Abstract
Collapsin response mediator protein 4 (CRMP4) is a molecular marker for immature neurons but only limited information is available on the spatiotemporal gene expression changes of Crmp4 in the developing rodent. In the present study, the variation of CRMP4 mRNA expression in the mouse brain was investigated from postnatal day (PD) 0 (the day of birth) to adulthood by in situ hybridization. The hybridization signals were broadly detected on PD0 and regional changes in expression during development were noted. Expression patterns of CRMP4 mRNA were classified into the following three types: (i) signals that were strongest on PD0 or PD7, weak or undetectable on PD14, and absent in adulthood: this pattern was observed in most brain areas; (ii) signals that were first detected on PD0 or PD7 and persisted into adulthood: this pattern was seen in the dentate gyrus and subventricular zone of the olfactory bulb (OB); and (iii) signals that were strongest on PD0 and decreased gradually with age but were still detectable in adulthood: this pattern was identified for the first time in the mitral cell layer of the OB. Analysis using quantitative real-time RT-PCR confirmed higher expression of CRMP4 mRNA in the OB than in other adult brain regions. The persistence of CRMP4 mRNA in the adult OB, including the mitral cell layer, suggests the possibility of both neurogenetic and non-neurogenetic functional roles of CRMP4 in this region.
Keywords: brain, collapsin response mediator protein 4, gene expression, in situ hybridization, mitral cell layer, mouse, olfactory bulb, postnatal development
Introduction
The development of a complex nervous system relies on the proliferation of neuronal precursor cells and the differentiation, migration, and extension of axons to appropriate synaptic targets. Many extra- and intracellular proteins contributing to this development have been discovered to date. For example, collapsin, a member of the semaphorin family, is involved in axonal pathfinding during neural development by inhibiting growth-cone extension. Collapsin response mediator protein (CRMP)-62, now known as CRMP2, was identified as the molecule required for the signal transduction of collapsin (Goshima et al. 1995). Five members of CRMPs (CRMP1–CRMP5) were subsequently identified in rats and mice (CRMP1–CRMP4; Wang & Strittmatter, 1996; CRMP5; Fukada et al. 2000; Inatome et al. 2000).
Recent studies indicate that CRMPs mediate signal transduction pathways related to the development of not only axons, but also of dendrites and spines. For example, CRMP1 regulates spine development through the phosphorylation of cyclin-dependent kinase 5 (Yamashita et al. 2007). CRMP2 regulates axon growth through phosphorylation by glycogen synthase kinase-3β (GSK-3β) and dendritic field organization (Cole et al. 2004; Uchida et al. 2005; Fang et al. 2011; Yamashita & Goshima, 2012). CRMP3 is required for dendritic organization in the hippocampal CA1 region as well as long-term potentiation in this area (Quach et al. 2008). CRMP4 regulates myelin-dependent axon outgrowth through the interaction with GSK-3β as well as dendrite bifurcation of hippocampal pyramidal neurons (Alabed et al. 2007, 2010; Niisato et al. 2012). CRMP5 is involved in the regulation of dendritic development in the cerebellar Purkinje cells through BDNF-TrkB signaling (Yamashita et al. 2011). All of the above-mentioned studies indicate the critical role of CRMPs in the developmental organization of normal neural networks.
Despite the significant role played by CRMPs during neural network formation, only a few studies have reported the spatiotemporal variation of CRMP expression over the whole brain (for CRMP3, Quach et al. 2000; for CRMP5 in the forebrain, McLaughlin et al. 2008). Instead, most studies on CRMPs have focused on specific neurons in distinct areas, such as Purkinje cells in the cerebellum, hippocampal CA1 pyramidal neurons or neurons in the cerebral cortex (Yoshimura et al. 2005; Ip et al. 2011; Yamashita et al. 2011; Niisato et al. 2012). Despite the lack of comprehensive information about variation in the regional distribution of CRMP4 over time, CRMP4 has traditionally been used as an immature neuron marker (Quinn et al. 1999; Seki, 2002). However, CRMP4 immunoreactivity has been demonstrated in the differentiated neurons of various regions of the adult rat brain (Nacher et al. 2000). Therefore, in the present study, to provide more detailed information on the spatiotemporal expression changes of CRMP4 mRNA, we examined the gene expression of Crmp4 in the whole brain from PD0 to adulthood by using in situ hybridization.
Materials and methods
Animals
Pregnant and juvenile mice (C57BL/6N) were purchased from Charles River, Japan. All experiments were conducted according to the Guidelines for the Care and Use of Experimental Animals of the University of Tokyo. On the day of birth (postnatal day 0, PD0), PD7, PD14, and 8 weeks after birth (adult), two or three male mice at each age were deeply anesthetized with pentobarbital and transcardially perfused with saline followed by 4% paraformaldehyde in 0.1 m phosphate buffer (PB, pH 7.3). After the brains were quickly removed and further fixed with 4% paraformaldehyde in PB overnight at 4 °C, they were rinsed with phosphate-buffered saline (PBS); passed through RNase-free graded sucrose solutions of 10, 20, and 30% in PB for 8–12 h (for each step) at 4 °C; and frozen in optimal cutting temperature (OCT) compound (Sakura, Tokyo, Japan). For quantitative real-time RT-PCR, 8-week-old male C57BL/6N mice (n = 4) were deeply anesthetized with pentobarbital. Immediately after cervical dislocation, their brains were removed. The olfactory bulb (OB), hippocampus, and cerebellum were immediately cut from the brain under a stereomicroscope and soaked separately in RNAlater (Ambion, Austin, TX, USA). The tissue pieces were homogenized and stored at −80 °C.
Probe generation
The cDNA containing 1792 bp of Rattus norvegicus CRMP4 (GenBank: AF389425.1) was generously donated by Prof. Y. Goshima (Yokohama City University Graduate School of Medicine, Yokohama, Japan). One microgram of cDNA was incubated overnight with XbaI (Roche Diagnostics, Tokyo, Japan) or HindIII (Roche Diagnostics) in 1 × SuRE/Cut Buffer H (Roche Diagnostics) at 37 °C to prepare template DNA for generation of antisense probe or sense probe. To generate an antisense or sense probe labeled with digoxigenin, the template DNA was incubated for 2 h at 37 °C with T7 (for antisense probe) or T3 (for sense probe) RNA polymerase (Roche Diagnostics) in 20 μL transcription mixture containing 1 μg template DNA, 2 μL transcription buffer (10× ), 2 μL DIG RNA labeling mix (Roche Diagnostics), 0.5 μL ribonuclease inhibitor (Toyobo, Tokyo, Japan), and 2 μL appropriate RNA polymerase. The reaction was stopped by heating for 15 min at 65 °C. Then, 2.5 μL LiCl (4 m), 2 μL EDTA (0.2 m, pH 8.0), and 75 μL absolute EtOH were added to the solution and incubated overnight at −20 °C. After centrifugation and removal of the supernatant, the pellet was dissolved in 50% formamide and stored at −20 °C.
In situ hybridization
Serial frontal cryosections (20-μm-thick) of the brain from the OB to the medulla oblongata were cut using a cryostat (CM-3050-S, Leica Microsystems) and thaw-mounted on MAS-coated glass slides (Matsunami, Osaka, Japan). A series of every 5th section of each brain from two or three males at each stage (PD0, PD7, PD14, and adult) was stained with cresyl violet (Muto, Tokyo, Japan) to determine the cytoarchitectonics of neuroanatomical structures. Another series of every 5th section was used for in situ hybridization to detect the expression of CRMP4 mRNA. After endogenous alkaline phosphatase (AP) activity was quenched with 0.2 m HCl, the sections were postfixed with 4% paraformaldehyde. Sections were then washed with PBS, treated with 1 μg mL−1 proteinase K (Promega, Tokyo, Japan) at 37 °C, and soaked again in 4% paraformaldehyde. After several washes, the sections were acetylated with 0.25% (v/v) acetic anhydride (Wako, Osaka, Japan) in 0.1 m triethanolamine (Sigma Aldrich, Tokyo, Japan). After washing with PBS, the sections were incubated with prehybridization solution [50% formamide, 3× saline sodium citrate (SSC), 0.12 m PB pH 7.4, 1× Denhardt solution (Sigma Aldrich), 120 mg mL−1 tRNA (Sigma Aldrich), 0.1 mg mL−1 calf thymus sperm DNA (Invitrogen, Tokyo, Japan), 10% dextran sulfate (Wako)] for 2 h at 50 °C. After denaturation of the DIG-labeled RNA probe in prehybridization buffer (0.1 mg mL−1) for 3 min at 85 °C, the solution was applied to the sections and incubated overnight at 50 °C. The sections were washed with 2× SSC containing 50% formamide at 50 °C, immersed with TNE (10 mm Tris-HCl, 0.5 m NaCl, and 1 mm EDTA) for 10 min at 37 °C and then processed using 20 μg mL−1 RNaseA (Sigma Aldrich) in TNE for 30 min at 37 °C. The sections were soaked again in TNE followed by washing with 2× SSC and 0.5× SSC at 50 °C. After a rinse in DIG-1 buffer (100 mm Tris-HCl, 150 mm NaCl, and 0.1% Tween 20), the sections were immersed in 1.5% blocking reagent (Roche Molecular Biochemicals, Mannheim, Germany) in DIG-1 buffer. After washing with DIG-1 buffer, the sections were incubated with an AP-conjugated anti-DIG antibody (1 : 1000; Roche Diagnostics). The sections were then washed with DIG-1 buffer, treated with DIG-3 buffer [100 mm Tris-HCl (pH 9.5), 100 mm NaCl, and 50 mm MgCl2], and treated with a chromogen solution (0.3375 mg mL−1 4-nitroblue tetrazolium chloride, 0.175 mg mL−1 5-bromo-4-chloro-3-indoyl-phosphate in DIG-3 buffer) until a visible signal was detected. The reaction was stopped by adding a reaction stop solution [10 mm Tris-HCl (pH 8.0) and 1 mm EDTA (pH 8.0)]. Sequentially, sections were dehydrated, embedded with Entellan New (Merck, Tokyo, Japan), and observed under a microscope (AXIO Imager A1; Zeiss, Jena, Germany).
Image analysis after in situ hybridization
Qualitative analysis of labeled images was performed on both hybridized sections, and the adjacent sections were stained with cresyl violet for optimal anatomical resolution according to a reference atlas for the developing mouse brain (Atlas of the Developing Mouse Brain). The relative CRMP4 mRNA labeling intensities were assessed for each brain area and visually scored on a qualitative scale of − (no staining detected), +/− (very weak), + (weak), ++ (moderate), and +++ (strong) according to the intensity of the hybridization signals.
Mirror-image sections subjected to immunohistochemical and in situ hybridization
Coronal mirror-image frozen sections (20-μm-thick) of the OB were prepared from another adult mouse (8 weeks after birth). The pairs of these sections were stained either by in situ hybridization as stated above or by immunohistochemical staining for polysialylated neural-cell-adhesion molecule (PSA-NCAM). For the latter staining, the sections were incubated with mouse monoclonal anti-PSA-NCAM antibody (Millipore; ×400) as the primary antibody and reacted with Alexa Fluor 488-conjugated anti-mouse IgG (Molecular Probes, Eugene, OR; ×200) as the secondary antibody. The nuclei were labeled with Hoechst 33258 (Wako).
Quantitative real-time RT-PCR
Total RNA was extracted from adult tissue pieces immersed in RNAlater by RNeasy kit (Qiagen, Valencia, CA, USA), and then converted to cDNA using a Reverse Transcription kit (Qiagen). The synthesized cDNA was used as a template in the subsequent PCR. Quantitative real-time RT-PCR analysis of mRNA was performed using a Thermal Cycler Dice Real Time System TP800 (Takara Bio) according to the manufacturer's protocol. The following mouse-specific primers were used: crmp4: forward 5′-GGTACAGAGCCTCAGCAAGG-3′, reverse 5′-TTATCCCCATTTCCAGCATC-3′, β-actin: forward 5′-GCTACAGCTTCACCACCACA-3′, reverse 5′-TCTCCAGGGAGGAAGAGGAT-3′. The amplification was achieved in a 20-μL volume containing 1 μL cDNA, 200 nm of each primer pair, and SYBR Premix Extaq (Takara Bio). The gene expression levels of crmp4 were standardized using simultaneously measured values of β-actin.
Statistical analysis
In the quantitative real-time RT-PCR experiment, statistical differences were determined by one-way anovas followed by Tukey's HSD post hoc tests for multiple comparisons (P < 0.05).
Results
The hybridization signals were clearly observed as black dots in sections of the brain removed from mice on PD0, when the sections were hybridized with the antisense probe (Fig. 1A). In contrast, the sense probe produced no specific signals on the adjacent section (Fig. 1B). The results obtained from the in situ hybridization study are summarized in Table 1. Details of development-related changes in CRMP4 mRNA expression are shown in the following brain regions.
Fig. 1.
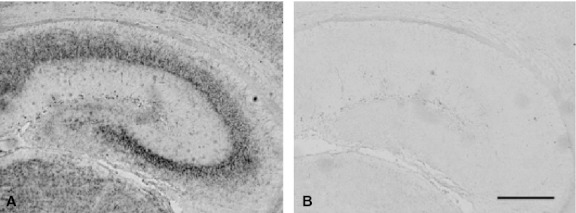
Specificity of anti-sense probe for detection of CRMP4 mRNA. In situ hybridization was performed on sections of the mouse brain using DIG-labeled antisense (A) and sense (B) probes for CRMP4 mRNA. Scale bar: 300 μm.
Table 1.
Spatiotemporal expression changes of CRMP4 mRNA in the brain
| Region | PD0 | PD7 | PD14 | Adult | Region | PD0 | PD7 | PD14 | Adult |
|---|---|---|---|---|---|---|---|---|---|
| Forebrain | Paratenial thalamic nu | ++ | + | − | − | ||||
| Main olfactory bulb | Centre-median thalamic nu | ++ | + | − | − | ||||
| Glomerular layer | +/− | ++ | +/− | +/− | Rhomboid nu | ++ | + | − | − |
| Mitral cell layer | +++ | ++ | + | + | Reuiens thalamic nu | ++ | + | − | − |
| Granule cell layer | − | − | − | − | Anteroventral thalamic nu | ++ | + | − | − |
| Subependymal zone | − | + | + | + | Reticular thalamic nu | ++ | + | − | − |
| Anterior olfactory nu | ++ | +++ | +/− | − | Ventral posterior complex | ++ | + | − | − |
| Accessory olfactory bulb | Ventral antero-lateral complex | ++ | + | − | − | ||||
| Glomerular layer | − | − | + | − | Ventral medial nu | ++ | + | − | − |
| Mitral cell layer | +++ | ++ | + | + | Parafascicular nu | ++ | ++ | − | − |
| Granule cell layer | − | − | − | − | Submedial nu | ++ | + | − | − |
| Olfactory tubercle | + | +/− | − | − | Lateral posterior nu | ++ | + | − | − |
| Cortical regions | Central lateral nu | ++ | ++ | − | − | ||||
| Primary motor cortex | ++ | ++ | − | − | Paracentral nu | ++ | ++ | − | − |
| Secondary motor cortex | ++ | ++ | − | − | Geniculate complex | ++ | ++ | − | − |
| Cingulate cortex | ++ | ++ | − | − | Posterior complex | ++ | ++ | − | − |
| Granular insular cortex | ++ | ++ | − | − | Hypothalamus | ||||
| Lateralorbital cortex | + | + | − | − | Paraventricular nu | + | + | − | − |
| Piriform cortex | ++ | +++ | + | − | Superchiasmatic nu | + | +/− | − | − |
| Endopiriform nu | + | + | − | − | Arcuate nu | ++ | +/− | − | − |
| Insular cortex | ++ | ++ | − | − | Supraoptic nu | + | +/− | − | − |
| prelimbic cortex | ++ | ++ | − | − | Mamillary nu | ++ | + | − | − |
| Dorsal peduncular cortex | ++ | ++ | − | − | Preoptic nu | ||||
| Infralimbic cortex | ++ | ++ | − | − | Ventmed preoptic nu | + | +/− | − | − |
| Somatosensory cortex | ++ | ++ | − | − | Magnocell preoptic nu | ++ | +/− | − | |
| Primary somatosensory cortex | ++ | ++ | − | − | Ventrat preoptic nu | +/− | +/− | − | − |
| Subiculum cortex | ++ | ++ | − | − | Subthalamic nu | ++ | +/− | − | − |
| Tenia tecta | Nu lateral olfactory tract, layer 2 | ++ | ++ | − | − | ||||
| Dorsal tenia tecta | ++ | ++ | − | − | Peduncular part lat hy | + | +/− | − | − |
| Ventral tenia tecta | ++ | ++ | − | − | Midbrain | ||||
| Striatum | + | + | − | − | Tectum | ||||
| Hippocampus | Superior colliuculus | + | + | − | − | ||||
| CA1 | +++ | +++ | − | − | Inferior colliuculus | + | + | − | − |
| CA2 | +++ | +++ | − | − | Tegmentus | ||||
| CA3 | +++ | +++ | − | − | Periaqueductal gray | ++ | + | − | − |
| Dentate gyrus | + | +++ | +/− | +/− | Red nu, magnocell part | ++ | + | − | − |
| Islands of Callejo, major | + | + | − | − | Substantia nigra | ++ | + | − | − |
| Septum | Interpedunc fossa | ++ | +/− | − | − | ||||
| Septohippocampal nu | ++ | ++ | − | − | Hindbrain | ||||
| Medial septal nu | ++ | ++ | − | − | Pons | ||||
| Lat septal nu, intermed | + | +/− | − | − | Pontine nuclei | ++ | ++ | − | − |
| Lat septal nu, dorsal part | ++ | + | − | − | Lateral lemniscus | ++ | +/− | − | − |
| Lat septal nu, vent part | + | + | − | − | Median raphe nu | + | + | − | − |
| Paralambdoid septal nu | + | − | − | − | Reticulotegmental nu pons | + | +/− | − | − |
| Lambdoid septal zone | + | − | − | − | Dorsal raphe | ++ | +/− | − | − |
| Amygdala | ++ | ++ | − | − | Laterodorsal tegmental nu | ++ | +/− | − | − |
| Bed nucleus | Cerebellum | ||||||||
| Principal nu | ++ | + | − | − | Cortex | + | + | − | − |
| Strial extension | ++ | +/− | − | − | Medial cerebellar nu | ++ | + | − | − |
| Thalamus | Interposed cerebellar nu | ++ | + | − | − | ||||
| Habenular nu | + | + | − | − | Medulla oblongata | ||||
| Paraventricular thalamic nu | ++ | ++ | − | − | Propositus nu | + | +/− | − | − |
| mediodorsal thalamic nu | ++ | ++ | − | − | Vestibular nu | + | +/− | − | − |
| Anterodorsal thalamic nu | ++ | ++ | − | − | Hypoglossal nu | + | + | − | − |
| Laterodorsal thalamic nu | ++ | ++ | − | − | Gigantocellular reticular nu | + | +/− | − | − |
| Reticluoustrial nu | ++ | ++ | − | − | nu X | + | + | − | − |
| Anteromedial thalamic nu | ++ | + | − | − | Inf olive | + | + | − | − |
nu, nucleus.
Forebrain
Olfactory bulb
On PD0, hybridization signals of strong intensity (+++) were detected in the mitral cell layer (MCL) of the main OB (MOB) (arrowheads in Fig. 2A and A′), whereas very weak and scattered signals were observed in the glomerular layer (arrows in Fig. 2A and A′).
Fig. 2.
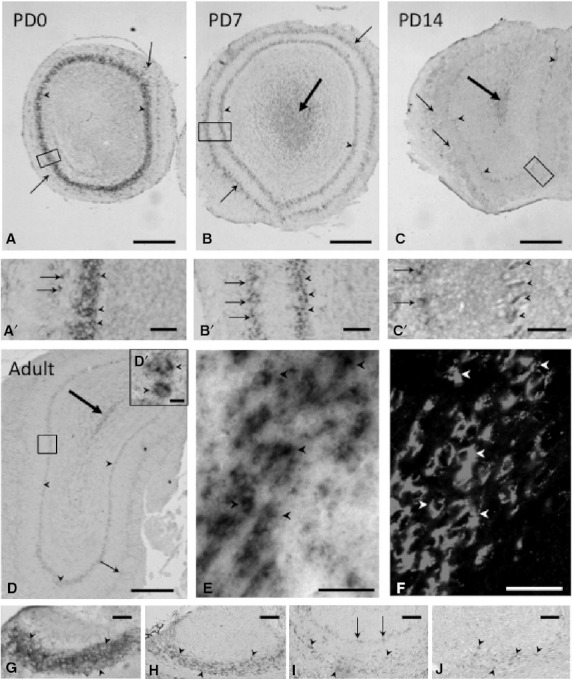
Expression and localization patterns of CRMP4 mRNA in the MOB and AOB during postnatal development. (A–D, A′–D′) Representative images of hybridization signals in the MOB on PD0 (A, A′), PD7 (B, B′), and PD14 (C, C′) and in adults (8 weeks old) (D, D′). (A′–D′) Higher magnification of delineated areas in A–D. Arrowheads and arrows indicate the MCL and glomerular layer, respectively. Big arrows in B, C, and D indicate hybridization signals in the subependymal zone. (E,F) Pictures of mirror-image sections of the subependymal zone in the adult OB. In situ hybridization signals of CRMP4 mRNA (E) were colocalized with PSA-NCAM immunoreactivity (F) in the same cells (arrowheads). (G–J) Representative images of hybridization signals in the AOB on PD0 (G), PD7 (H), and PD14 (I) and in adults (J). Arrowheads and arrows in G, H, I, and J indicate the MCL and glomerular layer, respectively. Scale bars: 400 μm (A–D), 100 μm (A′–B′ and G–H), 50 μm (C′,E–F and I–J), 10 μm (D′).
On PD7, the signals in the MCL of the MOB were weaker (arrowheads in Fig. 2B and B′), although those in the glomerular layer were stronger (small arrows in Fig. 2B and B′), as compared with those on PD0. The intensity of the MCL (moderate) on PD7 was almost the same as that of the glomerular layer on the same day. In addition, weak signals were broadly observed in the central part of the MOB – the subependymal layer – on PD7 (big arrow in Fig. 2B).
On PD14, the hybridization signals in the MCL and glomerular layer of the MOB were weak and very weak, respectively (arrowheads and small arrows indicate the MCL and glomerular layer, respectively, in Fig. 2C and C′). Cells with weak signals were observed in the subependymal layer of the MOB (big arrow in Fig. 2C).
In adults, weak signals were detected in the MCL of the MOB (arrowheads in Fig. 2D and D′). Signals in the glomerular layer of the MOB (small arrows in Fig. 2D) were very weak and scattered, but still detectable. In the subependymal layers, cells showing weak signals were clearly detected in adults (big arrow in Fig. 2D). Similar to characteristics of migrating neuroblasts, these cells were bipolar with extended leading and trailing processes and formed elongated cell aggregates referred to as ‘chains’. Because neuronal progenitor cells generated from the subventricular zone (SVZ) are known to migrate in the center of the OB, the rostral migratory stream (RMS) (Doetsch et al. 1997; Sawamoto et al. 2006), we examined whether these cells expressing CRMP4 mRNA were immature neurons in the RMS. We stained pairs of consecutive mirror-image sections by two different methods – in situ hybridization and immunohistochemistry with antibody against PSA-NCAM, an immature neuronal marker (Fig. 2E and F, respectively). The cells expressing CRMP4 mRNA (black arrowheads in Fig. 2E) corresponded to the PSA-NCAM-positive cells (white arrowheads in Fig. 2F), confirming that CRMP4 mRNA-expressing cells were immature neurons in the RMS.
On PD0, intense hybridization signals were observed in the MCL of the accessory OB (AOB) (arrowheads in Fig. 2G), although signals were not detectable in the glomerular layer. On PD7, signals in the MCL of the AOB were weaker (arrowheads in Fig. 2H) and still undetectable in the glomerular layer. However, on PD14, the signals in both the MCL and glomerular layer of the AOB were weak but detectable (Fig. 2I). In adults, weak signals were still detected in the MCL of the AOB (arrowheads in Fig. 2J).
Anterior olfactory nucleus
In the anterior olfactory nucleus (AON), a substructure of the olfactory areas located within the rostral portion of the cerebral cortex, hybridization signals of moderate intensity were found on PD0 (white arrow in Fig. 3A). Signals in the AON were quite strong on PD7 (white arrow in Fig. 3B), very weak on PD14 (white arrow in Fig. 3C), and almost undetectable in adults (Fig. 3D). As observed in the subependymal layer of the MOB, in the central part of the AON that constitutes a part of the RMS, no specific signals were observed on PD0 (Fig. 3A), but weak signals were first found on PD7 and detectable thereafter (big arrows in Fig. 3C, D and D′). The intensity of signals in the MCL in the rostral portion of the cerebral cortex was the strongest on PD0 (arrowheads in Fig. 3A), gradually decreased on PD7 and PD14 (arrowheads in Fig. 3B,C), and was no longer detected in adults (Fig. 3D). The intensity of signals in the glomerular layer was stronger on PD7 (small arrows in Fig. 3B) than on PD0 (small arrows in Fig. 3A), weaker on PD14 (small arrows in Fig. 3C), and undetectable in adults (Fig. 3D).
Fig. 3.
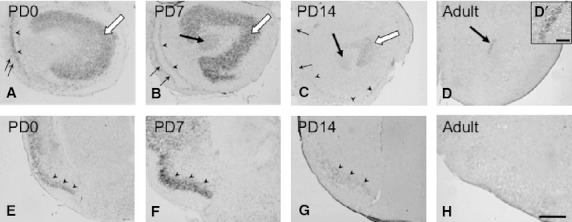
Expression and localization patterns of CRMP4 mRNA in the AON (A–D) and piriform cortex (E–H) during postnatal development. (A–D) Representative images of hybridization signals in the AON on PD0 (A), PD7 (B), and PD14 (C) and in adults (D). White arrows in A, B, and C indicate signals in the AON. Arrowheads and arrows in A, B, and C indicate the MCL and glomerular layer, respectively. Big arrows in B–D indicate signals in the central part of the AON. D′ corresponds to a higher magnification of the central part of the AON (a big arrow in D). (E–H) Representative hybridization signals (arrowheads) in the piriform cortex on PD0 (E), PD7 (F), and PD14 (G) and in adults (H). Scale bars: 400 μm (A–H), 100 μm (D′).
Piriform cortex
Moderate expression of CRMP4 mRNA was found in the piriform cortex on PD0 (arrowheads in Fig. 3E). The signals were quite strong on PD7 (arrowheads in Fig. 3F), weak on PD14 (arrowheads in Fig. 3G), and undetectable in adults (Fig. 3H).
Hippocampus
Intense hybridization signals were detected in the CA1–CA3 regions of the hippocampus on PD0 and PD7 (arrows in Fig. 4A,B) and nearly undetectable on PD14 and in adults (Fig. 4C,D). Conversely, in the dentate gyrus, signals were weak on PD0 (arrowheads in Fig. 4A), quite strong on PD7 (arrowheads in Fig. 4B), and weak again on PD14 (arrowheads in Fig. 4C). In adults, very weak scattered signals were found in the granule cell layer of the dentate gyrus (arrowheads in Fig. 4D and an arrow in high-magnification image).
Fig. 4.
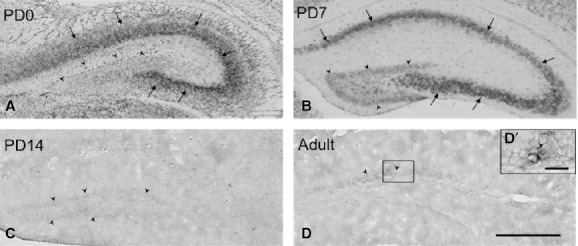
Expression and localization patterns of CRMP4 mRNA in the hippocampus during postnatal development (A–D). Representative images of hybridization signals in the dentate gyrus (arrowheads) and CA1–CA3 (arrows) on PD0 (A), PD7 (B), and PD14 (C) and in adults (D). Frame D′ corresponds to a higher magnification of a granule cell (an arrowhead) delineated in D. Scale bars: 400 μm (A–D); 20 μm (D′).
Cerebral cortex
In the somatosensory cortex of the cerebrum, hybridization signals of moderate intensity were widely observed in the cortical plate (layer II–IV) on PD0 (Fig. 5A). In addition to the signals in the cortical plate, subplate cells were positively stained (arrowheads in Fig. 5A). On PD7, moderately intense signals were found in layers II/III and V, and weak signals were distributed in layer VI (Fig. 5B). These signals were undetectable on PD14 (Fig. 5C) and in adults (data not shown). The distribution pattern described above was generally observed in other regions of the cerebral cortex.
Fig. 5.
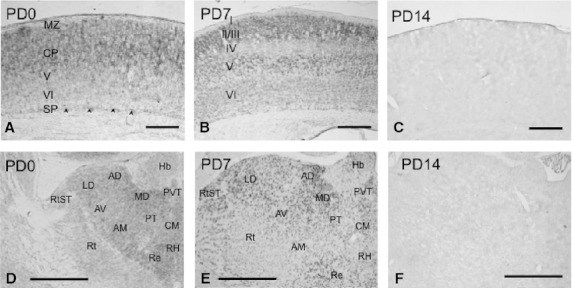
Expression and localization patterns of CRMP4 mRNA in the somatosensory cortex (A–C) and thalamus during postnatal development (D–F). (A–C) CRMP4 mRNA expression was detected in the cortical plate and subplate (arrowheads) on PD0 (A). The hybridization signals were observed in layers II/III, V, and VI on PD7 (B) of the somatosensory cortex. Signals were then undetectable after PD14. (D–E) CRMP4 mRNA expression was detected in many thalamic nuclei on PD0 (D) and PD7 (E), whereas it was not detectable after PD14 (F). Hb, habenular nucleus; PVT, paraventricular thalamic nucleus; MD, mediodorsal thalamic nucleus; PT, paratenial thalamic nucleus; CM, centromedian thalamic nucleus; AD, anterodorsal thalamic nucleus; AV, anteroventral thalamic nucleus; LD, laterodorsal thalamic nucleus; Re, reuniens thalamic nucleus reuniens thalamic nucleus; AM, anteromedial thalamic nucleus; RtST, reticulostrial nucleus; Rt, reticular thalamic nucleus. Scale bars: 200 μm (A), 400 μm (B,C), 500 μm (D–F).
Thalamus and hypothalamus
In the thalamus, hybridization signals of moderate intensity were detected in most areas of the thalamus on PD0, even though signals were weak in the habenular nucleus (Fig. 5D). On PD7, signals of moderate intensity were found in some nuclei, including the paraventricular thalamic nucleus (PVT) and mediodorsal thalamic nucleus (MD) (Fig. 5E). In contrast, in some nuclei, such as the paratenial thalamic nucleus (PT) and centromedian thalamic nucleus (CM), signals became weaker on PD7 than on PD0 (Fig. 5F). Signals in the thalamus were nearly undetectable on PD14 (Fig. 5F) and in adults (pictures not shown). Signals of moderate or weak intensities were broadly observed on PD0 in the hypothalamus (Fig. 6A). The signals were then moderate or became weaker on PD7 (Fig. 6B) and were undetectable on PD14 (Fig. 6C).
Fig. 6.
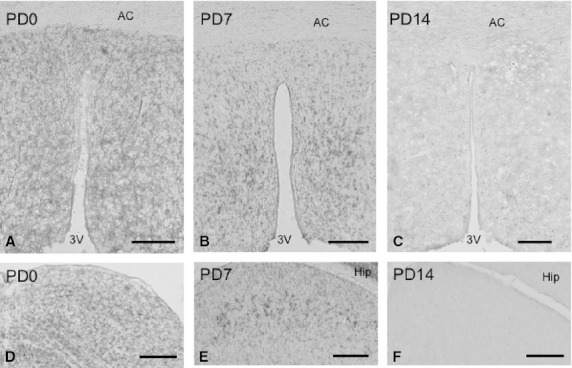
Expression and localization patterns of CRMP4 mRNA in the hypothalamus (A–C) and tectum during postnatal development (D–F). Representative images of hybridization signals in the hypothalamus and the superior colliculus on PD0 (A and D), PD7 (B and E), and PD14 (C and F). AC, 3V, and Hip indicate the anterior commissure, third ventricle, and hippocampus, respectively. Scale bars: 200 μm (A–C and F), 250 μm (D,E).
Midbrain
Signals of weak intensity were observed in the superior (Fig. 6D) and inferior colliculi on PD0. The signals were detected on PD7 (Fig. 6E) but were not observed after PD14 (Fig. 6F). In contrast, signals in tegmental nuclei, such as the red nucleus and substantia nigra, were stronger on PD0 than on PD7, and were undetectable after PD14 (Table 1).
Hindbrain
Cerebellum, pons, and medulla oblongata
Weak signals were broadly detected in the cerebellar cortex on PD0 (Fig. 7A). The intensity of signals in the external granule layer (EGL) and internal granule layer (IGL) was unchanged on PD7, whereas signals were not subsequently observed in the molecular layer (ML) and Purkinje cell layer (PCL) (Fig. 7B). Signals were almost undetectable on PD14 and in adults (Fig. 7C). In other regions of the hindbrain, such as the cerebellar, pontine, and medullary nuclei, the intensity of the signals was moderate or weak on PD0, unchanged or weaker on PD7, and undetectable after PD14 (Table 1).
Fig. 7.
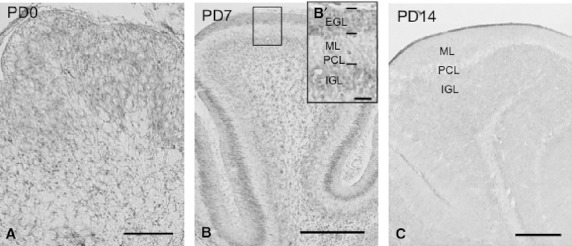
Expression and localization patterns of CRMP4 mRNA in the cerebellar cortex (A–C) during postnatal development. The insert in B is a higher magnification image of the delineated region in B. EGL, ML, PCL, and IGL indicate the external granule layer, molecular layer, Purkinje cell layer, and internal granule layer, respectively. Scale bars: 400 μm (A), 800 μm (B,C), 100 μm (B′).
Quantitative real-time RT-PCR
Relative expression levels of CRMP4 mRNA were compared among samples removed from the OB, hippocampus, and cerebellum in adults using quantitative real-time RT-PCR (Fig. 8). Expression of CRMP4 mRNA was significantly higher in the OB than in the hippocampus or cerebellum.
Fig. 8.
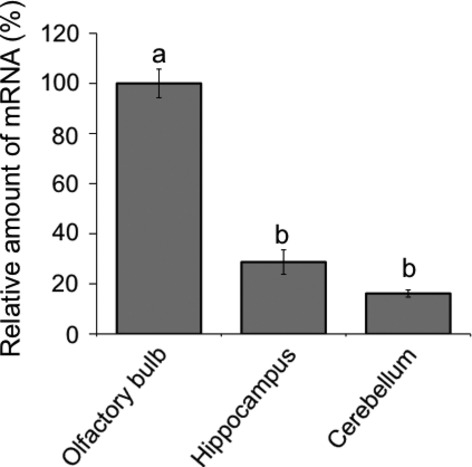
Quantitative evaluation of CRMP4 mRNA expression in the OB, hippocampus, and cerebellum. Relative CRMP4 mRNA levels were determined by quantitative real-time RT-PCR in extracts from each brain area of adults. The expression levels of CRMP4 mRNA were normalized to β-actin RNA. Different superior letters indicate statistical significance (P < 0.05) using one-way anovas followed by Tukey's HSD post hoc tests for multiple comparisons.
Discussion
In the present study, we comprehensively examined the spatiotemporal expression of CRMP4 mRNA in the mouse brain during postnatal development by in situ hybridization. According to expression patterns of CRMP4 mRNA (Table 1), the brain regions could be classified into the following three types: (i) the regions where hybridization signals were most intensive on PD0 or PD7, weak or undetectable on PD14, and undetectable in adults; (ii) the regions known as adult neurogenetic areas, the dentate gyrus and SVZ/RMS (subependymal zone of the OB in Table 1), where strong or weak signals were detected on PD7, and sparse or weak signals were detected in adults; (iii) the MCL of the OB, where strong signals on PD0 declined on PD7 and PD14, but were still detectable in adults. Relatively high expression of CRMP4 mRNA in the adult OB was also confirmed by quantitative real-time RT-PCR analysis.
This is the first study to clarify the regional variability of CRMP4 mRNA expression in the mouse brain from PD0 to adulthood. In a previous study, Northern blot and in situ hybridization analyses showed that the expression of CRMP4 mRNA in various areas of the rat brain is strong during embryonic stages, peaks around the first postnatal week, and is downregulated in the adult stage (Wang & Strittmatter, 1996). Other studies have demonstrated CRMP4 mRNA expression in immature neurons in adult neurogenetic brain regions (Wang & Strittmatter, 1996; Seki, 2002). Thus, our results concerning the first two classifications listed above are consistent with those of previous findings. Additionally, we have provided evidence for the third type, expressing CRMP4 mRNA in mature MCL of the OB for the first time, providing negative information regarding CRMP4 as an immature neuron marker.
Recent studies have demonstrated that CRMP4 has a regulatory function in the cytoskeletal dynamics of neuronal cells to control axonal growth. CRMP4 interacts with GAP-43 (Kowara et al. 2007) to regulate F-actin bundling (Rossienbroich et al. 2005) and mediates myelin-associated inhibitor (MAI)-dependent axonal inhibition via RhoA (Alabed et al. 2007, 2010). CRMP2 and CRMP4 are involved in the determination of neuronal polarity and axonal elongation as they bind tubulin heterodimers (Fukata et al. 2002; Yoshimura et al. 2005; Arimura & Kaibuchi, 2007). Furthermore, CRMP4 suppresses apical dendrite bifurcation of CA1 pyramidal neurons in the mouse hippocampus (Niisato et al. 2012). In the current study, the age-related variability of CRMP4 mRNA hybridization signals in the brain support these functional roles of CRMP4. In addition, after PD14, CRMP4 mRNA was expressed in the granule cell layer of the dentate gyrus and subependymal layer of the OB (Table 1). It has been reported that neurogenesis is maintained in adulthood in the subgranule zone of the dentate gyrus (Bayer, 1982b; Bayer et al. 1982a), and Seki (2002) previously demonstrated that CRMP4 and PSA-NCAM were colocalized in newly generated immature granule cells in the dentate gyrus of older rats. The subependymal zone of the OB was another brain region in the current findings where CRMP4 mRNA was expressed in adults (Table 1). Immunoreactive response to PSA-NCAM antibody indicates that those cells with hybridization signals in the subependymal layer were immature migratory neurons in the RMS, which is a specialized migratory route in the brain from the SVZ to the OB. Therefore, expression of CRMP4 mRNA in the adult neurogenetic regions and migratory route support the previous usage of CRMP4 as an indicator of immature neurons. In the present study, however, we also found the expression of CRMP4 mRNA in the MCL of the OB from PD0 to adult stage.
There are only a few studies on the expression of CRMP4 mRNA in rodent OB. Wang & Strittmatter (1996) reported that the OB has average hybridization signals on PD1, but they investigated neither the labeled layers of the OB nor their developmental changes. Tsim et al. (2004) examined the mRNA expression of CRMPs in the OB of female, male, and neonatal testosterone propionate-treated female rats during postnatal development and indicated significant age-related reduction but not hormonal effects on the transcription of the Crmp4 gene. Therefore, this is the first report to show the persistent expression of Crmp4 in the MCs of the OB in neonates and adults. The MC is one of the projection neurons in the OB that transfers signals of smell information from the olfactory nerve to the olfactory cortex. Their dendrites are extended to the glomerular cell layer and make synaptic contacts with axons of olfactory receptor neurons (ORNs) (Vassar et al. 1994; Mombaerts et al. 1996). The ORNs are located in the olfactory mucosa of the upper parts of the nasal cavity and are capable of regeneration throughout life, i.e. new synapses are generated between ORNs and MCs in adults (Beites et al. 2005). Furthermore, MCs form reciprocal synapses with granule cells or periglomerular cells, both of which are inhibitory neurons regulating the signal transduction of smell. Newly divided cells in the SVZ migrate via the RMS to the OB, where they differentiate into granule and periglomerular cells, and form reciprocal synapses with MCs (Lois & Alvarez-Buylla, 1993; Luskin, 1993). Thus, MCs maintain strong synapse plasticity throughout life. Recent studies also indicate synaptic functions of CRMPs (Brittain et al. 2009; Chi et al. 2009; Wang et al. 2010; Yamashita & Goshima, 2012). In the present study, sustained expression of CRMP4 mRNA in MCs from PD0 to the adult stage suggests the involvement of CRMP4 in synapse plasticity between MCs and ORNs, and/or between MCs and interneurons (either granule or periglomerular cells).
There have been some reports showing that CRMP4 protein is expressed in many regions in the adult rat brain, including both neurogenetic and non-neurogenetic regions (Geschwind et al. 1996; Nacher et al. 2000). Nacher et al. (2000) precisely examined CRMP4 expression by means of immunohistochemistry in young adult rats. Similar to the present results in adults, they found CRMP4-immunopositive cells in newly generated neurons, which included cells in the dentate granular layer and the SVZ cells migrating through the RMS to the OB (Nacher et al. 2000). However, they also showed immunoreactive CRMP4 in many other regions of the adult rat brain in which there is no record of adult neurogenesis or neuronal migration, e.g. in neurogliaform neurons in the piriform cortex, in neurons of the cerebral cortex, and in many processes and cells in the hypothalamus. In the OB, they found a few small differentiating neurons in the granular and external plexiform layers and the strongly labeled afferent projection from the olfactory nerve in the glomeruli of the MOB. In addition, small CRMP4-immunoreactive cells, suggested to be of oligodendroglial lineage, were seen throughout the rat brain (Nacher et al. 2000). The presence of these immunoreactive cells in non-neurogenetic regions is inconsistent with the results obtained in the current study. Cnops et al. (2006) also detected CRMP4-immunopositive neurons throughout cortical layers II to VI of adult cat visual cortex. In addition, Nacher et al. (2002) distinguished scattered CRMP4-positive neurons in adult lizard cerebral cortex. Conversely, many other researchers have failed to localize CRMP4 immunoreactivity in adult human (Gaetano et al. 1997; Kerfoot et al. 1999) and rat brain (Minturn et al. 1995; Wang & Strittmatter, 1996; Liu et al. 2003). These discrepancies among studies in the localization of CRMP4-immunoreactive cells and CRMP4 mRNA signals may be due to interspecies differences, antibody differences, and sensitivity differences between in situ hybridization and immunohistochemistry. Difference in CRMP4 translation and/or degradation rates may also account for these discrepancies in adult animals.
Concluding remarks
In the present study, we specifically demonstrated the expression changes in CRMP4 mRNA and confirmed that CRMP4 was expressed in immature neurons in the developing and adult brain. Furthermore, we found that CRMP4 mRNA was expressed in the adult MCL of the OB, where a high level of synaptic plasticity is retained even in adults. CRMPs are considered early effector proteins of neurodegenerative diseases (Taghian et al. 2012), and some reports have suggested the relationship between Alzheimer's disease and CRMPs (Uchida et al. 2005; Petratos et al. 2008). Our report provides basic information for a better understanding of the developmental, physiological, and neurodegenerative roles of CRMP4.
Acknowledgments
We would like to thank Prof. Goshima for providing us with a cDNA probe for Crmp4 in situ hybridization analysis. This study was supported by a Grant-in-Aid for Scientific Research (C) (KAKENHI). Grant number: 22500315 (to R. Ohtani-Kaneko).
Author contributions
A. Tsutiya and R. Ohtani-Kaneko designed the research, analyzed data, and wrote the paper; A. Tsutiya performed the research.
References
- Alabed YZ, Pool M, Ong Tone S, et al. Identification of CRMP4 as a convergent regulator of axon outgrowth inhibition. J Neurosci. 2007;27:1702–1711. doi: 10.1523/JNEUROSCI.5055-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabed YZ, Pool M, Ong Tone S, et al. GSK3 beta regulates myelin-dependent axon outgrowth inhibition through CRMP4. J Neurosci. 2010;30:5635–5643. doi: 10.1523/JNEUROSCI.6154-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura N, Kaibuchi K. Neuronal polarity: from extracellular signals to intracellular mechanisms. Nat Rev Neurosci. 2007;8:194–205. doi: 10.1038/nrn2056. [DOI] [PubMed] [Google Scholar]
- Bayer SA. Changes in the total number of dentate granule cells in juvenile and adult rats: a correlated volumetric and 3H-thymidine autoradiographic study. Exp Brain Res. 1982b;46:315–323. doi: 10.1007/BF00238626. [DOI] [PubMed] [Google Scholar]
- Bayer SA, Yackel JW, Puri PS. Neurons in the rat dentate gyrus granular layer substantially increase during juvenile and adult life. Science. 1982a;216:890–892. doi: 10.1126/science.7079742. [DOI] [PubMed] [Google Scholar]
- Beites CL, Kawauchi S, Crocker CE, et al. Identification and molecular regulation of neural stem cells in the olfactory epithelium. Exp Cell Res. 2005;306:309–316. doi: 10.1016/j.yexcr.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Brittain JM, Piekarz AD, Wang Y, et al. An atypical role for collapsin response mediator protein 2 (CRMP-2) in neurotransmitter release via interaction with presynaptic voltage-gated calcium channels. J Biol Chem. 2009;284:31375–31390. doi: 10.1074/jbc.M109.009951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi XX, Schmutzler BS, Brittain JM, et al. Regulation of N-type voltage-gated calcium channels (Cav2.2) and transmitter release by collapsin response mediator protein-2 (CRMP-2) in sensory neurons. J Cell Sci. 2009;122:4351–4362. doi: 10.1242/jcs.053280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cnops L, Hu TT, Burnat K, et al. Age-dependent alterations in CRMP2 and CRMP4 protein expression profiles in cat visual cortex. Brain Res. 2006;1088:109–119. doi: 10.1016/j.brainres.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Cole AR, Knebel A, Morrice NA, et al. GSK-3 phosphorylation of the Alzheimer epitope within collapsin response mediator proteins regulates axon elongation in primary neurons. J Biol Chem. 2004;279:50176–50180. doi: 10.1074/jbc.C400412200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F, García-Verdugo JM, Alvarez-Buylla A. Cellular composition and three-dimensional organization of the subventricular germinal zone in the adult mammalian brain. J Neurosci. 1997;17:5046–5061. doi: 10.1523/JNEUROSCI.17-13-05046.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang WQ, Ip JP, Li R, et al. Cdk5-mediated phosphorylation of Axin directs axon formation during cerebral cortex development. J Neurosci. 2011;31:13613–13624. doi: 10.1523/JNEUROSCI.3120-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukada M, Watakabe I, Yuasa-Kawada J, et al. Molecular characterization of CRMP5, a novel member of the collapsin response mediator protein family. J Biol Chem. 2000;275:37957–37965. doi: 10.1074/jbc.M003277200. [DOI] [PubMed] [Google Scholar]
- Fukata Y, Itoh TJ, Kimura T, et al. CRMP-2 binds to tubulin heterodimers to promote microtubule assembly. Nat Cell Biol. 2002;4:583–591. doi: 10.1038/ncb825. [DOI] [PubMed] [Google Scholar]
- Gaetano C, Matsuo T, Thiele CJ. Identification and characterization of a retinoic acid-regulated human homologue of the unc-33-like phosphoprotein gene (hUlip) from neuroblastoma cells. J Biol Chem. 1997;272:12195–12201. doi: 10.1074/jbc.272.18.12195. [DOI] [PubMed] [Google Scholar]
- Geschwind DH, Kelly GM, Fryer H, et al. Identification and characterization of novel developmentally regulated proteins in rat spinal cord. Dev Brain Res. 1996;97:62–75. doi: 10.1016/s0165-3806(96)00132-0. [DOI] [PubMed] [Google Scholar]
- Goshima Y, Nakamura F, Strittmatter P, et al. Collapsin-induced growth cone collapse mediated by an intracellular protein related to UNC-33. Nature. 1995;376:509–514. doi: 10.1038/376509a0. [DOI] [PubMed] [Google Scholar]
- Inatome R, Tsujimura T, Hitomi T, et al. Identification of CRAM, a novel unc-33 gene family protein that associates with CRMP3 and protein- tyrosine kinase(s) in the developing rat brain. J Biol Chem. 2000;275:27291–27302. doi: 10.1074/jbc.M910126199. [DOI] [PubMed] [Google Scholar]
- Ip JP, Shi L, Chen Y, et al. α2-chimaerin controls neuronal migration and functioning of the cerebral cortex through CRMP-2. Nat Neurosci. 2011;15:39–47. doi: 10.1038/nn.2972. [DOI] [PubMed] [Google Scholar]
- Kerfoot C, Vinters HV, Mathern GW. Cerebral cortical dysplasia: giant neurons show potential for increased excitation and axonal plasticity. Dev Neurosci. 1999;21:260–270. doi: 10.1159/000017405. [DOI] [PubMed] [Google Scholar]
- Kowara R, Ménard M, Brown L, et al. Co-localization and interaction of DPYSL3 and GAP43 in primary cortical neurons. Biochem Biophys Res Commun. 2007;363:190–193. doi: 10.1016/j.bbrc.2007.08.163. [DOI] [PubMed] [Google Scholar]
- Liu PC, Yang ZJ, Qiu MH, et al. Induction of CRMP-4 in striatum of adult rat after transient brain ischemia. Acta Pharmacol Sin. 2003;24:1205–1211. [PubMed] [Google Scholar]
- Lois C, Alvarez-Buylla A. Proliferating subventricular zone cells in the adult mammalian forebrain can differentiate into neurons and glia. Proc Natl Acad Sci U S A. 1993;90:2074–2077. doi: 10.1073/pnas.90.5.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luskin MB. Restricted proliferation and migration of postnatally generated neurons derived from the forebrain subventricular zone. Neuron. 1993;11:173–189. doi: 10.1016/0896-6273(93)90281-u. [DOI] [PubMed] [Google Scholar]
- McLaughlin D, Vidaki M, Karagogeos D. Localization of CRMP5 mRNA by in situ hybridisation during development of the mouse forebrain. Neurosci Lett. 2008;432:117–120. doi: 10.1016/j.neulet.2007.12.011. [DOI] [PubMed] [Google Scholar]
- Minturn JE, Fryer HJ, Geschwind DH, et al. TOAD-64, a gene expressed early in neuronal differentiation in the rat, is related to unc-33, a C. elegans gene involved in axon outgrowth. J Neurosci. 1995;15:6757–6766. doi: 10.1523/JNEUROSCI.15-10-06757.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, et al. Visualizing an olfactory sensory map. Cell. 1996;87:675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- Nacher J, Rosell DR, McEwen BS. Widespread expression of rat collapsin response-mediated protein 4 in the telencephalon and other areas of the adult rat central nervous system. J Comp Neurol. 2000;424:628–639. doi: 10.1002/1096-9861(20000904)424:4<628::aid-cne5>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Nacher J, Soriano S, Varea E, et al. CRMP-4 expression in the adult cerebral cortex and other telencephalic areas of the lizard Podarcis hispanica. Brain Res Dev Brain Res. 2002;139:285–294. doi: 10.1016/s0165-3806(02)00589-8. [DOI] [PubMed] [Google Scholar]
- Niisato E, Nagai J, Yamashita N, et al. CRMP4 suppresses apical dendrite bifurcation of CA1 pyramidal neurons in the mouse hippocampus. Dev Neurobiol. 2012 doi: 10.1002/dneu.22007. doi: 10.1002/dneu.22007 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Petratos S, Li QX, George AJ, et al. The β-amyloid protein of Alzheimer's disease increases neuronal CRMP-2 phosphorylation by a Rho-GTP mechanism. Brain. 2008;131:90–108. doi: 10.1093/brain/awm260. [DOI] [PubMed] [Google Scholar]
- Quach TT, Mosinger B, Jr, Ricard D, et al. Collapsin response mediator protein-3/unc-33-like protein-4 gene: organization, chromosomal mapping and expression in the developing mouse brain. Gene. 2000;242:175–182. doi: 10.1016/s0378-1119(99)00528-4. [DOI] [PubMed] [Google Scholar]
- Quach TT, Massicotte G, Belin MF, et al. CRMP3 is required for hippocampal CA1 dendritic organization and plasticity. FASEB J. 2008;22:401–409. doi: 10.1096/fj.07-9012com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn CC, Gray GE, Hockfield S. A family of proteins implicated in axon guidance and outgrowth. J Neurobiol. 1999;41:158–164. [PubMed] [Google Scholar]
- Rosslenbroich V, Dai L, Baader SL, et al. Collapsin response mediator protein-4 regulates F-actin bundling. Exp Cell Res. 2005;310:434–444. doi: 10.1016/j.yexcr.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Sawamoto K, Wichterle H, Gonzalez-Perez O, et al. New neurons follow the flow of cerebrospinal fluid in the adult brain. Science. 2006;311:629–632. doi: 10.1126/science.1119133. [DOI] [PubMed] [Google Scholar]
- Seki T. Expression patterns of immature neuronal markers PSA-NCAM, CRMP-4 and NeuroD in the hippocampus of young adult and aged rodents. J Neurosci Res. 2002;70:327–334. doi: 10.1002/jnr.10387. [DOI] [PubMed] [Google Scholar]
- Taghian K, Lee JY, Petratos S. Phosphorylation and cleavage of the family of collapsin response mediator proteins may play a central role in neurodegeneration after CNS trauma. J Neurotrauma. 2012;29:1728–1735. doi: 10.1089/neu.2011.2063. [DOI] [PubMed] [Google Scholar]
- Tsim TY, Wong EY, Leung MS, et al. Expression of axon guidance molecules and their related genes during development and sexual differentiation of the olfactory bulb in rats. Neuroscience. 2004;123:951–965. doi: 10.1016/j.neuroscience.2003.10.024. [DOI] [PubMed] [Google Scholar]
- Uchida Y, Ohshima T, Sasaki Y, et al. Semaphorin3A signalling is mediated via sequential Cdk5 and GSK3β phosphorylation of CRMP2: implication of common phosphorylating mechanism underlying axon guidance and Alzheimer's disease. Genes Cells. 2005;10:165–179. doi: 10.1111/j.1365-2443.2005.00827.x. [DOI] [PubMed] [Google Scholar]
- Vassar R, Chao SK, Sitcheran R, et al. Topographic organization of sensory projections to the olfactory bulb. Cell. 1994;79:981–991. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- Wang LH, Strittmatter SM. A family of rat CRMP genes is differentially expressed in the nervous system. J Neurosci. 1996;16:6197–6207. doi: 10.1523/JNEUROSCI.16-19-06197.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Brittain JM, Wilson SM, et al. Emerging roles of collapsin response mediator proteins (CRMPs) as regulators of voltage-gated calcium channels and synaptic transmission. Commun Integr Biol. 2010;3:172–175. doi: 10.4161/cib.3.2.10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita N, Goshima Y. Collapsin response mediator proteins regulate neuronal development and plasticity by switching their phosphorylation status. Mol Neurobiol. 2012;45:234–246. doi: 10.1007/s12035-012-8242-4. [DOI] [PubMed] [Google Scholar]
- Yamashita N, Morita A, Uchida Y, et al. Regulation of spine development by semaphorin3A through cyclin-dependent kinase 5 phosphorylation of collapsin response mediator protein 1. J Neurosci. 2007;27:12546–12554. doi: 10.1523/JNEUROSCI.3463-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita N, Mosinger B, Roy A, et al. CRMP5 (collapsin response mediator protein 5) regulates dendritic development and synaptic plasticity in the cerebellar Purkinje cells. J Neurosci. 2011;31:1773–1779. doi: 10.1523/JNEUROSCI.5337-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura T, Kawano Y, Arimura N, et al. GSK-3β regulates phosphorylation of CRMP-2 and neuronal polarity. Cell. 2005;120:137–149. doi: 10.1016/j.cell.2004.11.012. [DOI] [PubMed] [Google Scholar]


