Abstract
Alteromonas macleodii is a marine gammaproteobacterium with widespread distribution in temperate or tropical waters. We describe three genomes of isolates from surface waters around Europe (Atlantic, Mediterranean and Black Sea) and compare them with a previously described deep Mediterranean isolate (AltDE) that belongs to a widely divergent clade. The surface isolates are quite similar, the most divergent being the Black Sea (BS11) isolate. The genomes contain several genomic islands with different gene content. The recruitment of very similar genomic fragments from metagenomes in different locations indicates that the surface clade is globally abundant with little effect of geography, even the AltDE and the BS11 genomes recruiting from surface samples in open ocean locations. The finding of CRISPR protospacers of AltDE in a lysogenic phage in the Atlantic (English Channel) isolate illustrates a flow of genetic material among these clades and a remarkably wide distribution of this phage.
The genus Alteromonas, and the species A. macleodii, were described in one of the first large scale studies of aerobic marine bacterial isolates in 19721. The first isolates obtained from coastal waters of Oahu (Hawaii) were described as motile by unsheathed polar flagella, capable of growing on a minimal medium with glucose, and requiring sodium ion (a trait shared by most autochthonous marine bacteria). This microbe grows very quickly on marine agar (colonies are fully developed after 24 hours incubation at room temperature) and has been commonly isolated from many marine samples2. The abundance and distribution of A. macleodii was not fully understood, however, until PCR amplified 16S rDNA genes were sequenced directly from marine samples. A. macleodii closely related 16S rRNA genes were detected in Mediterranean waters from the deep-chlorophyll maximum (ca. 50 m deep) and from deeper waters (400 m deep) when the large-size fraction (>2 µm) was analyzed. rDNA sequences closely associated to A. macleodii were also recovered as an important part of the total biomass from confinement and mesocosm experiments in Mediterranean bacterioplankton3,4. Later studies using rRNA internal transcribed spacer (ITS) sequencing and hybridization of DNA samples collected from several marine samples from around the world5 indicated that A. macleodii cells represent a significant fraction of the bacterial population associated with particles or aggregates (2–5 µm filters) in temperate or tropical waters with average temperatures above 10°C. This temperature limitation precluded the presence of this microbe in any deep water samples with the exception of the Mediterranean, where the deep water mass never gets below 12°C. Analysis of isolates from the deep Mediterranean revealed differences in ITS and house-keeping genes that suggested the presence of a “Deep-Ecotype” (DE) in the Mediterranean6.
The genome of one A. macleodii AltDE isolate obtained from 1000 m deep in the South Adriatic was determined, and its comparison with a draft genome of the type strain A. macleodii ATCC 27126 (AltATCC, a surface isolate from Hawaii) led to the conclusion that the AltDE isolate was not a real deep dweller7. Specifically the presence in the genome of a photolyase gene indicated that the microbe is sometimes exposed to UV light. The AltDE isolate also lacked a characteristic loop in the 16S rRNA that has been found in all piezophilic gamma-proteobacteria8,9. On the other hand, many features of the genome such as more microaerophilic adaptations (hydrogenases, nitrate reductase and microaerophilic respiratory chains) were reminiscent of life even more committed to the particle associated lifestyle. This prompted the authors to propose that this microbe is really a larger particle ecotype that, due to their faster sinking rate, is largely found at deeper waters when the water mass temperature remains warm enough for the microbe to proliferate.
Two recent metatranscriptomic studies provide new insights into the lifestyle of A. macleodii10,11. Microcosm experiments were carried out to monitor the changes in transcript populations, in a water sample from 75 m deep in the central Pacific gyre (near Hawaiian Ocean Time-Series, HOT station). In one experiment, water was amended with dissolved organic matter (DOM) concentrated from the same environment, and in a second experiment water from a deeper sample (700 m) was added to the surface one to simulate the fertilization that happens when an upwelling or water mixing event takes place. The A. macleodii population increased in both experiments, from undetectable to ca. 10% of the population during the study period of 25 hours. The majority of A. macleodii associated reads in both treatments were dominated by genotypes which shared ~98% nucleotide identity with AltATCC. AltDE-like sequences were also detected, although at much lower abundance. This data illustrate by a totally different methodology the relevance of this microbe as an r-strategist that blooms under conditions of sudden increase in the availability of resources (probably DOM released or exudated by the phytoplankton)12,13,14.
Given the importance of A. macleodii strains that cluster with AltATCC (surface ecotype) and the lack of finished genomes from this group, we have fully sequenced and assembled three new genomes characterized by MLSA as belonging to this group. The origin of the three surface marine isolates sequenced has been described before6. Briefly, A. macleodii AD45 (AD45) comes from waters of a fish farm near Valencia, Spain15, A. macleodii 673 (673) comes from the L4 long-term coastal monitoring station in the Western English Channel16, and A. macleodii BS11 (BS11) was isolated from the Black Sea near the Karadag reservation off the Crimean peninsula (Ukraine). We also compared the new genomes to that of the previously sequenced AltATCC and AltDE. Our findings reveal striking genomic similarity among the three geographically distinct isolates and suggest that the surface isolates could be more physiologically diverse than AltDE.
Results
Phenotypic features
The general growth characteristics of the isolates were determined to compare the phenotypes (Supplementary Table 1 and 3; Supplementary Figures 1). All three A. macleodii isolates described here had a generation time of ca. 2.4 hours at 25°C and could grow at salinities from 1% to 18% with optimal growth achieved between 2 and 12% in marine salts. In the case of BS11 the salinity limit was lower (0.25%). Also small differences were found for the upper limit (20% for AltDE and 18% for AD45 and BS11). The wide salinity growth range observed for all three isolates is a remarkable euryhaline response. Similarly, the members of the genus Halomonas that are routinely found in the ocean35 and our own (unpublished data) are among the most euryhaline microbes known. It is possible that compatible solute accumulation renders these microbes capable of growing over a wide range of salinities that are never found in their natural environment. A similar situation was found with the growth temperature, where all strains could grow between 10 and 43°C even though ocean temperatures rarely exceed 30°C. Both phenomena could be a reflection of the resilient physiology of these microbes.
General features of the genome sequences
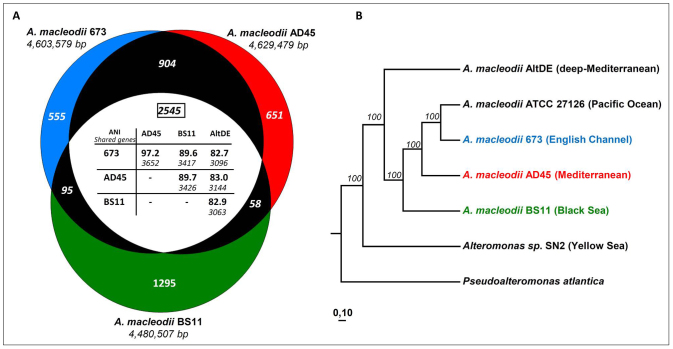
The general features of surface isolates (673, AD45 and BS11), the previously assembled genome of the deep isolate (AltDE), and the draft genome of the type strain AltATCC are shown in Table 1. To assess the presence of plasmids, PFGE was carried out with the three strains (plus AltDE). Only one plasmid of 45 Kb was found in AD45. It was subsequently assembled from the raw sequence data, and its structure confirmed by PCR. One circular chromosome was finally assembled for all the three strains with an average size of ca. 4.5 Mb. A total of 2545 orthologous genes were found between the three surface isolates genomes, AD45, 673 and BS11 and its average nucleotide identity (ANI) is shown in Figure 1 (panel A). The English Channel isolate 673 and the Mediterranean isolate AD45 both exhibited high ANI to the type strain AltATCC isolated from Hawaii (98.4% and 97.2%, respectively). These three surface isolates represent a closely associated clade with a large set of shared genes (close to 90% of ORFs detected). The Black Sea isolate BS11 appears more divergent with an ANI slightly below 90% and a much smaller set of shared genes (ca. 75%). In contrast, they all represent a very different clade from the deep Mediterranean isolate AltDE (Figure 1) which exhibited an ANI (ca. 82% with all the others) that is borderline consistent with it belonging to a different species of the same genus28,36. However the number of shared genes is in the same range as with the Black Sea isolate. The maximum likelihood tree generated from a concatenate of 67 house-keeping genes (Supplementary table 2) provided further support for the relationships suggested by the ANI data (Figure 1panel A).
Table 1. General features of the genomes.
| A. macleodii strain | 673 | BS11 | AD45 | AltDE | AltATCC |
|---|---|---|---|---|---|
| Size (bp) | 4 601 785 | 4 480 507 | 4 629 497 | 4 480 922 | 4 607 844 |
| % GC | 44,8 | 44,6 | 44,6 | 44,9 | 44,6 |
| Contigs | 1 | 1 | 1 | 1 | 716 |
| Total ORFs | 4 099 | 3 993 | 4 158 | 4 389 | 4 444 |
| rRNA operons | 5 | 5 | 5 | 5 | 5 |
| tRNAs | 75 | 69 | 71 | 63 | – |
| Hypothetical proteins | 544 | 551 | 577 | 362 | – |
| Function assigned | 3 555 | 3 442 | 3 587 | 4 027 | – |
| Plasmids | 0 | 0 | 1 (45 kbp) | 0 | – |
Figure 1. Relationships among the sequenced and reference strains.
(A) Scalar-Venn representation of shared genes among 673, AD45 and BS11. The average nucleotide identity (ANI) and the number of shared genes among the strains is indicated in the inset table. (B) Phylogenetic relationship (maximum-likelihood) among the available A. macleodii genomes derived from 67 concatenated housekeeping genes (Supplementary Table 2). Pseudoalteromonas atlantica T6c was used as an outgroup. Numbers on the branches represent the percentage of 100 bootstrap samples supporting the branch.
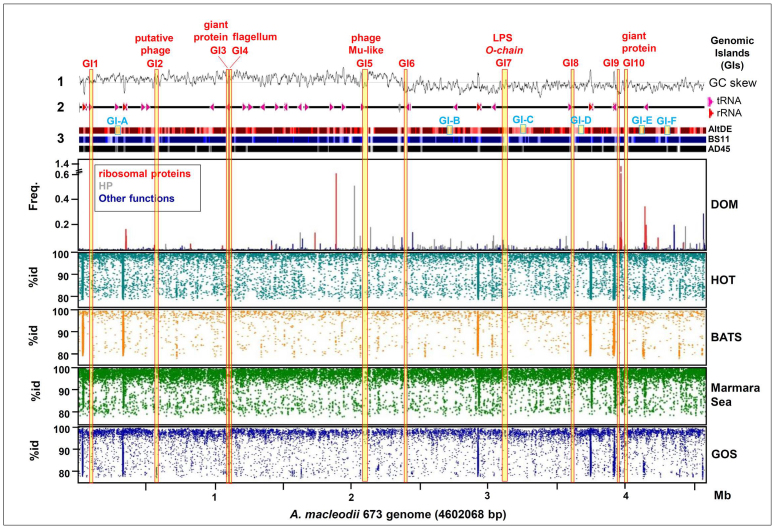
Synteny (Supplemetary Figure 2) was well preserved in all the strains (including AltDE). Only in the case of Alteromonas sp. strain SN237 there seems to be a large rearrangement from rRNA operon 1 to rRNA operon 5 (Supplementary Figure 3). However, within this context of highly conserved synteny there were genomic islands (GI) with differential gene content. GIs larger than 15 Kb are listed in Table 2 and highlighted in Figure 2 in which the genome of 673 was used as reference. The locations of GIs in the other genomes are shown in Supplementary Figures 2 and 4. GIs present in all the genomes could be identified as metagenomic islands38 as fragments that dramatically under-recruit (Figure 2), indicating the highly variable nature of these regions of the genomes. Often there were tRNAs and integrases or transposases flanking or within these GI. In addition, a variety of isolate-specific genes were scattered along the chromosome, with more than 50% contained in fragments of more than 5 Kb.
Table 2. Features of A. macleodii genomic islands.
| Associated to | Conserved | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GI | Start | End | Number of genes (hp*) | GC content (%) | Inferred character | tRNA | IS* | AD45 | BS11 | 673 | AltDE | |
| 1 | 81222 | 101697 | 18 (4) | 43,66 | — | Arg | IN* | • | ||||
| 2 | 560941 | 589307 | 25 (6) | 41,96 | Putative Phage | IN | • | |||||
| 673 | 3 | 1088580 | 1116505 | 8 (1) | 46,40 | Giant Protein | Arg | • | ||||
| 4 | 1134251 | 1152135 | 17 (2) | 42,92 | Flagellum | • | • | • | • | |||
| 5 | 2082298 | 2121603 | 47 (25) | 46,54 | Phage Mu | Leu | IN | • | ||||
| 6 | 2343436 | 2359304 | 14 (1) | 41,57 | — | • | ||||||
| 7 | 3118782 | 3158982 | 36 (9) | 40,26 | LPS O-chain | • | • | • | • | |||
| 8 | 3609548 | 3637119 | 27 (1) | 45,15 | Benzoate degradation | Leu | • | |||||
| 9 | 3959983 | 3974610 | 9 (4) | 36,51 | — | • | ||||||
| 10 | 4008301 | 4030719 | 5 (1) | 45,46 | Giant Protein | • | ||||||
| 1 | 78181 | 106449 | 19 (3) | 44,61 | — | Arg | • | |||||
| 2 | 266279 | 322009 | 64 (12) | 42,12 | Metal resistance | Phe | IN | • | • | • | ||
| BS11 | 3 | 1124500 | 1144989 | 16 (3) | 41,53 | Flagellum | • | • | • | • | ||
| 4 | 2279598 | 2304325 | 19 (3) | 41,23 | — | IN | • | |||||
| 5 | 2401542 | 2421598 | 29 (14) | 40,01 | — | TR* | • | |||||
| 6 | 2892357 | 2932344 | 52 (36) | 49,21 | Putative Phage | IN | • | |||||
| 7 | 3054010 | 3084688 | 27 (6) | 41,27 | LPS O-chain | • | • | • | • | |||
| 8 | 3507904 | 3530237 | 16 (4) | 44,37 | — | Leu | • | |||||
| 9 | 4330079 | 4370292 | 23(1) | 43,67 | Carbohydrates catabolism | • | ||||||
| 1 | 269912 | 303751 | 33 (6) | 40,43 | Metal resistance | Phe | IN | • | • | • | ||
| 2 | 879288 | 907000 | 15 (0) | 44,72 | Carbohydrates catabolism | • | ||||||
| AD45 | 3 | 1019221 | 1042766 | 15 (1) | 44,12 | Alginate degradation | ||||||
| 4 | 1154028 | 1169709 | 12 (1) | 41,72 | Flagellum | • | • | • | • | |||
| 5 | 1721883 | 1783016 | 41 (18) | 39,98 | — | IN | • | |||||
| 6 | 2538511 | 2554138 | 30 (15) | 37,42 | — | • | ||||||
| 7 | 2649619 | 2670481 | 24 (4) | 41,40 | Mercury resistance | • | • | |||||
| 8 | 3170033 | 3215337 | 33 (4) | 41,09 | LPS O-chain | • | • | • | • | |||
| 9 | 3700399 | 3721824 | 14 (3) | 45,54 | — | • | ||||||
| 10 | 3846400 | 3867991 | 23 (13) | 40,39 | Putative Phage | IN | • | |||||
| Deep-Surface* | A | 274551 | 311502 | 35 (3) | 45,11 | Chemotaxis | Phe | • | • | • | ||
| B | 2712110 | 2734217 | 20 (3) | 44,56 | Carbohydrates catabolism | • | • | • | ||||
| C | 3236155 | 3320545 | 21 (8) | 44,37 | — | • | • | • | ||||
| D | 3675224 | 3704363 | 22 (2) | 45,74 | Alcohol catabolism | • | • | • | ||||
| E | 4127396 | 4147382 | 19 (2) | 47,48 | Iron acquisition | • | • | • | ||||
| F | 4238710 | 4330900 | 73 (10) | 46,18 | — | • | • | • | ||||
IS* Insertion Sequence, IN* Integrase, TR* Transposase, HP* hypothetical protein, Deep-Surface* Genomic islands found in all the surface isolates but absent in the deep isolated AltDE.
Figure 2. Genomic islands and metagenome recruitment of the 673 genome.
The genome of 673 was used as reference to detect genomic islands absent in the other three available and largely synthenic genomes (AltDE, BS11 and AD45). From top to bottom: 1, GC skew, 2, location of tRNA and rRNA genes, 3, regions absent in each genome are indicated by discoloured bands as obtained from a genome atlas plot (http://www.cbs.dtu.dk/index.shtml); Yellow/blue squares and blue characters represent genomic islands (GI) larger than 15 Kb present in all the surface isolates but not in AltDE. GIs that are unique to 673 have been highlighted as vertical yellow/red rectangles and red characters, identified by the inferred function whenever possible (on top). The lower four panels indicate fragments recruited in different available metagenomes and their similarity to the homologous region of the 673 genome.
Strain specific GIs
O-chain of the Lipopolysaccharide (LPS)
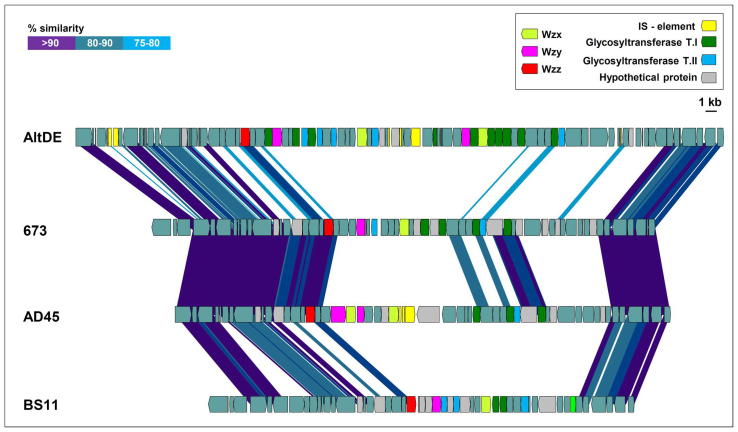
Comparative genomics has shown that some gene clusters are nearly always different when two closely related strains are sequenced39,40,41,42. Many of these differential islands are related to exposed structures and are used by phages or grazers as recognition targets43. In the case of Gram negative bacteria, the O-chain of the lipopolysaccharide (LPS) is an important component of the outer membrane and represents a paradigm of extreme variability among strains. It is a repeat-unit (between two and six sugar residues) polysaccharide extremely variable in the nature, order and linkage of the different sugars44. Originally, this diversity was interpreted as a mechanism of antigenic variation (hence the O-antigen designation). However, the variability in free living cells that are never exposed to any immune system indicates that this variability has other reasons to exist such as the above mentioned phage avoidance42,43. AltDE strain poses the longest O-chain cluster, followed by AD45, 673 and finally, BS11. The gene clusters are so different that only between the closely related AD45 and 673 could some homologous genes be found (Figure 3).
Figure 3. Schematic representation of the LPS O-chain cluster genes in A. macleodii AltDE, 673, AD45 and BS11 strains.
Wzy (O-antigen polymerase), Wzx (O-antigen flipase), Wzz (O-chain length determinant protein), Glycosyltransferase T.I (Glycosyltransferase type 1), Glycosyltransferase T.II (Glycosyltransferase type 2).
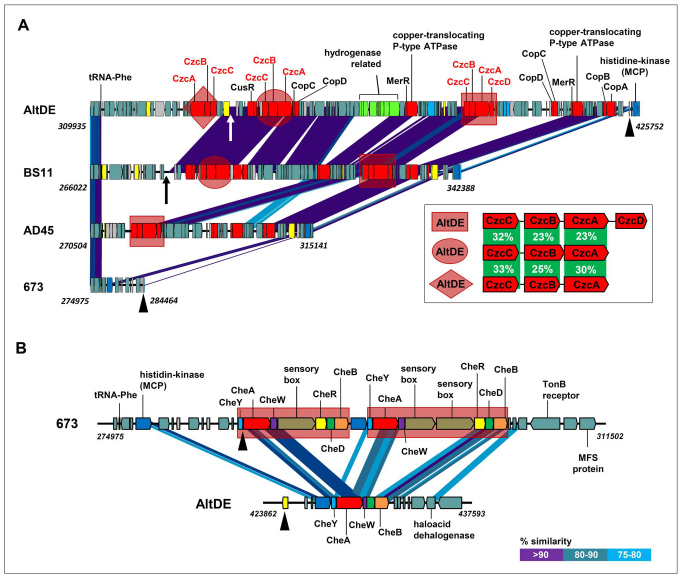
Exposed flagellar constituents and their glycosylation
Another GI that has been found to be different in all the strains is located inside the large gene cluster that codes for the synthesis of the flagellum. In all the A. macleodii strains sequenced the flagellar genes are located in a single large cluster (>70 genes). In the middle of this large cluster there is a set of genes that code for some exposed flagellar proteins such as FliC (flagellin) and genes that appear to be involved in glycosylation of the flagellar proteins. A similar flagellar glycosylation island has been described in Pseudomonas aeruginosa PAO145. These genes appear extremely diverse in each strain (Supplementary Figure 5). The LPS O-chain and the flagellar GIs are probably very important to the cell, this would explain why these regions are not associated to a tRNA or IS element that might produce frequent functional disruption.
Giant proteins
GI3 and GI10 in 673 are made up of a single gene that codes for a “giant protein”: 6388 and 5751 aminoacids respectively (Supplementary Figure 6). AltDE contains also a very different giant protein gene7. Giant proteins of this kind have been found in many bacterial genomes although their function remains elusive46. The protein of GI3 poses domains of “Bacterial Ig-like”47, which have been found in many surface proteins of bacteria and phages. The high variability of these proteins might indicate also a connection to phage recognition/evasion. Alternatively, they could act in the defense against predation by protists as has been described in Synechococcus WH810248.
Plasmid in AD45 and prophages
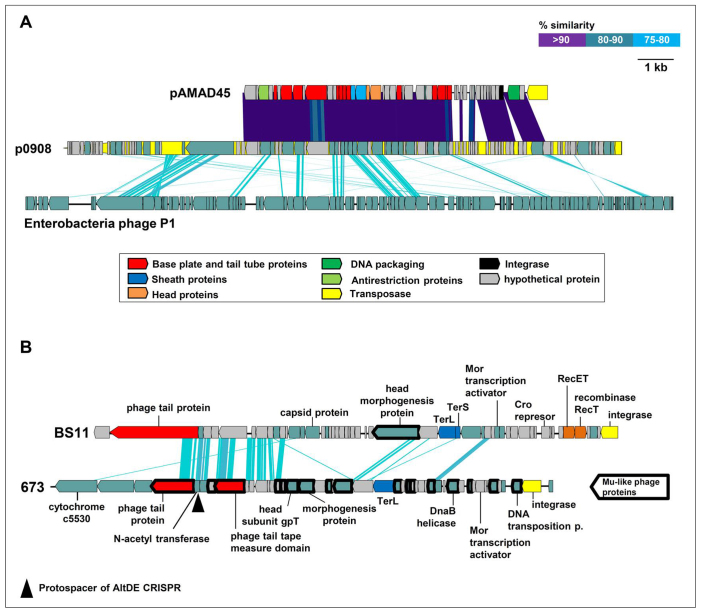
The only plasmid found in our collection of isolates was found in AD45 and has been named pAMBAS45. This 45 Kb plasmid with high GC content (49.5%) encodes 53 CDSs of which 43 showed high similarity to a p0908 81.4 Kb plasmid from a Vibrio sp. strain 0908 isolated from a salt marsh sediment49 (Figure 4 panel A). The Vibrio plasmid was characterized as a putative defective phage based on its similarity to the enterobacterial phage P1 but missing critical proteins for packaging and dispersal50. This could also be the case of pAMBAS45. The plasmid was extremely under-recruiting compared to the AD45 chromosome (data not shown) indicating that it is present only in some environmental lineages of AD45, and is not very prevalent in marine bacteria at large.
Figure 4. Plasmid and putative prophages found in the surface isolates.
(A) Schematic representations of the plasmids pAMAD45 of A. macleodii AD45, p0908 of Vibrio sp. 0908 and the enterobacterial (Escherichia coli) phage P1. (B) Schematic representations of the prophages found in A. macleodii BS11 and 673 strains. Arrows with thick borders are proteins similar to E. coli phage Mu. Black triangle indicates protospacer found in AltDE CRISPR.
Other typical unique islands in bacterial genomes are integrated lysogenic or defective phages. This seems to be the case of GI5 of 673 (39 Kbp) and GI6 of BS11 (40 Kbp) (Figure 4 panel B). The presence of a tape measure protein suggests that they could be long-tailed phages (Caudovirales). The conservation of the relative position of genes with similar annotation and the morphogenesis genes with similarities that range between 34 to 51% indicate that both putative prophages could be family-related. Besides, the 673 putative prophage has significant gene similarity and organization to Mu-like prophages present in marine γ-proteobacteria such as Fulvimarina pelagi HTCC2506 and Marinomonas sp. MED12151. Particularly revealing is the fact that the 673 Mu-like prophage possess a sequence identical to a clustered regularly interspaced short palindromic repeats (CRISPR) spacer present in AltDE. AltDE is the only A. macleodii sequenced genome with a CRISPR-Cas system. CRISPR act as a prokaryotic immune system and confer resistance to foreign DNA such as plasmids and phages52,53,54. Comparing the spacers of the CRISPR system of AltDE with the prophages in 673 and BS11 and the plasmid present in AD45, we found that the spacer 2 of the AltDE repeats is present at 100% identity in an N-acetyl-transferase gene in the 673 Mu-like prophage (see Figure 4 panel B). This finding confirms the prophage nature of this 673 island and suggests that this putative phage is geographically very widespread. Also, spacer 29 is present (with 4 mismatches) in a helicase placed in a conserved region of the three surface isolate genomes. In this area, there are 3 genes that are annotated as being of viral origin (a RNA chaperone, a DNA mismatch repair protein and a RNA polymerase sigma factor). They could have belonged to an inserted defective prophage in the common ancestor of the three strains that was also widely distributed. In any case, the sharing of phage sequences among strains that are so distant geographically, and in the case of AltDE, phylogenetically distant underscores the lack of barriers to genetic exchange within this diverse and widespread marine group.
Metal resistance
AltDE was shown to contain a large cluster of genes that code for metal resistance and a hydrogenase cluster7 located next to the single Phe-tRNA (Figure 5). This cluster contains a set of hydrogenase genes that have been the focus of recent interest being the most oxygen resistant hydrogenases described to date55,56,57. None of the surface isolates described here contains any of the hydrogenase components. However, the surface isolates genomes revealed a hypervariable region, different in each strain, and related to metal resistance. The way this GI has been increasing by successive insertions (or decreasing by deletions) can be visualized in Figure 5 panel A. The tRNA has obviously acted as an insertion target producing larger gene clusters with each insertion event. It is known that the conserved sequence of tRNA genes facilitate DNA integration and excision58. The simplest version of this region is present in 673, where the tRNA gene is next to the general chemotaxis region that is conserved in all the surface isolates (see below). The other strains, including AltDE contain many genes involved in heavy metal resistance. In AD45 there is a single cluster of genes that code for CzcA, CzcB and CzcC, the components of a heavy metal efflux pump important for resistance to cobalt, zinc and cadmium. Besides, the main components of the copper resistance operon, copA, copB, copC and copD59 and three other genes coding for efflux systems (RND, CDF and P-type ATPase) that serve also in the basic defense of the cell against heavy metals60 were also found. These genes are found in all the strains (except 673) with additional insertions of paralogous (see below) czc clusters. Thus, BS11 contained an additional set of czc genes. In this strain the duplication of part of the tRNA gene can still be detected as evidence of an integration event. Finally in AltDE there was another insertion that produced the tell-tale 3′ end tRNA gene fragment duplication and that carried a third set of the czc cassette (Figure 5 panel A). The comparison of all three AltDE czc cassettes reveals low similarity typically associated with paralogs (Figure 5, inset).
Figure 5. Variations in the metal resistance related island.
(A) tRNA-Phe related variable region (GIA in Figure 2). Regions highlighted by geometric figures (square, diamond or ellipse) show clusters of czc genes. The clusters highlighted by the same figure are identical in sequence. Arrows under BS11 and AltDE indicate the 3′ end of the tRNA gene section that is duplicated and represents a hallmark of an integration event. Inset in figure show the percentage of identity among the three paralogous sets of Czc clusters found in AltDE (or the two in BS11). Numbers on the left indicate the position in the genome of the Phe-tRNA first nucleotide. The numbers to the right are the positions of the 3′ end of the last gene represented at this end. (B) Region downstream of GIA (or just the Phe-tRNA gene in 673) that is common to all the surface isolates but very different in AltDE. The duplication of the major chemotaxis pathway in the surface isolates (only the one in 673 is indicated) is highlighted as brown rectangles. Black triangles indicate equivalent genomic locations in panel A and B.
Downstream from the metal-resistance region (or just the tRNA in the case of 673). The three surface isolates have a cluster of ca. 37 Kb that contains genes involved in chemotaxis (Figure 5 panel B). The chemotaxis genes include a co-transcribed histidine-kinase and response regulator typical of two component systems, and two sets of paralogous versions of the general chemotaxis pathway proteins CheA, CheB, CheD, CheR, CheW and CheY. The two versions of each gene were quite divergent with similarities ranging from 57% to 86%. Shine-Dalgarno sequences were present in all the genes upstream of the initiation codon indicating that all the copies could be expressed. In contrast, AltDE contains a single set of chemotaxis genes next to the metal-resistance-hydrogenase island whose genes are even more divergent (Figure 5 panel B). This redundant cluster in the surface isolates might be interpreted as reflecting a difference in the chemotactic behavior between the two clades. In the previous analysis of the AltDE genome compared to AltATCC7 a similar argument was suggested based on the much larger number of two component systems found in the AltATCC genome draft.
Specific GIs and genes in the Black Sea isolate BS11
The most divergent surface isolate BS11 contains a large set of 1295 unique genes not found in the other strains. Some of the differences appear associated to salinity (the main differential factor of the Black Sea). For example, all the strains have a Na+ /dicarboxylate symporter and a Na+/Pi cotransporter. However, while the genes of 673, AD45 and even AltDE are nearly identical (>90% similarity), the BS11 transporters were less than 40% similar to any of the others (Supplementary Figure 7). We found also BS11 specific differences in some of the genes involved in the Na+/H+ antiporters. Na+/H+ antiporters are membrane proteins that play a major role in pH and Na+ homeostasis in bacterial cells61. In 673, AD45 and AltDE we found five genes that code for three different antiporters, one for NhaA, one for NhaB and three that code for different versions of NhaC (possibly paralogs) with less than 40% similarity. BS11 lacks the most divergent (e.g., not found in E. coli) of the three NhaC genes. The absence of this specific paralogous NhaC only in BS11 might be associated to the lower salinity and therefore, less demanding Na+ extruding activity in the brackish environment of the Black Sea. Although the salinity growth range of BS11 is only slightly different from the other isolates (see above), there was a significant decrease in the NaCl requirements. Besides, the maximum growth rate exhibited in the laboratory is not always representative of the performance in the natural habitat.
We also found, in BS11, eleven genes coding for different families of glycosyl-hydrolases while only four and five such genes were found in the genomes of 673 and AD45 respectively. Nine of the eleven hydrolytic enzyme sequences in BS11 exhibits a predicted signal peptide which implies that these proteins may be involved in the processing of an extracellular biopolymer. In BS11 most of these genes are found together in the 40 Kb GI9 (Supplementary Figure 4) in which there were also two sugar transporters. In the same GI, several genes encoding proteins involved in the degradation of xylan were also present, like a gene coding for an α-L-arabinofuranosidase that is part of the hydrolytic system that carries out the hydrolysis62. Xylan is highly complex polysaccharide that is found in some algae. Growth experiments confirmed that BS11 is the only surface isolate degrading xylan.
Recruitment from metagenomes and metatranscriptomes
We have studied the recruitment of the A. macleodii genomes described here compared to all the published marine metagenomes. These include the water columns at the Bermuda Atlantic Time Series (BATS) station and at the HOT station31, which are representative of Northern central gyres of the Atlantic and Pacific Oceans and of very oligotrophic offshore waters. Also the different collections of the Global Ocean Survey (GOS)33 and the deep Marmara Sea metagenome (1000 m deep)32. BLASTN comparisons of the environmental reads against a local database constructed with the five genomes were performed and only the hits over a cut off 98% nucleotide identity in 90% of the length of the metagenomic read (see Materials and Methods) were considered as fragments recruited by the isolate genome. This high similarity threshold allowed us to identify the individual strains’ preferential recruitment from each environmental marine collection (Figure 6).To be able to compare the results, we normalized the numbers of hits against the genomes and the database sizes. Some representative genomes of marine bacteria were used also for comparison: Candidatus Pelagibacter ubique strains HTCC 1062 and HTCC 7211, Prochlorococcus marinus MIT9301 and Glaciecola sp. 4H-7+YE-5 (a close relative of A. macleodii). Yersinia pestis Z176003 and Escherichia coli K12-DH10B genomes were used as negative controls.
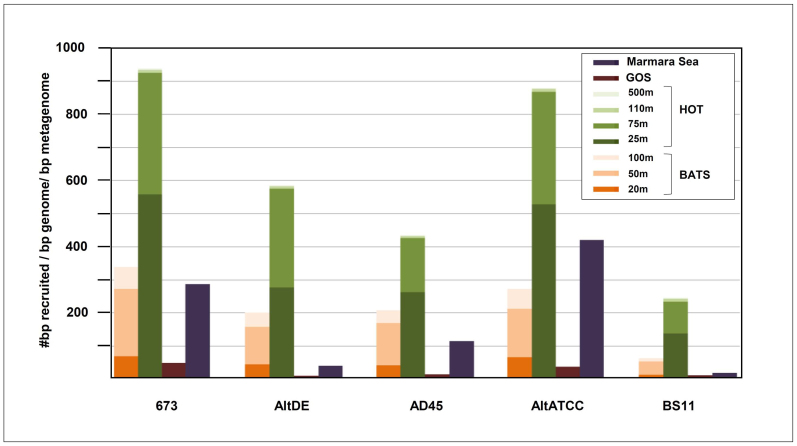
Figure 6. Recruitment of the genomes from reference metagenomes.
A. macleodii available genomes' relative recruitment of metagenomic reads at 98% identity and 90% coverage from some marine reference metagenomes (HOT, BATS GOS and Marmara).
Negative controls recruited very few hits confirming the specificity of the similarity threshold used (Supplementary Figure 8) while the marine genomes recruited abundantly. P. marinus MIT9301 recruited the most in the HOT dataset. The Candidatus P. ubique genomes recruited an order of magnitude less, probably because both come from much colder waters63. As expected10, 673 and AltATCC recruited especially well in the surface HOT samples (25 and 75 m deep), all depths combined each of these genomes recruited only about seven times less that P. marinus MIT9301 and nearly 10 times more than Candidatus P. ubique HTCC 7211. In slightly colder BATS waters (20°C compared to 25°C of the HOT station sample) A. macleodii seems to be less predominant. Both locations are oligotrophic with similar rates of primary production and carbon export, but BATS experiences stronger seasonal mixing and nutrient supply compared with HOT and at the sampling time (near the end of summer stratification) the surface layer in BATS was probably less productive.
In general, the Mediterranean strains (AltDE and AD45) and BS11 are less predominant in all the collections tested, reflecting perhaps some kind of endemism in such closed seas. However, the presence of AltATCC and 673 in the deep Marmara and also the presence of BS11 in the HOT sample indicate that geography is not the main factor of the unequal distribution of the strains. Although the Marmara sample comes from a 1000 m deep sample, the deep water mass of this sea is mainly fed by surface Mediterranean waters, compensating the surface outflow of lighter water from the Black sea32. These would explain why AltATCC, a surface isolate, and not the deep particle associated AltDE strain are the most abundant there. Although the total number of hits found in the GOS databases was larger than in the other datasets, the relative frequency decreased dramatically. This is because the majority of the GOS samples were recovered from waters that are too shallow (1–10 m), while A. macleodii appears to be more abundant in deeper waters (20–75 m)64.
We also compared our genomes against the DOM and deep sea water (DSW) metatranscriptomes10,65, both also from the HOT station. Again, in both metatranscriptomes, the 673, AltATCC and AD45 genomes recruited about double each than the BS11 or AltDE genomes (data not shown). As found before66 some genes from GIs (two hypothetical proteins found in 673, GI10 and GI7) are found among the more expressed genes.
Discussion
A. macleodii is a remarkably euryhaline and eurythermal bacterium, and its relatively large and complex genome reflects such an adaptable phenotype. All the available data indicate that A. macleodii is a typical r-strategist (investing most energy in multiplying fast) that takes advantage of its relatively large cell and genome size to exploit intensively localized (in time or space) nutrient rich micro-niches, such as when a bloom of phytoplankton occurs or when a nutrient rich particles become available. Its reliance on fast growth to compete efficiently might help explain its absence in waters where water temperature is low and prevents rapid growth. Obviously the large cells required to carry these large genomes cannot compete in normal conditions with the streamlined K-strategist of the Candidatus P. ubique type in the diluted, purely planktonic lifestyle. However, whenever a concentrated pool of organic matter is available, they can multiply more rapidly than any other heterotrophic prokaryote and reach high population densities as has been previously described11. One trait that is missing in the Alteromonadaceae is the potential to use light as energy source, by rhodopsins or bacteriochlorophyll dependent mechanisms, an extremely common feature in marine prokaryotes67. On the other hand, they seem to be highly specialized in the use of polymers and their degradation products68.
The comparison of the new genomes to the deep Mediterranean isolate AltDE, supports most of the previous conclusions6. The surface isolates have more genes involved in environmental sensing (for example histidine kinases) or chemotaxis, indicating a more demanding lifestyle in terms of the variability of their niche. Probably a significant part of their life cycle is spent free-living or commuting between particles compared to their larger-particle-associated counterparts. They also have more metabolic diversity in terms of degradation of organic compounds, particularly the Black Sea isolate. Also, the metabolic pathways of the surface isolates seem more dependent on purely aerobic metabolism, lacking all the nitrate reductase clusters present in AltDE7. All surface isolates lack the CRISPR-Cas system, possibly making them more susceptible to phages, and putative lysogenic phages have been found in the surface strains (e.g., Mu-like phage in 673), while no positively identified prophage could be found in AltDE.
The isolates from surface of the Mediterranean and the English Channel are remarkably similar to the type strain isolated from Hawaii. This together with the recruitment at high threshold similarity from available metagenomes that cover many different marine habitats has shown that the surface isolates clade represented by the Hawaiian AltATCC, English Channel 673 and Mediterranean AD45 isolates are indeed ubiquitous in temperate-tropical waters. They are more abundant at medium photic zone depths (25–100 m) as previously described64 which explains why they appear less prevalent in surface dominated datasets like the GOS. The unexpected finding of an AltDE- CRISPR protospacer69 in a putative lysogenic phage in the genome of 673 indicates the sharing of phages and hence potential genetic exchange between the two most divergent clades of A. macleodii, not to mention the wide distribution of this phage geographically speaking. Overall, our data are strongly against this microbe showing any geographic distribution pattern, other than the temperature associated limitations i.e. absence of all the strains we have studied in cold regions.
As seems to be the rule when comparing genomes of closely related Gram-negative strains some differential flexible genomic islands appeared related to surface-exposed structures42. The most typical being the cluster coding for glycosyltransferases and other enzymes involved in the synthesis of the O-chain of the LPS. This is probably related to this polysaccharide being the recognition target for many phages70. Interestingly a similar level of variation was found in the giant protein and the flagellum glycosylation Island what might be also related to their potential recognition by phages48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71. A very peculiar genomic island that was found to be different in all the isolates is the one apparently involved in metal resistance. It appears that strains acquire different czc gene cassettes (efflux pumps important for metal resistance) by successive insertions at a tRNA gene, with the numbers of czc cassettes varying from zero to three. This is clearly a highly variable region of the genome that also contained in AltDE the hydrogenase genes and at least one cluster of the general chemotaxis pathway proteins. The importance of this type of heavy metal efflux mechanisms for these microbes is elusive, but it is possible that the fast growth rate that they display under abundant nutrient supply, might increase the import of toxic products by the cell, such as heavy metals that are known to concentrate in marine particulate organic matter72.
As expected from previous MLSA work, and from the peculiar characteristics of its place of isolation, the Black Sea isolate (BS11) was markedly different from the other two surface isolates in overall sequence similarity and in differential gene content. Some differences appear to be associated to the lower salinity of this water body. Thus, one Na+/H+ antiporter found in 673, AD45 and even in AltDE was missing in BS11 which might reflect the fact that the lower salinity of the Black Sea requires less Na+ transporting capabilities. Also the sodium driven transporters found in BS11 were very different in sequence from the other A. macleodii isolates. In addition, phenotypically BS11 was the strain that could grow at the lowest salinity. All these data seem to indicate that BS11 is really adapted to a brackish environment like the Black Sea. However, the very strict recruitment analysis carried out here indicated the presence of some close relatives to this strain even in the central Pacific gyre. Therefore, the characteristics that we are identifying as Black Sea adaptation might just reflect a genotype that is better suited to grow at lower salinities regardless of the location.
Methods
Sample collection and sequencing
All the strains sequenced here come from surface waters. Details of isolation and origin are provided in Ivars-Martinez et al6. DNA was extracted by phenol-chloroform as described in Neumann et al17 and checked for quality on a 1% agarose gel. The quantity was measured using Quant-iT® PicoGreen® dsDNA Reagent (Invitrogen). The DNA of each strain was pyrosequenced in 1/4 run of the Roche 454 GS-FLX system (GATC, Konstanz, Germany) and additionally, around 1 Gb were obtained for each of the strains using the Illumina GAIIx technology (Macrogen, Korea). The coverage for each of the genomes, the amount of bases sequenced and other related details are in Supplementary Table 1. Low quality regions were completely clipped using sff_extract (by Jose Blanca). Two different programs were used in the assembly, Geneious Pro 5.0.1 (with default parameters (http://www.geneious.com)) and MIRA18. Both results were compared for equal assemblies. Oligonucleotides designed from the sequence of the ends of assembled contigs and PCR amplicons were generated and sequenced in order to concatenate them.
Growth assays
Growth at different temperatures (5, 8, 10, 40, 42, 43 and 45°C) and different salinities (0, 0.10, 0.20, 0.25, 0.50, 1, 2, 3.5, 4, 6, 8, 10, 12, 14, 15, 16, 18, 19 and 20%) was measured using a microplate optical density reader FLUOstar Optima from BMG LABTECH GmbH (Offenburg, Germany) at 595 nm always in duplicate. Marine broth was used as the basal medium. To investigate heavy metals resistance, the minimum inhibitory concentration for zinc acetate and mercury chloride were found by detecting growth visually in two-fold dilutions of the metals in marine broth. To measure maximum growth rate, cultures grown in Erlenmeyer flasks with orbital shaking (200 rpm) and incubated at 20°C. Growth was measured by the optical density at 595 nm.
Pulse field gel electrophoresis (PFGE)
To assess the presence of plasmids, PFGE was carried out with AltDE, AD45, 673 and BS11 strains. PFGE of the genomic DNA of the strains was performed in a contour-clamped homogeneous electric field (CHEF) system on a CHEF-DR III device (Bio-Rad Laboratories, Hercules, Calif.) with 1% agarose gels and modified 0.5x TBE buffer (45 mM Tris, 45 mM boric acid, 0.1 mM EDTA) at 14°C. Pulse time ramps and run times were varied to identify different plasmid conformations19,20. A lambda ladder PFGE marker (New England Biolabs) was used as a molecular size marker. After electrophoresis, the gels were stained with ethidium bromide (Sigma Co., St. Louis, Mo.), destained in distilled water, and photographed over an UV transilluminator.
Gene prediction, annotation and bioinformatics analysis
Gene prediction and Annotation of the new genomes and the formerly sequenced AltDE7 was carried out using the ISGA pipeline (http://isga.cgb.indiana.edu/). In addition, all predicted genes were compared to the NCBI nr protein database using BLASTP (e-value 10−5) and all hits were examined manually. ORFs smaller than 100 bp and without significant homology to other proteins were not considered. To confirm the presence of domains in the predicted proteins the hmmpfam program of the HMMER package21 (e-value 10-5) was used. BioEdit software was used to manipulate the sequences22. KEGG database was also used to analyze metabolic pathways23. tRNAs were identified using tRNAscan-SE24. GC content was calculated using the EMBOSS tool geecee25 and the GC-skew using the Oligoweb interface (http://insilico.ehu.es/oligoweb/). Reciprocal BLASTN and TBLASTX searches between the genomes were carried out, to identify of regions of similarity, insertions and rearrangements. Artemis v.926 and Artemis Comparison Tool ACTv.1227 were used for interactive visualization of genome comparisons. ANI (Average Nucleotide Identity) was calculated as defined in Ref.28 using a minimum cut-off of 50% identity and 70% of the length of the query gene. Sequences were aligned using MUSCLE version 3.629 and ClustalW30 and edited manually as necessary. The Venn-diagram in Figure 1 (panel A) was done with Venn Diagram Plotter (version March 289, 2010). Gene similarities are always provided as amino acid identities of the predicted peptides unless otherwise indicated. The maximum likelihood tree shown in Figure 1 (panel B) was created using an artificial concatenate of nucleotide sequences of 67 housekeeping genes using the PHYLIP package (version 3.69). The final consensus tree was created using MEGA (version 4.0.2). Values of 100 bootstrap replicates are indicated at branching points for those cases where they were > 50. The tree was rooted using Pseudoalteromonas atlantica T6c as outgroup. The genes used are listed in Supplementary Table 2.
Recruitments of environmental collections
Recruitment plots of the genomes were carried out against some available marine metagenomes31,32,33. BLASTN34 was carried out between a database formed by all the A. macleodii genomes (673, AltDE, AD45, AltATCC and BS11) and the environmental databases. A restrictive cut-off of 98% of identity in 90% of the length of the environmental read was established to guarantee that only similarities at the level of nearly identical microbes were counted. If a hit had the same score value for more than one genome, it was taken counted as a hit for both strains. Less stringent parameters were used to construct the plots of Figure 2 (second, third and fourth panels), where the cut-off used was 70% of identity in 50% of the length of the metagenomic read. The numbers of hits were normalized against the genomes and the database sizes. As controls, similar recruitment experiments were carried out for the marine bacteria: Candidatus Pelagibacter ubique strains HTCC1062 and HTCC7211, Prochlorococcus marinus MIT9301 and Glaciecola sp. 4H-7+YE-5 (a close relative of A. macleodii). Yersinia pestis Z176003 and Escherichia coli K12-DH10B genomes were used as negative controls. For the metatranscriptome recruitment of 673 strain in the DOM database10 showed in Figure 2 (first panel), we counted the number of bp recruited for each CDSs and normalized by the size of the gene.
Accession numbers
The sequences have been deposited in NCBI under the BioProject number PRJNA65407 for A. macleodii 673 PRJNA65405 for A. macleodii AD45 and PRJNA65401 for A. macleodii BS11.
Author Contributions
FRV conceived the work. MLP, ABMC, AGo and RG analyzed the data. Experiments were carried out by MLP, AGo, OO, AGh. FRV, MLP, and ABMC wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Nikole Kimes for helpful suggestions and for critically reading the manuscript. This work was supported by projects MAGYK (BIO2008-02444), MICROGEN (Programa CONSOLIDER-INGENIO 2010 CDS2009-00006), CGL2009-12651-C02-01 from the Spanish Ministerio de Ciencia e Inovación, DIMEGEN (PROMETEO/2010/089) and ACOMP/2009/155 from the Generalitat Valenciana. FEDER funds supported this project. ABMC was supported by CONSOLIDER-INGENIO 2010 CDS2009-00006. RG was supported by a Juan de la Cierva scholarship from the Spanish Ministerio de Ciencia e Inovación.
References
- Baumann L., Baumann P., Mandel M. & Allen R. D. Taxonomy of aerobic marine eubacteria. J Bacteriol 110, 402–429 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd M. M., Tang J., Kane M. & Emerson D. Captured diversity in a culture collection: case study of the geographic and habitat distributions of environmental isolates held at the american type culture collection. Appl Environ Microbiol 71, 2813–2823 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukall R. et al. Sulfitobacter mediterraneus sp. nov., a new sulfite-oxidizing member of the alpha-Proteobacteria. Int J Syst Bacteriol 49 Pt 2, 513–519 (1999). [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez J., Acinas S. G., Massana R. & Rodriguez-Valera F. Prevalence and microdiversity of Alteromonas macleodii-like microorganisms in different oceanic regions. Environ Microbiol 4, 42–50 (2002). [DOI] [PubMed] [Google Scholar]
- Schafer H., Servais P. & Muyzer G. Successional changes in the genetic diversity of a marine bacterial assemblage during confinement. Arch Microbiol 173, 138–145 (2000). [DOI] [PubMed] [Google Scholar]
- Ivars-Martinez E. et al. Biogeography of the ubiquitous marine bacterium Alteromonas macleodii determined by multi-locus sequence analysis (MLSA). Molecular Ecology, 17, 4092–4106 (2008). [DOI] [PubMed] [Google Scholar]
- Ivars-Martinez E. et al. Comparative genomics of two ecotypes of the marine planktonic copiotroph Alteromonas macleodii suggests alternative lifestyles associated with different kinds of particulate organic matter. Isme J 2, 1194–1212 (2008). [DOI] [PubMed] [Google Scholar]
- Lauro F. M. & Bartlett D. H. Prokaryotic lifestyles in deep sea habitats. Extremophiles (2007). [DOI] [PubMed] [Google Scholar]
- Lauro F. M., Chastain R. A., Blankenship L. E., Yayanos A. A. & Bartlett D. H. The unique 16S rRNA genes of piezophiles reflect both phylogeny and adaptation. Appl Environ Microbiol 73, 838–845 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarren J. et al. Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc Natl Acad Sci U S A 107, 16420–16427 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., McCarren J. & DeLong E. F. Transcriptional responses of surface water marine microbial assemblages to deep-sea water amendment. Environ Microbiol 14, 191–206 (2012). [DOI] [PubMed] [Google Scholar]
- Romera-Castillo C., Sarmento H., Alvarez-Salgado X. A., Gasol J. M. & Marrase C. Net production and consumption of fluorescent colored dissolved organic matter by natural bacterial assemblages growing on marine phytoplankton exudates. Appl Environ Microbiol 77, 7490–7498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y. et al. Differing growth responses of major phylogenetic groups of marine bacteria to natural phytoplankton blooms in the western North Pacific Ocean. Appl Environ Microbiol 77, 4055–4065 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemb O., West N., Bourrain M., Godon J. J. & Lebaron P. Effect of a transient perturbation on marine bacterial communities with contrasting history. J Appl Microbiol 109, 751–762 (2010). [DOI] [PubMed] [Google Scholar]
- Pujalte M. J., Sitja-Bobadilla A., Alvarez-Pellitero P. & Garay E. Carriage of potencially fish -pathogenic bacteria in sparus aurata cultured inMediterranean fish farms. Dis Aquat Organ 54, 119–126 (2003). [DOI] [PubMed] [Google Scholar]
- Southward A. J. et al. Long-term oceanographic and ecological research in the Western English Channel. Adv Mar Biol 47, 1–105 (2005). [DOI] [PubMed] [Google Scholar]
- Neumann B., Pospiech A. & Schairer H. U. Rapid isolation of genomic DNA from gram-negative bacteria. Trends Genet 8, 332–333 (1992). [DOI] [PubMed] [Google Scholar]
- Chevreux B., Wetter T. & Suhai S. Genome Sequence Assembly Using Trace Signals and Additional Sequence Information Computer Science and Biology: Proceedings of the German Conference on Bioinformatics (GCB). 99, 45–56 (1999).
- Hightower R. C., Metge D. W. & Santi D. V. Plasmid migration using orthogonal-field-alternation gel electrophoresis. Nucleic Acids Res 15, 8387–8398 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew M. K., Hui C. F., Smith C. L. & Cantor C. R. High-resolution separation and accurate size determination in pulsed-field gel electrophoresis of DNA. 4. Influence of DNA topology. Biochemistry 27, 9222–9226 (1988). [DOI] [PubMed] [Google Scholar]
- Eddy S. R. A probabilistic model of local sequence alignment that simplifies statistical significance estimation. PLoS Comput Biol 4, e1000069 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser 41, 95–98 (1999). [Google Scholar]
- Kanehisa M., Goto S., Kawashima S., Okuno Y. & Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res 32(Database issue), D277–280 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. M. & Eddy S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25, 955–964 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P., Longden I. & Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16, 276–277 (2000). [DOI] [PubMed] [Google Scholar]
- Rutherford K. et al. Artemis: sequence visualization and annotation. Bioinformatics 16, 944–945 (2000). [DOI] [PubMed] [Google Scholar]
- Carver T. J. et al. ACT: the Artemis Comparison Tool. Bioinformatics 21, 3422–3423 (2005). [DOI] [PubMed] [Google Scholar]
- Konstantinidis K. T. & Tiedje J. M. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A 102, 2567–2572 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G. & Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22, 4673–4680 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman M. L. & Chisholm S. W. Ecosystem-specific selection pressures revealed through comparative population genomics. Proc Natl Acad Sci U S A 107, 18634–18639 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaiser A., Zivanovic Y., Moreira D. & Lopez-Garcia P. Comparative metagenomics of bathypelagic plankton and bottom sediment from the Sea of Marmara. ISME J 5, 285–304 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch D. B. et al. The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol 5, e77 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye J. Z., Sylvan J. B., Edwards K. J. & Baross J. A. Halomonas and Marinobacter ecotypes from hydrothermal vent, subseafloor and deep-sea environments. FEMS Microbiol Ecol 75, 123–133 (2011). [DOI] [PubMed] [Google Scholar]
- Konstantinidis K. T. & Tiedje J. M. Towards a genome-based taxonomy for prokaryotes. J Bacteriol 187, 6258–6264 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H. M. et al. Complete genome sequence of the polycyclic aromatic hydrocarbon-degrading bacterium Alteromonas sp. strain SN2. J. Bacteriol 193, 4292–4293 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasic L. et al. Metagenomic islands of hyperhalophiles: the case of Salinibacter ruber. BMC Genomics 10, 570 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman M. L. et al. Genomic islands and the ecology and evolution of Prochlorococcus. Science 311, 1768–1770 (2006). [DOI] [PubMed] [Google Scholar]
- Cuadros-Orellana S. et al. Genomic plasticity in prokaryotes: the case of the square haloarchaeon. Isme J 1, 235–245 (2007). [DOI] [PubMed] [Google Scholar]
- Legault B. A. et al. Environmental genomics of Haloquadratum walsbyi in a saltern crystallizer indicates a large pool of accessory genes in an otherwise coherent species. BMC Genomics 7, 171 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Valera F. et al. Explaining microbial population genomics through phage predation. Nat Rev Microbiol 7, 828–836 (2009). [DOI] [PubMed] [Google Scholar]
- Avrani S., Wurtzel O., Sharon I., Sorek R. & Lindell D. Genomic island variability facilitates Prochlorococcus-virus coexistence. Nature 474, 604–608 (2011). [DOI] [PubMed] [Google Scholar]
- Samuel G. & Reeves P. Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr Res 338, 2503–2519 (2003). [DOI] [PubMed] [Google Scholar]
- Arora S., Bangera M., Lory S. & Ramphal R. A. genomic island in Pseudomonas aeruginosa carries the determinants of flagellin glycosylation. Proc Natl Acad Sci U S A 98: 9342–9347 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reva O. & Tummler B. Think big--giant genes in bacteria. Environ Microbiol 10, 768–777 (2008). [DOI] [PubMed] [Google Scholar]
- Fraser J. S., Yu Z., Maxwell K. L. & Davidson A. R. Ig-like domains on bacteriophages: a tale of promiscuity and deceit. J Mol Biol 359, 496–507 (2006). [DOI] [PubMed] [Google Scholar]
- Strom S. L., Brahamsha B., Fredrickson K. A., Apple J. K. & Rodriguez A. G. A giant cell surface protein in Synechococcus WH8102 inhibits feeding by a dinoflagellate predator. Environ Microbiol (2011). [DOI] [PubMed] [Google Scholar]
- Cook M. A., Osborn A. M., Bettandorff J. & Sobecky P. A. Endogenous isolation of replicon probes for assessing plasmid ecology of marine sediment microbial communities. Microbiology 147(Pt 8), 2089–2101 (2001). [DOI] [PubMed] [Google Scholar]
- Hazen T. H., Dongying W., Eisen J. A. & Sobecky P. A. Sequence characterization and comparative analysis of three plasmids isolated from environmental Vibrio spp. Appl. Environ. Microbiol. 73, 7703–7710 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H. Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas? ISME J 2, 579–589 (2008). [DOI] [PubMed] [Google Scholar]
- Barrangou R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007). [DOI] [PubMed] [Google Scholar]
- Deveau H., Garneau J. E. & Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol 64, 475–493 (2010). [DOI] [PubMed] [Google Scholar]
- Garneau J. E. et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71 (2010). [DOI] [PubMed] [Google Scholar]
- Weyman P. D. et al. Heterologous expression of Alteromonas macleodii and Thiocapsa roseopersicina [NiFe] hydrogenases in Synechococcus elongatus. PLoS One 6, e20126 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyman P. D., Smith H. O. & Xu Q. Genetic analysis of the Alteromonas macleodii [NiFe]-hydrogenase. FEMS Microbiol Lett. 322, 180–187 (2011). [DOI] [PubMed]
- Weyman P. D. et al. Heterologous expression of Alteromonas macleodii and Thiocapsa roseopersicina [NiFe] hydrogenases in Escherichia coli. Microbiology 157(Pt 5), 1363–1374 (2011). [DOI] [PubMed] [Google Scholar]
- Hacker J. & Kaper J. B. Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol 54, 641–679 (2000). [DOI] [PubMed] [Google Scholar]
- Cooksey D. A. Molecular mechanisms of copper resistance and accumulation in bacteria. FEMS Microbiol Rev 14, 381–386 (1994). [DOI] [PubMed] [Google Scholar]
- Nies D. H. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev 27, 313–339 (2003). [DOI] [PubMed] [Google Scholar]
- Padan E. & Schuldiner S. Na+/H+ antiporters, molecular devices that couple the Na+ and H+ circulation in cells. J. Bioenerg. Biomembr. 25, 647–669 (1993). [DOI] [PubMed] [Google Scholar]
- Rahman A. K., Sugitani N., Hatsu M. & Takamizawa K. A role of xylanase, alpha-L-arabinofuranosidase, and xylosidase in xylan degradation. Can J Microbiol 49, 58–64 (2003). [DOI] [PubMed] [Google Scholar]
- Giovannoni S. J. et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309, 1242–1245 (2005). [DOI] [PubMed] [Google Scholar]
- Acinas S. G., Anton J. & Rodriguez-Valera F. Diversity of free-living and attached bacteria in offshore Western Mediterranean waters as depicted by analysis of genes encoding 16S rRNA. Appl Environ Microbiol 65, 514–522 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarren P., Hall M. L. & Whitehead L. The Chemical Tuning of a Weak Zinc Binding Motif for Histone Deacetylase Using Electronic Effects. Chem Biol Drug Des. (2012). [DOI] [PubMed] [Google Scholar]
- Frias-Lopez J. et al. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci U S A 105, 3805–3810 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton R. J. et al. Genome characteristics of a generalist marine bacterial lineage. ISME J 4, 784–798 (2010). [DOI] [PubMed] [Google Scholar]
- Varbanets L. D. et al. [The Black Sea bacteria--producers of hydrolytic enzymes]. Mikrobiol Z 73, 9–15 (2011). [PubMed] [Google Scholar]
- Makarova K. S. et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9, 467–477 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz C. R. & Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem 71, 635–700 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskella B., Taylor T. B., Bates J. & Buckling A. Using experimental evolution to explore natural patterns between bacterial motility and resistance to bacteriophages. ISME J 5, 1809–17 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig P., Palanques A., Sanchez-Cabeza J. A. & Masqué P. Heavy metals in particulate matter and sediments in the southern Barcelona sedimentation system (North-western Mediterranean). Marine Chemistry 63, 311–329 (1999). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information