Abstract
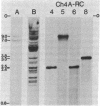
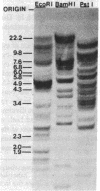
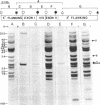
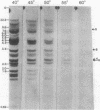
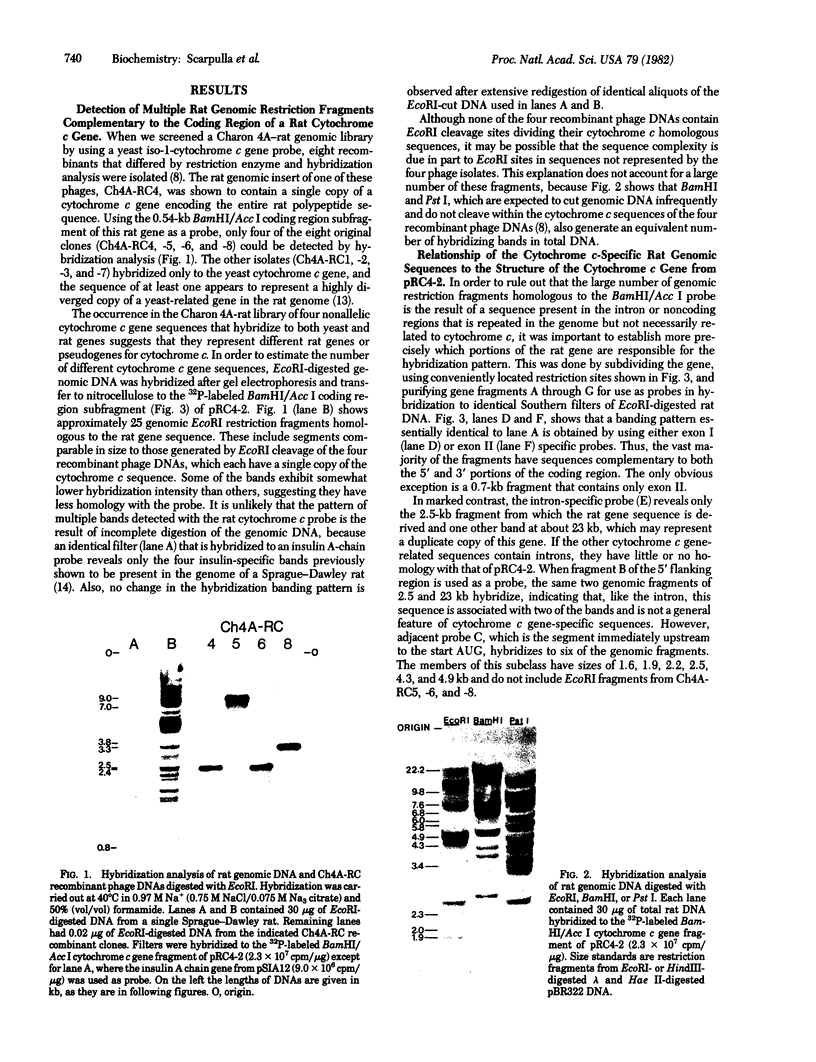
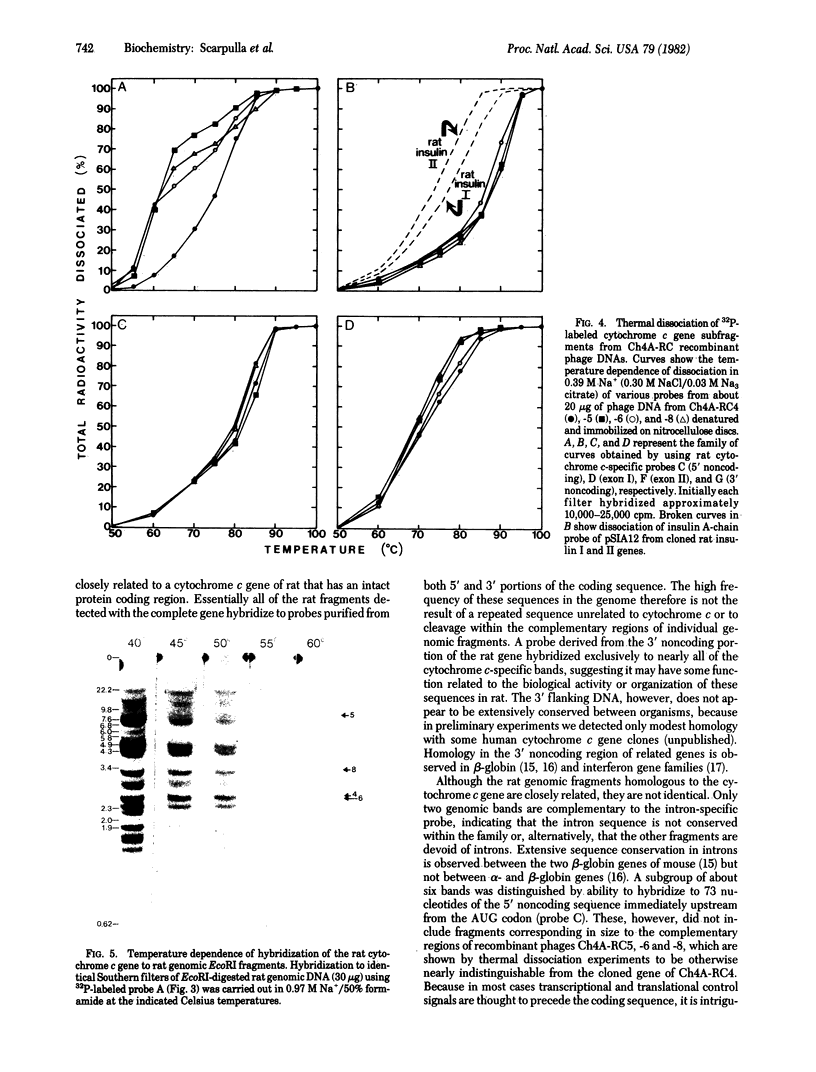
We use a rat cytochrome c gene that we previously isolated and determined the sequence of to estimate the number of related sequences present in the rat genome. Approximately 25 different EcoRI restriction endonuclease fragments from total rat DNA hybridize to the gene of known structure. Four of these correspond to homologous sequences present in four different lambda Charon 4A-rat cytochrome c recombinants previously isolated. Intact or nearly intact genes appear to reside on almost all of the genomic fragments, because they hybridize strongly to gene subfragments representing both 5' and 3' portions of the coding sequence as well as to 3' noncoding DNA that is found specifically associated with the coding region. A subgroup of about six of the fragments also shares homology within the 73 nucleotides immediately preceding the AUG codon. An intron-specific probe reveals only the EcoRI fragment from which it was derived and one other genomic fragment. On the basis of the temperature of complete dissociation of the coding region probe in 0.75 M NaCl/0.075 M Na3 citrate/50% (vol/vol) formamide, the 25 fragments are separable into three stringency classes of 40-50 degrees C, 50-55 degrees C, and 55-60 degrees C. The latter, high-stringency group of about seven fragments includes those cloned in the recombinant phage isolates, whose regions homologous to cytochrome c are shown to differ from the purified gene of known sequence by an amount equivalent to about 2% mismatched bases. Families of cytochrome c gene-related sequences are also found in the genomes of several other mammals, including humans.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blin N., Stafford D. W. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976 Sep;3(9):2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Cordell B., Bell G., Tischer E., DeNoto F. M., Ullrich A., Pictet R., Rutter W. J., Goodman H. M. Isolation and characterization of a cloned rat insulin gene. Cell. 1979 Oct;18(2):533–543. doi: 10.1016/0092-8674(79)90070-9. [DOI] [PubMed] [Google Scholar]
- Firtel R. A. Multigene families encoding actin and tubulin. Cell. 1981 Apr;24(1):6–7. doi: 10.1016/0092-8674(81)90494-3. [DOI] [PubMed] [Google Scholar]
- Goeddel D. V., Leung D. W., Dull T. J., Gross M., Lawn R. M., McCandliss R., Seeburg P. H., Ullrich A., Yelverton E., Gray P. W. The structure of eight distinct cloned human leukocyte interferon cDNAs. Nature. 1981 Mar 5;290(5801):20–26. doi: 10.1038/290020a0. [DOI] [PubMed] [Google Scholar]
- Hennig B. Change of cytochrome c structure during development of the mouse. Eur J Biochem. 1975 Jun 16;55(1):167–183. doi: 10.1111/j.1432-1033.1975.tb02149.x. [DOI] [PubMed] [Google Scholar]
- Jacq C., Miller J. R., Brownlee G. G. A pseudogene structure in 5S DNA of Xenopus laevis. Cell. 1977 Sep;12(1):109–120. doi: 10.1016/0092-8674(77)90189-1. [DOI] [PubMed] [Google Scholar]
- Konkel D. A., Maizel J. V., Jr, Leder P. The evolution and sequence comparison of two recently diverged mouse chromosomal beta--globin genes. Cell. 1979 Nov;18(3):865–873. doi: 10.1016/0092-8674(79)90138-7. [DOI] [PubMed] [Google Scholar]
- Lacy E., Maniatis T. The nucleotide sequence of a rabbit beta-globin pseudogene. Cell. 1980 Sep;21(2):545–553. doi: 10.1016/0092-8674(80)90492-4. [DOI] [PubMed] [Google Scholar]
- Margoliash E., Schejter A. Cytochrome c. Adv Protein Chem. 1966;21:113–286. doi: 10.1016/s0065-3233(08)60128-x. [DOI] [PubMed] [Google Scholar]
- Montgomery D. L., Leung D. W., Smith M., Shalit P., Faye G., Hall B. D. Isolation and sequence of the gene for iso-2-cytochrome c in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Jan;77(1):541–545. doi: 10.1073/pnas.77.1.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NUNNIKHOVEN R. Amino acid composition and some other properties of yeast cytochrome c in comparison with horse-heart cytochrome c. Biochim Biophys Acta. 1958 Apr;28(1):108–119. doi: 10.1016/0006-3002(58)90436-0. [DOI] [PubMed] [Google Scholar]
- Nishioka Y., Leder A., Leder P. Unusual alpha-globin-like gene that has cleanly lost both globin intervening sequences. Proc Natl Acad Sci U S A. 1980 May;77(5):2806–2809. doi: 10.1073/pnas.77.5.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka Y., Leder P. The complete sequence of a chromosomal mouse alpha--globin gene reveals elements conserved throughout vertebrate evolution. Cell. 1979 Nov;18(3):875–882. doi: 10.1016/0092-8674(79)90139-9. [DOI] [PubMed] [Google Scholar]
- Orgel L. E., Crick F. H. Selfish DNA: the ultimate parasite. Nature. 1980 Apr 17;284(5757):604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Maniatis T. The structure of a human alpha-globin pseudogene and its relationship to alpha-globin gene duplication. Cell. 1980 Sep;21(2):537–544. doi: 10.1016/0092-8674(80)90491-2. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Scarpulla R. C., Agne K. M., Wu R. Isolation and structure of a rat cytochrome c gene. J Biol Chem. 1981 Jun 25;256(12):6480–6486. [PubMed] [Google Scholar]
- Sherman F., Stewart J. W. Genetics and biosynthesis of cytochrome c. Annu Rev Genet. 1971;5:257–296. doi: 10.1146/annurev.ge.05.120171.001353. [DOI] [PubMed] [Google Scholar]
- Sherman F., Taber H., Campbell W. Genetic determination of iso-cytochromes c in yeast. J Mol Biol. 1965 Aug;13(1):21–39. doi: 10.1016/s0022-2836(65)80077-8. [DOI] [PubMed] [Google Scholar]
- Smith M., Leung D. W., Gillam S., Astell C. R., Montgomery D. L., Hall B. D. Sequence of the gene for iso-1-cytochrome c in Saccharomyces cerevisiae. Cell. 1979 Apr;16(4):753–761. doi: 10.1016/0092-8674(79)90091-6. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Vanin E. F., Goldberg G. I., Tucker P. W., Smithies O. A mouse alpha-globin-related pseudogene lacking intervening sequences. Nature. 1980 Jul 17;286(5770):222–226. doi: 10.1038/286222a0. [DOI] [PubMed] [Google Scholar]
- Wallace R. B., Shaffer J., Murphy R. F., Bonner J., Hirose T., Itakura K. Hybridization of synthetic oligodeoxyribonucleotides to phi chi 174 DNA: the effect of single base pair mismatch. Nucleic Acids Res. 1979 Aug 10;6(11):3543–3557. doi: 10.1093/nar/6.11.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]