, such as identifying splicing events and allelic differences. Paired-end sequencing, in which both ends of each RNA fragment are sequenced, is particularly effective for discovering alternatively spliced mRNA isoforms. In this way, even though only short stretches of cDNA are sequenced, it is possible to map pairs of fragments over a longer span. In cancer gene expression studies, this approach allows for the detection of fusion genes that may have special relevance to disease (6). For example, Chinnaiyan's group (5) used paired-end RNA-seq to detect BCR-ABL1 and TMPRSS2-ERG fusion genes as well as several novel ETS gene fusion events in prostate tumors. Allele-specific expression analysis using NGS raises the prospect of integrating population-based discoveries with laboratory-based experiments (8), linking the association of genetic variants with disease to the molecular biology basis of disease.
The choice of NGS protocol used for mRNA analysis can be crucial to the success of an experiment. The RNA-seq protocol that is most similar to expression microarray experiments is polyA-select, strand-specific, single-end sequencing (9). An NGS-centered experimental design optimized for one aspect of biology will probably not be optimal for others. Designing an experiment to understand one key biological question is likely to be more successful than generating data with multiple objectives in mind. For example, NGS using paired-end sequencing effectively identifies splicing events (10), allowing for a survey of isoforms but usually not a quantitative comparison of isoform abundance between conditions. Splicing information comes at a cost because paired-end sequencing is more expensive per read than single-end sequencing. If the objective is to identify differentially expressed genes and to use the abundance information on these genes to understand a biological process, the better choice is single-end sequencing. Compatibility of newly generated data with archival data, which permits better data reuse, is another consideration. In this respect, RNA-seq is not different from platform considerations in expression microarrays.
Instead of sequencing mRNA from the whole transcript, it is possible to reduce the complexity of the sequence population by targeting sequence near restriction enzyme recognition sites. This is the principle of Serial Analysis of Gene Expression (SAGE)-seq. SAGE-seq offers several advantages over RNA-seq: SAGE-seq does not suffer from the gene length bias and at the same sequencing depth quantifies rare transcripts more accurately (11). In addition, SAGE-seq can be carried out with smaller sample aliquots. On the other hand, SAGE-seq contains less information than RNA-seq techniques in terms of detection of allele-specific transcription and splice variants.
Global Run-On (GRO)-seq, a high-throughput adaptation of the nuclear run-on experiment, is an assay that measures the orientation and density of transcriptionally competent RNA polymerase, serving as a proxy for the rate of RNA transcription rather than mRNA concentration (12, 13). This technique is important in dissecting different aspects of the RNA life cycle (14), in genome-wide studies of transcriptional regulation (12), and in the development of quantitative mathematical models linking transcription factor (TF)-binding events and transcription. A time course of GRO-seq in estrogen-stimulated breast cancer cells, when viewed together with estrogen receptor (ER) chromatin immunoprecipitation (ChIP)-seq data, confirmed several features of ERα transcription regulation that had been discovered by microarray measurements of mRNA concentration (15). Interestingly, analysis of binding and transcription indicated that ER binding was more strongly associated with GRO-seq than with microarray measurements. Moreover, this time course revealed rapid and heterogeneous dynamics of hormone-induced RNA transcription that could not be detected through measurements of mRNA concentration. GRO-seq has also been used to detect enhancer RNA, nascent noncoding RNA at putative enhancer regions (16). These enhancer RNA may serve as good markers of enhancers that are interacting with promoters to induce RNA transcription.
A drawback of the GRO-seq technique is that the nuclear run-on assay is carried out ex vivo. An alternative approach to measuring the rate of RNA production is to use in vivo mechanisms to label the nucleotides that have been incorporated into newly transcribed RNA. In the 4-thiouridine-seq technique, 4-thiouridine is used as the label. Total RNA is separated through thiol-specific biotinylation into previously accumulated RNA and newly transcribed RNA that is assayed using RNA-seq (17). In a study of the response of immune dendritic cells to pathogens, 4-thiouridine RNA was compared with total RNA (18) to impute transcription rate and degradation rate, and the former was found to be the main determinant of total RNA levels.
Some applications of NGS were carried out exclusively using microarray technologies in the past. Although NGS approaches do offer advantages to microarrays, depending on the objectives of the study, microarrays may, in some cases, still be the better technology choice. When studying gene expression changes in annotated genes, the standardization in manufacturing, extensive experience with protocols in core laboratories, and well-established computational approaches make microarrays a good choice. Furthermore, large databases of gene expression microarray experiments can be exploited in combination with emerging genomic data sets to help interpret new biological data.
NGS in the form of RNA-seq has several advantages over microarrays in its greater coverage of transcripts, treatment of alternative splicing, and its potential for greater dynamic range and accuracy. RNA-seq is sometimes claimed to be a superior platform due to its digital nature; the result of NGS can be counts of short sequence reads that map to annotated transcripts. However, there is no intrinsic advantage to having discrete numbers rather than continuous ones. Most importantly, NGS technologies do not eliminate biology-related sources of variance that are usually greater than the technical sources in both microarray and NGS. Consequently, many of the experimental considerations that are important in microarray studies also apply to NGS, in particular the need for biological replicates, suitable controls, and measures to reduce batch effects and enable the measurement of such effects (19).
Transcription Factor Cistrome Mapping
Gene expression profiling provides invaluable information about which genes are associated with conditions, disease outcome, or phenotype. However, the key to understanding how mRNA levels are controlled lies in the elucidation of transcription regulation by transcription factors (TFs). A large number of drugs, including contraceptives, antiinflammatories, and cancer treatments, modulate transcription by targeting nuclear receptors. Understanding transcriptional regulation would contribute to our understanding of which patients will respond to drug therapies, the optimization of these therapies, and avenues for treatment by drug combinations.
ChIP-seq is a broadly used technology that can be effective in profiling genome-wide TF-binding sites, provided a good antibody and sufficient biological material are available. A fundamental question that arises in the analysis of any ChIP-seq dataset is why binding sites are found in some regions of the genome but not others. Although ChIP-seq experiments have shown that TFs bind in a highly tissue- and condition-specific manner (20–23), they have also shown that within a cell type, multiple TFs often share common binding loci. Cooperation between TFs are likely to be a central cause of this phenomenon. One TF may facilitate the binding of another through a variety of mechanisms, including the following: 1) heterodimerization of factors that bind DNA together through heterodimer DNA sequence recognition (24, 25); 2) physical interaction between the primary bound factor and the second factor, where only the primary factor is bound to the DNA (26, 27); 3) stabilization of the nucleosome position, exposing nucleotides for binding by the second factor (28); 4) cooperative effects in which several TFs, binding close to each other but not physically interacting, compete with a nucleosome for occupancy of a genomic locus (29, 30); 5) recruitment of chromatin remodelers that reorganize nucleosome positioning or composition in a way that allows the second factor to bind to DNA (31); and 6) recruitment of factors that modify the tails of histones in a way that facilitates binding at the locus (32).
Recent studies suggest that TF binding is a complex dynamic process where many of the above processes play essential roles (33–35). Although, taken in isolation, TF DNA-binding motifs have little power to predict in vivo binding sites, motif analysis of ChIP-seq data can provide important clues to how binding sites are defined. For example, motif analysis of the liver X receptor (LXR) cistrome by the Glass group showed PU.1 and AP-1 motifs to be enriched, along with the LXR motif (32). PU.1 and CCAAT/enhancer-binding protein (C/EBP) ChIP-seq and gene knockdown experiments in macrophage precursors demonstrated how PU.1 and C/EBP support the binding of each other, creating chromatin that later enables LXR binding in mature macrophages. This study exemplifies how a combination of facilitating mechanisms involving several TF interactions over the developmental history of a cell lineage can be fundamental to understanding the definition of one cistrome (32). Numerous other nuclear receptor ChIP-seq studies have found nuclear receptor binding to occur in conjunction with other TF that facilitate their binding: for example, forkhead box A1 (FOXA1) with ER and androgen receptor (AR) (15, 36), AP-1 with glucocorticoid receptor (GR) (37), and C/EBP with peroxisome proliferator-activated receptor-γ (38).
Although motif analyses can be used to discover factors that collaborate to define TF-binding sites, such analyses are often confounded by factors such as GC content, repetitive DNA elements, and binding hotspots. Matching controls is one way of accounting for confounders; control sites are selected in such a way that they sample a similar distribution to the TF cistrome in terms of GC content, distance from the transcription start site (TSS), and other factors. Another way of dealing with this issue is to exploit the high accuracy of ChIP-seq in defining the precise locations of TF-binding sites. Significance of association between TF binding and a motif can be assessed using statistics on the position of the motif relative to the predicted location of TF binding as implemented in the BINOCh software (39). Because multiple members of a TF family often have similar DNA-binding motifs, complementary methods need to be used to narrow down the target. Work by the Brown laboratory (40) revealed the important role of FOXA1 in defining a large subset of ER-binding sites in the MCF7 breast cancer cell line. This association was initially realized through a motif analysis of ER ChIP-chip data that revealed forkhead motifs to be strongly associated with the ER cistrome. FOXA1 was identified as the primary forkhead candidate because it was the most highly expressed forkhead and most highly correlated with ER in panels of breast cancer gene expression microarray data.
Genome-wide ChIP-chip and ChIP-seq experiments have shown that usually there is no clear one-to-one relationship between TF-binding sites and regulated genes. There are usually many more TF-binding sites than regulated genes, most of which lie far from any TSS. Some speculate that many of these distant sites might be nonfunctional and exist simply because of the combinatorial sequence possibilities that inevitably arise in a large genome. Although some studies fail to identify an association between genome-wide gene expression and TF binding, we find using a simple model that TF binding is usually statistically associated with regulation, even if not highly predictive. In our model, the regulatory potential of each TF-binding site is a decreasing function of the genomic distance between the site and the TSS, and the regulatory effect on the TSS is a sum of the influence of the individual sites (41).
Frequently, when a TF is activated, there are genes that are differentially expressed without any TF-binding site being detected sufficiently close to the TSS to have a regulatory effect. One explanation is that not all the regulatory sites have been determined by ChIP-seq. Another is that the gene is regulated by secondary factors. A third is that the cell line of use has genomic structural variations not considered when ChIP-seq reads are mapped to the reference genome. Last but not least, although a gene may have no TF-binding site near the TSS in genome sequence space, specific three-dimensional interactions may be bringing the regulatory enhancers closer to the gene promoters over a long genomic distance. The opposite also occurs; certain genes have one or more TF-binding sites near the TSS and ought to be regulated, but are not. The status of the promoter, functionality of the observed binding sites, or repressive factors may all contribute to this. ChIP-seq predictions of regulated genes can be highly specific at low levels of sensitivity; however, much needs to be done to understand genome-wide regulation. As data accumulate, it will become necessary to incorporate all relevant TF-binding data into quantitative statistical models before drawing conclusions about the role of a particular factor. Otherwise, it will be hard to understand how new data add to the old. Computational genome-wide models will become indispensible to nuclear receptor biology.
A broad range of tools for ChIP-seq data analysis is available via the cistrome web interface at http://cistrome.org/ap/ (42). This allows users to carry out many of the common ChIP-seq data analysis tasks such as determining where sites lie relative to genomic annotations and the degree of overlap between TF cistromes.
Epigenome Profiling
Posttranslational histone modifications have been correlated with diverse functional categories of genomic loci. For example, H3K4me3 is found mainly at the promoters of actively transcribed genes (43, 44), H3K27me3 is associated with genes repressed by polycomb complex proteins (45), and acetylation marks are often linked to actively transcribed genes (43). H3K4me1 and -2 are strongly associated with the binding of many TFs and are more predictive of TF binding than TF DNA motifs themselves (46). A broad sense of genome-wide chromatin state in a particular cell type can be obtained by profiling a small set of histone modifications (47). Because histone modifications are often highly correlated with each other and antibodies vary greatly in efficacy (44, 48), the most effective strategy for delineating epigenomic domains requires careful selection of target modifications and antibodies, starting with H3K4me3 for active promoters, H3K27me3 for polycomb repressed domains, and H3K4me1/2 for enhancers. Although nuclear receptors are recruited to the genome at regions enriched in specific histone modifications, they also recruit histone-modification enzymes that modify the epigenome in a targeted way. FOXA1 binds genome-wide to H3K4me2-enriched regions in prostate and breast cancer cell line-specific regions to facilitate ER and AR binding (49).
In addition to helping identify TF-binding sites, some histone modifications may be used as a readout of enzymatic activity to determine TF-binding sites that actively regulate genes. For example, the presence of H3K27ac at putative enhancer sites in a variety of cell types has been shown to boost the association of transcribed genes with those binding sites (50, 51). p300 and CREB-binding protein histone acetyltransferases are known to deposit this histone modification (44, 51) and may also help identify active enhancers (52, 53). Although H3K27ac appears to be generic, associating with a broad spectrum of active enhancers, some histone modifications appear to be more TF specific. For example, marks of coactivator-associated arginine methyltransferase 1 activity, including H3R17me2, appear to be highly associated with ER binding in MCF7 and to discriminate between active and inactive ERα enhancers (49). O'Malley and colleagues (54) carried out a genome-wide ChIP-seq study of steroid receptor cofactor steroid receptor coactivator 3 binding. The results showed that steroid receptor coactivator 3 binds frequently in association with ER and that the ER sites with the cofactor are significantly more active than those without. It is puzzling why these factors are recruited to some sites and not others. Studies of the genome-wide location of histone-modifying enzymes can add further insight into nuclear receptor activity. Elegant studies by Lazar (55) and Evans (56) groups used ChIP-seq to demonstrate the involvement of histone deacetylase histone deacetylase-3 and Rev-erbα in controlling the circadian rhythm of hepatic metabolism.
High-resolution analysis of nucleosome occupancy in the vicinity of TF-binding sites has shown that TF-binding sites frequently occur in nucleosome-free regions (21, 46). Nucleosome occupancy data near TF-binding sites can be most efficiently obtained by micrococcal nuclease digestion followed by H3K4me2 or H3K27ac ChIP-seq. In an experiment examining changes in H3K4me2 nucleosome occupancy before and after testosterone stimulation (46), we observed a pattern in which nucleosomes flanking the binding site were stabilized, whereas a nucleosome destabilization was observed in the vicinity of the binding site itself. This method allows for the inference of TF-binding events from nucleosome occupancy data and TF-binding DNA motifs (21, 57, 58).
Deoxyribonuclease (DNase) I-hypersensitive sites (DHS) are short regions of the genome that are highly sensitive to deoxyribonuclease I enzymatic cleavage. Such sites frequently occur at loci that are bound by TFs and provide a universal map of chromatin sites accessible to TF binding. Together with DNA motif analysis, DHS can be used to infer the binding sites of specific TFs (59, 60). DHS studies have shown that there is a high degree of overlap between genome-wide GR, C/EBP, and peroxisome proliferator-activated receptor-γ binding and DHS sites (61). The differences in the degree of overlap between DHS sites and TF-binding sites may be due to data quality and sequencing depth, although differences in DHS intrinsic to the TF-chromatin interaction are also possible. DHS can also provide insight into the biology of TF binding. For example, DHS and ChIP-seq experiments carried out by the Hager group (62) showed that, although GR had previously been recognized as a factor that is capable of binding condensed chromatin, in a mouse mammary epithelial line, DNase I-accessible chromatin is evident before GR activation and predetermined the majority of activated GR-binding sites. Follow-up work, including motif analysis, gene knockdown, and ChIP-seq demonstrates the importance of AP-1 in defining these open chromatin regions (37).
In addition to being used to characterize DNA as accessible or inaccessible, DHS can be used as a quantitative measure of chromatin accessibility. Using changes in DHS between hormone-stimulated and unstimulated conditions, we were able to accurately identify ER- and AR-binding sites (63). DHS is an efficient way of characterizing the static and dynamic aspects of the chromatin landscape and complements both TF-binding and histone-mark data.
DNA itself is subject to epigenetic modifications in the form of 5-methyl-cytosine and 5-hydroxy-methyl-cytosine (64). DNA methylation plays crucial roles in normal transcription regulation in development and aberrant gene expression in cancer (65, 66). NGS techniques for profiling genome-wide DNA methylation include bisulfite sequencing (MethylC-seq) (67) and reduced-representation bisulfite sequencing (68), immunoprecipitation of methylated DNA [MeDIP-seq (69), and MBD-seq (70)], and DNA cleavage using methylation-specific restriction enzymes followed by fragment size selection (71, 72). Each of these techniques has its own cost, coverage and quantitative characteristics and should be chosen to address specific hypotheses (73).
Interactions in Three Dimensions
Several studies have revealed that chromosomes form loops in a site-specific way (74–77). These observations are based on the chromosome conformation capture (3C) technique in which chromatin that has been fragmented in vivo is ligated so as to reveal DNA fragments that are in close proximity in three-dimensional space even if distant in genomic sequence space (78). The 3C technique is a locus-specific technology that has been generalized to assay multiple interactions in 4C (79), 5C (79–81), 6C (82, 83), and Hi-C assays (84, 85). Hi-C is a NGS-based assay that can, in principal, be used to detect all genomic interactions. In practice, Hi-C coverage of interactions in the human genome is sparse using current NGS technologies, and the resulting maps of genomic interactions are still of low resolution.
Selecting only the genome-wide regions that are of special interest is a way of ameliorating the genome size problem. A technique called ChIA-PET does this by examining interactions between sites associated with a particular TF or histone modification, using ChIP to target specific protein-DNA complexes. NGS is used to sequence paired-end tags (PET). Mapping the resulting ChIA-PET sequences to the reference genome reveals relationships between chromosomal regions brought into close proximity by the interaction of the ChIP target factor (7). ChIA-PET is useful for regulatory studies but needs to be interpreted carefully. The method is dependent on ChIP enrichment that is highly variable between sites. Although ChIA-PET uses ChIP to enrich for a specific TF, this does not necessarily mean that the interaction is directly mediated by that TF. It is possible that other proteins are responsible for the formation of stable looping interactions.
Looping is likely to be stochastic and may differ between cell types and conditions. An observation that is consistently made in looping experiments is that the frequency of interaction decays with distance between sites, and strong interactions become less frequent between sites that are remote from each other. Genomic distance between sites has been well characterized and is largely consistent with three-dimensional interactions and is therefore a baseline on which three-dimensional interactions data can elaborate. Although isolated cases of distant three-dimensional interactions have been observed to be important for transcriptional regulation, the generality of this looping phenomenon and its importance in regulating genome-wide transcription remain unanswered questions.
Future Directions
NGS technologies have provided unprecedented experimental opportunities for probing the mechanisms that regulate transcription. Hypothesis-driven experiments using NGS approaches will deepen our understanding of key molecular interactions. Gene regulation, however, involves numerous factors that interact in quantitative, time- and context-specific ways. Genome-wide data on chromatin status, TF binding, and gene expression in a broad array of cell types and conditions will be needed to develop models describing the roles of the large number of factors known to influence transcription. Technological innovation, systematic data organization, process automation, and protocol optimization can all be used to increase the generation and utility of NGS data. Although DNA-sequencing technologies are steadily improving throughput in terms of sequencing depth, sample processing is currently a bottleneck in data generation. Extensive use of robotic systems, batch processing, and multiplexing as well as techniques to reduce required starting material will be needed to provide sufficient data to model this complex biological process.
Better data management, dissemination, and analysis methods will be needed to make use of current and future NGS data. As part of a project to organize TF ChIP-seq data, we have created a nuclear receptor cistrome database (http://cistrome.org/NR_Cistrome/) (41). Making NGS data available to the research community is important, because genome-wide data from one study can provide useful insights in other studies. To improve the use of publicly available NGS data, low-level analysis pipelines need to be put in place to systematically assess data quality and to summarize data in a consistent way. If done well, this will enable data sets to be meaningfully compared and integrated. In addition, mechanisms need to be put in place to allow users to search for data sets by flexible criteria.
Effective use of genomic technologies requires close collaboration between computational and experimental biologists. A common model of collaboration is one in which an experimental plan is developed and executed by an experimental biologist before consulting a computational biologist. Although useful findings can emerge in this way, a better model of collaboration is one in which experimental and computational biologists are involved at every step. This requires computational biologists to understand some details of the genomic techniques and the biological questions of interest. It also requires experimental biologists to have an appreciation of the statistical and computational modeling issues that may arise. We propose that experiments be performed in a staged manner. For example, if several genomic experiments are proposed, the most important one should be conducted first, and some data analysis should be done to determine whether information acquired at this stage might change the overall experimental design. Staging may also be used to explicitly define genome-wide follow-up experiments. To elucidate the complexities of nuclear receptor biology and its impact on disease, genomic and computational technologies need to be used in synergy with human understanding.
Acknowledgments
This work was supported by the United States National Institutes of Health (HG4069) and the National Natural Science Foundation of China (31028011).
Disclosure Summary: C.A.M. and Q.T. have nothing to declare. X.S.L. is a shareholder of Bristol-Myers Squibb, Abbott Laboratories, and Johnson & Johnson BMS, ABT, and JNJ stocks (total less than $30K) and served on the scientific advisory board of GenPathway from 2009–2010.
NURSA Molecule Pages†:
Ligands: 17β-estradiol | Progesterone | Dexamethasone | Dihydrotestosterone | 1α,25-dihydroxyvitamin D3.
Annotations provided by Nuclear Receptor Signaling Atlas (NURSA) Bioinformatics Resource. Molecule Pages can be accessed on the NURSA website at www.nursa.org.
- AP-1
- Adaptor protein complex 1
- AR
- androgen receptor
- C/EBP
- CCAAT/enhancer-binding protein
- ChIP
- chromatin immunoprecipitation
- DHS
- DNase I-hypersensitive sites
- DNase
- deoxyribonuclease
- ER
- estrogen receptor
- FOXA1
- forkhead box A1
- GR
- glucocorticoid receptor
- GRO
- Global Run-On
- LXR
- liver X receptor
- NGS
- next-generation sequencing
- RNA-seq
- mRNA sequencing
- SAGE
- Serial Analysis of Gene Expression
- TSS
- transcription start site.
References
- 1. Oshlack A, Wakefield MJ. 2009. Transcript length bias in RNA-seq data confounds systems biology. Biol Direct 4:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Young MD, Wakefield MJ, Smyth GK, Oshlack A. 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cheung E, Kraus WL. 2010. Genomic analyses of hormone signaling and gene regulation. Annu Rev Physiol 72:191–218 [DOI] [PubMed] [Google Scholar]
- 4. Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, Sam L, Barrette T, Palanisamy N, Chinnaiyan AM. 2009. Transcriptome sequencing to detect gene fusions in cancer. Nature 458:97–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maher CA, Palanisamy N, Brenner JC, Cao X, Kalyana-Sundaram S, Luo S, Khrebtukova I, Barrette TR, Grasso C, Yu J, Lonigro RJ, Schroth G, Kumar-Sinha C, Chinnaiyan AM. 2009. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci USA 106:12353–12358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Campbell PJ, Stephens PJ, Pleasance ED, O'Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, Teague JW, Menzies A, Goodhead I, Turner DJ, Clee CM, Quail MA, Cox A, Brown C, Durbin R, Hurles ME, Edwards PA, Bignell GR, Stratton MR, Futreal PA. 2008. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet 40:722–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, Chew EG, Huang PY, Welboren WJ, Han Y, Ooi HS, Ariyaratne PN, Vega VB, Luo Y, Tan PY, Choy PY, Wansa KD, Zhao B, Lim KS, Leow SC, Yow JS, et al. 2009. An oestrogen-receptor-α-bound human chromatin interactome. Nature 462:58–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rozowsky J, Abyzov A, Wang J, Alves P, Raha D, Harmanci A, Leng J, Bjornson R, Kong Y, Kitabayashi N, Bhardwaj N, Rubin M, Snyder M, Gerstein M. 2011. AlleleSeq: analysis of allele-specific expression and binding in a network framework. Mol Syst Biol 7:522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628 [DOI] [PubMed] [Google Scholar]
- 10. Katz Y, Wang ET, Airoldi EM, Burge CB. 2010. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 7:1009–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu ZJ, Meyer CA, Choudhury S, Shipitsin M, Maruyama R, Bessarabova M, Nikolskaya T, Sukumar S, Schwartzman A, Liu JS, Polyak K, Liu XS. 2010. Gene expression profiling of human breast tissue samples using SAGE-Seq. Genome Res 20:1730–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL. 2011. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell 145:622–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Core LJ, Waterfall JJ, Lis JT. 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322:1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. 2011. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev 25:742–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat Genet 38:1289–1297 [DOI] [PubMed] [Google Scholar]
- 16. Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, Glass CK, Rosenfeld MG, Fu XD. 2011. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474:390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dölken L, Ruzsics Z, Rädle B, Friedel CC, Zimmer R, Mages J, Hoffmann R, Dickinson P, Forster T, Ghazal P, Koszinowski UH. 2008. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA 14:1959–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rabani M, Levin JZ, Fan L, Adiconis X, Raychowdhury R, Garber M, Gnirke A, Nusbaum C, Hacohen N, Friedman N, Amit I, Regev A. 2011. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat Biotechnol 29:436–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hansen KD, Wu Z, Irizarry RA, Leek JT. 2011. Sequencing technology does not eliminate biological variability. Nat Biotechnol 29:572–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, Chen Z, Beroukhim R, Wang H, Lupien M, Wu T, Regan MM, Meyer CA, Carroll JS, Manrai AK, Jänne OA, Balk SP, Mehra R, Han B, Chinnaiyan AM, Rubin MA, True L, Fiorentino M, Fiore C, Loda M, Kantoff PW, Liu XS, Brown M. 2009. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell 138:245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Verzi MP, Shin H, He HH, Sulahian R, Meyer CA, Montgomery RK, Fleet JC, Brown M, Liu XS, Shivdasani RA. 2010. Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev Cell 19:713–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Krum SA, Miranda-Carboni GA, Lupien M, Eeckhoute J, Carroll JS, Brown M. 2008. Unique ERα cistromes control cell type-specific gene regulation. Mol Endocrinol 22:2393–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lupien M, Meyer CA, Bailey ST, Eeckhoute J, Cook J, Westerling T, Zhang X, Carroll JS, Rhodes DR, Liu XS, Brown M. 2010. Growth factor stimulation induces a distinct ERα cistrome underlying breast cancer endocrine resistance. Genes Dev 24:2219–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu VC, Delsert C, Andersen B, Holloway JM, Devary OV, Näär AM, Kim SY, Boutin JM, Glass CK, Rosenfeld MG. 1991. RXRβ: a coregulator that enhances binding of retinoic acid, thyroid hormone, and vitamin D receptors to their cognate response elements. Cell 67:1251–1266 [DOI] [PubMed] [Google Scholar]
- 25. Kliewer SA, Umesono K, Mangelsdorf DJ, Evans RM. 1992. Retinoid X receptor interacts with nuclear receptors in retinoic acid, thyroid hormone and vitamin D3 signalling. Nature 355:446–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Owen GI, Richer JK, Tung L, Takimoto G, Horwitz KB. 1998. Progesterone regulates transcription of the p21(WAF1) cyclin-dependent kinase inhibitor gene through Sp1 and CBP/p300. J Biol Chem 273:10696–10701 [DOI] [PubMed] [Google Scholar]
- 27. Johansson-Haque K, Palanichamy E, Okret S. 2008. Stimulation of MAPK-phosphatase 1 gene expression by glucocorticoids occurs through a tethering mechanism involving C/EBP. J Mol Endocrinol 41:239–249 [DOI] [PubMed] [Google Scholar]
- 28. Shim EY, Woodcock C, Zaret KS. 1998. Nucleosome positioning by the winged helix transcription factor HNF3. Genes Dev 12:5–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adams CC, Workman JL. 1995. Binding of disparate transcriptional activators to nucleosomal DNA is inherently cooperative. Mol Cell Biol 15:1405–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Steger DJ, Workman JL. 1997. Stable co-occupancy of transcription factors and histones at the HIV-1 enhancer. EMBO J 16:2463–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Becker PB, Hörz W. 2002. ATP-dependent nucleosome remodeling. Annu Rev Biochem 71:247–273 [DOI] [PubMed] [Google Scholar]
- 32. Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38:576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Beato M, Vicent GP. 2012. Impact of chromatin structure and dynamics on PR signaling. The initial steps in hormonal gene regulation. Mol Cell Endocrinol 357:37–42 [DOI] [PubMed] [Google Scholar]
- 34. Zaret KS, Carroll JS. 2011. Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25:2227–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Biggin MD. 2011. Animal transcription networks as highly connected, quantitative continua. Dev Cell 21:611–626 [DOI] [PubMed] [Google Scholar]
- 36. Wang Q, Li W, Liu XS, Carroll JS, Jänne OA, Keeton EK, Chinnaiyan AM, Pienta KJ, Brown M. 2007. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell 27:380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Biddie SC, John S, Sabo PJ, Thurman RE, Johnson TA, Schiltz RL, Miranda TB, Sung MH, Trump S, Lightman SL, Vinson C, Stamatoyannopoulos JA, Hager GL. 2011. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol Cell 43:145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert CJ, Jr, Liu XS, Lazar MA. 2008. PPARγ and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev 22:2941–2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Meyer CA, He HH, Brown M, Liu XS. 2011. BINOCh: binding inference from nucleosome occupancy changes. Bioinformatics 27:1867–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122:33–43 [DOI] [PubMed] [Google Scholar]
- 41. Tang Q, Chen Y, Meyer C, Geistlinger T, Lupien M, Wang Q, Liu T, Zhang Y, Brown M, Liu XS. 2011. A comprehensive view of nuclear receptor cancer cistromes. Cancer Res 71:6940–6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y, Shin H, Wong SS, Ma J, Lei Y, Pape UJ, Poidinger M, Chen Y, Yeung K, Brown M, Turpaz Y, Liu XS. 2011. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol 12:R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39:311–318 [DOI] [PubMed] [Google Scholar]
- 44. Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40:897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448:553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, Mieczkowski P, Lieb JD, Zhao K, Brown M, Liu XS. 2010. Nucleosome dynamics define transcriptional enhancers. Nat Genet 42:343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473:43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823–837 [DOI] [PubMed] [Google Scholar]
- 49. Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. 2008. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132:958–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470:279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA 107:21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chan HM, La Thangue NB. 2001. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci 114:2363–2373 [DOI] [PubMed] [Google Scholar]
- 53. Das C, Lucia MS, Hansen KC, Tyler JK. 2009. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 459:113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lanz RB, Bulynko Y, Malovannaya A, Labhart P, Wang L, Li W, Qin J, Harper M, O'Malley BW. 2010. Global characterization of transcriptional impact of the SRC-3 coregulator. Mol Endocrinol 24:859–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA. 2011. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331:1315–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, Liddle C, Auwerx J, Downes M, Panda S, Evans RM. 2012. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 485:123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hoffman BG, Robertson G, Zavaglia B, Beach M, Cullum R, Lee S, Soukhatcheva G, Li L, Wederell ED, Thiessen N, Bilenky M, Cezard T, Tam A, Kamoh B, Birol I, Dai D, Zhao Y, Hirst M, Verchere CB, Helgason CD, Marra MA, Jones SJ, Hoodless PA. 2010. Locus co-occupancy, nucleosome positioning, and H3K4me1 regulate the functionality of FOXA2-, HNF4A-, and PDX1-bound loci in islets and liver. Genome Res 20:1037–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, Rosen ED. 2010. Comparative epigenomic analysis of murine and human adipogenesis. Cell 143:156–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pique-Regi R, Degner JF, Pai AA, Gaffney DJ, Gilad Y, Pritchard JK. 2011. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res 21:447–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee BK, Sheffield NC, Gräf S, Huss M, Keefe D, Liu Z, London D, McDaniell RM, Shibata Y, Showers KA, Simon JM, Vales T, Wang T, Winter D, Zhang Z, Clarke ND, Birney E, Iyer VR, Crawford GE, Lieb JD, Furey TS. 2011. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res 21:1757–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Siersbæk R, Nielsen R, John S, Sung MH, Baek S, Loft A, Hager GL, Mandrup S. 2011. Extensive chromatin remodelling and establishment of transcription factor 'hotspots' during early adipogenesis. EMBO J 30:1459–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. 2011. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet 43:264–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. He HH, Meyer CA, Chen MW, Jordan VC, Brown M, Liu XS. 2012. Differential DNase I hypersensitivity reveals factor-dependent chromatin dynamics. Genome Res 22:1015–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324:930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bird A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev 16:6–21 [DOI] [PubMed] [Google Scholar]
- 66. Feinberg AP, Vogelstein B. 1983. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 301:89–92 [DOI] [PubMed] [Google Scholar]
- 67. Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462:315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454:766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jacinto FV, Ballestar E, Esteller M. 2008. Methyl-DNA immunoprecipitation (MeDIP): hunting down the DNA methylome. Biotechniques 44:35, 37, 39 passim [DOI] [PubMed] [Google Scholar]
- 70. Serre D, Lee BH, Ting AH. 2010. MBD-isolated genome sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res 38:391–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. 2009. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol 27:361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, Turecki G, Delaney A, Varhol R, Thiessen N, Shchors K, Heine VM, Rowitch DH, Xing X, Fiore C, Schillebeeckx M, Jones SJ, Haussler D, Marra MA, Hirst M, Wang T, Costello JF. 2010. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466:253–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y, Olshen A, Ballinger T, Zhou X, Forsberg KJ, Gu J, Echipare L, O'Geen H, Lister R, Pelizzola M, Xi Y, Epstein CB, Bernstein BE, Hawkins RD, Ren B, Chung WY, et al. 2010. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol 28:1097–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. 2002. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell 10:1453–1465 [DOI] [PubMed] [Google Scholar]
- 75. Murrell A, Heeson S, Reik W. 2004. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet 36:889–893 [DOI] [PubMed] [Google Scholar]
- 76. Spilianakis CG, Flavell RA. 2004. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol 5:1017–1027 [DOI] [PubMed] [Google Scholar]
- 77. Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR. 2007. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J 26:2041–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Dekker J, Rippe K, Dekker M, Kleckner N. 2002. Capturing chromosome conformation. Science 295:1306–1311 [DOI] [PubMed] [Google Scholar]
- 79. Zhao Z, Tavoosidana G, Sjölinder M, Göndör A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R. 2006. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet 38:1341–1347 [DOI] [PubMed] [Google Scholar]
- 80. Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. 2006. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet 38:1348–1354 [DOI] [PubMed] [Google Scholar]
- 81. Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, Green RD, Dekker J. 2006. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res 16:1299–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. 2005. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet 37:31–40 [DOI] [PubMed] [Google Scholar]
- 83. Tiwari VK, Cope L, McGarvey KM, Ohm JE, Baylin SB. 2008. A novel 6C assay uncovers Polycomb-mediated higher order chromatin conformations. Genome Res 18:1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS. 2010. A three-dimensional model of the yeast genome. Nature 465:363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Shopland LS, Lynch CR, Peterson KA, Thornton K, Kepper N, Hase J, Stein S, Vincent S, Molloy KR, Kreth G, Cremer C, Bult CJ, O'Brien TP. 2006. Folding and organization of a contiguous chromosome region according to the gene distribution pattern in primary genomic sequence. J Cell Biol 174:27–38 [DOI] [PMC free article] [PubMed] [Google Scholar]