Abstract
Nucleocytoplasmic transport is mediated by nuclear pore complexes (NPCs), enormous assemblies composed of multiple copies of ∼30 different proteins called nucleoporins. To unravel the basic scaffold underlying the NPC, we have characterized the species-specific scaffold nucleoporin Nup37 and ELY5/ELYS. Both proteins integrate directly via Nup120/160 into the universally conserved heptameric Y-complex, the critical unit for the assembly and functionality of the NPC. We present the crystal structure of Schizosaccharomyces pombe Nup37 in complex with Nup120, a 174-kDa subassembly that forms one of the two short arms of the Y-complex. Nup37 binds near the bend of the L-shaped Nup120 protein, potentially stabilizing the relative orientation of its two domains. By means of reconstitution assays, we pinpoint residues crucial for this interaction. In vivo and in vitro results show that ELY5 binds near an interface of the Nup120–Nup37 complex. Complementary biochemical and cell biological data refine and consolidate the interactions of Nup120 within the current Y-model. Finally, we propose an orientation of the Y-complex relative to the pore membrane, consistent with the lattice model.
Keywords: macromolecular assemblies, structural biology
Compartmentalization of the genetic material into the nucleus, and thus physical separation of transcription and translation, is the hallmark feature that distinguishes eukaryotes from prokaryotes. The nucleus is enclosed by the nuclear envelope (NE), a double-membrane layer composed of the inner nuclear membrane (INM) and outer nuclear membrane (ONM), separated by the perinuclear space (1). Passage across the NE occurs through circular openings, or pores, where INM and ONM fuse. The pores are lined with massive protein assemblies, called nuclear pore complexes (NPCs) (2, 3). Principally conserved in all eukaryotes, NPCs are estimated to be ∼50 MDa in size and composed of ∼30 different proteins (nucleoporins or Nups), which are arranged in multiple copies around a central eightfold rotational axis (4). Apart from the well-known transport function (5–7), NPCs participate in a number of additional cellular processes, including nuclear organization, cell cycle regulation, chromatin maintenance, and DNA repair (8).
To understand the molecular mechanisms underlying these different functions, elucidating the structure of the NPC is necessary. Recent cryoelectron tomographic studies of the intact NPC embedded in the NE reached a resolution of 6 nm, revealing general features of the assembly (9). The NPC scaffold appears porous and lattice-like, and it has a central framework that is roughly twofold symmetrical across the NE plane. The symmetry breaks at the pore periphery, where distinct features, cytoplasmic filaments and the nuclear basket, respectively, are visible. Although these tomographic studies are informative, the resolution afforded by X-ray crystallography is needed to study the molecular details of the NPC. The enormous size of the NPC and its partially dynamic character require that these studies focus for now on the subcomplexes into which nucleoporins are organized (4).
The large body of data on the subcomplexes that build the NPC points toward a model in which two large multimeric subcomplexes form the main, stable architectural scaffold of the NPC. These two subcomplexes are the 7- to 10-membered Y-complex and the 5-membered Nic96 complex. Although an approximate model of the Nic96 complex has just begun to emerge (10, 11), more is known about the essential Y-complex (also referred to as the Nup84 complex in yeast or Nup107–Nup160 complex in vertebrates) (12, 13). Seven conserved proteins, yeastNup120/humanNup160, Nup133, Nup84/107, Nup85, Nup145C/96, Sec13, and Seh1, form the eponymous structure composed of two short arms and one extended stalk, which are connected at a central hub (14, 15). A recently determined ∼35-Å resolution structure from negative-stain EM has allowed a tentative fit of its constituents (15).
In addition to its conserved core of seven proteins, the Y-complex contains up to three additional proteins, Nup37, Nup43, and ELYS, depending on the organism (16, 17). Although a specific function for both Nup37 and Nup43 is still unknown, their protein architecture (18), as well as their stable integration into the NPC (19), suggests that they are scaffolding units (4). Thus, they may be responsible for some of the difference observed between NPCs from different organisms (9). ELYS, initially described in Xenopus egg extracts (17), is thought to initiate postmitotic NPC assembly by recruiting the Y-complex to chromatin (20). Although ELYS is a large, multidomain protein in metazoa with a length of ∼2,000- to 2,500 aa, a distant homolog, ELY5, is found in many fungi, although curiously not in budding yeast. ELY5, which is only ∼300 aa in length, may have reduced functionality compared with vertebrate ELYS/Mel-28.
Here, we show that the integration of Nup37 and ELY5/ELYS into the Y-complex is mediated through a network of interactions involving Nup120/160. We present the crystal structure of Schizosaccharomyces pombe Nup120 in complex with Nup37 and describe a biochemical analysis of the binding interface. We show by in vivo localization and in vitro reconstitution assays that S. pombe ELY5 is incorporated into the nuclear pore scaffold through interactions with Nup120 and Nup37. We suggest roles for the species-specific architectural nucleoporins and describe how our data fix the orientation of the Y-complex relative to the pore membrane.
Results
Nup120/160 Is the Binding Platform for Integrating Nup37 and ELY5/ELYS into the Y-Complex.
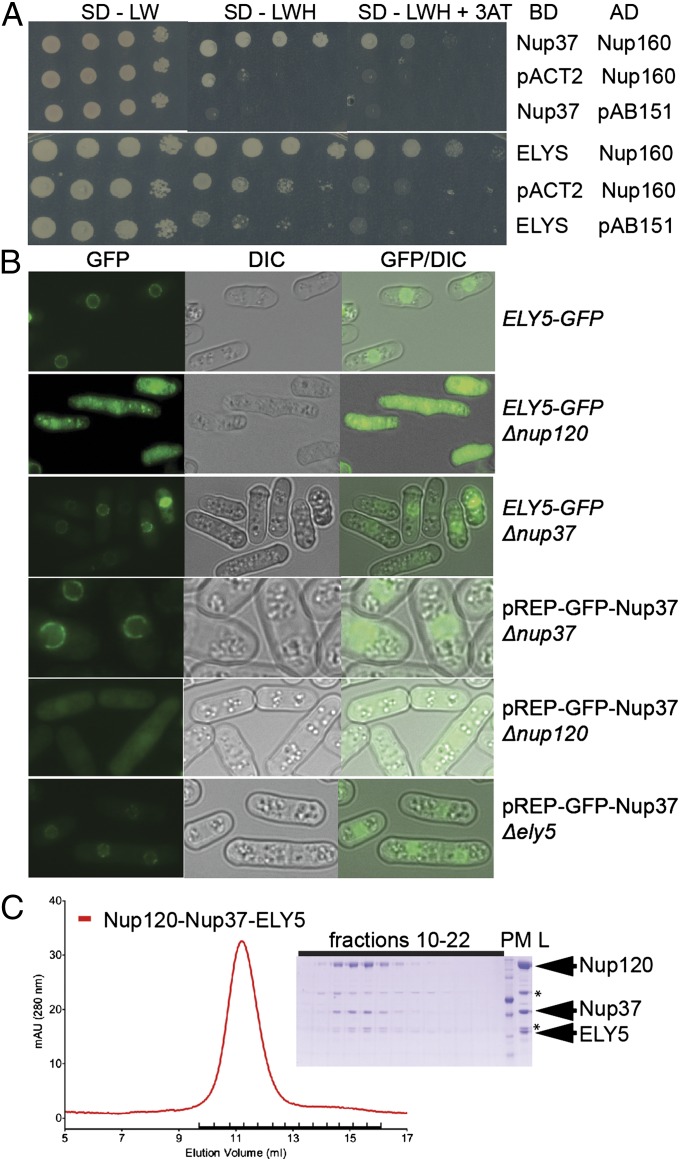
To investigate the direct binding partner of Nup37 and ELYS within the Y-complex, we used a yeast two-hybrid (Y2H) approach with human proteins (Fig. 1). Nup160 emerged as the direct interacting partner of Nup37 and ELYS (Fig. 1A).
Fig. 1.
Nup120/160 recruits Nup37 and ELY5/ELYS to the NPC. (A) Y2H interactions between human nucleoporins. Plasmids expressing Lex4 DNA-binding domain (BD) and Gal4 activation domain (AD) fusion constructs of nucleoporins were transformed into a yeast strain, and 10-fold serial dilutions were spotted on synthetic dropout (SD)-Leu-Trp (LW) and SD-Leu-Trp-His (LWH) plates supplemented with 3-amino-1,2,4-triazole (3AT). As a negative control, a combination of the fusion construct and the empty plasmid (pAB151, pACT2) was used. (B) In vivo localization of S. pombe Nup37 and ELY5. Both GFP nucleoporins become distributed throughout the cell in a Δnup120 background (second and fifth rows). GFP-tagged ELY5 and Nup37 are properly targeted to the NPC in Δnup37 and Δely5 S. pombe cells (third and sixth rows). (Magnification: 1,000×.) (C) ELY5 forms a complex with Nup120-Nup37 in a gel filtration experiment. Selected fractions (elution volume: 9.85–16.35 mL), marked with a black bar on the chromatogram, were analyzed by SDS/PAGE. The asterisk (*) denotes common impurities. L, loaded proteins; PM, protein marker.
We validated the Y2H data by in vivo localization studies in S. pombe (Fig. 1) using GFP-tagged proteins. GFP nucleoporins in the WT strain showed characteristic nuclear rim staining (Fig. 1B and Table S1). A Δnup120 strain with a genomic copy of ELY5-GFP revealed fluorescence signal diffusely dispersed throughout the cytoplasm (Fig. 1B), indicating that ELY5 is not recruited to the NPC. Similarly, GFP-Nup37 did not localize properly to the nuclear rim in the Δnup120 strain. WT localization of ELY5 was restored after expressing either plasmid-borne, full-length Nup120 or Nup120 α-helical domain (residues 546–1,136; Fig. S1). The localization defects of ELY5 as well as Nup37 were characteristic of Δnup120 because they were not reproduced in a Δnup37 or Δely5 strain, respectively (Fig. 1B).
We also performed binding studies with purified proteins in vitro. Full-length Nup120 and Nup37, overexpressed in Escherichia coli, formed a stable complex as judged by size exclusion chromatography. Full-length ELY5 was overexpressed as a GFP-fusion product under the nmt1 promoter in S. pombe. Ni-NTA beads saturated with Nup120, Nup37, or Nup120–Nup37 complex were incubated with GFP-ELY5 in S. pombe extract. ELY5 binds to beads when Nup120 or Nup120–Nup37 complex is present (Fig. S1D).
To test whether Nup37 affects the Nup120–ELY5 interaction in S. pombe, we generated yeast strains expressing plasmid-encoded, flag-tagged Nup120 and genomically encoded GFP-tagged ELY5 and affinity-purified the Nup120–ELY5 complex from WT and Δnup37 strains. We found that Nup120 pulled down ELY5 from both strains but that the efficiency was higher in the WT (Fig. S1E), indicating that Nup37 contributes to ELY5 binding.
To map the region of ELY5 that mediates the interaction with the nuclear pore, we examined ELY5 truncations (Fig. S1C). Localization in S. pombe shows that removal of the 26 C-terminal residues from ELY5 does not eliminate binding to the NPC but that N-terminal and further C-terminal truncations of ELY5 compromised the association (Fig. S1). Using ELY51–272 overexpressed in E. coli, we could detect ELY51–272-Nup120–Nup37 complex formation in a gel filtration experiment (Fig. 1C).
Our combined biochemical and in vivo findings support a model in which Nup37 binds directly to Nup120 and the complex recruits ELY5.
Structure Determination of the Nup120–Nup37 Complex.
We obtained crystals of a Nup120–Nup37 complex that adventitiously included an additional E. coli protein as determined by SDS/PAGE of the dissolved crystals. The contaminant was identified as E. coli glutamate dehydrogenase (ecGDH) by mass spectrometric analysis.
The crystals diffracted to 7.0 Å, and the structure was solved iteratively using a combination of molecular replacement (MR) and single isomorphous replacement with anomalous dispersion phasing (Fig. 2, Fig. S2, and Table S2; details are provided in SI Materials and Methods).
Fig. 2.
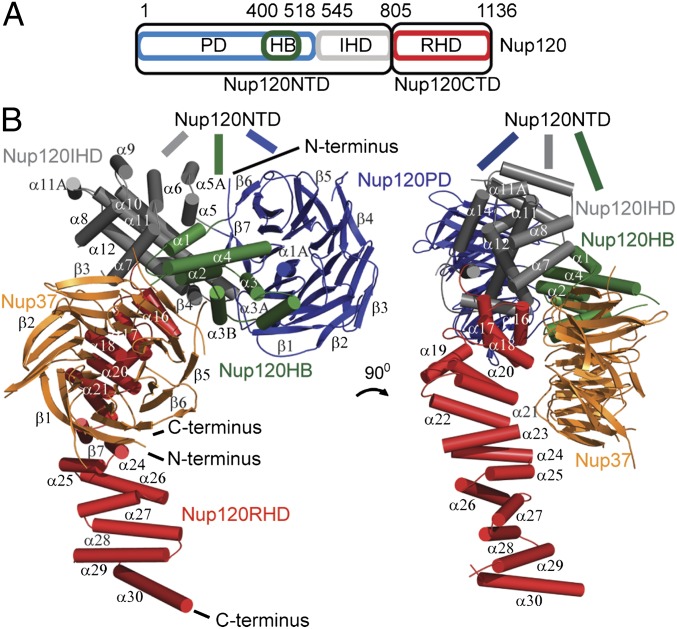
Structural analysis of the Nup120–Nup37 complex. (A) Domain structure of Nup120. The propeller domain (PD, blue) with the helical bundle insertion (HB, green) and the irregular α-helical domain (IHD, gray) make the 91 kDa Nup120 NTD. The 39 kDa Nup120 CTD is built from a regular α-helical domain (RHD, red). (B) Cartoon representation of the Nup120-Nup37 heterodimer. The complex comprises full-length nucleoporins with Nup120 colored as in A and Nup37 colored in orange. The β-strands are drawn as arrows, and the α-helices are drawn as cylinders. The two views are rotated by 90° with respect to each other. Nup37 binds close to the bend of the L-shaped Nup120 molecule.
We initially obtained an MR solution for ecGDH, which we had separately determined at 2.0 Å (Table S3). These phases were used to find seven tantalum-cluster sites in a derivative (Fig. S2A). Combined MR–single-wavelength anomalous dispersion (SAD) phases, together with density modification, resulted in an interpretable electron density map that outlined the molecular arrangement within the asymmetrical unit (asu). The asu contains two Nup120–Nup37 complexes and one molecule of ecGDH (Fig. S2A). We could dock two copies of the published N-terminal Nup120 fragment from Saccharomyces cerevisiae [Protein Data Bank (PDB) ID codes 3HXR and 3F7F] into the electron density. At this stage, it became obvious that the C-terminal ∼170 residues of Nup120 are somewhat flexible, because they adopt slightly different positions in the two copies in the asu (Fig. S2E). Thus, a truncated Nup120ΔC (residues 1–961)–Nup37 complex formed better ordered crystals that diffracted to 4.4 Å (Table S2). We built the 4.4-Å structure using selenomethionine (SeMet)-SAD phases and assigned the sequence of Nup120. Nup37 modeling in the complex was aided by a 2.6-Å apo-Nup37 structure solved and refined separately (Table S4). The refined 4.4-Å structure was used to generate the best density map for the 7.0-Å, full-length Nup120–Nup37 complex, which then also enabled building and at least tentative assignment of the sequence of the entire complex, including the C-terminal ∼170 residues of Nup120.
Structure of the Nup120–Nup37 Complex.
Nup120 is an L-shaped molecule composed of two principal domains, a ∼91-kDa N-terminal domain (NTD) and a ∼39-kDa C-terminal domain (CTD) (Fig. 2). The NTD of S. pombe Nup120 closely resembles the S. cerevisiae homolog described recently (21, 22). The two Nup120 NTD structures, which have 23% sequence identity, superpose with an r.m.s.d. of 2.6 Å. The NTD begins with a seven-bladed β-propeller domain (PD), interrupted by six helices inserted between blades 6 and 7. These helices, together with an additional helix (1A), make an unusual and characteristic helical bundle (HB) insertion. In S. pombe, the bundle has seven helices, but it only has four helices in S. cerevisiae. The NTD is completed by an irregular α-helical domain (IHD), which has extensive contact with both the PD and HB. The core of the IHD is formed by two long hydrophobic α-helices (helix α11 and helix α12), which pack against each other, whereas the remaining α-helices wrap around this central stalk and bury it in the hydrophobic core. The entire NTD is a compact, rigid domain.
The CTD emerges in a sharp 90° angle from the NTD and forms an elongated α-helical stack composed of 16 α-helices (α15–α30) (Fig. 2B). Fourteen α-helices are arranged in seven antiparallel α-helical pairs reminiscent of the HEAT-repeat motif. Exceptions are the fifth α-helix (α19), which is positioned in an extended loop, and the terminal helix α30, which caps the α-helical stack. The HEAT repeats are stacked on top of one another in a staggered fashion. The antiparallel α-helices vary in length as well as in the twist angle between each α-helical pair, causing the domain to have a slightly curved shape.
As illustrated in Fig. 2B, Nup37 binds close to the elbow of the L-shaped Nup120, likely stabilizing the relative positions of the two Nup120 domains. Nup37 is a seven-bladed β-propeller with a diameter of ∼50 Å and a thickness of ∼30 Å (Fig. S3). Blade 7 shows a characteristic velcro-closure arrangement of strands (23). The loops are named according to which of the four strands (A–D) in a blade they connect. The DA and BC loops that define the top surface of the β-propeller are short, except for the elongated connection between strands 4D and 5A. The long loops at the bottom surface of Nup37 are characteristic structural elements in an otherwise fairly canonical seven-bladed β-propeller. Some of these insertions are not conserved in the shorter vertebrate homologs (Fig. S3A).
The elaborate interface between Nup120 and Nup37 buries a surface area of ∼4,100 Å2. It has a mixture of van der Waals, polar, and charged interactions. The HB insertion in the Nup120 β-propeller cradles the side surface of Nup37 formed by blades 3 and 4, whereas the top of the Nup37 β-propeller abuts and interacts with the first four α-helical pairs of the CTD of Nup120 (Fig. 2 and Fig. S2C). The difference between the two Nup120–Nup37 complexes in the crystallographic asu is largely limited to the last eight α-helices of Nup120, which do not interact with Nup37 and are slightly rotated with respect to each other (Fig. S2E). We cocrystallized the Nup120–Nup37 complex with ecGDH, which helps in forming crystal contacts. We have also crystallized the Nup120–Nup37 complex in the absence of ecGDH, but we could only record lower resolution data (Fig. S2D and Tables S5 and S6). Superposing this ecGDH-free complex onto the two Nup120–Nup37 complexes in the 7.0-Å crystal form shows that ecGDH has no influence on the overall molecular arrangement within the complex (Fig. S2E).
The flexibility seen in the CTD is typical for many HEAT-repeat proteins and often important for their function (24). The random conical tilt EM reconstruction of the assembled Y-complex showed conformational flexibility at the tentatively inferred position for Nup120 (15). Our structure suggests that the flexibility of Nup120 results predominantly from movement within the α-helical stack of the CTD. The small interface between the NTD and CTD suggests that the two domains might pivot with respect to each other, particularly in the absence of Nup37.
Comparison of Nup120 with Other Scaffold Components of the NPC.
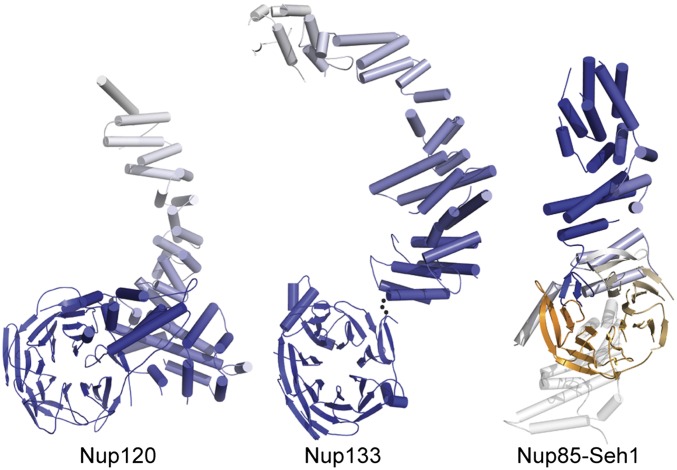
Scaffold nucleoporins can be categorized according to their tertiary structure elements. Structure prediction suggested that Nup157/170, Nup133, and Nup120 are particularly similar because they are all composed of an N-terminal PD and a C-terminal α-helical repeat domain (25). Although the similarity between Nup157/170 and Nup133 has been experimentally confirmed (26), Nup120 is distinctly different (Fig. 3). It cannot be meaningfully superposed on Nup157/170 or Nup133. The main differences are that the N-terminal β-propeller is integrated into the NTD of Nup120 as described above, whereas in Nup157/170 and Nup133, it forms a separate, flexible attached domain. The α-helical portion of Nup120 is divided into three parts: HB, IHD, and regular α-helical domain (RHD). Although the RHD is a α-helical, reasonably canonical, repeat domain, the IHD is idiosyncratic, and we have not detected topological similarity to it in other proteins. Distinct from both the Nup120 and Nup133 folds is the ancestral coatomer element (ACE1) fold, which was described by Brohawn et al. (27) and is found in four additional scaffold nucleoporins. ACE1 is a ∼65-kDa tripartite (crown, trunk, and tail) α-helical domain that has a characteristic fold-back arrangement of its constituent α-helical stack, a feature not observed in Nup120. Some ACE1 proteins, such as Sec16, Sec31, Nup85, and Nup145C, bind β-propellers in trans, but the relative orientation of both elements is, again, quite different from that found in Nup120.
Fig. 3.
Nup120 is structurally distinct from other nucleoporins, composed of a β-propeller domain combined with an α-helical domain. Full-length Nup120 (Left), Nup133 (Center), and Nup851–545 (Right) are gradient-colored blue to white from the N terminus to the C terminus. The structures of the Nup133 β-propeller domain and α-helical domain were determined separately, and are oriented arbitrarily to one another. The ACE1 protein Nup85 in complex with Seh1 (gradient-colored orange to white) exhibits a distinct fold-back structure not observed in Nup120 or Nup133. The Nup85 tail module was modeled and is shown transparently in white.
Analysis of the Nup120–Nup37 Binding Interface.
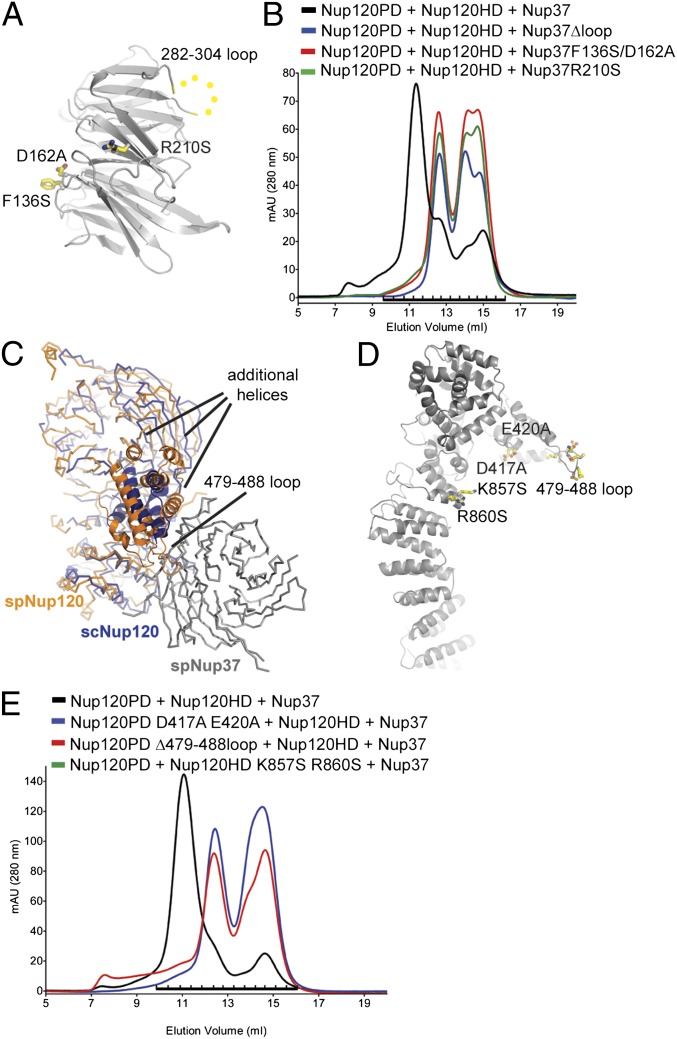
To confirm correct sequence assignment in the crystal structure of the complex, we designed surface mutations in Nup37 and Nup120 that should retain the protein structure but abolish binding of the partner protein (Fig. 4 and Fig. S4). As an assay, we examined the formation of a stable ternary complex between Nup37; the PD of Nup120, including the inserted HB (Nup120PD, residues 1–545); and the complete α-helical portion of Nup120 (Nup120HD, residues 546–1,136). The two Nup120 fragments bind to each other only in the presence of Nup37 (Fig. 4B and Fig. S4A).
Fig. 4.
Mutations in Nup37 or Nup120 abolish complex formation. (A) Cartoon representation of Nup37 with elements critically involved in Nup120 binding highlighted in yellow. (B) Gel filtration analysis of Nup37 mutants binding to Nup120 domains shows that the designed mutations abolish the interaction. Complex formation, indicated by an elution peak at ∼11 mL as shown for WT Nup37, is no longer observed for any of the mutants (Fig. S4D). (C) Ribbon representation of superposed Nup120 NTDs from S. cerevisiae and S. pombe. Differences in the HB region, important for Nup37 binding, are marked. A cartoon representation is used for HB insertion. Nup37 is depicted in the ribbon representation and colored in gray. (D) Cartoon representation of Nup120, with elements critically involved in Nup37 binding highlighted in yellow. (E) Binding data with mutants in Nup120. Gel filtration profiles indicate that the designed mutations in Nup120 abolish interaction with Nup37 (Fig. S4E).
We first probed the interaction of Nup37 with the HB insertion of Nup120 (Fig. S4 B and C). Residue R210, situated in blade 4 of Nup37 (Fig. 4A), interacts with the Nup120 loop connecting the first two α-helices in the HB insertion (residues 410–418). Mutation of the residue to serine abolished Nup120 binding (Fig. 4B). Complementary mutation of Nup120 (Nup120PD D417A E420A) also abolished complex formation (Fig. 4E and Fig. S4E). Residues 282–304 build the Nup37 3CD loop, which is disordered in the apo-Nup37 structure but becomes partially ordered on Nup120 binding. Deletion of the Nup37 3CD loop (residues 282–304) abolished complex formation (Fig. 4B). Likewise, deletion of the contacting loop in Nup120 (residues 479–488; Fig. 4 C and D) hindered Nup37 binding (Fig. 4E).
We then tested the Nup37–Nup120–CTD interaction. Two point mutants in Nup37, F136S within the 2D3A loop and D162A in the 3BC loop, respectively, abolish binding to the second HEAT repeat (α17 and α18) in the Nup120–CTD interaction (Fig. 4B). Reciprocally, a double mutation of the Nup120–CTD interaction surface exposed residues R860 and K857 to serine also disrupted Nup37 binding (Fig. 4E).
Interactions Within the Y-Complex.
Biochemical characterization of the S. cerevisiae Y-complex and its components has shown that Nup120, Nup145C-Sec13, and Nup85-Seh1 self-assemble into a heteropentameric complex in vitro (14). Further binding assays have established that the tail domains of Nup145C and Nup85 are required for the integration into the Y-complex (27). In the current model, the C terminus of Nup120 is central to the hub structure of the Y-complex, bridging Nup85 and Nup145C.
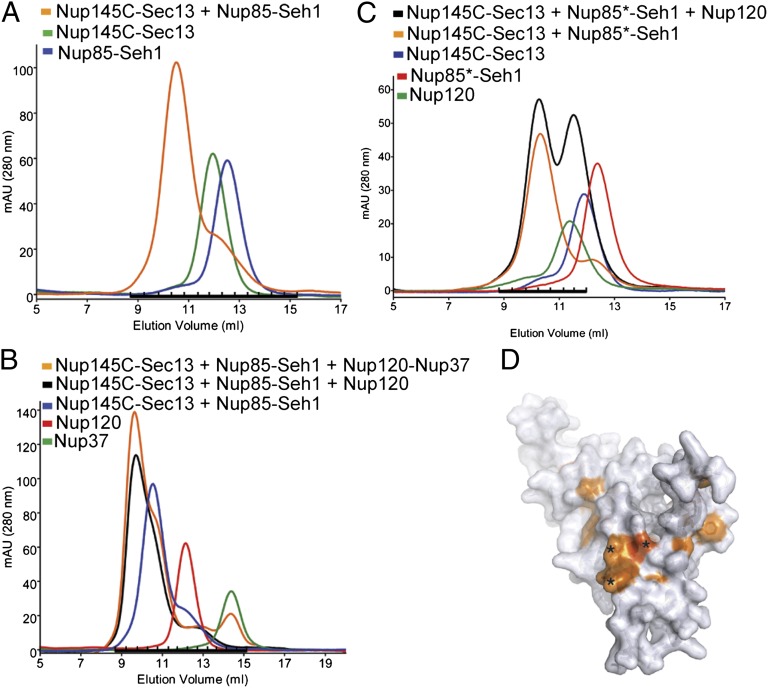
To demonstrate that Nup37 is a stable component of the Y-complex, we reconstituted the heterohexameric Nup145C–Sec13–Nup85–Seh1–Nup120–Nup37 complex from purified components in our biochemical assay (Fig. 5 and Fig. S5). Contrary to previous data, we were able to form a tetrameric complex between Nup145C-Sec13 and Nup85-Seh1 in the absence of Nup120 (Fig. 5A and Fig. S5C). This result indicates direct interaction between Nup145C-Sec13 and Nup85-Seh1 and leads us to modify the current Y-model (Fig. 6).
Fig. 5.
Interaction analysis of the S. pombe Y-complex. (A) S. pombe Nup85-Seh1 stably interacts with Nup145C-Sec13, as judged by the formation of a higher molecular-weight species using size exclusion chromatography (Fig. S5C). (B) Nup120 binds to preformed Nup85-Seh1-Nup145C-Sec13 tetramer. Addition of Nup37 to this pentameric complex results in the formation of a stable hexameric complex (Fig. S5D). (C) Nup85 binds Nup120 and Nup145C via its tail module. Binding of the heterodimeric Nup85–Seh1 complex with triple mutation L632D, V632D, and Y636N in the Nup85 tail module (marked as Nup85*-Seh1) to the Y-complex was analyzed by gel filtration. Nup85*-Seh1 binds the Nup145C-Sec13 heterodimer, but incorporation of Nup120 into the tetrameric complex is abolished (Fig. S7B). (D) Threading model of the Nup85 tail module with mapped sequence conservation colored from white to dark orange. Black stars denote the position of the triple mutation used in this study.
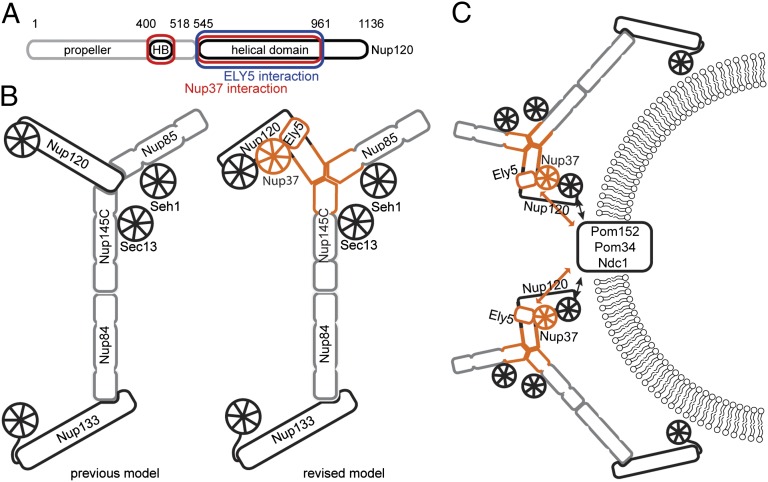
Fig. 6.
Refined assembly model for the Y-complex. (A) Nup120 regions involved in Nup37 and ELY5 binding are boxed in red and blue, respectively. The domain structure of Nup120 with PD in gray and α-helical regions in black is shown. (B) Relative positions and interactions between the proteins in the Y-complex are shown; the previous consensus model (Left) and the modified model based on the data presented here (Right) are shown. The hub, where the two arms and the stalk of the Y-complex cojoin, has mutual interactions between all extensions rather than Nup120 forming an exclusive tether. The additional proteins Nup37 and ELY5 are recruited via Nup120. Proteins belonging to the ACE1 class are colored in gray. (C) Model of the interaction network within the Nup120–Nup37–ELY5 complex. Black solid arrows depict direct protein–protein interactions, whereas orange arrows indicate proximity to the pore membrane proteins proposed by our work.
To map the interaction site of Nup120 with Nup85 and Nup145C, we used truncation mutants. In S. cerevisiae, the Nup120–NTD interaction (residues 1–757) does not interact stably with either Nup145C or Nup85 (21). Our S. pombe Nup120ΔC construct is substantially longer (residues 1–961), but it still does not bind the heterotetrameric Nup145C–Sec13–Nup85–Seh1 complex (Fig. S5E). The addition of Nup37 to Nup120ΔC also does not restore binding to the tetrameric complex (Fig. S5F). Furthermore, we were unable to detect nuclear rim staining for GFP-labeled Nup120ΔC in in vivo localization studies (Fig. S5B). Thus, we conclude that the region of Nup120 required for its incorporation into the Y-complex lies within the C-terminal ∼170 residues. Nup120C (residues 965–1,136) is the most conserved part of Nup120 (Fig. S6); it incorporates efficiently into NPCs in vivo (Fig. S5B) and contains the binding site for both Nup85-Seh1 and Nup145C-Sec13, as shown by in vitro binding (Fig. S5G).
A previous study has shown that the tail modules of the ACE1 proteins Nup145C and Nup85 are necessary for interaction with Nup120 (27). In contrast, when the heptameric Y-complex was purified from S. cerevisiae and its 3D structure was analyzed by EM, a different model was proposed, in which the crown region of Nup85, rather than the tail, contributes to the interactions within the hub (15). To distinguish between the two orientations of Nup85-Seh1 within the Y-complex, we performed further studies. The crystal structure of the Nup85–Seh1 complex does not include the tail module of Nup85 (27). We therefore used MODELLER (28) to create a threading model of the S. pombe Nup85 tail module using the crystal structure of Nic96 as a template (29) (Fig. 5D). A few residues in the penultimate helix are hydrophobic, conserved, and surface-exposed in our model, and we hypothesized that they make the binding sites for Nup145C or Nup120 (Fig. S7). We mutated the sequence L632VNVY636 to DDNVN, introducing charges and replacing tyrosine with a smaller amino acid, asparagine. The overall fold of Nup85 was not perturbed by this modification, because the protein binds Seh1 and behaves indistinguishably from WT as judged by gel filtration. The mutations in the penultimate helix did not abolish binding to Nup145C-Sec13 (Fig. 5C), but the tetrameric Nup85–Seh1–Nup145C–Sec13 complex no longer bound Nup120 (Fig. 5C). Thus, our biochemical data show that the tail module of Nup85 interacts with the tail region of Nup145C and the CTD of Nup120 (Fig. 6).
Discussion
We have characterized the integration of Nup37 and ELY5/ELYS into the NPC and established that their binding partner is Nup120/160 (Fig. 6 A and B). Nup37 and ELY5/ELYS are stable architectural nucleoporins that occur, independent of one another, in a subset of eukaryotes (30).
A central question regarding the NPC scaffold is the orientation of the Y-complex relative to the pore membrane and to the other components of the NPC. Two very different models have been proposed. In the “head-to-tail” model, eight Y-complexes form a closed ring around the central channel (22, 31, 32). In the “lattice” model, Y-complexes are oriented “head-to-head” along the positive curvature of the pore membrane, with Nup133 facing outward (27, 33) (Fig. 6C). We show here by in vivo fluorescence microscopy that Nup37 and ELY5 require Nup120 for proper NPC integration in S. pombe. In Aspergillus nidulans, Nup37 and ELY5 are required to maintain Y-complex association with transmembrane nucleoporins (TM-Nups) during semiopen mitosis (34). Together, these data strongly suggest that Nup37 and ELY5/ELYS bridge between TM-Nups and the Y-complex through Nup120/160 (Fig. 6C). Therefore, our data allow us to position the Nup120/160 arm of the Y-complex toward the pore membrane. This orientation is also consistent with previous data showing Nup120/160 interactions with the TM-Nups Pom152 and Pom121 in yeast and humans, respectively (35). Based on our lattice model with the stalk of the Y-complex orientated along the positive curvature of the pore membrane (33) and the assumption that the Y-complex is relatively flat (14, 15), the deduced membrane-proximal Nup120/160 position automatically orients Nup85 toward the central channel (Fig. 6C).
It can be envisioned that both the head-to-tail and lattice models can orient Nup120/160 toward the pore membrane and Nup85 toward the pore channel. Our structure of full-length Nup120 shows, however, that the residues previously suggested to be critical for a direct interaction of Nup120 with Nup133 in the head-to-tail model (22) are not conserved in S. pombe (Fig. S6). Specifically, residue D641 in S. cerevisiae, proposed to be critical for direct scNup133 binding (22), is conserved neither in S. pombe nor in metazoans. Therefore, the head-to-tail model is either specific to S. cerevisiae or the presumed Nup120–Nup133 interaction is mediated very differently in the various species. We think that both alternatives are rather unlikely.
In summary, the present work, together with previous contributions from many laboratories, narrows down the possibilities for assembly of the architectural scaffold of the NPC. It shows that even at the level of the basic scaffold, species-specific differences occur that may result in substantially different NPC assemblies. It remains to be determined how these differences translate into function.
Materials and Methods
The details of protein expression, purification, crystallization, structure determination, protein interaction analysis, and in vivo studies are provided in SI Materials and Methods. In short, the proteins were expressed in E. coli using a pET-Duet vector modified to contain 3C-cleavable N-terminal 6 × His-SUMO tag. Recombinant proteins were purified using several chromatographic techniques. The Nup37 structure was solved by SAD, and the ecGDH structure was solved by MR. The Nup120–Nup37 complex structure was solved using a combination of MR and SAD phasing. Data collection and refinement statistics are summarized in Tables S2–S6. Nup37, Nup1201–967-Nup37, Nup120-Nup37-ecGDH, and ecGDH are deposited in the PDB with ID codes 4FHL, 4FHM, 4FHN, and 4FCC, respectively.
Supplementary Material
Acknowledgments
We thank staff at beamlines 24-ID-C/-E at Argonne National Laboratory for assistance with data collection, especially K. R. Rajashankar for help with obtaining tantalum phases; D. Kim for providing access to the fluorescence microscope; V. Doye for providing the Δnup120 S. pombe strain; E. Spear and M. Halic for advice on yeast experiments; and N. Leksa, A. Ulrich, G. Kabachinski, M. Halic, and T. Maier for critically reading the manuscript. This work was supported by National Institutes of Health Grant GM077537 (to T.U.S.), a Pew Scholar Award (to T.U.S.), and a Croatian Science Foundation fellowship (to S.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4FHL, 4FHM, 4FHN, and 4FCC).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205151109/-/DCSupplemental.
References
- 1.Güttinger S, Laurell E, Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol. 2009;10(3):178–191. doi: 10.1038/nrm2641. [DOI] [PubMed] [Google Scholar]
- 2.Onischenko E, Weis K. Nuclear pore complex-a coat specifically tailored for the nuclear envelope. Curr Opin Cell Biol. 2011;23:293–301. doi: 10.1016/j.ceb.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hetzer MW, Wente SR. Border control at the nucleus: Biogenesis and organization of the nuclear membrane and pore complexes. Dev Cell. 2009;17:606–616. doi: 10.1016/j.devcel.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brohawn SG, Partridge JR, Whittle JRR, Schwartz TU. The nuclear pore complex has entered the atomic age. Structure. 2009;17:1156–1168. doi: 10.1016/j.str.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cook AG, Conti E. Nuclear export complexes in the frame. Curr Opin Struct Biol. 2010;20:247–252. doi: 10.1016/j.sbi.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 6.Rodríguez-Navarro S, Hurt E. Linking gene regulation to mRNA production and export. Curr Opin Cell Biol. 2011;23:302–309. doi: 10.1016/j.ceb.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Xu D, Farmer A, Chook YM. Recognition of nuclear targeting signals by Karyopherin-β proteins. Curr Opin Struct Biol. 2010;20:782–790. doi: 10.1016/j.sbi.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: Bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol. 2010;11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- 9.Frenkiel-Krispin D, Maco B, Aebi U, Medalia O. Structural analysis of a metazoan nuclear pore complex reveals a fused concentric ring architecture. J Mol Biol. 2010;395:578–586. doi: 10.1016/j.jmb.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 10.Amlacher S, et al. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell. 2011;146:277–289. doi: 10.1016/j.cell.2011.06.039. [DOI] [PubMed] [Google Scholar]
- 11.Theerthagiri G, Eisenhardt N, Schwarz H, Antonin W. The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. J Cell Biol. 2010;189:1129–1142. doi: 10.1083/jcb.200912045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walther TC, et al. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 2003;113:195–206. doi: 10.1016/s0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- 13.Harel A, et al. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell. 2003;11:853–864. doi: 10.1016/s1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- 14.Lutzmann M, Kunze R, Buerer A, Aebi U, Hurt E. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 2002;21:387–397. doi: 10.1093/emboj/21.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kampmann M, Blobel G. Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex. Nat Struct Mol Biol. 2009;16:782–788. doi: 10.1038/nsmb.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loïodice I, et al. The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol Biol Cell. 2004;15:3333–3344. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rasala BA, Orjalo AV, Shen Z, Briggs S, Forbes DJ. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc Natl Acad Sci USA. 2006;103:17801–17806. doi: 10.1073/pnas.0608484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol. 2004;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- 20.Franz C, et al. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep. 2007;8(2):165–172. doi: 10.1038/sj.embor.7400889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leksa NC, Brohawn SG, Schwartz TU. The structure of the scaffold nucleoporin Nup120 reveals a new and unexpected domain architecture. Structure. 2009;17:1082–1091. doi: 10.1016/j.str.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seo H-S, et al. Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex. Proc Natl Acad Sci USA. 2009;106:14281–14286. doi: 10.1073/pnas.0907453106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaudhuri I, Söding J, Lupas AN. Evolution of the β-propeller fold. Proteins. 2008;71:795–803. doi: 10.1002/prot.21764. [DOI] [PubMed] [Google Scholar]
- 24.Kappel C, Zachariae U, Dölker N, Grubmüller H. An unusual hydrophobic core confers extreme flexibility to HEAT repeat proteins. Biophys J. 2010;99:1596–1603. doi: 10.1016/j.bpj.2010.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Devos D, et al. Simple fold composition and modular architecture of the nuclear pore complex. Proc Natl Acad Sci USA. 2006;103:2172–2177. doi: 10.1073/pnas.0506345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whittle JR, Schwartz TU. Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element. J Biol Chem. 2009;284:28442–28452. doi: 10.1074/jbc.M109.023580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, Schwartz TU. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science. 2008;322:1369–1373. doi: 10.1126/science.1165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eswar N, Eramian D, Webb B, Shen MY, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2008;426:145–159. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 29.Jeudy S, Schwartz TU. Crystal structure of nucleoporin Nic96 reveals a novel, intricate helical domain architecture. J Biol Chem. 2007;282:34904–34912. doi: 10.1074/jbc.M705479200. [DOI] [PubMed] [Google Scholar]
- 30.Neumann N, Lundin D, Poole AM. Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS ONE. 2010;5:e13241. doi: 10.1371/journal.pone.0013241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandez-Martinez J, et al. Structure-function mapping of a heptameric module in the nuclear pore complex. J Cell Biol. 2012;196:419–434. doi: 10.1083/jcb.201109008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kampmann M, Atkinson CE, Mattheyses AL, Simon SM. Mapping the orientation of nuclear pore proteins in living cells with polarized fluorescence microscopy. Nat Struct Mol Biol. 2011;18:643–649. doi: 10.1038/nsmb.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brohawn SG, Schwartz TU. Molecular architecture of the Nup84-Nup145C-Sec13 edge element in the nuclear pore complex lattice. Nat Struct Mol Biol. 2009;16:1173–1177. doi: 10.1038/nsmb.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu HL, De Souza CPC, Osmani AH, Osmani SA. The three fungal transmembrane nuclear pore complex proteins of Aspergillus nidulans are dispensable in the presence of an intact An-Nup84-120 complex. Mol Biol Cell. 2009;20:616–630. doi: 10.1091/mbc.E08-06-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitchell JM, Mansfeld J, Capitanio J, Kutay U, Wozniak RW. Pom121 links two essential subcomplexes of the nuclear pore complex core to the membrane. J Cell Biol. 2010;191:505–521. doi: 10.1083/jcb.201007098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.