Abstract
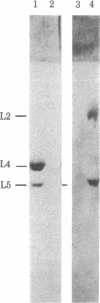
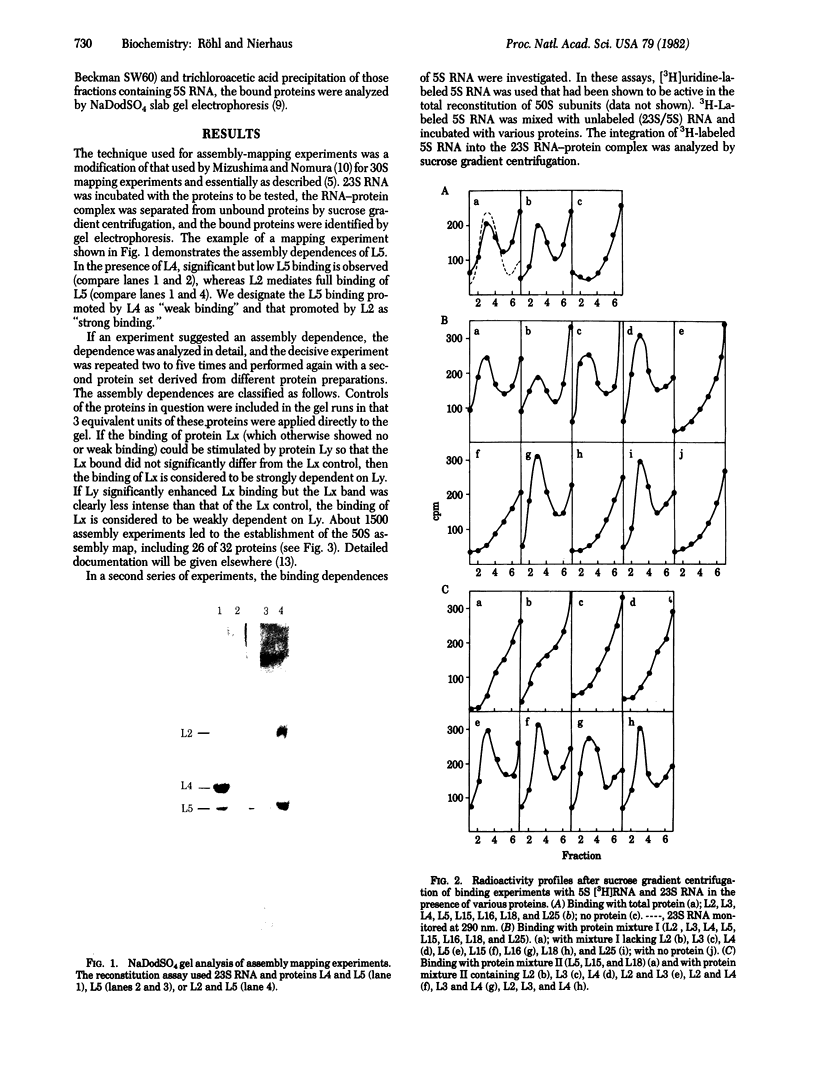
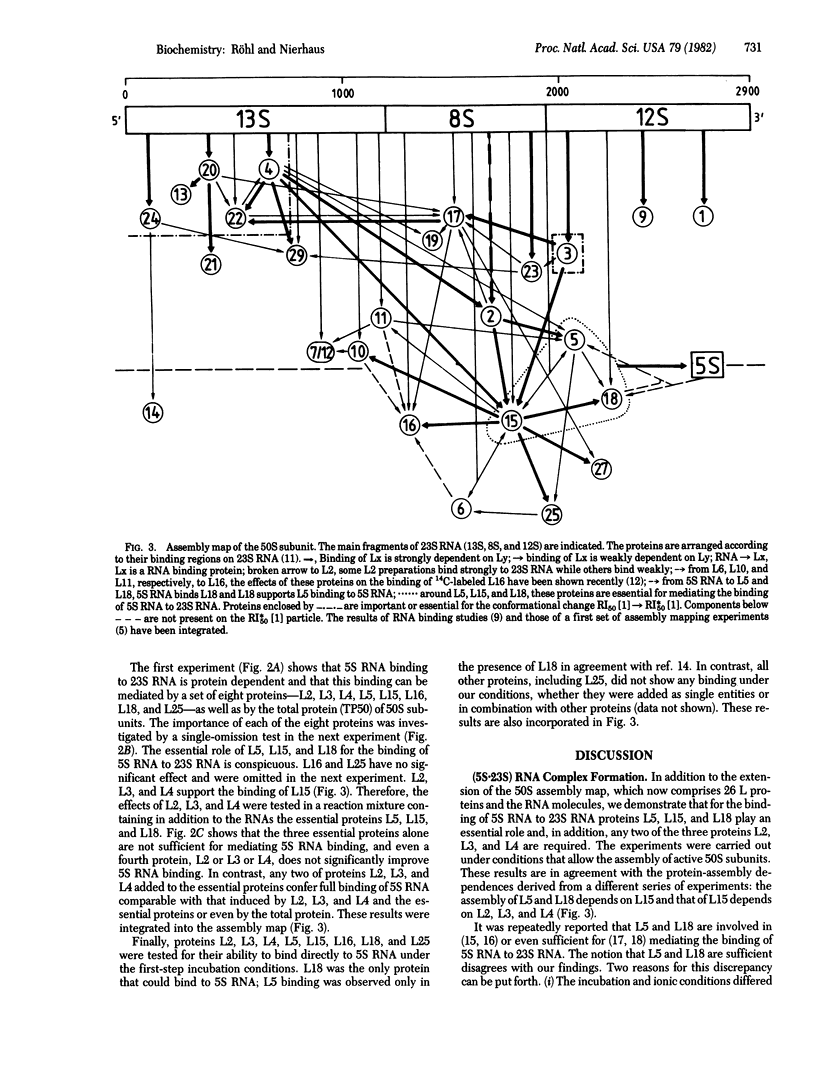
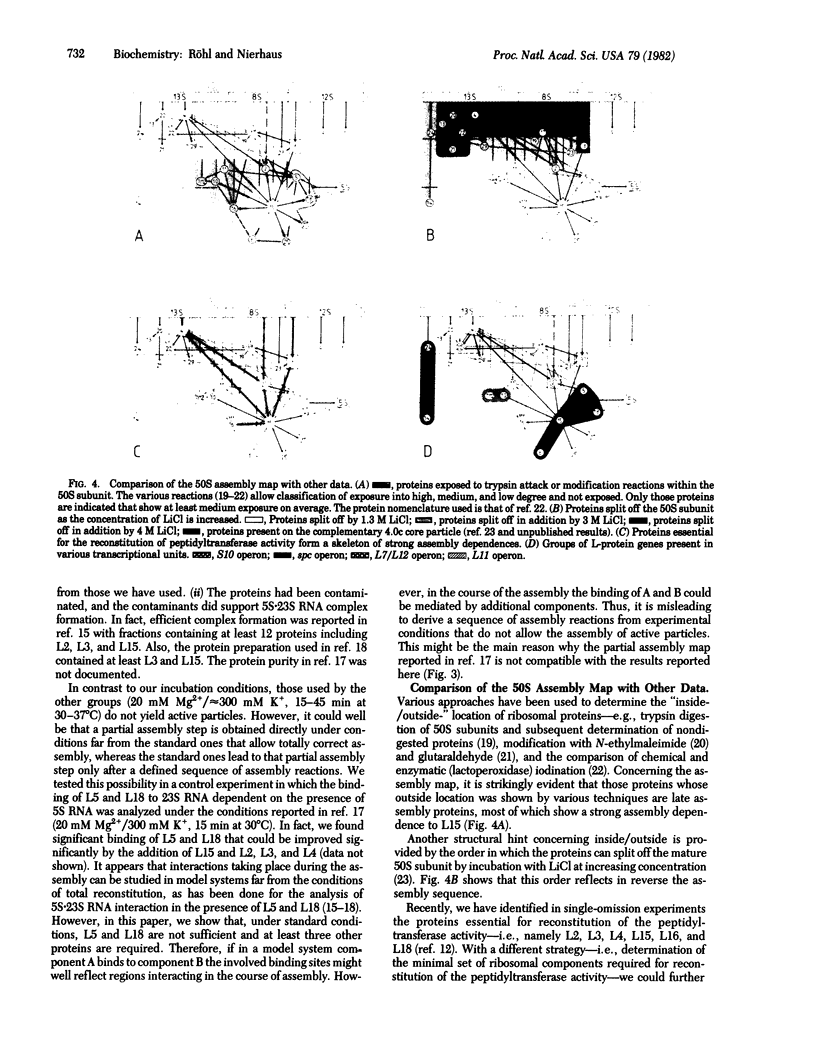
Stoichiometric amounts of ribosomal proteins and RNA derived from the 50S subunit reconstitute to fully active particles under the conditions of a two-step incubation procedure. After the first incubation, all components are found in a particle that is activated in the second incubation [Dohme, F. & Nierhaus, K. H. (1976) J. Mol. Biol. 107, 585-599]. Here we describe the assembly dependences of the ribosomal components in the first incubation. Assembly dependence is the requirements of one protein that, before it binds, another must be first built into the ribosome. After incubation of 23S RNA and the proteins under observation, the mixture was subjected to sucrose gradient analysis. The RNA-protein complex was precipitated with trichloroacetic acid and the proteins were identified by NaDodSO4 gel electrophoresis. The assembly dependences of 26 proteins could be elucidated. In a second series of experiments, the incorporation of 3H-labeled 5S RNA in the 23S-protein complex was analyzed. It was found that L5, L15, and L18 are absolutely required for 5S RNA incorporation. In addition, two of the three proteins L2, L3, and L4 are needed, in excellent agreement with the protein dependences. The data are summarized in an assembly map. Comparison with other data shows a structural domain at the 5' end of 23S RNA around protein L20 combining all proteins essential in the early assembly. All the proteins essential for the reconstitution of the peptidyltransferase protein form a skeleton of strong assembly dependences. Finally, L proteins whose genes are present in large transcriptional units on the chromosome depend on each other during assembly.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Crichton R. R., Wittmann H. G. Ribosomal proteins. XXIV. Trypsin digestion as a possible probe of the conformation of Escherichia coli ribosomes. Mol Gen Genet. 1972;114(2):95–105. doi: 10.1007/BF00332780. [DOI] [PubMed] [Google Scholar]
- Dean D., Yates J. L., Nomura M. Escherichia coli ribosomal protein S8 feedback regulates part of spc operon. Nature. 1981 Jan 1;289(5793):89–91. doi: 10.1038/289089a0. [DOI] [PubMed] [Google Scholar]
- Dohme F., Nierhaus K. H. Role of 5S RNA in assembly and function of the 50S subunit from Escherichia coli. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2221–2225. doi: 10.1073/pnas.73.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohme F., Nierhaus K. H. Total reconstitution and assembly of 50 S subunits from Escherichia coli Ribosomes in vitro. J Mol Biol. 1976 Nov 15;107(4):585–599. doi: 10.1016/s0022-2836(76)80085-x. [DOI] [PubMed] [Google Scholar]
- Fuenteun J., Monier R., Garrett R., Le Bret M., Le Pecq J. B. Effect of 50 S subunit proteins L5, L18 and L25 on the fluorescence of 5 S RNA-bound ethidium bromide. J Mol Biol. 1975 Apr 25;93(4):535–541. doi: 10.1016/0022-2836(75)90245-4. [DOI] [PubMed] [Google Scholar]
- Gray P. N., Garrett R. A., Stoffler G., Monier R. An attempt at the identification of the proteins involved in the incorporation of 5-S RNA during 50-S ribosomal subunit assembly. Eur J Biochem. 1972 Jul 24;28(3):412–421. doi: 10.1111/j.1432-1033.1972.tb01927.x. [DOI] [PubMed] [Google Scholar]
- Hampl H., Schulze H., Nierhaus K. H. Ribosomal components from Escherichia coli 50 S subunits involved in the reconstitution of peptidyltransferase activity. J Biol Chem. 1981 Mar 10;256(5):2284–2288. [PubMed] [Google Scholar]
- Homann H. E., Nierhaus K. H. Ribosomal proteins. Protein compositions of biosynthetic precursors and artifical subparticles from ribosomal subunits in Escherichia coli K 12. Eur J Biochem. 1971 May 28;20(2):249–257. doi: 10.1111/j.1432-1033.1971.tb01388.x. [DOI] [PubMed] [Google Scholar]
- Isono K., Kitakawa M. Cluster of ribosomal protein genes in Escherichia coli containing genes for proteins S6, S18, and L9. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6163–6167. doi: 10.1073/pnas.75.12.6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahan L., Kaltschmidt E. Glutaraldehyde reactivity of the proteins of Escherichia coli ribosomes. Biochemistry. 1972 Jul 4;11(14):2691–2698. doi: 10.1021/bi00764a022. [DOI] [PubMed] [Google Scholar]
- Litman D. J., Cantor C. R. Surface topography of the Escherichia coli ribosome enzymatic iodination of the 50S subunit. Biochemistry. 1974 Jan 29;13(3):512–518. doi: 10.1021/bi00700a017. [DOI] [PubMed] [Google Scholar]
- Marquardt O., Roth H. E., Wystup G., Nierhaus K. H. Binding of Escherichia coli ribosomal proteins to 23S RNA under reconstitution conditions for the 50S subunit. Nucleic Acids Res. 1979 Aug 10;6(11):3641–3650. doi: 10.1093/nar/6.11.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima S., Nomura M. Assembly mapping of 30S ribosomal proteins from E. coli. Nature. 1970 Jun 27;226(5252):1214–1214. doi: 10.1038/2261214a0. [DOI] [PubMed] [Google Scholar]
- Moore P. B. Reaction of N-ethyl maleimide with the ribosomes of Escherichia coli. J Mol Biol. 1971 Aug 28;60(1):169–184. doi: 10.1016/0022-2836(71)90456-6. [DOI] [PubMed] [Google Scholar]
- Moore P. B. The preparation of deuterated ribosomal materials for neutron scattering. Methods Enzymol. 1979;59:639–655. doi: 10.1016/0076-6879(79)59119-8. [DOI] [PubMed] [Google Scholar]
- Newberry V., Garrett R. A. The role of the basic N-terminal region of protein L18 in 5S RNA-23S RNA complex formation. Nucleic Acids Res. 1980 Sep 25;8(18):4131–4142. doi: 10.1093/nar/8.18.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierhaus K. H., Dohme F. Total reconstitution of 50 S subunits from Escherichia coli ribosomes. Methods Enzymol. 1979;59:443–449. doi: 10.1016/0076-6879(79)59106-x. [DOI] [PubMed] [Google Scholar]
- Prakash V., Aune K. C. Molecular interactions between ribosomal proteins: a study of the S6-S18 interaction. Arch Biochem Biophys. 1978 Apr 30;187(2):399–405. doi: 10.1016/0003-9861(78)90050-4. [DOI] [PubMed] [Google Scholar]
- Roth H. E., Nierhaus K. H. Assembly map of the 50-S subunit from Escherichia coli ribosomes, covering the proteins present in the first reconstitution intermediate particle. Eur J Biochem. 1980 Jan;103(1):95–98. doi: 10.1111/j.1432-1033.1980.tb04292.x. [DOI] [PubMed] [Google Scholar]
- Sieber G., Nierhaus K. H. Kinetic and thermodynamic parameters of the assembly in vitro of the large subunit from Escherichia coli ribosomes. Biochemistry. 1978 Aug 22;17(17):3505–3511. doi: 10.1021/bi00610a013. [DOI] [PubMed] [Google Scholar]
- Spierer P., Wang C. C., Marsh T. L., Zimmermann R. A. Cooperative interactions among protein and RNA components of the 50S ribosomal subunit of Escherichia coli. Nucleic Acids Res. 1979 Apr;6(4):1669–1682. doi: 10.1093/nar/6.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spierer P., Zimmermann R. A. Stoichiometry, cooperativity, and stability of interactions between 5S RNA and proteins L5, L18, and L25 from the 50S ribosomal subunit of Escherichia coli. Biochemistry. 1978 Jun 27;17(13):2474–2479. doi: 10.1021/bi00606a002. [DOI] [PubMed] [Google Scholar]
- Wystup G., Teraoka H., Schulze H., Hampl H., Nierhaus K. H. 50-S subunit from Escherichia coli ribosomes. Isolation of active ribosomal proteins and protein complexes. Eur J Biochem. 1979 Oct;100(1):101–113. doi: 10.1111/j.1432-1033.1979.tb02038.x. [DOI] [PubMed] [Google Scholar]