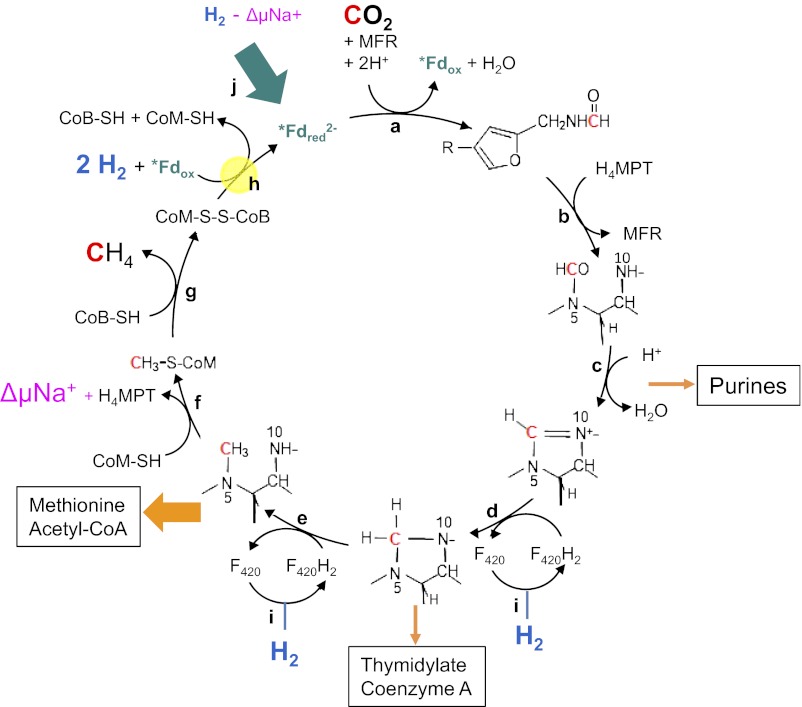
In 1988, Rouvière and Wolfe (1) suggested that methane formation from H2 and CO2 by methanogenic archaea could be a cyclical process. Indirect evidence indicated that the first step, the reduction of CO2 to formylmethanofuran, was somehow coupled to the last step, the reduction of the heterodisulfide (CoM-S-S-CoB) to coenzyme M (CoM-SH) and coenzyme B (CoB-SH). Over 2 decades passed until the coupling mechanism was unraveled in 2011: Via flavin-based electron bifurcation, the reduction of CoM-S-S-CoB with H2 provides the reduced ferredoxin (Fig. 1h) required for CO2 reduction to formylmethanofuran (2) (Fig. 1a). However, one question still remained unanswered: How are the intermediates replenished that are removed for the biosynthesis of cell components from CO2 (orange arrows in Fig. 1)? This anaplerotic (replenishing) reaction has recently been identified by Lie et al. (3) as the sodium motive force-driven reduction of ferredoxin with H2 catalyzed by the energy-converting hydrogenase EhaA-T (green arrow in Fig. 1).
Fig. 1.
Wolfe cycle of CO2 reduction to methane with 4 H2 in hydrogenotrophic methanogenic archaea. Orange arrows indicate biosynthetic reactions that remove intermediates (thickness of the arrows reflects the quantitative importance), the green arrow illustrates the anaplerotic reaction catalyzed by EhaA-T, and yellow highlighting represents the electron-bifurcating reaction. Reactions f and j are catalyzed by membrane-associated enzyme complexes. All other reactions are catalyzed by cytoplasmic enzymes. F420, coenzyme F420; *Fd, specific ferredoxin; H4MPT, tetrahydromethanopterin; MFR, methanofuran; ΔμNa+, electrochemical sodium ion potential. Enzymes: a, formylmethanofuran dehydrogenase; b, formylmethanofuran/H4MPT formyltransferase; c, methenyl-H4MPT cyclohydrolase; d, methylene-H4MPT dehydrogenase; e, methylene-H4MPT reductase; f, methyl-H4MPT/coenzyme M methyltransferase; g, methyl-coenzyme M reductase; h, electron-bifurcating hydrogenase–heterodisulfide reductase complex; i, F420-reducing hydrogenase; j, energy-converting hydrogenase catalyzing the sodium motive force-driven reduction of ferredoxin with H2. With Methanothermobacter marburgensis, it has been shown that one of the two C1 carbons in purines is derived from formate (C-2) and the other is derived from formyl-H4MPT/methenyl-H4MPT (C-8) (25).
Biological methane formation from H2 and CO2 (ΔG°′ = −131 kJ/mol) is not only a quantitatively important process but possibly one of the oldest (4). It involves such coenzymes as methanofuran, tetrahydromethanopterin (H4MPT), and CoM-SH as C1-unit carriers. These coenzymes were first thought to be unique to anaerobic methanogenic archaea (5) but were later found also in methanotrophic bacteria (6) and in some other microorganisms (7, 8). CO2 reduction to methane begins with its reduction to formylmethanofuran with reduced ferredoxin (Fdred2−) (9–11), which is regenerated by reduction of oxidized ferredoxin (Fdox) with H2 (12). The redox potential of the Fdox/Fdred2− couple of −500 mV is almost 200 mV more negative than that of the 2H+/H2 couple at a pH of 7 and the H2 partial pressure of 10 Pa prevailing in the anaerobic habitats of hydrogenotrophic methanogens (11). The endergonic reduction of ferredoxin with H2 must therefore somehow be coupled to one of the three downstream exergonic reactions.
The first exergonic reaction is the transfer of the methyl group from methyl-H4MPT to CoM-SH (ΔG°′= −30 kJ/mol), which is catalyzed by the membrane-associated and energy-conserving enzyme complex MtrA-H (13) (Fig. 1f). The reaction is associated with the build-up of an electrochemical sodium ion potential, which, in turn, drives both ATP synthesis via the membrane-associated AoA1-ATP synthase (not shown) and ferredoxin reduction with H2 via the membrane-associated and energy-converting hydrogenase complexes EhaA-T (Fig. 1j) and EhbA-Q (not shown) (11).
The second exergonic reaction is the reduction of methyl-coenzyme M with CoB-SH to methane and the heterodisulfide CoM-S-S-CoB (ΔG°′ = −30 kJ/mol) catalyzed by cytoplasmic methyl-coenzyme M reductase (Fig. 1g). As far as known, this reaction is not coupled with ferredoxin reduction or energy conservation (14, 15).
The third exergonic reaction is the reduction of CoM-S-S-CoB with H2 (ΔG°′ = −55 kJ/mol) catalyzed by a cytoplasmic hydrogenase–heterodisulfide reductase complex (Fig. 1h), which couples this reaction with the endergonic reduction of ferredoxin with H2 (ΔG°′ = +16 kJ/mol) and renders the methanogenic pathway cyclical (2). Coupling is via the newly discovered mechanism of flavin-based electron bifurcation (16).
Using elegant genetic experiments, Lie et al. (3) show that when the genes for six of the seven chromosomally encoded hydrogenases of Methanococcus maripaludis are deleted, this model organism can still grow on formate, but only when H2 is also present. Only the energy-converting hydrogenase EhaA-T appears to be essential. During growth on formate, the hydrogenase–heterodisulfide reductase complex (Fig. 1h) is substituted by an electron-bifurcating formate dehydrogenase–heterodisulfide reductase complex and the F420-reducing hydrogenase (Fig. 1i) is substituted by F420-reducing formate dehydrogenase. The H2 is used in substoichiometric amounts (< 0.2 mol of H2 per mol of CH4) that approximately comprise the amount of H2 required for autotrophic CO2 fixation, which suggests an anaplerotic role of EhaA-T.
Why is an anaplerotic reaction so important? It is because intermediates are continuously withdrawn from the cycle for the biosynthesis of purines (DNA and RNA); thymidylate (DNA); CoA; methionine (protein and S-adenosyl-methionine); and, quantitatively most important, acetyl-CoA (Fig. 1), which is synthesized from methyl-H4MPT, CO, and CoA in autotrophic methanogens (17). If reduced ferredoxin is not replenished, the cycle would come to a halt. The same would happen if electron bifurcation is imperfectly coupled (11). However, imperfect coupling appears not to be of quantitative importance, as evidenced by the relatively low H2 requirement during growth on formate of the M. maripaludis mutant lacking the six hydrogenases. This finding is another important result of the work of Lie et al. (3).
The elucidation of the cycle all began in 1977 with an experiment by Gunsalus and Wolfe (18), which showed that in cell extracts of Methanobacterium thermoautotrophicum strain ΔH (now Methanothermobacter thermoautotrophicus), the reduction of CO2 with H2 to methane proceeds only in the presence of catalytical amounts of methyl-coenzyme M. Later, it was found that methyl-coenzyme M could be substituted by catalytical amounts of CoM-S-S-CoB (19). This finding was reminiscent of the observation of Hans Adolf Krebs (20) that in liver slices, the formation of urea from ammonia and CO2 is stimulated by catalytical amounts of ornithine, which led to the discovery of the first metabolic cycle, the urea cycle. This was followed by Krebs’ elucidation of the tricarboxylic acid cycle for acetate oxidation. Later, Hans L. Kornberg (21) showed that anaplerotic reactions were necessary to replenish the intermediates of the Krebs cycle used for biosynthesis. With the anaplerotic reaction found by Lie et al. (3), the cycle of CO2 reduction to methane is finally complete. This methanogenic cycle is now referred to as the Wolfe cycle, in honor of Ralph S. Wolfe (22), who has been the motor driving its elucidation.
One can argue that the Wolfe cycle is not a cycle comparable to the Krebs cycle because the first and last steps are coupled via the electron carrier ferredoxin rather than by a carbon compound. Also, the Embden–Meyerhof pathway would be a cycle if formulated with ATP as an intermediate connecting the first step and the last step, namely, the hexokinase reaction and the pyruvate kinase reaction. However, the Wolfe cycle differs from the Embden–Meyerhof pathway in that the ferredoxin involved appears to be used mainly in the cycle, whereas the ATP of the Embden–Meyerhof pathway is also used elsewhere. Evidence for a specific ferredoxin comes from the finding that the energy-converting hydrogenase EhbA-Q, which catalyzes the sodium motive force-driven reduction of ferredoxin with H2, has a purely anabolic function and cannot substitute for the EhaA-T complex catalyzing the same reaction. This can be explained by assuming that the two hydrogenases prefer to use different ferredoxins. Indeed, in the genome of all methanogens analyzed, several genes for ferredoxins with quite different predicted structures are encoded (23). An alternative explanation is that ferredoxin pools are compartmentalized. The finding that the hydrogenase–heterodisulfide reductase complex (Fig. 1h) and the formylmethanofuran dehydrogenase complex (Fig. 1a) form super complexes points in this direction (24).
How widespread is the Wolfe cycle? Biochemical and genomic information indicates that with modifications, the cycle operates in all members of the Methanobacteriales, Methanopyrales, Methanococcales, Methanomicobiales, and Methanocellales that can grow on H2/CO2 and/or formate. Only the relatively few members of the Methanosarcinales that grow on H2 and CO2 are exceptions, although these members appear to contain all the genes required for the operation of the cycle. In Methanosarcina barkeri, for example, the reduction of CoM-S-S-CoB is catalyzed by two membrane-associated enzyme complexes and is associated with the build-up of an electrochemical proton potential, which, in turn, drives the reduction of the ferredoxin required for CO2 reduction to formylmethanofuran via an energy-converting hydrogenase (11). In M. barkeri during growth on H2 and CO2, the function of the energy-converting hydrogenase is therefore catabolic rather than anaplerotic.
Footnotes
The author declares no conflict of interest.
See companion article on page 15473.
References
- 1.Rouvière PE, Wolfe RS. Novel biochemistry of methanogenesis. J Biol Chem. 1988;263:7913–7916. [PubMed] [Google Scholar]
- 2.Kaster AK, Moll J, Parey K, Thauer RK. Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea. Proc Natl Acad Sci USA. 2011;108:2981–2986. doi: 10.1073/pnas.1016761108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lie TJ, et al. Essential anaplerotic role for the energy-converting hydrogenase Eha in hydrogenotrophic methanogenesis. Proc Natl Acad Sci USA. 2012;109:15473–15478. doi: 10.1073/pnas.1208779109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. Methanogens: Reevaluation of a unique biological group. Microbiol Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DiMarco AA, Bobik TA, Wolfe RS. Unusual coenzymes of methanogenesis. Annu Rev Biochem. 1990;59:355–394. doi: 10.1146/annurev.bi.59.070190.002035. [DOI] [PubMed] [Google Scholar]
- 6.Chistoserdova L, Vorholt JA, Thauer RK, Lidstrom ME. C1 transfer enzymes and coenzymes linking methylotrophic bacteria and methanogenic Archaea. Science. 1998;281:99–102. doi: 10.1126/science.281.5373.99. [DOI] [PubMed] [Google Scholar]
- 7.Chistoserdova L, et al. The enigmatic planctomycetes may hold a key to the origins of methanogenesis and methylotrophy. Mol Biol Evol. 2004;21:1234–1241. doi: 10.1093/molbev/msh113. [DOI] [PubMed] [Google Scholar]
- 8.Krishnakumar AM, et al. Getting a handle on the role of coenzyme M in alkene metabolism. Microbiol Mol Biol Rev. 2008;72:445–456. doi: 10.1128/MMBR.00005-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertram PA, Thauer RK. Thermodynamics of the formylmethanofuran dehydrogenase reaction in Methanobacterium thermoautotrophicum. Eur J Biochem. 1994;226:811–818. doi: 10.1111/j.1432-1033.1994.t01-1-00811.x. [DOI] [PubMed] [Google Scholar]
- 10.Vorholt JA, Thauer RK. The active species of ‘CO2’ utilized by formylmethanofuran dehydrogenase from methanogenic Archaea. Eur J Biochem. 1997;248:919–924. doi: 10.1111/j.1432-1033.1997.00919.x. [DOI] [PubMed] [Google Scholar]
- 11.Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. Methanogenic archaea: Ecologically relevant differences in energy conservation. Nat Rev Microbiol. 2008;6:579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- 12.Thauer RK, et al. Hydrogenases from methanogenic archaea, nickel, a novel cofactor, and H2 storage. Annu Rev Biochem. 2010;79:507–536. doi: 10.1146/annurev.biochem.030508.152103. [DOI] [PubMed] [Google Scholar]
- 13.Gottschalk G, Thauer RK. The Na(+)-translocating methyltransferase complex from methanogenic archaea. Biochim Biophys Acta. 2001;1505(1):28–36. doi: 10.1016/s0005-2728(00)00274-7. [DOI] [PubMed] [Google Scholar]
- 14.Thauer RK. Biochemistry of methanogenesis: A tribute to Marjory Stephenson. Microbiology. 1998;144:2377–2406. doi: 10.1099/00221287-144-9-2377. [DOI] [PubMed] [Google Scholar]
- 15.Scheller S, Goenrich M, Boecher R, Thauer RK, Jaun B. The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane. Nature. 2010;465:606–608. doi: 10.1038/nature09015. [DOI] [PubMed] [Google Scholar]
- 16.Buckel W, Thauer RK. 2012. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na(+) translocating ferredoxin oxidation. Biochim Biophys Acta, 10.1016/j.bbabio.2012.07.002.
- 17.Berg IA, et al. Autotrophic carbon fixation in archaea. Nat Rev Microbiol. 2010;8:447–460. doi: 10.1038/nrmicro2365. [DOI] [PubMed] [Google Scholar]
- 18.Gunsalus RP, Wolfe RS. Stimulation of CO2 reduction to methane by methylcoenzyme M in extracts Methanobacterium. Biochem Biophys Res Commun. 1977;76:790–795. doi: 10.1016/0006-291x(77)91570-4. [DOI] [PubMed] [Google Scholar]
- 19.Bobik TA, Wolfe RS. Activation of formylmethanofuran synthesis in cell extracts of Methanobacterium thermoautotrophicum. J Bacteriol. 1989;171:1423–1427. doi: 10.1128/jb.171.3.1423-1427.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krebs HA. Reminiscences and Reflections. Oxford: Clarendon Press; 1981. pp. 51–60. [Google Scholar]
- 21.Kornberg HL. Anaplerotic sequences in microbial metabolism. Angew Chem Int Ed. 1965;4:558–565. [Google Scholar]
- 22.Wolfe RS. My kind of biology. Annu Rev Microbiol. 1991;45:1–35. doi: 10.1146/annurev.mi.45.100191.000245. [DOI] [PubMed] [Google Scholar]
- 23.Kaster AK, et al. 2011. More than 200 genes required for methane formation from H2 and CO2 and energy conservation are present in Methanothermobacter marburgensis and Methanothermobacter thermautotrophicus. Archaea, 10.1155/2011/973848.
- 24.Costa KC, et al. Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase. Proc Natl Acad Sci USA. 2010;107:11050–11055. doi: 10.1073/pnas.1003653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schick M, et al. Biosynthesis of the iron-guanylylpyridinol cofactor of [Fe]-hydrogenase in methanogenic archaea as elucidated by stable-isotope labeling. J Am Chem Soc. 2012;134:3271–3280. doi: 10.1021/ja211594m. [DOI] [PubMed] [Google Scholar]