Summary
One of the advantages of nanotechnology is the feasibility to construct therapeutic particles carrying multiple therapeutics with defined structure and stoichiometry. The field of RNA nanotechnology is emerging. However, controlled assembly of stable RNA nanoparticles with multiple functionalities which retain their original role is challenging due to refolding after fusion. Herein, we report the construction of thermodynamically stable X-shaped RNA nanoparticles to carry four therapeutic RNA motifs by self-assembly of reengineered small RNA fragments. We proved that each arm of the four helices in the X-motif can harbor one siRNA, ribozyme, or aptamer without affecting the folding of the central pRNA-X core, and each daughter RNA molecule within the nanoparticle folds into their respective authentic structures and retains their biological and structural function independently. Gene silencing effects were progressively enhanced as the number of the siRNA in each pRNA-X nanoparticles gradually increased from one to two, three, and four. More importantly, systemic injection of ligand-containing nanoparticles into the tail-vein of mice revealed that the RNA nanoparticles remained intact and strongly bound to cancers without entering the liver, lung or any other organs or tissues, while remaining in cancer tissue for more than 8 h.
Keywords: Nanotechnology, Nanobiotechnology, Nanomotor, DNA packaging motor, Bacteriophage phi29, RNA nanotechnology, RNA nanoparticle, RNA therapeutics
Introduction
Living organisms possess wide assortments of elegant nanomachines, patterned arrays and highly structured macromolecules performing diverse biological functions. Macromolecules of DNA, RNA and proteins have intrinsically defined features at the nanometer scale and can serve as powerful building blocks for the bottom-up fabrication of biomimetic nanostructures and nanodevices [1,2]. RNA is a particularly attractive candidate for such applications [3—8], since it can be designed and manipulated with a level of simplicity characteristic of DNA, while possessing a versatile flexibility in structure and function similar to some properties of proteins [9]. Simple chemical modifications such as, 2′-Fluoro (2′F) can generate RNAs resistant to degradation without changing its folding into appropriate 3D structure, while retaining authentic biological and enzymatic functions [10,11].
There are many types of RNA molecules that could potentially be utilized for nanotechnology-based therapy such as small interfering RNAs [12—14], ribozymes [15—17], RNA aptamers [18,19], riboswitches [20,21], and miRNAs [22—24]. Although the methods for gene silencing with high efficacy and specificity have been achieved in vitro, the effective delivery of RNA to specific cells in vivo remains challenging. The development of a safe, efficient, specific and nonpathogenic nanodevice for the delivery of multiple therapeutic RNAs is in high demand. RNA nanotechnology holds great potential in this regard: (1) Homogeneous RNA nanoparticles can be manufactured with high reproducibility and known stoichiometry, thus avoiding unpredictable side effects or nonspecific toxicity associated with heterogeneous structures. (2) Using the bottom up approach, RNA nanoparticles can be assembled harboring multiple therapeutic, reporter and/or targeting payloads for synergetic effects [12]. (3) Cell type-specific gene targeting can be achieved via simultaneous delivery and detection modules which reduces off-target toxicity and lowers the concentration of the drug administered, thus reducing the side effects of the therapeutics. (4) RNA nanoparticle size typically ranges from 10—50 nm, an optimal size for a non-viral vector as they are large enough to be retained by the body yet small enough to pass through the cell membrane via the cell surface receptors mediated endocytosis. The advantageous size has the potential to greatly improve the pharmacokinetics, pharmacodynamics, biodistribution, and toxicology profiles by avoiding non-specific cell penetration [25]. (5) Protein-free RNA nanoparticles with RNA aptamers as anti-receptors can yield superior specificity compared to protein anti-receptors while displaying lower antibody-inducing activity. This will provide an opportunity for repeated administration and treatment of chronic diseases. (6) RNA nanoparticles are treated as chemical drugs rather than biological entities, which might facilitate FDA approval.
However, one of the challenges in this emerging field of RNA nanotechnology is the relative instability of the nanoparticles without covalent modifications or cross-linking, resulting in the dissociation at ultra low concentrations in vivo after systemic injection. This has seriously hindered the delivery efficiency and therapeutic applications of RNA nanoparticles.
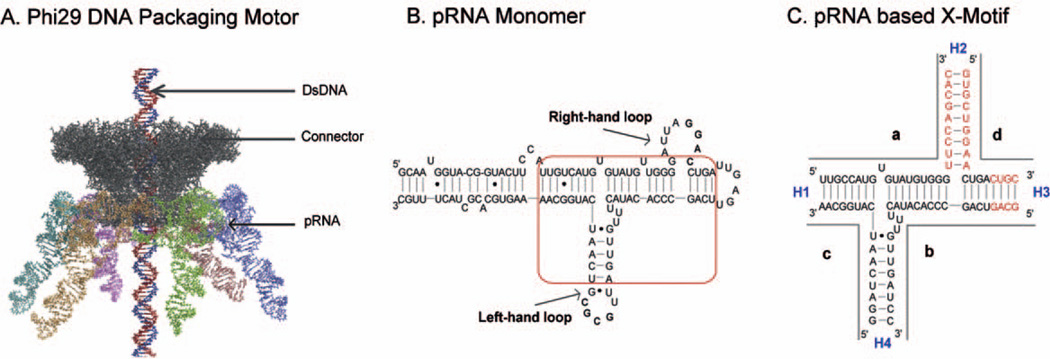
The feasibility of RNA nanotechnology in disease therapy has been exemplified in the phi29 pRNA therapeutic system [11—15,17,26]. The DNA packaging motor of bacteriophage phi29 (Fig. 1A) is geared by a hexameric pRNA ring [27—29], which contains two functional domains [30,31]. The central domain of each pRNA subunit contains two interlocking loops, denoted as the right- and left-hand loops (Fig. 1B) that can be reengineered to form dimers or trimers via hand-in-hand interactions [32—36]. The helical DNA packaging domain is located at the 5′/3′ paired ends [31,37]. The two domains are connected by a three-way junction (3WJ) region [11]. We have recently demonstrated that the 3WJ region extracted from the pRNA can be assembled from three pieces of RNA oligos to construct trivalent RNA nanoparticles [11]. In this communication, we demonstrate that the centerfold domain of the pRNA could be engineered to form a X-shaped motif (Fig. 1C), which was thermodynamically stable, resistant to denaturation by 8 M urea and remained intact at ultra-low concentrations. Incubation of four RNA oligos each carrying one of the four small RNA molecules, siRNA, receptor binding aptamer, or folate resulted in the formation of tetravalent RNA nanoparticles as potential therapeutic agents. We proved that each one of the four helices in the X-motif can serve as a sticky-end to link one siRNA or other therapeutic molecules without affecting the folding of the central pRNA-X core. Systemic injection of ligand-containing nanoparticles into the tail-vein of mice revealed that the RNA nanoparticles remained intact and strongly bound to cancers without entering the liver, lung or any other vital organs or tissues.
Figure 1.
Sequence and secondary structure of phi29 DNA-packaging RNA (pRNA). (A) Illustration of the phi29 packaging motor geared by hexameric pRNA ring (cyan, orange, green, blue, brown, and purple structures). (B) Sequence of pRNA monomer Ab′. The central domain for constructing the pRNA-X is boxed. (C) The core of the pRNA-X domain composed of four RNA oligos (a, b, c, and d). Helical segments are represented as H1, H2, H3 and H4. The additional bases used to construct the pRNA-X motif in helices H2 and H3 are marked in red. Ab′ indicates non-complementary loops [27].
Results and discussion
Length and sequence requirements of the helical stem regions of the pRNA for the assembly of the X-motif
The pRNA-X motif was constructed by (1) opening the right-hand loop of pRNA to insert 9 base pairs, thereby forming a double helical segment (Helix H2), and (2) extending the H3 helix by 4 base pairs (Fig. 1C). The length of the helices H1, H3 and H4 were 8 base pairs, respectively, while H2 was 9 base pairs long. Although 6 base pairs are sufficient for the assembly of the junction domain, 8 base pairs are necessary to keep the junction domain stable under strongly denaturing conditions [11].
Thermodynamically stable properties displayed by the pRNA X-motif
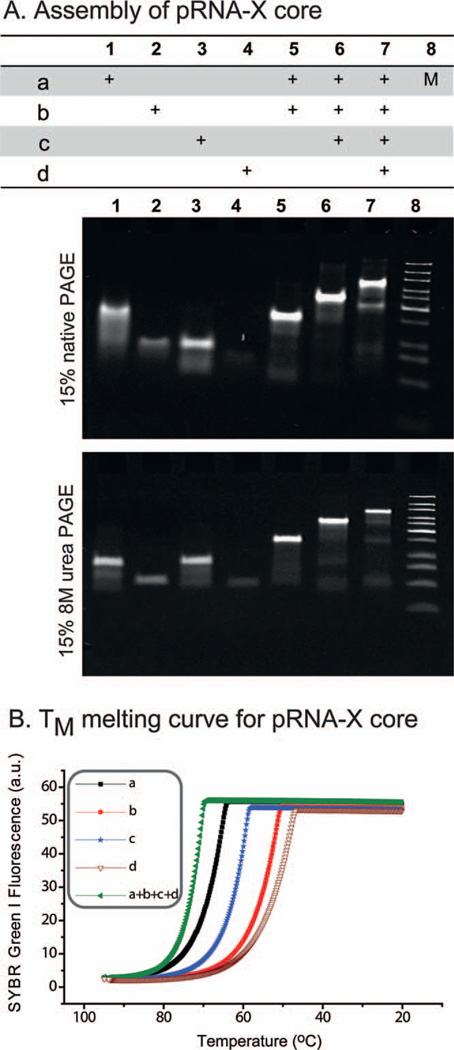
The pRNA-X motif was assembled by mixing four RNA oligos, denoted a, b, c, and d in stoichiometric ratio at room temperature. The affinity and efficiency of assembly was investigated by both gel shift assays and melting experiments conducted under the physiological buffer TMS in the presence of 5 mM magnesium and 100 mM sodium chloride at pH 7.6. The gel-shift assays demonstrate that if one or two strands are omitted (lanes 1—6), they have a faster migration rate compared to the pRNA-X-core (lane 7) (Fig. 2A, top). The core remained stable in 8 M urea (Fig. 2A, bottom), thereby demonstrating its stable properties. Melting experiments displayed a very smooth, high-slope temperature dependent melting curve indicating that the four oligos of the pRNA-X core (TM of 62.7 ± 3.2 °C) have a higher affinity to interact favorably compared to any of the individual component strands (Fig. 2B). The robust attributes of the 3WJ core have already been demonstrated using competition assays at different temperatures and in presence of 0—8 M urea [11].
Figure 2.
Assembly and stability of the pRNA-X core. In the table, ‘+’ indicates the presence of the RNA oligo in samples of the corresponding lanes M: DNA ladder. (A) 15% native PAGE and 8 M denaturing PAGE gels showing the step-wise assembly of the pRNA-X core. (B) Melting curves for the individual pRNA-X strands (magenta, red, blue, and black) and the pRNA-X core (green).
Self-assembly of stable RNA nanoparticles harboring small RNA molecules linked to the X-motif
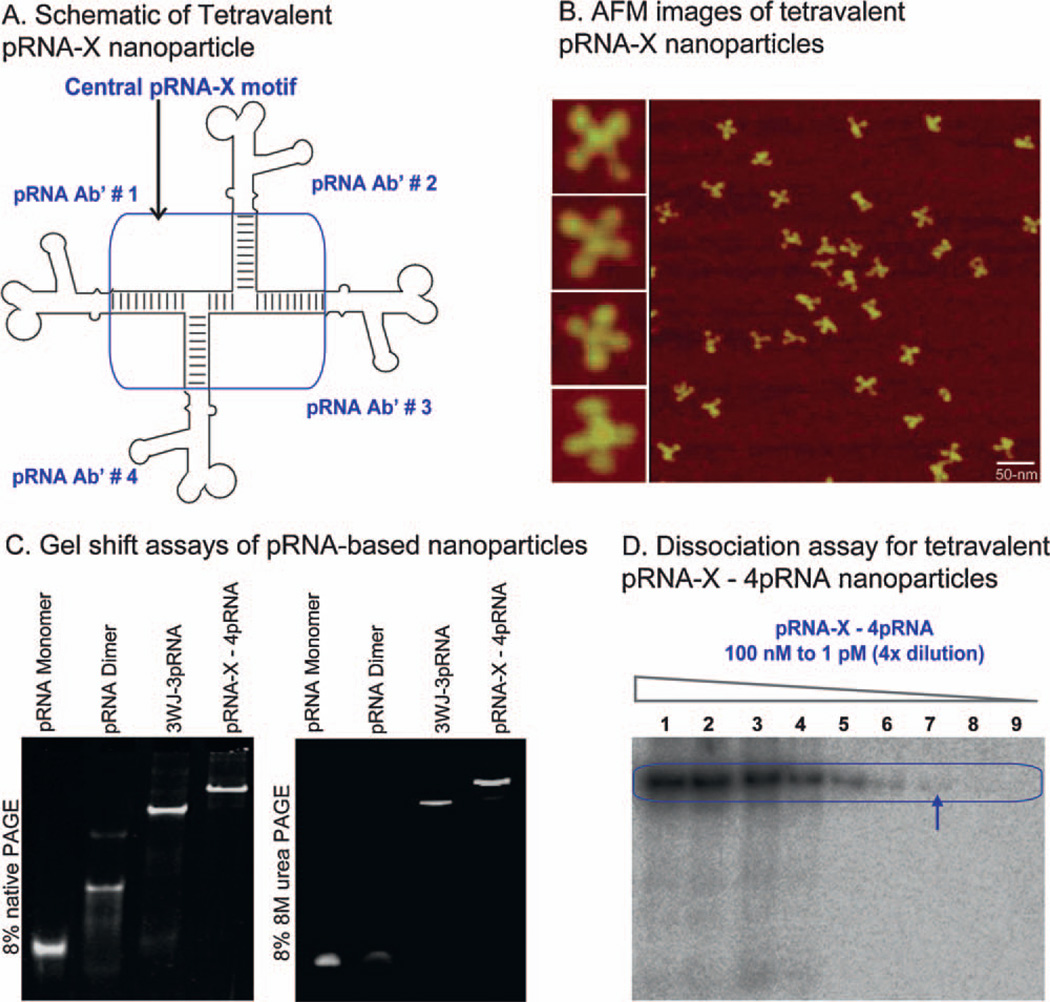
Extension of the phi29 pRNA at the 3′-end does not affect the folding of pRNA global structure [11,34]. Accordingly, the sequences of the four RNA oligos a, b, c, and d were placed at the 3′-end of the pRNA monomer, Ab′. Mixing of the four resulting pRNA chimeras at equimolar concentrations led to the assembly of X-shaped branched nanoparticles harboring one pRNA at each branch. AFM images confirmed the formation of larger RNA complexes with four-branches (Fig. 3A and B), which were consistent with gel shift assays comparing monomer (Ab′), dimer (Aa′), 3WJ-3pRNA [11] and pRNA-X—4pRNA (Fig. 3C). The dimer (Aa′) formed via hand-in hand interactions of interlocking loops is only stable up to 2 M urea, while the 3WJ—3pRNA and the pRNA-X—4pRNA are both stable in presence of 8 M urea. The pRNA-X—4pRNA nanoparticles can also be co-transcribed and self-assembled in one step during transcription with high yield.
Figure 3.
Construction of tetravalent pRNA-X nanoparticles harboring monomeric pRNA at each branch. (A) Schematic of pRNA-X—4pRNA constructs, (B) corresponding AFM images, (C) 8% native (left) and denaturing (right) PAGE gel, (D) dissociation assay for the pRNA-X—4pRNA constructs by 4-fold serial dilution with [32P] pRNA-X—4pRNA (lanes 1—9) from 100 nM to 1 pM. Arrow indicates the lowest detectable concentration of 25 pM. Scale bar: 50 nm.
As a candidate therapeutic RNA nanoparticle, the pRNA-X constructs with four branches harboring multi-module functionalities have to remain intact after systemic delivery, where it will exist at ultra low concentrations due to dilution by circulating blood. To assay the dissociation, [α-32P] labeled pRNA-X nanoparticles were serially diluted to extremely low concentrations (100 nM to 1 pM). The concentration for dissociation was below the detection limit of the [32P]-labeling technology; at 25 pM in TMS buffer, the lowest detectable concentration, the pRNA-X nanoparticles showed no signs of dissociation (Fig. 3D).
Construction of a variety of therapeutic RNA nanoparticles using the X-motif as scaffold
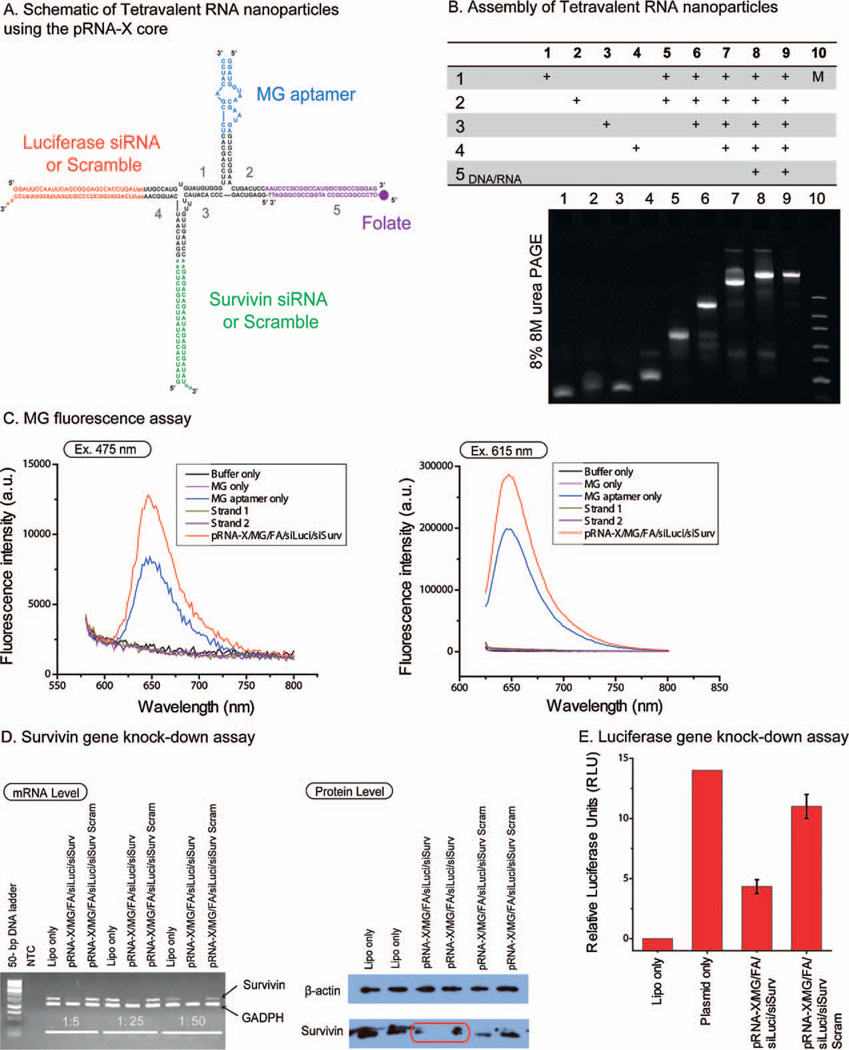
Tetravalent RNA nanoparticles were constructed using the pRNA-X motif as a scaffold by incorporating four functional modules: MG (malachite green dye, triphenylmethane) aptamer, luciferase siRNA (siLuci), survivin siRNA (siSurv) and folate (FA) (Fig. 4A), denoted [pRNA-X/MG/FA/siLuci/siSurv] or corresponding scramble siRNA control. The presence of the functional moieties did not interfere with the formation of the pRNA-X core and the tetravalent complex assembled with high affinity (Fig. 4B). The purified constructs (lane 9, Fig. 4B) were stable in absence of magnesium and remained intact under strongly denaturing conditions, even after the incorporation of functionalities. In the next sections, we evaluated whether the incorporated RNA moieties in the pRNA-X nanoparticles retain their original folding and functionalities.
Figure 4.
Construction of multi-module RNA nanoparticles harboring MG (malachite green) aptamer, folate, luciferase siRNA, and survivin siRNA. (A) Schematic and sequences of the tetravalent pRNA-X constructs. (B) Step-wise assembly of RNA nanoparticles using the pRNA-X as scaffold with functionalities assayed by 8% denaturing urea PAGE. In the table, ‘+’ indicates the presence of the RNA strands in samples of the corresponding lanes. (C) Functional assay of the MG aptamer incorporated in pRNA-X nanoparticles. MG fluorescence was measured using excitation wavelengths 475 and 615-nm. (D) Target gene knock-down effects of survivin siRNA showed by RT-PCR (GADPH is the endogenous control) on mRNA level and Western Blot assay (β-actin bands served as loading control) on protein level. (E) Dual-luciferase assay for target gene knock-down of luciferase gene. The relative firefly luciferase activity reflects the level of luciferase gene expression and is obtained by normalizing firefly luciferase activity using the internal control renilla luciferase activity. Error bars represent s.d. (N = 3).
Assessment of MG fluorescence
MG binding aptamer [38] was used as model system for structure and function verification. Free MG is not fluorescent by itself, but emits fluorescent light after binding to the aptamer. Fused MG-binding aptamer retained its capacity to bind MG, as revealed by its fluorescence emission (Fig. 4C). The fluorescence is comparable to optimized positive controls and therefore confirming that the MG aptamer assembled from two strands of the pRNA-X after incorporation into the RNA nanoparticles.
Targeted gene silencing assay in cancer cell model
Two pRNA-X nanoparticles were constructed for assaying the gene silencing effects harboring: (1) folate and survivin siRNA [pRNA-X/MG/FA/siLuci/si/Surv]; (2) folate and survivin siRNA scramble control [pRNA-X/MG/FA/siLuci/siSurv Scram]. After 48-h transfection, both reverse transcription-PCR (RT-PCR) assayed on mRNA level and western blot assayed on protein expression confirmed reduced survivin gene expression level of [pRNA-X/MG/FA/siLuci/si/Surv] nanoparticles compared to the scramble control (Fig. 4D). The mechanism of siRNA release from the pRNA nanoparticles is by Dicer processing, as established previously [14].
Targeted gene silencing of luciferase
Dual-luciferase reporter system was used to quantitatively measure the gene silencing effects of the pRNA-X constructs harboring the siRNA targeting firefly luciferase gene [39] (Fig. 4A). The relative luciferase activity was used to reflect the expression level of firefly luciferase gene by normalizing the firefly luciferase activity with the internal control, renilla luciferase activity. The results indicated that [pRNA-X/MG/FA/siLuci/si/Surv] nanoparticles displayed ~70% decrease in firefly luciferase gene expression (Fig. 4E).
Cell binding and entry of pRNA-X nanoparticles
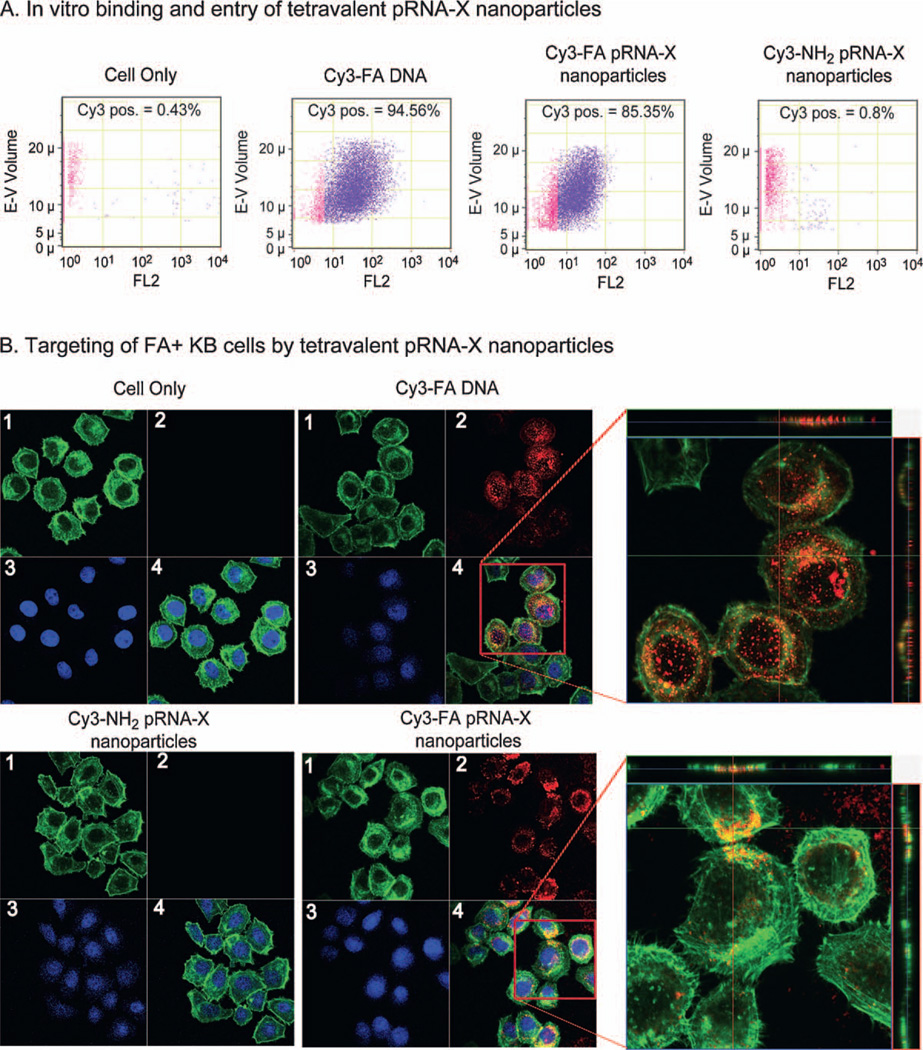
Folate was incorporated in the pRNA-X nanoparticles to serve as a cancer cell delivery agent via folate receptor-mediated endocytosis [11,14,40]. Fluorescent pRNA-X nanoparticles with folate conjugated into one of the branches of the pRNA-X complex were tested for cell binding efficiency. pRNA-X harboring FA and Cy3 labels [Cy3—pRNA-X/MG/FA/siLuci/si/Surv] served as the test sample, while the negative control harbored NH2 and Cy3 labels [Cy3—pRNA-X/MG/NH2/siLuci/si/Surv]. Flow cytometry (Fig. 5A) and confocal imaging indicated a strong binding of the RNA nanoparticles and efficient entry into the targeted cells, as demonstrated by the excellent co-localization and overlap of the fluorescent pRNA-X nanoparticles (red) and cytoplasma (green) (Fig. 5B).
Figure 5.
Binding and entry of tetravalent pRNA-X nanoparticles into targeted cells. (A) Flow cytometry revealed that [pRNA-X/MG/FA/siLuci/si/Surv] nanoparticles bound and specifically entered cells. Positive and negative controls were Cy3—FA—DNA and Cy3[pRNA-X/MG/NH2/siLuci/si/Surv] (without FA), respectively. (B) Confocal images showed targeting of folate receptor positive (FA+) KB cancer cells by the co-localization (overlap, 4) of cytoplasm (green, 1) and fluorescent RNA nanoparticles (red, 2) (magnified, right panel). Blue represents nuclei, 3.
Gene silencing effects were progressively enhanced as the number of siRNA in each pRNA-X nanoparticles increased gradually from one, two three to four
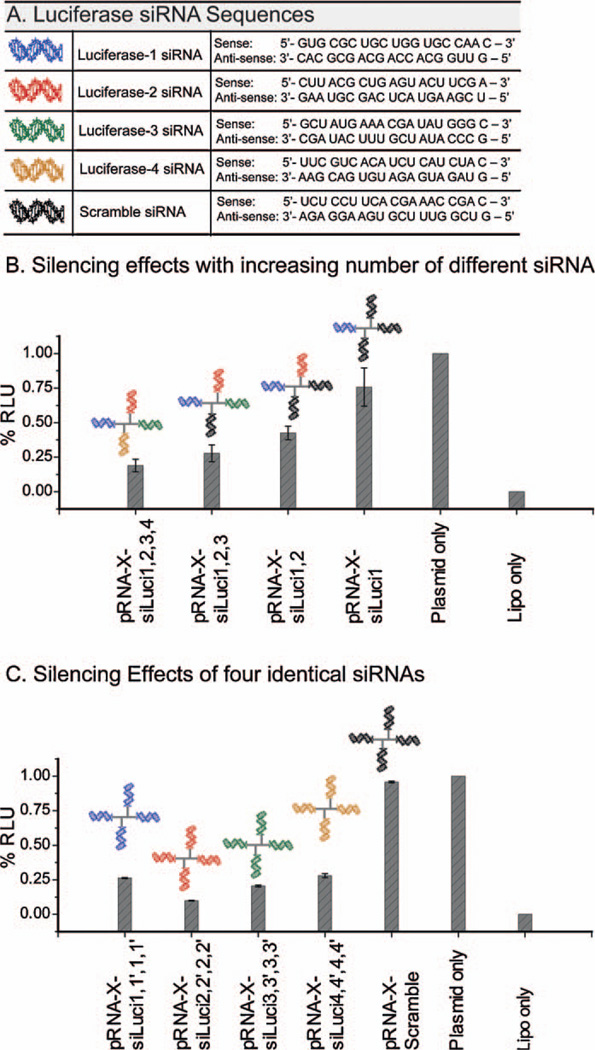
Tetravalent pRNA-X complexes were constructed harboring multiple luciferase siRNAs to assay for enhanced gene silencing effects. Dual-luciferase reporter system was used to quantitatively measure the gene silencing effects. For all the constructs, the total concentration of RNA was kept constant at 1.25 nM. The target sites on the luciferase gene for the four siRNAs (Fig. 6A) were located at 153—173, 196—216, 498—518, and 846—869 positions, as published in the literature [41,42]. The incorporation of four identical siRNA sequences compromised the assembly of the X-motif due to self-folding of the complementary sequences of the respective siRNAs. To facilitate the assembly, the siRNA sequences were reversed (denoted with a prime, such as siLuci-1′) at alternate helical branch locations. The reversed sequences had no impact on the functionality of the siRNA.
Figure 6.
Construction of tetravalent pRNA-X nanoparticles harboring multiple siRNA for enhanced gene silencing effects. (A) Sequences and notations of siRNA used in tetravalent constructs. Blue: siLuci-1 and 1′; red: siLuci-2 and 2′; green: siLuci-3 and 3′; orange: siLuci-4 and 4′; black: control siRNA [42]. (B) and (C) Quantification of Luciferase gene expression: Effects of increasing number of different Luciferase siRNAs (siLuci-1, 2, 3 and 4) (A); and four identical siRNA constructs (siLuci-1, 2, 3 or 4) incorporated in the pRNA-X motif (B). RLU, relative luciferase units; siLuci-1′, 2′ 3′ and 4′ represent reversed siRNA sequences for siLuci-1, 2, 3 and 4, respectively. Error bars represent s.d. (N = 3).
Silencing effects with increasing number of different luciferase siRNA
As the number of different luciferase siRNAs were gradually increased in the pRNA-X motif, progressive increase in silencing effects were observed as follows, ~25% (for 1 siRNA; 3 scramble siRNA), 57% (for 2 siRNA; 2 scramble siRNA), 72% (for 3 siRNA; 1 scramble siRNA), and 81% (for 4 siRNA) (Fig. 6B).
Silencing effects of four identical luciferase siRNAs
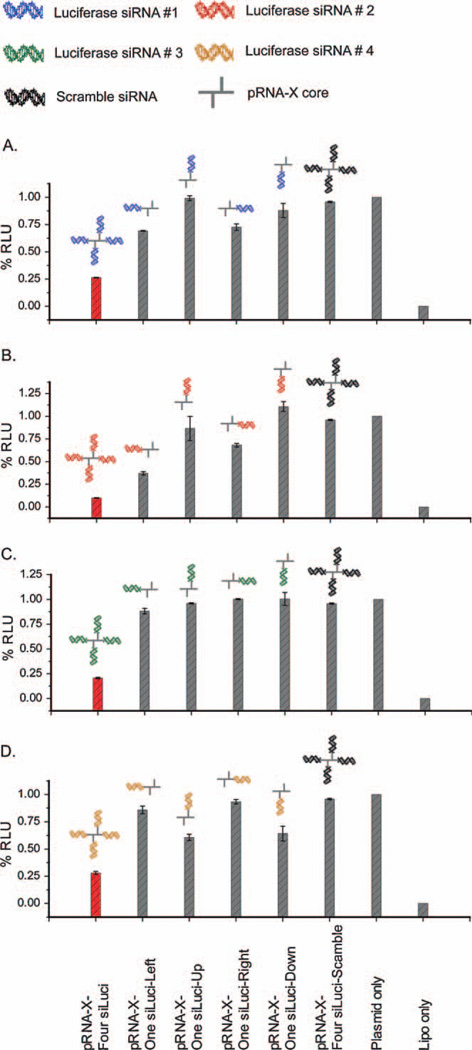
Significant silencing effects were observed in presence of four identical siRNAs fused to the pRNA-X motif, compared to a single siRNA as follows, ~74% (for four siLuci-1), ~90% (for four siLuci-2), ~80% (for four siLuci-3), and ~72% (for four siLuci-4) (Fig. 6C). For comparison, we constructed the X-motif harboring a single siRNA (either siLuci-1 or 2 or 3 or 4) at helical locations H1, H2, H3 or H4, respectively (Fig. 7A—D). The silencing effects of the four different siRNAs increased following the trend, siLuci-2 > siLuci-1 ≈ siLuci-4 > siLuci-3. The functionality of the siRNA was comparable at each of the arms of the X-motif. The data demonstrated that greatly enhanced effects were observed in presence of four identical siRNAs compared to a single siRNA (Fig. 7A—D).
Figure 7.
Comparison of gene silencing effects for a single vs. multiple siRNAs incorporated in the pRNA-X motif. (A—D) Four identical siRNA (siLuci-1, A; siLuci-2, B; siLuci-3, C; siLuci-4, D) compared with a single siRNA harbored at each pRNA-X motif arm; RLU: relative luciferase units; siLuci-1′, 2′ 3′ and 4′ represent reversed siRNA sequences for siLuci-1, 2, 3 and 4, respectively. Error bars represent s.d. (N = 3).
Silencing effects of pRNA-X harboring four luciferase siRNAs versus single standard siRNA
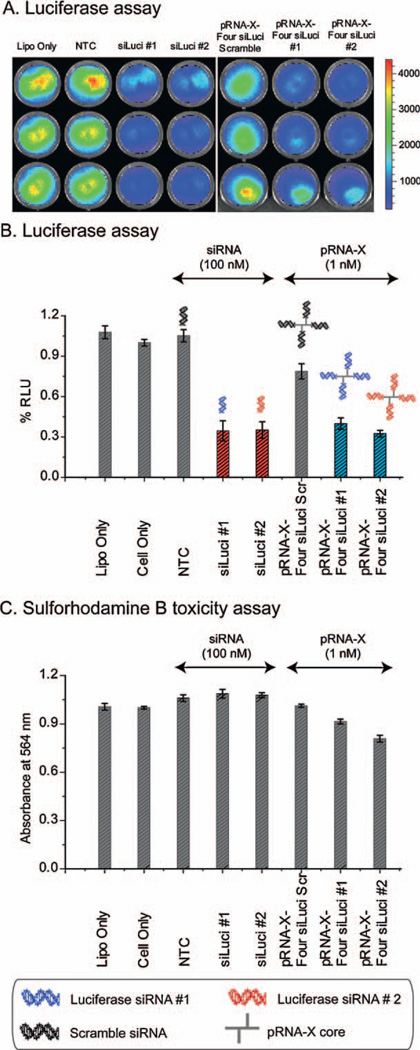
Firefly luciferase reporter was chosen as knockdown target for in vitro pRNA-X treatment. Two siRNA sequences (siLuci-1 and 2) were selected and tested in HT29 colon cancer cells with stable luciferase expression. To demonstrate that the pRNA-X system provides a superior delivery vehicle for siRNA, a direct comparison of the in vitro knock-down efficacy compared to standard siRNA (siLuciferase-1 and 2) was carried out (Fig. 8A and B). A 1% concentration of pRNA-X nanoparticles with four siRNA modules can achieve the same silencing effects (~60—70% decrease compared to scramble controls) as the siRNA by itself. Absence of nonspecific luciferase inhibition was confirmed by sulforhodamine B assay (Fig. 8C).
Figure 8.
(A) and (B) siRNA and pRNA-X nanoparticles were transfected into HT29 GFP-Luc cells with Lipofectamine 2000 (siRNA#1 and 2: 100 nM; pRNA-X: 1 nM). (C) Effect of pRNA transfection on the cytotoxicity in HT29 cells. Growth inhibition was measured after treatment of cells with pRNA for 24 h in 96-well plates. Cell numbers were quantitated by staining with sulforhodamine B and expressed relative to cells treated with Lipofectamine alone. RLU, relative luciferase units. Error bars represent s.d. (N = 3).
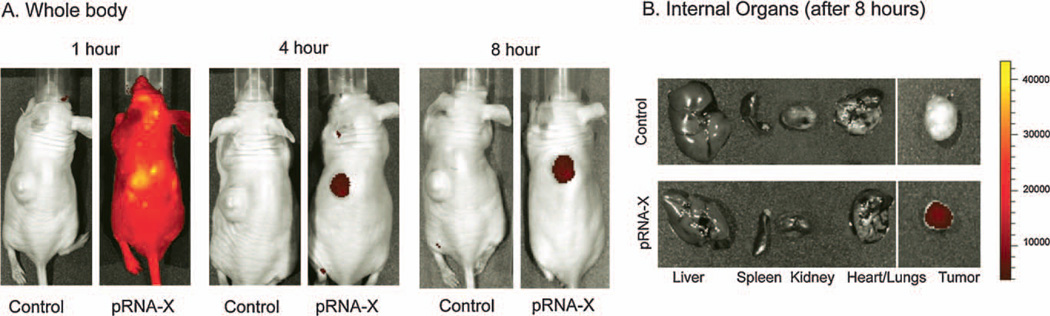
In vivo targeting of RNA nanoparticles to cancer xenograft by systemic injection
To confirm the chemical and thermodynamic stability of the X-shaped RNA nanoparticles for specific cancer targeting in vivo, RNA nanoparticles were constructed with one of the four RNA fragments carrying the folate to serve as a ligand for binding to the cancer cells, and another RNA fragment carrying the fluorescent dye Alexa-647. The nanoparticles were systemically injected (i.e., tail vein injections) into athymic nude mice bearing KB cells (folate receptor positive) xenografts in the subcutaneous flanks. Whole body imaging (at time points, 1-h, 4-h and 8-h) were carried out after intravenous administration of FA—pRNA-X—Alexa647 nanoparticles. The pRNA-X nanoparticles strongly bound to the tumor xenografts within 4-h (Fig. 9). The animals were sacrificed at 8-h time point and organ imaging revealed that fluorescence was localized specifically in the tumor and was not detected in other organs of the body, indicating that the particles are not trapped in the liver, lungs, kidneys or other tissues or organs after systemic injection (Fig. 9B). The results were confirmed to be reproducible by three independent labs with five trials. Together, these findings suggest a very selective targeting of the pRNA nanoparticles to tumors and not to normal tissues which would make this delivery system highly efficacious for future clinical applications.
Figure 9.
In vivo binding and targeting of pRNA-X nanoparticles. (A) The pRNA-X (harboring Folate and Alexa-647) nanoparticles specifically targeted folate-receptor positive tumor xenografts upon systemic administration in nude mice, as revealed by whole body imaging (A), and internal organ imaging (B). Control: PBS treated mice. Scale bar: fluorescent intensity.
Conclusions
We demonstrated that the central domain of bacteriophage phi29 motor pRNA can be engineered into a stable X-motif to carry four functional modules in the absence of metal ions. The resulting nanoparticles were thermodynamically stable, and resistant to dissociation under strongly denaturing conditions or at ultra-low concentrations. Incubation of four RNA oligos representing therapeutic functional motifs resulted in the self-assembly of tetravalent RNA nanoparticles as potential therapeutic agents. Progressive enhancement of gene silencing effects were observed when the number of siRNA in each pRNA-X nanoparticles increased from one to two, three and four. The results demonstrated that a wide range of therapeutic RNA molecules targeting cancer and viral infected cells can potentially be fused to the multivalent pRNA-X motif to achieve enhanced silencing effects.
Experimental procedures
In vitro synthesis and purification of pRNA
The pRNA were synthesized by enzymatic methods as described previously [31]. RNA oligos were synthesized chemically by IDT (Iowa). RNAs were purified by 8 M urea 8% PAGE. The corresponding bands were excised under UV shadow and eluted from the gel over 4 h at 37 °C in the elution buffer (0.5 M NH4OAc, 0.1 mM EDTA, 0.1% SDS, and 0.5 mM MgCl2) followed by ethanol precipitation overnight at −20 °C (2.5 volume of 100% ethanol and 1/10 volume of 3 M NaOAc). The precipitate was pelleted by centrifugation (16500 × g, 30 min), washed with 70% ethanol, and dried by speed vacuum. Finally, the RNA dried pellet was rehydrated in 0.05% DEPC treated water and stored at −20 °C.
Construction of multi-module RNA nanoparticles
The sequences for each of the RNA strands a, b, c, and d were added to the 3′-end of each 117-nt pRNA-Ab′ and synthesized using two primers: 3′-end primer encoding the a, b, c, and d, respectively, and a common 5′-end primer and the Ab′ template. The template was then used in transcription mixture and the pRNA was then purified in 8% urea PAGE gel in TBM buffer, as described previously [11].
The sequences for the luciferase siRNA, survivin siRNA, malachite green (MG) binding aptamer and folate labeled RNA were rationally designed with the sequences of the strands a, b, c, and d, respectively (Fig. 4). Multi-module nanoparticles pRNA-X/MG aptamer/Folate/luciferase siRNA and survivin siRNA, denoted [pRNA-X/MG/FA/siLuci/siSurv] or scramble control, denoted [pRNA-X/MG/FA/siLuci/siSurv Scram] were assembled from five individual fragments including a 26-nt folate labeled RNA (Trilink) or folate-DNA strand (synthesized in house). The individual RNA strands (fragments 1, 2, 3 and 4, Fig. 4A) were transcribed from DNA template amplified by PCR. Fluorescent dyes were labeled on one RNA strand by using the Label IT® siRNA Tracker Intracellular Localization Kit, Cy3™ (Mirus Bio LLC). The five RNA strands were mixed after purification in TMS buffer at equal molar ratio and then heated up to 80 °C for 5 min, followed by slow cooling to 4 °C. The assembled nanoparticles were then purified from 8% native PAGE gel.
Dilution assay to test dissociation at extremely low concentrations
The stability of the tetravalent pRNA-X—4pRNA nanoparticles were evaluated by radiolabel assays. Purified [32P] pRNA-X—4pRNA complexes were serially diluted from 100 nM to 1 pM in TMS buffer, and then loaded onto 8% native PAGE gel for autoradiograph.
Melting experiments
The melting experiments were conducted by monitoring the fluorescence of the pRNA-X component strands using the LightCycler® 480 Real-Time PCR System (Roche). 1× SYBR Green I dye (Invitrogen) (emission 465—510 nm), which binds double-stranded nucleic acids, but not single-stranded ones was used for all the experiments. The respective RNA oligonucleotides were mixed at room temperature in physiological TMS buffer. The pRNA-X core strands were slowly cooled from 95 °C to 20°C at the ramping rate of 0.11 °C/s. Data was analyzed by the LightCycler® 480 Software using the first derivative of the melting profile. The TM value represents the mean and standard deviation from 4 independent experiments.
Flow cytometry analysis of folate mediated cell binding
Human cervical cancer Hela cells [American Type Culture Collection (ATCC)] were maintained in folate-free RPMI-1640 medium (Gibco), then trypsinized and rinsed with PBS (137 mM NaCl, 2.7 mM KCl, 100 mM Na2HPO4, 2 mM KH2PO4, pH 7.4). 200 nM Cy3 labeled pRNA-X complexes harboring folate [Cy3—pRNA-X/MG/FA/siLuci/si/Surv], and folate-free control [Cy3—pRNA-X/MG/NH2/siLuci/si/Surv] were each incubated with 2 × 105 KB cells at 37 °C for 1 h. After washing with PBS, the cells were resuspended in PBS buffer. Flow cytometry (Beckman Coulter) was used to observe the cell binding efficacy of the Cy3—pRNA-X nanoparticles.
Confocal microscopy
Hela cells were grown on glass coverslides in folate free medium overnight. Cy3 labeled pRNA-X complexes harboring folate [Cy3—pRNA-X/MG/FA/siLuci/si/Surv], and folate-free control [Cy3—pRNA-X/MG/NH2/siLuci/si/Surv] were each incubated with the cells at 37 °C for 2 h. After washing with PBS, the cells were fixed by 4% paraformaldehyde and stained by Alexa Fluor® 488 phalloidin (Invitrogen) for cytoskeleton and TO-PRO®-3 iodide (642/661) (Invitrogen) for nucleus. The cells were then assayed for binding and cell entry by Zeiss LSM 510 laser scanning confocal microscope.
Malachite green (MG) aptamer fluorescence assay
The pRNA-X tetravalent RNA nanoparticles [pRNA-X/MG/FA/siLuci/si/Surv] harboring MG binding aptamer (100 nM) [38] was mixed with MG (2 µM) in binding buffer containing 100 mM KCl, 5 mM MgCl2, and 10 mM HEPES (pH 7.4) and incubated at room temperature for 30 min (Fig. 4C). The fluorescence was measured using a fluorospectrometer (Horiba Jobin Yvon; SPEX Fluolog-3), excited at 475 nm (scanning from 540 to 800 nm for emission) and 615 nm (scanning from 625 to 800 nm for emission).
Assay for the silencing of genes in cancer cell model
Two pRNA-X nanoparticles were constructed for assaying the gene silencing effects harboring: (1) folate and survivin siRNA [pRNA-X/MG/FA/siLuci/si/Surv]; (2) folate and Survivin siRNA scramble control [pRNA-X/MG/FA/siLuci/siSurv Scram].
Hela cells were transfected with 25 nM of the individual pRNA-X complexes using Lipofectamine 2000 (Invitrogen). After 48-h treatment, cells were collected and target gene silencing effects were assessed by both RT-PCR and Western blot assays.
Cells were processed for total RNA using illustra RNAspin Mini kits (GE Healthcare, Buckinghamshire, UK). The first complementary DNA strand was synthesized on mRNA (500 ng) from Hela cells using SuperScript III First-Strand Synthesis System (Invitrogen) according to manufacturer’s instruction. PCR was performed using GoTaq Flexi DNA polymerase (Promega). Reactions were carried out in a final volume of 25 µL which contained complementary DNA from first-strand synthesis (the cDNA template was 1:5, 1:25 and 1:50 diluted respectively), 1× GoTaq Flexi colorless buffer, 2.5 mmol/L Mg2+, 0.2 mmol/L deoxynucleoside triphosphates, 0.2 µmol/L of each primer, and 0.02 U/µL GoTaq Flexi DNA polymerase. The PCR condition was 95 °C for 5 min then 25 cycles of 94 °C for 1 min, 55 °C for 1 min and 72 °C for 1 min, followed by 72 °C for 10 min.
Primers for human GAPDH [43] and survivin [44] are:
GAPDH left: 5′-ACGGATTTGGTCGTATTGGGCG-3′;
GAPDH right: 5′-CTCCTGAAGATGGTGATGGAA-3′;
Survivin left: 5′-GCATGGGTGCCCCGACGTTG-3′;
Survivin right: 5′-GCTCCGGCCAGAGGCCTCAA-3′.
Cells were rinsed and harvested in lysis buffer. Protein concentrations were determined and equal amounts of proteins were loaded onto a 15% PAGE. Membranes were blocked, incubated with primary antibody to survivin and β-actin (R&D Systems, Minneapolis, MN), and conjugated to a secondary antibody (Sigma—Aldrich, St. Louis, MO). Membranes were then blotted by ECL kit (Millipore, Billerica, MA) and exposed to film.
Assay for the silencing of genes in cancer cell model
Dual-luciferase assays were used to test the potential of the pRNA-X complex in escorting siRNA delivered into cells. For dual-luciferase assays, Hela cells were seeded in 24-well plates. Gene silencing assays were performed by co-transfecting [pRNA-X/MG/FA/siLuci/si/Surv] with both plasmid pGL3 and pRL-TK (Promega, Madison, WI) coding for firefly and renilla luciferase, respectively. The latter served as an internal control to normalize the luciferase data (Dual-Luciferase Reporter Assay System; Promega). Cells were washed once with PBS and lysed with passive lysis buffer. The plates were shaken for 15 min at room temperature. 20 µL of lysate were added to 100 µL of luciferase assay reagent (LAR II) and firefly luciferase activity was measured. Upon addition of 100 µL of Stop & Glo Reagent, control measurements of renilla luciferase activity were then obtained. The data was then normalized with respect to the renilla activity for determining the average ratio of firefly to renilla activity over several trials.
HT29 colon cancer cells, that express GFP and firefly luciferase (HT29 GFP-Luc), were plated at 10,000 cells/well in a 96-well black plate (Corning Life Sciences; Tewksbury, MA) overnight and then transfected with control siRNA (NTC), siLuci#1, and siLuci#2 using Lipofectamine (Invitrogen). The siRNAs and pRNA-X nanoparticles were transfected into HT29 GFP-Luc cells at 100 nM and 1 nM each, correspondingly. d-Luciferin potassium salt (Research Products International Corp, Mount Prospect, IL) was dissolved in sterile PBS to make a stock solution of 10 mg/mL; the cell culture media was removed before addition of 100µL of a 150 µg/mL solution of d-luciferin in PBS. Plates were incubated at 37 °C for 5 min before imaging. The IVIS Spectrum system was used for in vitro imaging of cells in 96-well plates. For quantification of the detected light, regions of interest were drawn by using Living Image 3.1 software, and the photon counts per second from each well were recorded and plotted. Cell numbers were quantified by staining with sulforhodamine B assay (Geno Technology, St. Louis, MO) as described previously [45].
AFM imaging
For all samples, specially modified mica surfaces (APS mica) were used. The APS mica was obtained by incubation of freshly cleaved mica in 167 nM 1-(3-aminopropyl)silatrane. The details of APS mica surface modification is described elsewhere [46,47]. The RNA samples were diluted with 1× TMS buffer to a final concentration of 3—5 nM. Then, the droplet of samples (5—10 µL) was immediately deposited on APS mica. After 2 min incubation on the surface, excess samples were washed with DEPC treated water and dried under a flow of Argon gas. AFM images in air were acquired using MultiMode AFM NanoScope IV system (Veeco/Digital Instruments, Santa Barbara, CA) operating in tapping mode. Two types of AFM probes were used for tapping mode imaging in air: (1) regular tapping Mode Silicon Probes (Olympus from Asylum Research, Santa Barbara, CA) with a spring constant of about 42 N/m and a resonant frequency between 300 and 320 kHz. (2) Non-contact NSG01 _DLC probes (K-Tek Nanotechnology, Wilsonville, OR) with a spring constant of about 5.5 N/m and a resonance frequency between 120 and 150 kHz.
Animal trial: in vivo targeting of tumor xenograft by systemic injection of pRNA-X nanoparticles
Male athymic nudenu/nu (6—8 weeks old) mice were obtained from Taconic (Hudson, NY) and housed in clean, pathogen-free rooms in an environment with controlled temperature (27 °C), humidity, and a 12 h light/dark cycle. All animal experiments were approved by the Institutional Animal Care and Use Committee at University of Kentucky and were conducted in accordance with guidelines issued by the National Institutes of Health for the care of laboratory animals. Mice were fed a folate-free diet (Harlan Laboratories; Indianapolis, IN) for a total of 2 weeks before the experiment and injected with KB cells (3 × 106 cells per mouse in 100 µL PBS) subcutaneously. When the tumors grew to about 500 mm3, the mice were anesthetized using isoflurane gas (2% in oxygen at 0.6 L/min flow rate) and injected intravenously through the tail vein with a single dose of 3 µg of pRNA per gram of body weight of 2′-F U/C modified Folate-Alexa647-labeled pRNA-X nanoparticle in 300 µL of PBS. Whole-body imaging (Ex-Max 650 nm/Em-Max 668 nm) was carried out at 1-h, 4-h and 8-h after pRNA administration on IVIS Spectrum station (Caliper Life Sciences; Hopkinton, MA). Composite images obtained were comprised of black and white digital photos with an overlay of images reflecting fluorescent activity. The density map, measured as photons/second/cm2/steradian (p/s/cm2/sr), were created using the Living Image 3.1 (Caliper Life Sciences; Hopkinton, MA) software and represented as a color gradient centered at the maximal spot. Following CO2 asphyxiation at 8-h after pRNA administration, the tumors, liver, heart, lung and kidney of the mice were dissected and individually imaged.
Acknowledgements
The research was supported by NIH Grants EB003730 and CA151648 to P.G. AFM images were obtained at Nanoimaging Core Facility supported by NIH SIG program, and UNMC Program of ENRI. We thank Dr. Zhenqi Zhu from Dr. Malak Kotb’s Lab at University of Cincinnati for help with the TM assays. PG is a cofounder of Kylin Therapeutics, Inc., and Biomotor and Nucleic Acid Nanotechnology Development Corp. Ltd.
Biographies

Dr. Farzin Haque, Ph.D., is a research assistant professor in the University of Kentucky College of Pharmacy, Department of Pharmaceutical Sciences. He received his B.A. degree in biochemistry and mathematics (2004) from Lawrence University and a Ph.D. degree in chemistry (2008) from Purdue University. He held a postdoctoral appointment (2009—2011) at the University of Cincinnati, with Professor Peixuan Guo. Dr. Haque’s scholarly interest broadly focuses on nanoscience and nanotechnology in biology and medicine. These include, RNA nanotechnology — construction of RNA nanoparticles for therapeutic and diagnostic applications; and nanopore-based technology for single molecule detection and sensing of chemicals and biopolymers.

Dr. Dan Shu, M.D., is a research associate professor in the University of Kentucky College of Pharmacy, Department of Pharmaceutical Sciences. She received her M.D. degree from Tongji Medical University, China in 1990. She worked as a research scientist at Purdue University Cancer Research Center, and Nanobiomedical Center, University of Cincinnati prior to joining the University of Kentucky. Dr. Shu’s research is focused on RNA Nanotechnology-use RNA as a nanotechnology platform to construct RNA nanoparticles with functional modalities for therapeutics, targeting, and diagnostics; and single molecule biophysical studies, such as FRET to understand the structure—function relation for individual biomolecules.

Dr. Yi Shu, Ph.D., is currently a postdoctoral fellow at College of Pharmacy, University of Kentucky. She obtained her B.S. from Lanzhou University, China (2004), M.S. from China CDC (2007), and Ph.D. from Nanobiotechnology Center at College of Engineering and Applied Science, University of Cincinnati (2012). She has a broad training in biology, immunology, virology, and nanobiotechnology. Her recent research is focused on applying bacteriophage phi29 packaging RNA (pRNA) as building blocks for bottom-up assembly of various RNA based nano-delivery platforms (RNA nanotechnology). The constructed RNA nanoparticles are utilized to deliver therapeutics into specific targets for potential cancer and viral disease treatment.

Dr. Luda S. Shlyakhtenko, Ph.D., is a research associate professor in the Department of Pharmaceutical Sciences, and the co-director of Nano-Imaging Core Facility at the College of Pharmacy, University of Nebraska Medical Center. Her work during past two decades on the study of structure, dynamics and interactions of various bimolecular systems was based on the use of atomic force microscopy (AFM) as an imaging and probing tool. She developed several immobilization approaches, such as silatrane surface chemistry to study the structures of different protein/DNA complexes, unusual DNA and RNA structures, the structures of proteins, macromolecular ensembles, and different types of carriers for drug delivery.

Dr. Piotr Rychahou, M.D., is an assistant professor of Markey Cancer Center and Surgery Department at University of Kentucky. He received his M.D. degree from Belarusian State Medical University in 2002 and M.M.S from University of Texas Medical Branch in 2007. His current interests involve studying mechanisms regulating colorectal cancer metastasis, colorectal cancer metastasis models and colorectal cancer metastases treatment with nanotechnology.

Dr. B. Mark Evers, M.D., Since 2009, he has served as Director of Markey Cancer Center; professor and vice-chair, Department of Surgery; Markey Cancer Foundation Endowed Chair; and physician-in-chief, Oncology Service Line, at the University of Kentucky. He earned his M.D. from the University of Tennessee and performed his General Surgery Residency at the University of Louisville. After completing a Gastrointestinal Physiology Fellowship at the University of Texas Medical Branch, he joined that faculty. During his 20 years there he filled many roles, including the Robertson-Poth Distinguished Chair in General Surgery, Director of the Sealy Center for Cancer Cell Biology, and Director of the UTMB Comprehensive Cancer Center. Dr. Evers has received more than 30 awards and held several leadership positions in national organizations. His many research projects, which include a SPORE and NIH MERIT awards, have been continuously funded by the NIH for the past 20 years. He holds three patents, has authored more than 300 publications, and serves on several editorial and review boards.

Dr. Peixuan Guo, Ph.D., is William Farish Endowed Chair in Nanobiotechnology, Markey Cancer Center, professor of College of Pharmacy at University of Kentucky, and director of NIH/NCI Cancer Nanotechnology Platform Partnership Program: “RNA Nanotechnology for Cancer Therapy”. He obtained his Ph.D. from University of Minnesota, and postdoctoral training at NIH, joined Purdue University in 1990, was tenured in 1993, became a full Professor in 1997, and was honored as a Purdue Faculty Scholar in 1998. He constructed phi29 DNA-packaging motor, discovered phi29 motor pRNA, pioneered RNA nanotechnology, incorporated phi29 motor channel into lipid membranes for single-molecule sensing with potential for high-throughput dsDNA sequencing. He is a member of two prominent national nanotech initiatives sponsored by NIH, NSF, NIST, and National Council of Nanotechnology, director of one NIH Nanomedicine Development Center from 2006 to 2011. His work was featured hundreds of times over radio or TV such as ABC, NBC, newsletters NIH, NSF, MSNBC, NCI, ScienceNow, etc. http://www.eng.uc.edu/nanomedicine/peixuanguo.html
Footnotes
Authors contributions
P.G. conceived, designed and led the project; F.H., D.S., and Y.S., designed and conducted the experiments; L.S. obtained the AFM images. P.G., F.H., D.S., and Y.S. analyzed the data and co-wrote the manuscript. R.P and B.M.E carried out the xenograft and animal imaging.
References
- 1.Niemeyer CM. Trends Biotechnol. 2002;20:395. doi: 10.1016/s0167-7799(02)02022-x. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt OG, Eberl K. Nature. 2001;410:168. doi: 10.1038/35065525. [DOI] [PubMed] [Google Scholar]
- 3.Studnicka GM, Rahn GM, Cummings IW, Salser WA. Nucleic Acids Res. 1978;5:3365. doi: 10.1093/nar/5.9.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yap NL, Rao VB. J. Mol. Biol. 1996;263:539. doi: 10.1006/jmbi.1996.0597. [DOI] [PubMed] [Google Scholar]
- 5.Zuker M. Science. 1989;244:48. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 6.Jaeger JA, SantaLucia JJ, Tinoco IJ. Annu. Rev. Biochem. 1993;62:255. doi: 10.1146/annurev.bi.62.070193.001351. [DOI] [PubMed] [Google Scholar]
- 7.Pleij CW, Bosch L. Meth. Enzymol. 1989;180:289. doi: 10.1016/0076-6879(89)80107-7. [DOI] [PubMed] [Google Scholar]
- 8.Correll CC, Freeborn B, Moore PB, Steitz TA. Cell. 1997;91:705. doi: 10.1016/s0092-8674(00)80457-2. [DOI] [PubMed] [Google Scholar]
- 9.Guo P. Nat. Nanotechnol. 2010;5:833. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J, Guo S, Cinier M, Shlyakhtenko L, Shu Y, Chen C, et al. ACS Nano. 2010;5:237. doi: 10.1021/nn1024658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shu D, Shu Y, Haque F, Abdelmawla S, Guo P. Nat. Nanotechnol. 2011;6:658. doi: 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khaled A, Guo S, Li F, Guo P. Nano Lett. 2005;5:1797. doi: 10.1021/nl051264s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo S, Tschammer N, Mohammed S, Guo P. Hum. Gene Ther. 2005;16:1097. doi: 10.1089/hum.2005.16.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo S, Huang F, Guo P. Gene Ther. 2006;13:814. doi: 10.1038/sj.gt.3302716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoeprich S, Zhou Q, Guo S, Qi G, Wang Y, Guo P. Gene Ther. 2003;10:1258. doi: 10.1038/sj.gt.3302002. [DOI] [PubMed] [Google Scholar]
- 16.Sarver NA, Cantin EM, Chang PS, Zaia JA, Ladne PA, Stephens DA, et al. Science. 1990;247:1222. doi: 10.1126/science.2107573. [DOI] [PubMed] [Google Scholar]
- 17.Liu H, Guo S, Roll R, Li J, Diao Z, Shao N, et al. Cancer Biol. Ther. 2007;6:697. doi: 10.4161/cbt.6.5.3962. [DOI] [PubMed] [Google Scholar]
- 18.Ellington AD, Szostak JW. Nature. 1992;355:850. doi: 10.1038/355850a0. [DOI] [PubMed] [Google Scholar]
- 19.Gold L. Harvey Lect. 1995;91:47. [PubMed] [Google Scholar]
- 20.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Nature. 2004;428:281. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 21.Mulhbacher J, St-Pierre P, Lafontaine DA. Curr. Opin. Pharmacol. 2010;10:551. doi: 10.1016/j.coph.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Chen Y, Zhu X, Zhang X, Liu B, Huang L. Mol. Ther. 2010;18:1650. doi: 10.1038/mt.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, et al. Proc. Natl. Acad. Sci. U. S. A. 2010;107:6328. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye X, Liu Z, Hemida M, Yang D. PLoS One. 2011;6:e21215. doi: 10.1371/journal.pone.0021215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdelmawla S, Guo S, Zhang L, Pulukuri S, Patankar P, Conley P, Trebley J, et al. Mol. Therapy. 2011;19:1312. doi: 10.1038/mt.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang HM, Su Y, Guo S, Yuan J, Lim T, Liu J, Guo P, et al. Antiviral Res. 2009;83:307. doi: 10.1016/j.antiviral.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C, Zhang C, Guo P. RNA. 1999;5:805. doi: 10.1017/s1355838299990350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao F, Zhang H, Guo P. Nucleic Acids Res. 2008;36(20):6620. doi: 10.1093/nar/gkn669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shu D, Zhang H, Jin J, Guo P. EMBO J. 2007;26:527. doi: 10.1038/sj.emboj.7601506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reid RJD, Bodley JW, Anderson D. J. Biol. Chem. 1994;269:5157. [PubMed] [Google Scholar]
- 31.Zhang CL, Lee C-S, Guo P. Virology. 1994;201:77. doi: 10.1006/viro.1994.1267. [DOI] [PubMed] [Google Scholar]
- 32.Guo P, Erickson S, Anderson D. Science. 1987;236:690. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- 33.Guo P, Zhang C, Chen C, Trottier M, Garver K. Mol. Cell. 1998;2:149. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 34.Shu D, Moll WD, Deng Z, Mao C, Guo P. Nano Lett. 2004;4:1717. doi: 10.1021/nl0494497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen C, Sheng S, Shao Z, Guo P. J. Biol. Chem. 2000;275(23):17510. doi: 10.1074/jbc.M909662199. [DOI] [PubMed] [Google Scholar]
- 36.Shu D, Huang L, Hoeprich S, Guo P. J. Nanosci. Nanotechnol. 2003;3:295. doi: 10.1166/jnn.2003.160. [DOI] [PubMed] [Google Scholar]
- 37.Cairns J, Overbaugh J, Miller S. Nature. 1988;335:142. doi: 10.1038/335142a0. [DOI] [PubMed] [Google Scholar]
- 38.Baugh C, Grate D, Wilson C. J. Mol. Biol. 2000;301:117. doi: 10.1006/jmbi.2000.3951. [DOI] [PubMed] [Google Scholar]
- 39.Shu Y, Cinier M, Fox SR, Ben-Johnathan N, Guo P. Mol. Therapy. 2011;19:1304. doi: 10.1038/mt.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu Y, Low PS. Adv. Drug Deliv. Rev. 2002;54:675. doi: 10.1016/s0169-409x(02)00042-x. [DOI] [PubMed] [Google Scholar]
- 41.Miyagishi M, Sumimoto H, Miyoshi H, Kawakami Y, Taira K. J. Gene Med. 2004;6:715. doi: 10.1002/jgm.556. [DOI] [PubMed] [Google Scholar]
- 42.Nakashima Y, Abe H, Abe N, Aikawa K, Ito Y. Chem. Commun. (Camb.) 2011;47:8367. doi: 10.1039/c1cc11780g. [DOI] [PubMed] [Google Scholar]
- 43.Guha M, Plescia J, Leav I, Li J, Languino LR, Altieri DC. Cancer Res. 2009;69:4954. doi: 10.1158/0008-5472.CAN-09-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fulda S, Debatin KM. Cancer Res. 2004;64:337. doi: 10.1158/0008-5472.can-03-1656. [DOI] [PubMed] [Google Scholar]
- 45.Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, et al. J. Natl. Cancer Inst. 1990;82:1107. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 46.Shlyakhtenko LS, Gall AA, Filonov A, Cerovac Z, Lushnikov A, Lyubchenko YL. Ultramicroscopy. 2003;97:279. doi: 10.1016/S0304-3991(03)00053-6. [DOI] [PubMed] [Google Scholar]
- 47.Lyubchenko YL, Shlyakhtenko LS. Methods. 2009;47:206. doi: 10.1016/j.ymeth.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]