Abstract
The RtcB protein has recently been identified as a 3′-phosphate RNA ligase that directly joins an RNA strand ending with a 2′,3′-cyclic phosphate to the 5′-hydroxyl group of another RNA strand in a GTP/Mn2+-dependent reaction. Here, we report two crystal structures of Pyrococcus horikoshii RNA-splicing ligase RtcB in complex with Mn2+ alone (RtcB/ Mn2+) and together with a covalently bound GMP (RtcB-GMP/Mn2+). The RtcB/ Mn2+ structure (at 1.6 Å resolution) shows two Mn2+ ions at the active site, and an array of sulfate ions nearby that indicate the binding sites of the RNA phosphate backbone. The structure of the RtcB-GMP/Mn2+ complex (at 2.3 Å resolution) reveals the detailed geometry of guanylylation of histidine 404. The critical roles of the key residues involved in the binding of the two Mn2+ ions, the four sulfates, and GMP are validated in extensive mutagenesis and biochemical experiments, which also provide a thorough characterization for the three steps of the RtcB ligation pathway: (i) guanylylation of the enzyme, (ii) guanylyl-transfer to the RNA substrate, and (iii) overall ligation. These results demonstrate that the enzyme’s substrate-induced GTP binding site and the putative reactive RNA ends are in the vicinity of the binuclear Mn2+ active center, which provides detailed insight into how the enzyme-bound GMP is tansferred to the 3′-phosphate of the RNA substrate for activation and subsequent nucleophilic attack by the 5′-hydroxyl of the second RNA substrate, resulting in the ligated product and release of GMP.
Keywords: RNA repair, tRNA splicing, two-metal-ion catalysis
RNA ligases join two RNA strands whose ends are produced by specific RNases in many biological processes during tRNA processing/splicing, antiphage, or unfolded protein response (1–3). Most widely studied are the 5′-Phosphate (5′-P) RNA ligases (4–6) that specifically catalyze the nucleophilic attack of a free 3′-hydroxyl on an activated 5′-P. However, these ligases cannot directly join two RNA strands ending with a 2′,3′-cyclic phosphate (RNA>p) and with a 5′-hydroxyl group. Before ligation, these strands require the action of a polynucleotide kinase (forming a 5′-P) and a phosphoesterase (generating a free 3′-OH) (6). The joining mechanism of 5′-P polynucleotide ligases involves a 3′-hydroxyl and an activated 5′-P end for internucleotide phosphodiester bond formation (7, 8). The 5′-P activation enzymes include group I and II self-splicing introns, the spliceosomal apparatus, and DNA/RNA polymerases, as well as other nucleotidyl transferases (9–11). Similarly, the tRNAHis guanylyltransferase ligates the 3′-OH of GTP to the 5′-terminus of tRNA through its adenylylation-activated 5′-P; this gives the appearance of reverse polarity of nucleotide addition relative to normal RNA polymerases (12).
The 3′-Phosphate (3′-P) RNA ligase activities were identified three decades ago; they use a 3′-P as a donor for joining two RNAs without involvement of phosphorylation of the 5′-hydroxyl or dephosphorylation of the 2′,3′-cyclic phosphate (13, 14). Recently, enzymes with this activity were purified from Methanopyrus kandleri or HeLa cells (15, 16), and then assigned to the RtcB family. Cloned family members from Pyrococcus horikoshii (PH), Pyrobaculum aerophilum, Escherichia coli, and Homo sapiens (as the central component of a four-protein complex) have been shown to have ligase activity (15–18). E. coli RtcB is most active in the presence of Mn2+ and stimulated by GTP (17). Biochemical analysis revealed a covalent RtcB-GMP intermediate to an identified histidine and a mechanism reminiscent of Mg2+-dependent adenylylation of 5′-P DNA and T4 RNA ligases (17, 19, 20). Before the discovery of the RtcB activity a crystal structure of the apo-PH RtcB was solved, revealing a unique protein fold with a putative metal ion-binding site (21). This structure could not explain the divalent metal ion specificity, nor reveal the RNA and GTP binding sites. We report here two PH RtcB structures and present: (i) binding of two Mn2+ ions in the single active center, and (ii) H404 as the site of enzyme guanylylation catalyzed by the D65-H404 dyad (Fig. 1). Our additional biochemical analysis reveals a stable 3′-P guanylylated RNA intermediate as an activation step for the 3′-P ligation.
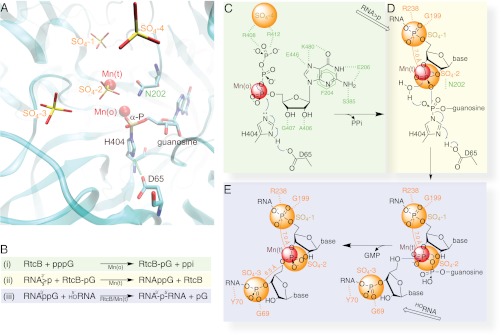
Fig. 1.
An overview of structure-derived reaction mechanism of the 3′-P RNA ligation. (A) Composite features of the two PH RtcB structures reported here with emphasis on the relationship of four sulfates and two Mn2+ ions with guanylylated H404 in the catalytic site in ribbons representation along with other highlighted residues, namely D65 and N202. (B) A summary of the RtcB-catalyzed pathway in three individual steps (described in the main text and schematized in C–E) as deduced from our structures and biochemistry. (C) The RtcB residues that specifically recognize the substrate GTP are shown in green. The putative PPi site coincides with SO4-4 (orange). The carboxylate group of D65 is hydrogen bonded to H404 Nδ1-H, thus enhancing the electron negativity of H404 Nε2 for nucleophilic attack on GTP’s α-phosphate that is in contact with the Mn2+ in octahedral geometry. (D) Coordination of tetrahedral Mn2+ allows binding the RNA strand ending with the 2′,3′-cyclic phosphate (RNA>p) superimposed with the last two phosphates at the SO4-1 and SO4-2 sites. (E) Positioning the HORNA strand with its first phosphate at the SO4-3 allows the optimal geometry of the 5′-hydroxyl for nucleophilic attack on the 3′-P activated terminus to form the internucleotide phosphodiester bond with the release of GMP.
Results and Discussion
RtcB Structures Have Two Mn2+ Ions at the Active Site and a Covalently Bound GMP.
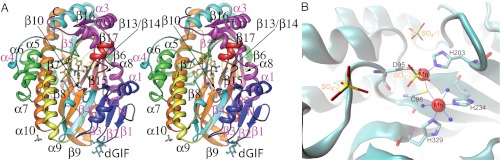
The overall structure of the RtcB/Mn2+ complex with four sulfate ions (SO4-1, SO4-2, SO4-3, and SO4-4) bound near the putative active site is similar to an earlier RtcB structure that lacked metal ions (21). The identity of the two bound Mn2+ was established in anomalous difference Fourier maps with peaks over 25 σ. One Mn2+, Mn(t), is in tetrahedral coordination with three conserved ligands (D95, C98, and H203) and a sulfate ion, SO4-2, which may mimic the 2′,3′-cyclic phosphate terminus of the RNA in the ligation reaction (Fig. 2). The second Mn2+, Mn(o), is in octahedral coordination also with three conserved ligands (C98, H234, and H329) and three water molecules. The occurrence of two different coordination states of the divalent metal ions and the involvement of the C98 thiol as a ligand explains the preference of Mn2+ over Mg2+. The sequential distance from SO4-1 to SO4-2 to SO4-3 is 7.0 and 6.5 Å (Fig. S1), respectively, which is consistent with the typical P-P distance of a single-stranded RNA (ssRNA). Indeed, a 5′-to-3′ ssRNA can be modeled with a reasonably good fit in the active site, whereas a modeled 3′-to-5′ ssRNA results in steric clashes of its bases with the enzyme. SO4-4 is 9.2 Å away from SO4-2 and its location is near to electron density found in the structure of the GTP-reacted RtcB/Mn2+-GMP/pyrophosphate (PPi) complex, which can be assigned as PPi (Figs. S1 and S2).
Fig. 2.
The Mn2+-bound RtcB complex. (A) Overview of the secondary structures of RtcB in rainbow colors from the N-to-C terminus in stereo. Active site residues, metal ions, four sulfate molecules, and (1,5-anhydro-d-glucose)-α1β2-(2,5-anhydro-l-fructose) (labeled as dGlF) are shown in balls-and-sticks. (B) Close-up view on the RtcB/Mn2+ active site. Residues cordinating the two Mn2+ ions in octahedral and tetrahedral geometry are highlighted. SO4-2 and three ordered water molecules serve as additional ligands.
A disaccharide with unusual linkage was tightly bound to the enzyme in the RtcB/Mn2+ complex. This disaccharide was structurally identified as α-(1,5-anhydro-d-glucopyranosyl)-(1,2)-β-(2,5-anhydro-d-fructofuranose) (Fig. S3).
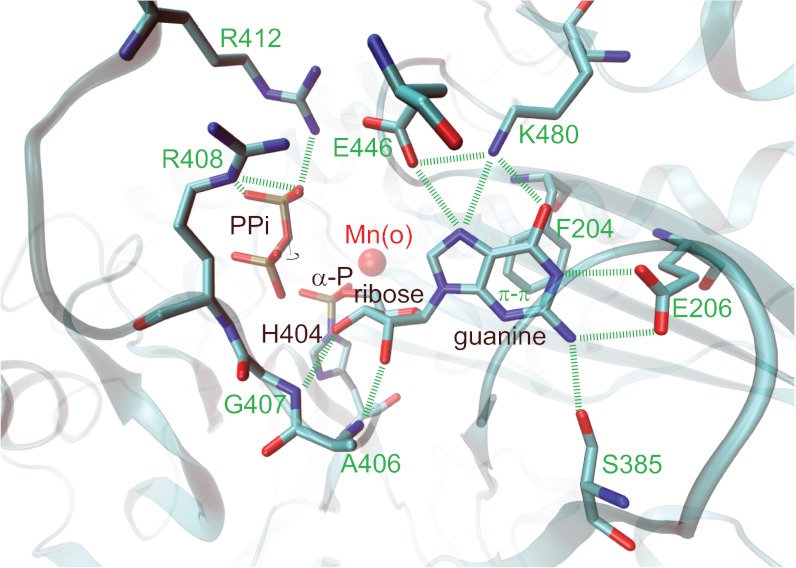
For determination of the second structure, the PH RtcB protein preparation was preincubated with Mn2+ and GTP before crystallization (SI Methods). Unbiased residual difference Fourier maps showed that RtcB has reacted with GTP to form a covalent link with Nε2 of H404 at its α-phosphate, forming a 5′-GMP-His (GPH) intermediate (Fig. 3 and Fig. S2). The GPH phosphate is in contact with N202 and Mn(o) in the active site (Fig. 3). All RtcB residues interacting with the G nucleobase are conserved, which include eight well-defined hydrogen bonds. At the Watson–Crick base-pairing face of the guanine, E206 and S385 of RtcB recognize the N1 and N2 hydrogen bond donors, and K480 recognizes the O6 hydrogen bond acceptor. In addition, E446 interacts with N7 of the guanine in which E446 has to be protonated, likely through interactions with three positively charged residues, K480, R408, and R412. The ligase also binds the O2′ and O3′ hydroxyls of the guanosine ribosyl moiety through hydrogen bonds to backbone amides of A406 and G407. Because the guanine is completely sandwiched in a hydrophobic cleft composed of the F204 side chain and the main chain of the P378-G379-S380-M381 residues (Fig. S2), most of the guanosine-recognition hydrogen bonds benefit from a hydrophobic effect.
Fig. 3.
GTP substrate recognition revealed from the RtcB-GMP/Mn2+ structure. RtcB has reacted with GTP to covalently bind GMP to H404. Annotated PPi (Fig. S2) interacts with R408 and R412. Residues specifically recognizing the guanine base and the ribose are indicated.
Substrate-Induced GTP-Binding Site and Guanylylation Active Site.
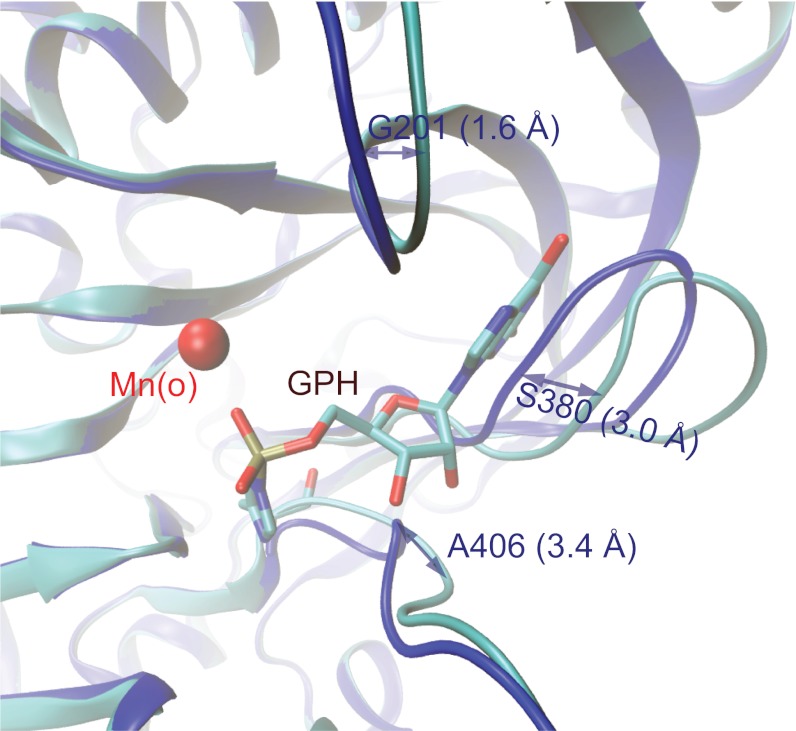
Structural comparison of the two RtcB complexes reported here reveals GTP-induced conformational changes that include repositioning of the β13 and β14 hairpins with the largest Cα displacement of 3.0 Å at S380 and an alignment of the D65-H404 dyad, which is important for RtcB guanylylation (Fig. 4). This induced-fit feature prevented us from predicting GTP binding and the guanylylation site at H404 from the RtcB/Mn2+ complex or from any previous structure (21). For formation of guanosine ribosyl O2′ and O3′ hydrogen bonds, the backbone amides of A406 and G407 were flipped with the maximal Cα displacement of 3.4 Å. In the Mn2+-bound RtcB complex, multiple conformations of the backbone peptide bonds in this region have also been observed. Moreover, the hydrophobic walls sandwiching the G nucleobase seen in the RtcB-GMP/Mn2+ complex are collapsed in the RtcB/Mn2+ complex, burying the two negatively charged residues E206 and E446 and a few ordered water molecules. Backbone conformational changes near H404-G405 are likely to be critical for the alignment of the D65-H404 dyad for activation of H404. In the RtcB/Mn2+ complex, D65 interacts with the backbone amide of G405 and does not align with H404. In the RtcB-GMP/Mn2+ complex, D65 has been reoriented so that it now interacts with Nδ1-H of H404 as well as its backbone amide. With the D65-H404 hydrogen bond (Fig. 1), D65 removes a proton from Nδ1 and makes Nε2 more nucleophilic for protein guanylylation in a mechanism reminiscent of the D-H catalytic dyad of RNase A (22).
Fig. 4.
Induced GTP-binding pocket as revealed from structural comparison. Superpositon of the RtcB/Mn2+ (blue) and RtcB-GMP/Mn2+ (turquois) structure shows opening of the cleft for the guanine base. The peptide backbone of A406 and G407 reorientate to allow amide interactions with the hydroxyl groups of the ribose.
The GTP-induced conformational changes extend to the metal ion binding site, suggesting possible multiple reaction steps in the single catalytic site. In the RtcB-GMP/Mn2+ complex, only one Mn2+ was observed at the equivalent location of Mn(o) in the RtcB/Mn2+ complex, and the second Mn2+ corresponding to Mn(t) along with bound SO4-2 (the 2′,3′-cyclic phosphate mimic for later steps) was missing. In both complexes, Mn(o) has the same three protein ligands (C98, H234, and H329); but in the RtcB-GMP/Mn2+ complex, this Mn2+ ion interacts with the nascent phosphate of GPH, implying that it may have a catalytic role in the guanylylation reaction, perhaps by stabilizing the transition state.
Enzymatic Identification of RtcB Reaction Intermediates.
To establish the catalytic importance of the structural findings described here, biochemical studies were undertaken to unravel the 3′-P RNA ligation mechanism catalyzed by this enzyme, particularly on the role of guanylylation (Fig. 5). We first defined the guanylylated enzyme, then the guanylylated RNA intermediate, and finally the overall ligation reaction in three steps (Fig. 1). For this purpose, a nonradioactive RNA substrate was incubated with α-[32P]GTP and RtcB, not only to label the enzyme, but also to catalyze the transfer of [32P]GMP from the enzyme-bound into the RNA-bound form (i.e., to capture the RNA-3′p-5′-[32P]-G intermediate) (Fig. 5). The radioactive [32P]GMP bound to the RNA migrated more slowly than the 21-nt RNA substrate, but was at the same position as that of an intermediate from an internally labeled RNA substrate synthesized by RtcB with nonradioactive GTP (Fig. 5). Hence, the RNA ligase-activity assay included not only the overall RNA ligation, but also the formation of the RNA intermediate. Together with the SDS/PAGE analysis that monitored the RtcB-[32P]GMP formation, these three activity assays, as outlined in Fig. 1B, were used to characterize RtcB variants with alterations in structurally-identified important amino acid residues (Table S1).
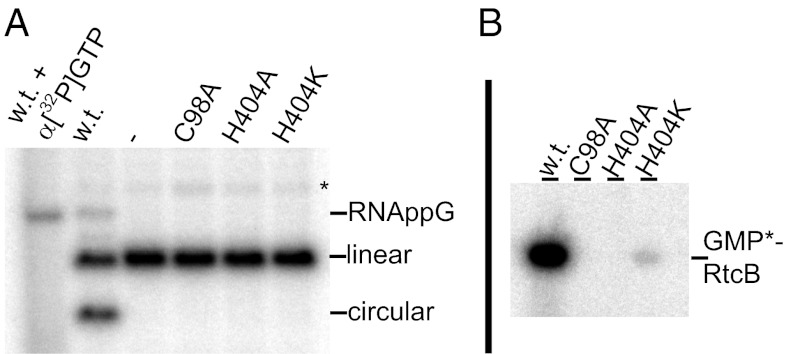
Fig. 5.
Activity assays for PH RtcB. (A) The 21-nt RNA with 5′-hydroxyl and 2′,3′-cyclic phosphate temini is incubated with WT or mutant RtcB proteins and separated on PAGE for PhosphorImager analysis. WT RtcB allows not only formation of the faster migrating circular RNA, but also cumulation of a slower migrating RNA. The same nonradioactive RNA incubated with α[32P]GTP identifies this species as guanylylated RNA intermediate. (B) WT and three RtcB variants are incubated with α[32P]GTP and separated on SDS PAGE for PhosphorImager analysis indicating protein guanylylation.
Enzymatic Characterization of RtcB Active Site Residues Using Site-Directed Mutagenesis.
All metal ion-binding residues in the active site are important for catalysis, as demonstrated by single substitutions with alanine that severely reduced the extent of overall RNA ligation (Table S1). C98 coordinates both Mn2+ ions as C98A abolishes all three enzymatic activities. D95 and H203 coordinate Mn(t) as D95A or H203A completely abolishes RNA ligation, even though they have only minor effects on protein guanylylation. H329 is a ligand for Mn(o). However, H329A remained active in RNA guanylylation, but produced just barely detectable amount of the guanylylated RtcB intermediate. These results suggest a possible subtle division of the two metal ions in individual steps of reaction [i.e., Mn(o) is important for protein guanylylation, but Mn(t) is essential for overall RNA ligation]. Based on coordination geometry, Zn2+ could substitute for Mn(t) (for RNA ligation reaction), but not for Mn(o) (for protein guanylylation). This finding may explain an unusual Zn2+-dependent Pyrobaculum RtcB RNA ligation activity independent of either ATP or GTP (15), because of preguanylylation before purification, a hypothesis that now can be tested. Furthermore, octahedrally coordinated Mn2+ was implicated in the adenylylation reaction from the crystal structure of E. coli RNA 3′-terminal phosphate cyclase RtcA in complex with ATP and Mn2+ (23).
The matched spacing of the observed sulfates on RtcB to the phosphates in ssRNA suggests possible RNA binding at these sulfate locations. RtcB variants with alterations at these sites were enzymatically characterized (Table S1). The R238A substitution at the SO4-1 site allows RtcB guanylylation but prevents the transfer of GMP to the 3′-terminal phosphate of the RNA substrate. Similarly, any disruptive substitutions at the SO4-3 site reduces the extent of overall ligation by as much as 50-fold, whereas protein guanylylation and RNA guanylylation that is independent of 3′-RNA binding to this site are unaffected. With experimentally confirmed binding sites for 5′ and 3′-RNAs, it is immediately apparent that the 2′,3′-cyclic phosphate must bind at the SO4-2 site at the binuclear metal ion center.
Our RtcB-GMP/Mn2+ structure shows that guanylylation at H404 is through the Pα-Nε2 bond formation when H404 Nδ1-H is aligned with D65 for deprotonation (Fig. 1). Consistent with this proposal, either H404A or D65A substitution completely abolishes RtcB’s protein guanylylation (Table S1). In an H404-D65 independent reaction, H404K restores some protein guanylylation activity but fails to carry out subsequent transfer of GMP to the RNA. As in any phosphoryl-transfer reaction involving concerted bond breakage and bond formation with inversion of the phosphoryl center, the bonds to be broken and formed have to be linearly aligned (9). Extrapolation of the newly formed Pα-Nε2 bond of GPH directs the orientation of the leaving PPi toward the Mn(t) site. Unfortunately, both PPi and Mn(t) were missing in the complex, presumably removed by centrifugation before crystallization when the PPi product precipitated with Mn2+ (SI Methods).
Interestingly, residual Fo-Fc difference Fourier maps have shown that a minor fraction of either unreacted RtcB/GTP/Mn2+ complex or the reacted RtcB/GMP/PPi/Mn2+ complex might exist in a given crystal where two phosphates of GTP or PPi occupied near the SO4-4 site. When we made substitutions near the SO4-4 site, such as R408A or R412A, the enzyme guanylylation was impaired, suggesting an important role of these residues in the catalytic cycle (Table S1). However, the precise functional role of this site remains to be further char3acterized because it is not aligned with the formed Pα-Nε2 bond opposite the phosphoryl center.
Enzyme Specificity: GTP vs. ATP, 3′-P Activation vs. 5′-P Activation, and Ligation vs. Hydrolysis.
Almost all residues in the induced GTP-binding pocket contribute to GTP recognition, as judged by disruptive effects of any substitutions on protein guanylylation as well as on the remaining two steps of RtcB’s reaction pathway (Table S1). All substituted residues are highly conserved, including E206, S385, and K480, which specifically recognize guanine through its hydrogen bond donor and acceptor pattern (Fig. 3). However, this finding presents a problem: namely, why the 3′-P RNA ligase activity purified from HeLa extracts appeared to be ATP-dependent in contrast to PH RtcB (24). Nevertheless, we found two homologous variations in the GTP-binding pocket between bacteria/archaea (F204 and P378) and eukaryotes (Y204 and G378). We wanted to know whether they were sufficient to confer the nucleotide specificity. Accordingly we made one F204Y/P378G double substitution in PH RtcB to mimic those found in eukaryotes, but unfortunately they failed to switch the requirement of activation cofactor from GTP to ATP (Table S1). Therefore, further studies on the human tRNA splicing ligase complex are necessary to understand the apparent ATP-dependent activity. There are a few probable reasons: (i) other subunits within the complex such as DDX1 may confer the ATP specificity (16); (ii) there might be a different ATP induced-fit mechanism; or (iii) the residual activity may result from preguanylylated RtcB that occurs before purification. If the third theory is the case, any inadvertent contamination of preguanylylated enzymes (or any other forms of preactivation) could be misleading during the enzymatic characterization process.
In the RtcB-GMP/Mn2+ complex (Fig. 3), N202 interacts with the α-phosphate of GMP, which will be attacked by the nucleophile oxygen of the 3′-P for 3′-P activation. This complex might be more relevant to the RNA guanylylation than to the protein guanylylation because the N202A substitution abolishes the RNA guanylylation, but not protein guanylylation (Table S1). Thus, some structural rearrangements may have already occurred after the protein guanylylation, which prevents us from defining the precise protein guanylylation mechanism. Nevertheless, this complex provides a clue as to why RtcB exclusively guanylylates the RNA 3′-P but not the RNA 5′-P (Fig. 1). During the guanylyl transfer from the protein to RNA, the attacking nucleophile must be located on the opposite side of the leaving group of H404 relative to the phosphoryl center [i.e., the phosphate that interacts with Mn(t) as revealed by SO4-2 in the RtcB/Mn2+ complex] (Fig. 1). Based on our RNA binding model, the phosphate of 3′-P of the 5′-RNA is in better aligned than that of 5′-P of the 3′-RNA. The alignment is defined by the orientation of an attacking oxygen atom on the phosphate relative to Pα of H404-GMP with the alignment angle for 3′-P of the 5′-RNA of 145° from SO4-1 to SO4-2 and to Pα of H404-GMP, and the angle for 5′-P of the 3′-RNA of 98° from SO4-3 to SO4-2 and to Pα of H404-GMP. Moreover, the attacking oxygen atom on this phosphate to the α-phosphate of H404-GMP also has the shortest interatomic distance when it is 3′-P of the 5′-RNA. In contrast, the RNA phosphate activation by T4 RNA ligase 1 is more promiscuous than PH RtcB, where T4 RNA ligase 1 prefers to adenylylate a 5′-P, but can also adenylylate 3′-P when 5′-P is not present (25). Similarly, E. coli RtcA can also adenylylate a 5′-P when a 3′-acceptor is missing (26). The promiscuity of phosphate activation by these enzymes is likely the result of a more symmetric nature of the formed phosphoanhydride bond relative to activation cofactors (AMP or GMP) than PH RtcB.
After 3′-P activation, the next step is the attack of the RNA 5′-hydroxyl on the activated RNA 3′-P for RNA ligation. However, the activation of the 3′-P by the phosphoanhydride bond to GMP and coordination to Mn(t) might also allow water as a nucleophile (Fig. 1). The hydrolytic product is RNA 3′-P, which is also a substrate for bacterial/archaeal RtcB, but not for the human RtcB complex. With the RNA>p ligase-enriched HeLa fraction (24, 27), only 30% of RNA>p led to the ligation product and the remaining 70% of RNA>p was converted to the inactive RNA 3′-P through the hydrolytic pathway. Thus, the 3′-terminal phosphate RNA cyclase is required to ensure efficient RNA>p ligation in eukaryotes. Interestingly, RtcA and RtcB copurified after three chromatographic steps during RNA>p ligase activity purification and during affinity purification of a 42-protein kinesin-cargo complex from mouse brain extracts (16, 28). The biological role of RtcA was previously overlooked either because it was redundant in the bacterial/archaeal RtcB system or because the RNA substrates produced by the Ire1 or the tRNA splicing endonucleases always contain the 2′,3′-cyclic phosphate termini.
Functional and Mechanistic Implications to Archaeal and Human RtcB.
Taking together all individual steps of the RtcB-catalyzed 3′-P RNA ligation reaction (Fig. 1), striking similarities of the 5′-P and the 3′-P ligation pathway emerge on the mechanistic level. Both enzymes covalently bind AMP or GMP to transfer them to the RNA-terminal phosphate of either the 5′- or 3′-end for activation, followed by nucleophilic attack by the RNA-encoded hydroxyl group from the opposite end to form a canonical 3′,5′-phosphodiester bond. Why is there a unique fold and active site of RtcB? This occurance might be because of the characteristic feature of RNAs having a 2′-hydroxyl and a 3′-hydroxyl next to one another with two possibilities of phosphodiester bond formation when 3′-P is activated, one intrastrand 2′,3′-cyclization and the other interstrand ligation, specifically catalyzed by RtcA and RtcB, respectively. Interestingly, archaea and animals have both 3′-P and 5′-P RNA ligation pathways, of which the 3′-P pathway has posttranslational regulations identified. For example, the phosphorylation site S439 of human RtcB (29) can be mapped onto the PH RtcB 415 position, next to the two essential R408 and R412 residues. This location coincides with regions involved in GTP-induced conformational changes, immediately suggesting a possible regulatory mechanism for the enzymatic activity. RtcB from the archaeon Thermococcus kodakarensis also has an unidentified tyrosine phosphorylation site (30). Additionally, in the Caenorhabditis elegans Parkinson disease model, RtcB (F16A11.2) depletion by RNAi was correlated with Parkinson symptoms and neurodegeneration, whereas RtcB overproduction resulted in neuroprotection (31). Thus, the tightly regulated eukaryotic RtcB pathway has more important functions than just resealing broken tRNAs.
In conclusion, our structural findings for PH RtcB with two Mn2+, four sulfates, and GMP bound are supported by a comprehensive biochemical analysis. This work brings together the features of a protein with unique sequence/fold, and the recently identified enzymatic activity of 3′-P RNA ligation. The active site has been established as the center for two Mn2+ ions with tetraheadral and octaheadral coordination geometry, thus explaining the Mn2+ specificity for the three-step reaction pathway. Three sulfates are bound by RtcB next to the binuclear Mn2+ center; their significance as RNA phosphate backbone binding sites is enzymatically validated in this work. The modeled 5′- to 3′-direction is confirmed by biochemical dissection of the protein’s individual enzymatic activities. The guanylylated H404 is in contact with one Mn2+ ion at the active site, and next to the proposed substrate RNA termini. These findings are summarized in a reaction scheme (Fig. 1); they now provide a framework for further characterization of the eukaryotic RtcB pathway.
Methods
RtcB-Coding Plasmid Constructs, Protein Expression and Purification, and Crystallization of RtcB Complexes.
Two extein-coding sequences of PH RtcB (PH1602) were assembled by overlap PCR extension, as previously described (21). The coding plasmids were modified to have a His6-tag for rapid protein purification, first with an additional 11-amino acid residue carboxyl-terminal His6-tag (for the first RtcB/Mn2+ structure, RtcB-1) (SI Methods) and then with a tobacco etch virus protease-cleavable amino-terminal His6-tag (for the second GTP-reacted RtcB/Mn2+ structure, RtcB-2) (SI Methods).
For the first RtcB complex, PH RtcB-1 was transformed into E. coli Rosetta (DE3) pLysS (Novagen). Cells were grown in LB with Studier auto-induction supplements NPS and 5052 (32) at 37 °C to an optical density of 0.8 and continued at 18 °C for 16 h. Harvested cells were disrupted in Ni-NTA lysis buffer [20 mM Tris-HCl, pH 8, 500 mM NaCl, 3 mM MnCl2, 10 mM imidazole, 10 mM β-mercaptoethanol (β-ME)] at 4 °C with Roche complete EDTA-free protease inhibitor mixture by lysozyme and sonication. Standard His6-tag and Superdex 200 gel-filtration procedures were applied to purify RtcB, which was then stored in final buffer, 10 mM Tris-HCl pH 8, 200 mM NaCl, 3 mM MnCl2, 10 mM β-ME. The RtcB/Mn2+ complex was crystallized using the sitting-drop vapor-diffusion method with 1.5 M (NH4)2SO4, 75 mM NaCl, and 0.1 M Hepes-NaOH pH 7.5 in reservoir solution.
For the second RtcB complex, PH RtcB-2 (SI Methods) was transformed into E. coli B834 (DE3)-plus-pRARE2. Cells were grown in LB medium containing 30 μg/mL kanamycin and 34 μg/mL chloramphenicol at 37 °C to an optical density of 0.5–0.6. Isopropyl-β-d-thiogalactopyranoside was then added to a final concentration of 1 mM and incubated for 5 h at 37 °C for induction. Harvested cells were disrupted using sonication and incubated for 30 min at 70 °C (heat denaturation) in buffer containing 50 mM Na-phosphate pH 8.0, 500 mM NaCl, 60 mM KCl, 5% (wt/vol) sucrose, and 5 mM β-ME before the cell debris was removed using centrifugation at 45,000 × g for 30 min. Standard His6-tag procedures were applied to purify RtcB (SI Methods) in final buffer, 20 mM Hepes-NaOH pH 7.5, 200 mM NaCl, 10% glycerol, and 1 mM DTT. For activation, 12 mg/mL RtcB was incubated with 12 mM MnCl2 at 80 °C for 15 min and centrifuged at 20,000 × g for 15 min at 20 °C to remove any precipitate. The cleared supernatant was incubated with GTP and MnCl2 for 40 min at 80 °C with the final concentrations: 10 mg/mL for RtcB and 10 mM for both MnCl2 and GTP. The reaction mixture was again centrifuged at 20,000 × g for 15 min to remove any new precipitate. The 10-mg/mL RtcB-GMP/Mn2+ complex was crystallized using the hanging-drop vapor-diffusion method at 20 °C for about 10 d when mixed with equal volume (2.9 μL) of reservoir solution containing 2 M Na-malonate pH 7.0, 20% glycerol, and 10 mM MnCl2.
X-Ray Diffraction, Data Collection, and Structure Determination.
For the RtcB/Mn2+ complex, crystals were cryoprotected with additional 50% (wt/vol) sucrose in the crystallization reservoir solution before freezing in liquid N2. Diffraction data were collected using synchrotron radiation sources at beam line 24ID-E, Northeast Collaborative Access Team, Advanced Photon Source, Argonne National Laboratory, Chicago, IL. For the RtcB-GMP/Mn2+ complex, crystals were directly picked from crystallization droplets and flash-frozen under a stream of liquid N2 gas at 100 K. Data were collected at beam line BL-1A, Photon Factory. Data were processed as summarized in Table S2. Starting with our previous apo-RtcB structure of 1UC2 (21), structure determination and refinement of the two RtcB complexes described here are summarized in Table S2.
Preparation of tRNA Splicing Intermediates and RNA Ligase-Activity Assays.
The tRNA splicing intermediates were obtained by T7 transcription of the intron-containing Archeuka pretRNA and cleaved with Mjan EndA, as described elsewhere (15). In vitro RNA ligase assays were carried out at 37 °C for 30 min in 50 mM Tris-HCl pH 7.4, 3 mM MnCl2, 0.1 mM GTP, 10 mM β-ME, with [32P]-labeled 40 fmol RNA substrate (40,000 cpm) and 5 μg PH RtcB-1 enzyme. The labeled RNA products were phenol-extracted, ethanol-precipitated, and separated using urea PAGE (12.5%), then visualized/quantified using PhosphorImager analysis (Fig. 5). For identification of a guanylylated RNA intermediate, nonradioactive tRNA splicing intermediates (400 ng) were incubated in 50 mM Tris-HCl pH 7.4, 3 mM MnCl2, 10 mM β-ME, 13 pmol α[32P]GTP, and 10 μg PH RtcB-1 for 20 min at 37 °C. The RNA products were phenol extracted, ethanol precipitated and separated by PAGE for PhosphorImager analysis (Fig. 5).
Supplementary Material
Acknowledgments
We thank Yong Xiong for inspiring discussions, the staff of the protein databank for defining the unusual sucrose, and the beamline staff at BL-1A of the High Energy Accelerator Research Organization Photon Factory for assistance in data collection. This work was supported in part by National Insititute of General Medical Sciences Grants GM022854 (to D.S.) and GM063276-05 (to W.H.K.); Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science 21370041 (to M.Y.) and 22870001 (to A.N.); and by grants from the Steitz Center for Structural Biology, Gwangju Institute of Science and Technology, Republic of Korea (to S.H.E. and J.W.). M.E. was a Feodor Lynen Fellow of the Alexander von Humboldt Foundation and A.N. was a Japan Society for the Promotion of Science Postdoctoral Fellow for Research Abroad.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4DWR and 4DWQ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213795109/-/DCSupplemental.
References
- 1.Popow J, Schleiffer A, Martinez J. Diversity and roles of (t)RNA ligases. Cell Mol Life Sci. 2012;69:2657–2670. doi: 10.1007/s00018-012-0944-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amitsur M, Levitz R, Kaufmann G. Bacteriophage T4 anticodon nuclease, polynucleotide kinase and RNA ligase reprocess the host lysine tRNA. EMBO J. 1987;6:2499–2503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sidrauski C, Cox JS, Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–413. doi: 10.1016/s0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- 4.Phizicky EM, Schwartz RC, Abelson J. Saccharomyces cerevisiae tRNA ligase. Purification of the protein and isolation of the structural gene. J Biol Chem. 1986;261:2978–2986. [PubMed] [Google Scholar]
- 5.Englert M, Beier H. Plant tRNA ligases are multifunctional enzymes that have diverged in sequence and substrate specificity from RNA ligases of other phylogenetic origins. Nucleic Acids Res. 2005;33:388–399. doi: 10.1093/nar/gki174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Englert M, Sheppard K, Gundllapalli S, Beier H, Söll D. Branchiostoma floridae has separate healing and sealing enzymes for 5′-phosphate RNA ligation. Proc Natl Acad Sci USA. 2010;107:16834–16839. doi: 10.1073/pnas.1011703107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pascal JM. DNA and RNA ligases: Structural variations and shared mechanisms. Curr Opin Struct Biol. 2008;18:96–105. doi: 10.1016/j.sbi.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 8.Tomkinson AE, Vijayakumar S, Pascal JM, Ellenberger T. DNA ligases: Structure, reaction mechanism, and function. Chem Rev. 2006;106:687–699. doi: 10.1021/cr040498d. [DOI] [PubMed] [Google Scholar]
- 9.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adams PL, et al. Crystal structure of a group I intron splicing intermediate. RNA. 2004;10:1867–1887. doi: 10.1261/rna.7140504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J. Inclusion of weak high-resolution X-ray data for improvement of a group II intron structure. Acta Crystallogr D Biol Crystallogr. 2010;66:988–1000. doi: 10.1107/S0907444910029938. [DOI] [PubMed] [Google Scholar]
- 12.Hyde SJ, et al. tRNA(His) guanylyltransferase (THG1), a unique 3′-5′ nucleotidyl transferase, shares unexpected structural homology with canonical 5′-3′ DNA polymerases. Proc Natl Acad Sci USA. 2010;107:20305–20310. doi: 10.1073/pnas.1010436107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filipowicz W, Shatkin AJ. Origin of splice junction phosphate in tRNAs processed by HeLa cell extract. Cell. 1983;32:547–557. doi: 10.1016/0092-8674(83)90474-9. [DOI] [PubMed] [Google Scholar]
- 14.Zofallova L, Guo Y, Gupta R. Junction phosphate is derived from the precursor in the tRNA spliced by the archaeon Haloferax volcanii cell extract. RNA. 2000;6:1019–1030. doi: 10.1017/s1355838200000613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Englert M, Sheppard K, Aslanian A, Yates JR, 3rd, Söll D. Archaeal 3′-phosphate RNA splicing ligase characterization identifies the missing component in tRNA maturation. Proc Natl Acad Sci USA. 2011;108:1290–1295. doi: 10.1073/pnas.1018307108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Popow J, et al. HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science. 2011;331:760–764. doi: 10.1126/science.1197847. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka N, Chakravarty AK, Maughan B, Shuman S. Novel mechanism of RNA repair by RtcB via sequential 2′,3′-cyclic phosphodiesterase and 3′-Phosphate/5′-hydroxyl ligation reactions. J Biol Chem. 2011;286:43134–43143. doi: 10.1074/jbc.M111.302133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desai KK, Raines RT. tRNA ligase catalyzes the GTP-dependent ligation of RNA with 3′-phosphate and 5′-hydroxyl termini. Biochemistry. 2012;51:1333–1335. doi: 10.1021/bi201921a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nandakumar J, Shuman S, Lima CD. RNA ligase structures reveal the basis for RNA specificity and conformational changes that drive ligation forward. Cell. 2006;127:71–84. doi: 10.1016/j.cell.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 20.Chakravarty AK, Subbotin R, Chait BT, Shuman S. RNA ligase RtcB splices 3′-phosphate and 5′-OH ends via covalent RtcB-(histidinyl)-GMP and polynucleotide-(3′)pp(5′)G intermediates. Proc Natl Acad Sci USA. 2012;109:6072–6077. doi: 10.1073/pnas.1201207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okada C, Maegawa Y, Yao M, Tanaka I. Crystal structure of an RtcB homolog protein (PH1602-extein protein) from Pyrococcus horikoshii reveals a novel fold. Proteins. 2006;63:1119–1122. doi: 10.1002/prot.20912. [DOI] [PubMed] [Google Scholar]
- 22.Schultz LW, Quirk DJ, Raines RT. His…Asp catalytic dyad of ribonuclease A: Structure and function of the wild-type, D121N, and D121A enzymes. Biochemistry. 1998;37:8886–8898. doi: 10.1021/bi972766q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chakravarty AK, Smith P, Shuman S. Structures of RNA 3′-phosphate cyclase bound to ATP reveal the mechanism of nucleotidyl transfer and metal-assisted catalysis. Proc Natl Acad Sci USA. 2011;108:21034–21039. doi: 10.1073/pnas.1115560108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perkins KK, Furneaux H, Hurwitz J. Isolation and characterization of an RNA ligase from HeLa cells. Proc Natl Acad Sci USA. 1985;82:684–688. doi: 10.1073/pnas.82.3.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hinton DM, Brennan CA, Gumport RI. The preparative synthesis of oligodeoxyribonucleotides using RNA ligase. Nucleic Acids Res. 1982;10:1877–1894. doi: 10.1093/nar/10.6.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chakravarty AK, Shuman S. RNA 3′-phosphate cyclase (RtcA) catalyzes ligase-like adenylylation of DNA and RNA 5′-monophosphate ends. J Biol Chem. 2011;286:4117–4122. doi: 10.1074/jbc.M110.196766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Filipowicz W, Konarska M, Gross HJ, Shatkin AJ. RNA 3′-terminal phosphate cyclase activity and RNA ligation in HeLa cell extract. Nucleic Acids Res. 1983;11:1405–1418. doi: 10.1093/nar/11.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: Isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 29.Molina H, Horn DM, Tang N, Mathivanan S, Pandey A. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci USA. 2007;104:2199–2204. doi: 10.1073/pnas.0611217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeon SJ, Fujiwara S, Takagi M, Tanaka T, Imanaka T. Tk-PTP, protein tyrosine/serine phosphatase from hyperthermophilic archaeon Thermococcus kodakaraensis KOD1: Enzymatic characteristics and identification of its substrate proteins. Biochem Biophys Res Commun. 2002;295:508–514. doi: 10.1016/s0006-291x(02)00705-2. [DOI] [PubMed] [Google Scholar]
- 31.Hamamichi S, et al. Hypothesis-based RNAi screening identifies neuroprotective genes in a Parkinson’s disease model. Proc Natl Acad Sci USA. 2008;105:728–733. doi: 10.1073/pnas.0711018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.