Abstract
Ultrasonics offers the possibility of developing sophisticated fluid manipulation tools in lab-on-a-chip technologies. Here we demonstrate the ability to shape ultrasonic fields by using phononic lattices, patterned on a disposable chip, to carry out the complex sequence of fluidic manipulations required to detect the rodent malaria parasite Plasmodium berghei in blood. To illustrate the different tools that are available to us, we used acoustic fields to produce the required rotational vortices that mechanically lyse both the red blood cells and the parasitic cells present in a drop of blood. This procedure was followed by the amplification of parasitic genomic sequences using different acoustic fields and frequencies to heat the sample and perform a real-time PCR amplification. The system does not require the use of lytic reagents nor enrichment steps, making it suitable for further integration into lab-on-a-chip point-of-care devices. This acoustic sample preparation and PCR enables us to detect ca. 30 parasites in a microliter-sized blood sample, which is the same order of magnitude in sensitivity as lab-based PCR tests. Unlike other lab-on-a-chip methods, where the sample moves through channels, here we use our ability to shape the acoustic fields in a frequency-dependent manner to provide different analytical functions. The methods also provide a clear route toward the integration of PCR to detect pathogens in a single handheld system.
Keywords: phononic crystal, surface acoustic waves, nucleic acid amplification test, mechanical cell lysis
Acoustic waves contain a mechanical energy that can be used to manipulate fluids, cells, and samples (1). A range of ultrasonic transducers have previously been developed, including those using surface acoustic wave (SAW) devices, as a practical solution to actuate fluids on microfluidic chips (2, 3). SAWs have the advantage that, despite using low powers, the energy is concentrated at the interface between the fluid and the substrate, enabling a range of fluid manipulations on a chip. Despite this ability to implement low power microfluidics, one potential disadvantage of using a SAW chip is the relatively high cost of the piezoelectric wafer. In an alternative configuration, the SAW can be coupled into a disposable superstrate (Fig. 1A) placed on the surface of the piezoelectric chip (4, 5), thus providing a low cost technology.
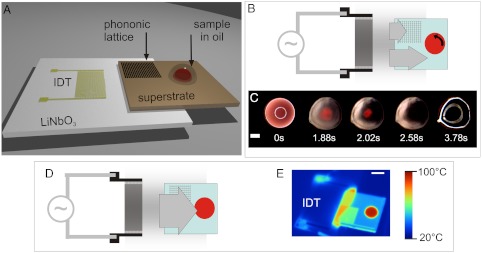
Fig. 1.
(A) Integrated lysis and PCR set up. Device architecture showing the IDT used to generate the SAW on the LiNbO3 piezoelectric wafer. The SAW is coupled into a superstrate. A microliter-sized droplet is positioned and coated with a layer of oil to prevent evaporation during processing. SI Appendix, Fig. S1 illustrates the interaction of the SAW with the drop. SI Appendix, Fig. S2 shows patterned hydrophilic tracks on the hydrophobic silicon superstrate, used to position the sample in place (3 mm disk, surrounded by a 200-μm wide annulus, 1 mm away). (B–E) Depending on the excitation frequency of the SAW, the square phononic lattice, machined into the superstrate, either filters or transmits the ultrasonic wave. Under conditions of SAW attenuation (9.5 MHz) the blood cells in the sample are lysed (B–C), whereas under transmission the ultrasonic wave causes heating (D–E) in a power-dependant manner. The lysis function occurs as a consequence of the propagation of the acoustic wave being hindered at a frequency within the stop-band of the phononic lattice, breaking the symmetry of the SAW and creating a rotational movement within the sample (B). This rotation results in shear flows that contribute to the disruption of the cell membrane. (C) The images are stills from SI Appendix, Movie S1 (9.5 MHz, 3.1 W) obtained through reflection microscopy. The first image (0 s) is taken before SAW actuation, whereas the last image (3.78 s) is taken after the SAW actuation has been turned off, to better show the increased transparency of the sample due to the disappearance of scattering cells after lysis. (Scale bar, 1 mm.) (D) Outside of the stop-band, the SAW propagates symmetrically and can be used to heat the sample. (E) The infrared image shows the drop at the denaturation temperature (95 °C, 1.3 W at 18 MHz). (Scale bar, 5 mm.)
Using such superstrates, we have recently demonstrated an alternative and improved method for performing complex fluid manipulations in which the ultrasonic waves are coupled into phononic lattices. Importantly, the functionality of such phononic structures is dependent upon the acoustic frequency. By using phononics to locally shape the acoustic fields and by switching between different ultrasonic wavelengths, we have designed tools capable of enabling different fluid manipulations on the disposable superstrate (5–7).
In this paper we show the implementation of nucleic acid based tests (NATs) on a microfluidic chip to demonstrate the potential of phononic technologies in diagnostics. Such tests usually require well-equipped laboratories with sample preparation apparatus, such as a centrifuge and thermal cyclers to carry out PCR. We now demonstrate that these apparatus can be replaced by microfluidic chips, using low cost phononic technology.
Microfluidics and lab-on-a-chip (LOC) devices have promised the development of portable point-of-care systems that could bring such complex assays to resource-limited settings in developing countries (8). In general, many of the techniques underpinning LOC involve a fluid being introduced into a device and being analyzed as it moves along a channel, with different functions being performed at different locations (9). In using phononics, we now propose a different paradigm, where the drop of sample remains stationary and the geometry of the phononic lattice and frequency of the SAW excitation combine to determine the functions available on the chip. By analogy with electronics, where a circuit is constructed by using different components, we propose to build complex bioassays by combining different phononic lattices, each of which delivers a unique diagnostic function to the stationary drop.
Our focus is on real-time PCR using conventional thermal cycling for the diagnosis of malaria, a disease which remains a significant burden on the health of individuals living in developing countries (10). Any strategy to curb the incidence of malaria and the consequent morbidity relies upon making efficient diagnostic identification of the blood stages of the aetiological agent that causes malaria, the unicellular protozoan parasites of the genus Plasmodium. As a model for the human pathogen, we worked with the rodent malaria parasite Plasmodium berghei in blood.
To access the appropriate genomic nucleic acids, the red blood cells and the parasite’s outer membranes both have to be disrupted. A typical off-chip cell lysis procedure (for blood or for other samples) involves numerous steps. These may include the use of lytic agents, enzymes, or detergents, which can inhibit subsequent analysis and have to be removed (11), most usually using centrifugation before further processing. Purification of the nucleic acids from the processed sample for analysis may also be required, a step known as enrichment. In existing NAT LOC devices these procedures have been integrated onto microfluidic chips, using the controlled mixing enabled by the behavior of liquids in the microdimensions (12, 13). Their reliance on lytic agents introduces significant dilutions of the sample/specimen that may compromise sensitivity and necessitates a cumbersome liquid-driving system to move the fluids around the chip, a concern for point-of-care applications. A number of other techniques have been developed for chemical-free lysis of cell samples in microfluidic platforms to overcome these limitations (14–19). Implementing such techniques has however been a particular difficulty in developing a fully integrated sample-to-answer solution for molecular diagnostics (20).
In this paper, we now show how SAW induced mechanical lysis of blood cells and of blood cells with malarial parasites within them, followed by real-time PCR detection of the parasite DNA using SAW as the heating mechanism, provide a method for implementing malarial detection on a disposable superstrate. By using a phononic lattice, both functions are carried out on the same platform (Fig. 1 B–E), switching between lysis and heating by changing the input frequency. Although only these two functions are combined here, the use of phononic lattices opens a wide range of applications and could be used to combine other SAW actuated techniques, such as the direction of drops for high-throughput analysis (21, 22) or nebulization (7).
Cell lysis using acoustic energy has previously been described either through harsh cavitation at high energies, or by using beads as crushing support (18). Proceeding differently now, we use SAW to create pressure waves and shear stresses, showing that both white blood cells and red blood cells can be lysed, as well as the parasites within, without cavitation or additional reagents introduced in the sample. SAW lysis efficiency was comparable to chemical and other mechanical lysis methods (17). Because of the mechanical friction that SAWs induce as they propagate, they can also be used for heating liquid samples. The input power can thereby serve as a method to cycle fluids between different temperatures (23). Here we adapt the virtual reaction chamber (VRC) described previously (24), covering a microliter-sized sample with a droplet of mineral oil (SI Appendix and Fig. 1A) to enable high temperature droplet processing (Fig. 1E) with SAW.
Thus, the same acoustic actuation mechanism can be used to lyse malaria-infected blood and amplify a specific sequence of the parasite’s genomic DNA via thermal cycle PCR. Fluorescence detection of the parasite-specific PCR product allows us to determine the level of parasitemia. In future the technique has the potential to discriminate between different species and drug-resistant organisms. This technology now provides us with a route toward a sensitive low cost, low power diagnostic tool to be deployed in developing countries for better infectious disease diagnostic.
Results and Discussion
The detection of parasites via their genetic material requires access to the nucleic acids within the parasites and the generation of a parasite-specific detectable signal. Here we perform these functions on a disposable SAW platform in microliter-sized drop samples, using phononic lattices to control the wave propagation and subsequently the functions enabled, in a frequency-dependent manner.
SAW Lysis.
SAWs were generated on a LiNbO3 piezoelectric wafer and coupled onto a phononic superstrate, via a thin layer of water-based gel. Upon reaching a droplet of liquid on the propagating surface, they are refracted as pressure waves inside the droplet at a specific angle depending on the speed of sound in both materials (2). By adjusting the power input in the device and the surface tensions at the droplet pinning contact line, different wave amplitudes give rise to different phenomena (3), from streaming at low powers to movement, jetting, and nebulization at high power. Here we make use of the pressure distribution inside the liquid when streaming is induced to create vortices. In their simplest states, these vortices are used for particle concentrations in the center of the droplet (25). We previously demonstrated that a phononic lattice, used as a frequency tuneable filter, could enable the centrifugation of blood cells within a drop on a disposable superstrate (5). We now further develop this technique and show a new function (Fig. 1 B–C), where at 9.5 MHz blood cells are first drawn into the center of the droplet at the beginning of the actuation (Fig. 1C after 1.88 s and SI Appendix, Movie S1) and then at increased powers, the conditions of pressures and shear stress at the center of the vortices mechanically disrupt them (Fig. 1C after 3.78 s). The result is cell lysis (SI Appendix, Movie S1), transforming the droplet from turbid (due to the presence of the blood cells, as shown in Fig. 1C 0 s) to translucent (as seen in Fig. 1C 3.78 s). We further investigated the phenomena using other cell types including the mammalian HL60 (SI Appendix, Fig. S3), MCF7 cells expressing a GFP-actin fusion (SI Appendix, Fig. S4 and Movie S2), and a trypanosome, Trypanosoma cyclops (SI Appendix, Fig. S5).
To understand the mechanism by which the cells lyse, we used the movement of beads to quantify the velocities of fluid flows within the drop at low power (milliwatt) and calculated the shear applied to the cells (SI Appendix, Figs. S6 and S7). These results were used to quantify the shears within the droplet, estimated as ca. 85 Pa under the conditions used for lysis (SI Appendix, Methods), sufficient for a mechanical disruptive effect on mammalian cells (26). Although previous literature suggests that shear forces at least an order of magnitude higher might be needed for the highly deformable RBC (27), we propose that it is a combination of these SAW-induced shear forces, coupled with the rapidly changing acoustic pressure fields and radiation pressures, which enables the rapid SAW lysis (which takes less than 3 s). Although the dissipation of acoustic energy in the droplet will generate heat, the temperature of blood droplets during the SAW lysis only rises to 38 °C, confirming that the lysis process was not due to an increase in temperature (28).
Lysis efficiency.
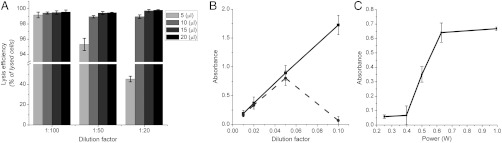
The extent of lysis of different biological samples was evaluated by counting the number of cells present at the end of the process and comparing it with the number of cells in an unprocessed sample. Fig. 2A shows the high rate of cell lysis for the conditions tested (> 98% for most conditions) and above 99.8% (± 0.16% standard deviation) for the optimized condition (20 μL samples at a power of 0.63 W). These results compare well with both chemical (cut off for efficient lysis at 99.5% in 18) and nonchemical protocols (17). Further, SI Appendix, Fig. S3 shows that cells were not lysed by the SAW at a lower exciting power (0.2 or 0.3 W), which caused only their concentration in the center of the droplet, whereas they were lysed efficiently when the vortices were created at 0.8 W (98.3% ± 1.4%). Similar results were obtained for different cell types [(SI Appendix, Fig. S5) shows the lysis of T. cyclops, a model of sleeping sickness].
Fig. 2.
Lysis efficiency (A) evaluated by reporting the number of cells lysed (percentage of original contents) for blood samples. Error bars are the estimated standard deviation over three samples. Diluted whole blood was processed for different volumes and different dilutions. It should be noted that the y axis is broken to show the results for 5 μL droplets of blood diluted 20 times (45%). (B–C) Absorbance of diluted blood samples (B, 540 nm—C, 414 nm), lysed chemically with Triton X-100 (squares, solid line) and using SAW (disks, dashed line). Droplets of 20 μL were processed on the SAW device for different dilutions of whole blood samples (B), and as a function of power for a 1∶50 diluted blood sample (dilution factor 0.02) (C). The SAW excitation was applied for 10 s.
Release of intracellular contents.
The availability of intracellular material in the solution after cell lysis on the SAW device was studied spectroscopically. The absorbance of the solutions at different wavelengths was used to evaluate the presence of haemoglobin (λ = 414 nm and 540 nm). Haemoglobin contained in the red blood cells is the most widely used marker of red blood cell lysis and spectroscopy is used routinely to evaluate its level in plasma as a diagnostic tool for haemolysis. Fig. 2B shows the level of haemoglobin in 20 μL samples of blood diluted with Triton X-100. For dilutions higher than 1∶20 (dilution factor < 0.05), the samples lysed with SAW or chemically were the same.
For more concentrated solutions, the efficiency dropped considerably, almost in a digital manner (Fig. 2B). The range of efficient dilutions can be explained by the fact that the formation of the vortices required for lysis was not possible at higher concentrations, where the blood cells formed clusters that disrupted the flow and prevented efficient streaming. Similarly, it was also noted that the lysis efficiency for drops of 5 μL decreased dramatically, which could be attributed to this same phenomena. By understanding the acoustic conditions for lysis, controlling the power of the SAW, the dilution of the sample and the volume it was possible to optimize conditions where intracellular contents are released, as shown in Fig. 2C, allowing the integration of blood lysis into a sequence of fluidic manipulation in a biological assay.
Amplification/detection of released DNA.
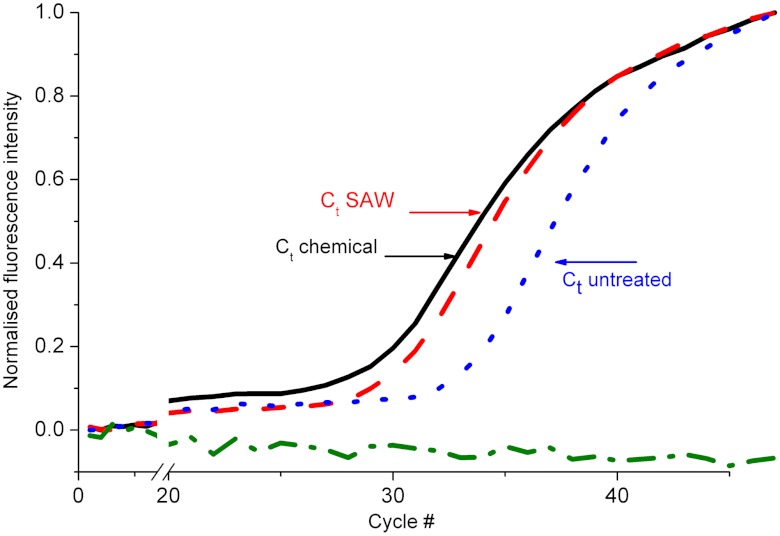
We evaluated the bioavailability of released analytes from cells lysed with SAW by performing a quantitative real-time PCR (qPCR) on genomic DNA from a blood sample. After lysis, the samples were processed on a LightCycler 480 with primers for the beta-actin (ActB) gene and the copy number present quantified (SI Appendix, Methods). Fig. 3 shows the fluorescence signal over the cycles for untreated whole blood (small dashed blue line), chemically lysed whole blood (full black line), SAW-lysed whole blood (red dashed line), and a negative control (deionised water, no template, green) processed identically. These results demonstrate the presence of genomic DNA in the samples lysed with SAW and its availability for successful PCR. Although the curve for untreated whole blood (blue) shows available DNA within the samples, as would be expected from dead cells or free circulating DNA, the signal arises much later than for the treated samples, evidenced by the higher threshold cycle (Ct) value for untreated whole blood.
Fig. 3.
Amplification of DNA released from diluted whole blood. Real-time PCR curves showing the intensity of fluorescence as a function of the amplification cycle for untreated blood (blue, dash), chemically lysed blood (black), SAW-lysed blood (red, long dash), and a control (no template, green, dash-dot). The x axis is broken to show more detail at the exponential phase. The cycle number Ct is the abscise of the secondary inflexion point (SI Appendix, Methods). Results show an increase in the amount of DNA in the samples after lysis in similar proportions for chemical or SAW-based lysis.
A quantification of the amount of DNA from the amplification curves indicated that a similar level of DNA was present in the samples lysed with SAW and in chemically lysed samples (Table 1). The differences may be explained by variability in handling, as the samples treated with SAW were recovered manually from the chip, pooled, and transferred to a tube before being processed for PCR, which may have resulted in sample loss.
Table 1.
Fold increase in the DNA copy number of ActB in qPCR reactions conducted on whole blood samples that were lysed using Triton X-100 or SAW
| Sample | Ratio ± SEM |
| Whole blood, untreated | 1.00 ± 0.00 |
| Whole blood, lysed chemically | 11.26 ± 1.40 |
| Whole blood, lysed with SAW | 6.84 ± 1.56 |
SAW PCR.
Here we show that heating induced by SAW propagation within the sample (Fig. 1 D and E) can be implemented as a thermocycler to perform real-time PCR on the same platform as that used for lysis, by switching the frequency, from F1(9.5 MHz) to frequency F2(18 MHz) and cycling the excitation power. We made use of the dissipation of acoustic energy in a liquid droplet that generates heat to increase the sample’s temperature. The cooling between two temperature steps was achieved through passive dissipation into a heat sink, placed below the LiNbO3 transducer (SI Appendix, Fig. S8 shows a schematic of the set up) as the excitation voltage is turned off.
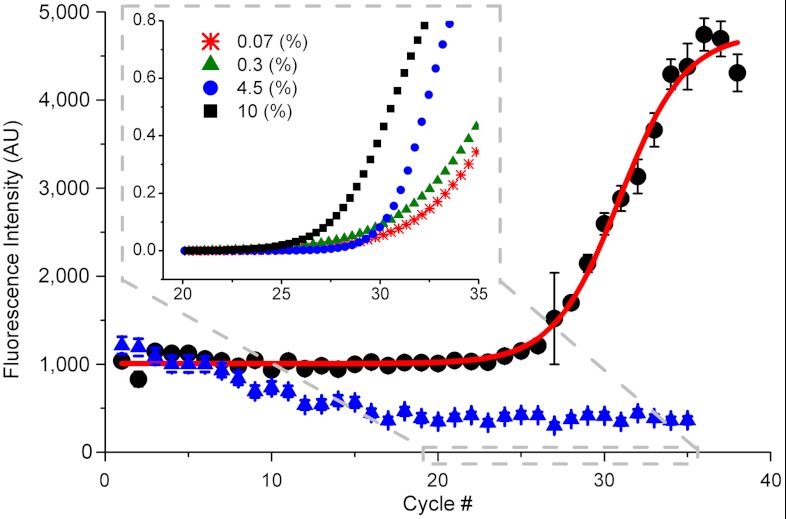
As a model for an infectious disease, we detected rodent-specific malaria parasites (P. berghei) in the blood of infected mice. After SAW lysis, the samples were mixed with a real-time PCR mix and the temperature cycled between 95 °C and 62 °C (SI Appendix, Methods and Fig. S9 for a temperature profile), by applying powers of 1.3 W and 0.3 W, respectively. Fig. 4 shows a typical amplification curve for an infected (4.5% parasitemia, black dots) and a noninfected mouse blood sample (blue triangle). The characteristic sigmoidal increase in fluorescence (red line) is indicative of successful PCR, whereas the noninfected sample only shows a decrease, which could be attributed to photobleaching.
Fig. 4.
SAW real-time PCR. Fluorescence intensity (arbitrary units) for each cycle showing the amplification of infected mouse blood at 4.5% parasitemia (black dots) but not of noninfected mouse blood (0%, blue triangle). The nonlinear fit (red line) is performed with a sigmoid function in Origin 8.0. Error bars are standard deviation over 1 s measurement. Inset shows a close up on the region where the signal overcomes the background for different parasitemia levels, 10% (black square), 4.5% (blue dot), 0.3% (green triangle), and 0.07% (red star). The fluorescence signal was normalized to 0–1 for easier comparison, showing an earlier rise for higher concentrations of parasites.
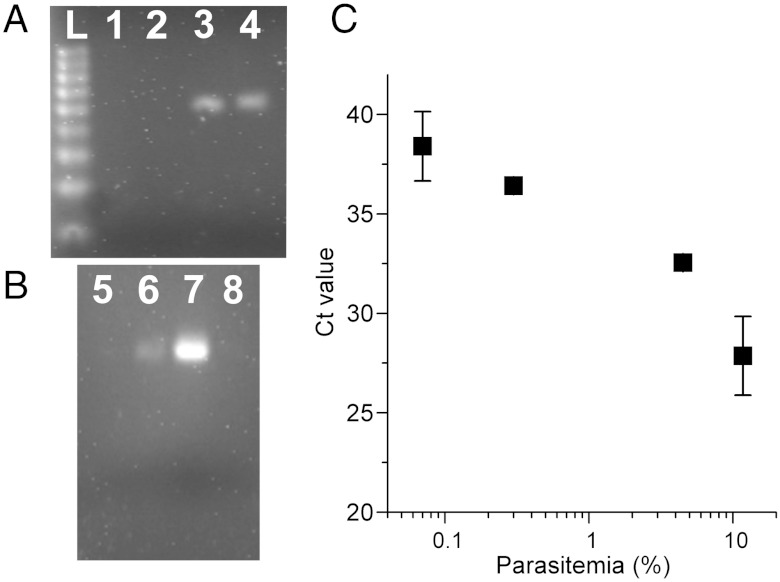
The amplification and specificity were confirmed by gel electrophoresis and were comparable with conventional PCR, carried out in a commercial thermo-cycler (Fig. 5 A and B). These results clearly demonstrate that the SAW lysis process is not only able to disrupt the cell membranes as shown for human blood (Fig. 2), but also gives access to the nucleic acids of the parasites within these cells.
Fig. 5.
Electrophoretic gel (Agarose) analysis of the amplicon from (A) the real-time PCR on SAW and (B) the conventional thermocycler. (L) DNA ladder 100 bp, (1, 2 and 5) noninfected mouse blood, (3, 4 and 6) infected mouse blood lysed with SAW, (7) infected mouse chemically isolated DNA, (8) no template. The gels show specific amplification of the predicted target DNA fragments at 64 bp, in agreement with conventional PCR. (C) The Ct value as a function of parasite concentration.
Fig. 4, Inset also shows that, as expected, as the concentration of parasites within the blood cells increases, so the amplification is detectable after fewer cycles. To evaluate this relationship we performed a threshold analysis (Fig. 5C), revealing a linear relationship over approximately two logs of the parasitemia.
The amplification over 40 cycles was carried within 45 min with a manual control of the temperature, although this time could be further reduced with an automated control to below 30 min. Including the lysis step, the whole assay would be complete under 35 min. This time is similar to that for the gold standard blood smear (60 min) and the rapid test kits (20 min) (29).
We have demonstrated the detection of parasite DNA at a parasitemia of 0.07% (a level equivalent to ca. 30 parasites in the PCR sample). This number is within the same order as current PCR methodologies [0.0001% or 1 parasite/μL of blood (30)] and better than routine smear diagnostic [500 parasite/μL (30)] and rapid tests [> 1,000/μL (29)] and corresponded to a 1 day incubation after mouse infection.
Although we selected a single locus from the P. berghei genome to assess this unique SAW-based PCR method, there is no doubt an optimization of the design of the primer set and PCR conditions for human malaria may improve the detection limit further. Here we have demonstrated an efficient and sensitive device for malaria, where the analysis may help to identify drug resistance, at the point-of-care (31), which is not possible with microscopy. The platform could, in future, also be developed toward the detection of other infectious organisms (trypanosoma) in other samples (such as tuberculosis in sputum), or RNA-based methodologies (viruses using reverse transcription PCR). The platform enables the whole analysis at minor costs and at low power. We estimate that the SAW PCR assay would require ca. 0.75 Wh, whereas a standard cell phone battery is able to produce ca. 4.9 Wh (1,320 mAh, 3.7 V), the equivalent of 6–7 assays. Similarly, the superstrate could be readily machined into lower cost materials, such as glass and, when scaled up, would not increase the overall cost. An integrated SAW-based device could be implemented on portable system to access remote areas in developing countries, thereby providing efficient diagnostics where it is needed.
Conclusions
We have demonstrated a SAW platform which we have implemented on a disposable superstrate patterned with a phononic lattice, to carry out the mechanical reagent-free lysis of microliter-sized droplets of diluted blood, followed by nucleic acid-based amplification and detection of parasite-specific sequences using real-time PCR. Acoustic and radiative pressure, when coupled with shear flows, induced by the interaction of the SAW with the phononic lattice, generated vortices in the liquid, which allowed controlled disruption of cells. Changing the excitation frequency resulted in efficient heating of the sample, as a consequence of dissipation of acoustic energy within the drop. Passive cooling and reheating enabled NAT. This technology not only demonstrates the ability to detect 0.00012% malaria parasitemia in blood on a low cost, low power platform, but also shows the potential that phononics has, when coupled with microfluidics, in providing a frequency-dependent toolbox for LOC in a stationary drop.
Materials and Methods
Sample preparation including phononic crystal design, cell culture, detailed experimental conditions for qPCR, gel electrophoresis, and calculations of shear are described in SI Appendix.
Device Fabrication.
The SAW was propagated on piezoelectric 128° Y-cut X-propagating 3-in LiNbO3 wafers. The devices consisted of 20 pairs of electrodes to form an interdigitated transducer (IDT) with a pitch of 200 μm, width of 100 μm, and an aperture of 10 mm, yielding a frequency of approximately 10 MHz for the propagating SAW. Over the duration of the project, different generations of IDTs were used. Although they had the same basic design, variations in the fabrication process yielded slightly different resonance frequencies between 9.4 and 9.7 MHz. The frequency of 18 MHz was excited as a harmonic on the same design. An example of the S11 parameter is shown in SI Appendix, Fig. S10.
For transmission microscopy, a silicon superstrate could not be used and a slanted IDT was fabricated to provide spatially controlled SAW propagation to enable the rotational momentum in the drop (32). Four-inch double-sided polished wafers were used. The transparent slanted electrode IDT contained 20 pairs of electrodes, with a pitch from 154 μm at the highest frequency (approximately 13 MHz) and 222 μm at the lowest frequency (approximately 9 MHz), with an aperture of 3 cm. The fingers width varied accordingly from 77 to 111 μm.
The phononic crystal superstrate are as described previously (5). Their fabrication is detailed in SI Appendix.
System Set Up.
The IDT was connected to an MXG Analog Signal Generator N5181A (Agilent Technologies) in conjunction with a Mini Circuits ZHL-5W-1, 5–500 MHz amplifier and a 3 A, ± 24 V dc power supply. The lysis was observed under a stereomicroscope (Leica MZ 12, with a ProgRes SpeedXTcore3 camera from Jenoptik) or a fast confocal microscope (Zeiss, LSM 5 Live) as mentioned in the text.
The superstrate was placed on top of the piezoelectric wafer and coupled with 2 μL of water-based gel spread manually (KY Jelly; Johnson and Johnson) in between, yielding a film approximately 50-μm thick.
The temperature was monitored using an infrared camera (FLIR i60; FLIR Systems) and the fluorescent signal was quantified using a photomultiplier tube (PMT, Ion Optix) mounted on a microscope with a 5× objective. SI Appendix, Fig. S8 provides a sketch of the set up.
Measuring Free Haemoglobin after Lysis.
Haemoglobin released from the red blood cells was quantified by measuring direct light absorption at λ = 414 nm and 540 nm (33). Although the standard methodology (American Society for Testing and Materials, 756∶2008) uses λ = 540 nm as the observation wavelength, it necessitates the intermediate step of adding a reagent to improve the signal, which also lyses the cells. To avoid the bias of an additional chemical lysis, we adopted a direct measurement.
A range of blood dilutions was processed on the SAW system. Six samples of 20 μL of each dilution were lysed at the power specified in the text, collected in their entirety (pooled) in an Eppendorf tube, and diluted five times. The extent of lysis was compared to a chemical method. The diluted blood samples were mixed (1∶1 vol/vol) with a solution of 6% (wt/wt) Triton X-100 (Sigma; T-9284) in PBS and agitated for 5 min. Finally a plasma sample was prepared by centrifuging the blood at 1,000 × g for 10 min.
All samples were centrifuged at 1,000 × g for 10 min prior to measurement in the spectrophotometer (Hitachi; U-2000), which was blanked with PBS. The absorbance for the chemically lysed samples is reported after subtracting the value for a solution of 3% Triton X-100.
Cell Counting.
The extent of lysis was also studied by counting the cells remaining intact after the SAW treatment. Different volumes of diluted blood were processed on the SAW system. After resuspension of the contents of the droplet, 10 μL of the solution was harnessed and inserted into a haemocytometer (Neubauer improved). The remaining cells were counted and the extent of lysis reported as a percentage with regards to the cell contents of the original solution. The error is the estimated standard deviation with a 94% confidence.
Other types of cells were also lysed and the lysis efficiency studied in a similar fashion (SI Appendix): HL60 cells at a concentration of 1 million cells/mL in PBS, trypanosomes (Cyclops) at a concentration of 3 million/mL.
PCR Detection of P. berghei using SAW-Lysed Mouse Blood.
Mice (Tyler’s outbred strain) were pretreated with phenylhydrazine (to increase the number of reticulocytes) two days prior to P. berghei infection. Mice were then infected by intraperitoneal injection with 200–500 μL of P. berghei-infected blood stock (stables made and cryopreserved at approximately 3%) at day 0 and subsequently blood samples were taken to determine parasitemia levels. Blood smears were stained with Giemsa staining and 10 fields containing on average 300 cells/field were counted. When desired levels of parasitemia were reached, blood was harvested by cardiac puncture into heparin tubes.
SAW methods were used to lyse diluted whole blood (1∶50 in PBS). For conventional PCR, 1 μL of the lysate was mixed in 20 μL PCR mix, which contained 2.4 μM forward primer, 2.4 μM reverse primer, and 1× Brilliant III Ultra Fast qPCR Master Mix (Agilent Technologies; 600880). This mix was used to amplify a 64 nt fragment derived from P. berghei-specific sequence PBANKA_142330 using primer 1 (aggtggaggtatgggaggaa) and primer 2 (cccatctcttccatctctgaa) over 30 cycles (95 °C for 15 s, 60 °C for 15 s, 72 °C for 15 s).
SAW PCR.
Five microliters of SAW lysate diluted three times in water was mixed with the PCR mix as above, including 0.15 μM fluorescent probe, leading to a 20 μL volume. Three microliters of the prepared mix was dispensed on the phononic superstrate hydrophilic spot and covered with 15 μL of mineral oil (Sigma; M3516). After an initial 5 min denaturing step at 95 °C (approximately 1.3 W), the temperature of the sample was cycled between 95 °C for 10 s and 62 °C (approximately 0.3 W) for 15 s, with a final extension step at 62 °C for 3 min. The fluorescence intensity was measured at the end of each 62 °C step. The temperature was monitored with the IR camera and the power adjusted manually. SI Appendix, Fig. S9 shows a typical temperature profile obtained with SAW. Electrophoresis is performed as described in SI Appendix, Methods in a 4% agarose gel prepared in 0.5× Tris/Borate/EDTA buffer and stained with ethidium bromide.
Mouse Malaria Ct Analysis.
The analysis was performed with the software OriginPro (v8 SR1; OriginLabs). First, the intensity was normalized between [0,1]. Then, using nonlinear curve fitting, the fluorescence signal was fitted to a 4-parameter sigmoid function and the inflexion point selected as the threshold cycle value.
Supplementary Material
ACKNOWLEDGMENTS.
The authors thank Dr. Lisa C. Ranford-Cartwright and Mrs. Elizabeth Peat (Institute of Biomedical and Life Sciences, Division of Infection and Immunity, Glasgow Biomedical Research Center, University of Glasgow) for the access to blood samples; Miss Abeer Syed (Biomedical Engineering, Glasgow University), and Professor Mike Barrett (Institute of Infection, Immunity and Inflammation, University of Glasgow) for the trypanosomes; Miss Mayuree Chanasakulniyom (Biomedical Engineering, Glasgow University) for the actin-GFP cells, Dr. Andrew Glidle (Biomedical Engineering, Glasgow University) for discussions on statistical analysis, and Mrs. Angela McBride (Institute of Infection Immunity and Inflammation, Glasgow University) for supplying malaria infected mouse blood. We acknowledge funding from RAdical SOlutions for Researching the proteome (RASOR), funded by Biotechnology and Biological Sciences Research Council and Engineering and Physical Sciences Research Council (EPSRC), EPSRC Grant EP/F 040857/01, The Bill and Melinda Gates Foundation (Grand Challenges Explorations), Scottish Enterprise (Proof-of-concept), Proxomics (EPSRC Grant EP/I017887/1). The Wellcome Trust (083811/Z/07/Z), and EU FP7 Network of Excellence EviMalar (242095).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206055109/-/DCSupplemental.
References
- 1.Friend J, Yeo LY. Microscale acoustofluidics: Microfluidics driven via acoustics and ultrasonics. Rev Mod Phys. 2011;83:647–704. [Google Scholar]
- 2.Achim W. Acoustically driven planar microfluidics. Superlattices Microstruct. 2003;33:389–396. [Google Scholar]
- 3.Yeo LY, Friend JR. Ultrafast microfluidics using surface acoustic waves. Biomicrofluidics. 2009;3:012002. doi: 10.1063/1.3056040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hodgson RP, Tan M, Yeo L, Friend J. Transmitting high power rf acoustic radiation via fluid couplants into superstrates for microfluidics. Appl Phys Lett. 2009;94:024102. [Google Scholar]
- 5.Wilson R, et al. Phononic crystal structures for acoustically driven microfluidic manipulations. Lab Chip. 2011;11:323–328. doi: 10.1039/c0lc00234h. [DOI] [PubMed] [Google Scholar]
- 6.Bourquin Y, Wilson R, Zhang Y, Reboud J, Cooper JM. Phononic crystals for shaping fluids. Adv Mater. 2011;23:1458–1462. doi: 10.1002/adma.201004455. [DOI] [PubMed] [Google Scholar]
- 7.Reboud J, et al. Nebulisation on a disposable array structured with phononic lattices. Lab Chip. 2012;12:1268–1273. doi: 10.1039/c2lc20705b. [DOI] [PubMed] [Google Scholar]
- 8.Yager P, Domingo GJ, Gerdes J. Point-of-care diagnostics for global health. Annu Rev Biomed Eng. 2008;10:107–144. doi: 10.1146/annurev.bioeng.10.061807.160524. [DOI] [PubMed] [Google Scholar]
- 9.Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 10.World Health Organization. World Malaria Report. WHO. 2010.
- 11.Wilson IG. Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sethu P, Anahtar M, Moldawer LL, Tompkins RG, Toner M. Continuous flow microfluidic device for rapid erythrocyte lysis. Anal Chem. 2004;76:6247–6253. doi: 10.1021/ac049429p. [DOI] [PubMed] [Google Scholar]
- 13.Chen X, Cui DF, Liu CC. Online cell lysis and DNA extraction on a microfluidic biochip fabricated by microelectromechanical system technology. Electrophoresis. 2008;29:1844–1851. doi: 10.1002/elps.200700551. [DOI] [PubMed] [Google Scholar]
- 14.Baek S, Min J, Park J-H. Wireless induction heating in a microfluidic device for cell lysis. Lab Chip. 2010;10:909–917. doi: 10.1039/b921112h. [DOI] [PubMed] [Google Scholar]
- 15.Lee DW, Cho Y-H. A continuous electrical cell lysis device using a low dc voltage for a cell transport and rupture. Sens Actuators B Chem. 2007;124:84–89. [Google Scholar]
- 16.Siegrist J, et al. Validation of a centrifugal microfluidic sample lysis and homogenization platform for nucleic acid extraction with clinical samples. Lab Chip. 2010;10:363–371. doi: 10.1039/b913219h. [DOI] [PubMed] [Google Scholar]
- 17.Carlo DD, Jeong K-H, Lee LP. Reagentless mechanical cell lysis by nanoscale barbs in microchannels for sample preparation. Lab Chip. 2003;3:287–291. doi: 10.1039/b305162e. [DOI] [PubMed] [Google Scholar]
- 18.Taylor MT, et al. Lysing bacterial spores by sonication through a flexible interface in a microfluidic system. Anal Chem. 2000;73:492–496. doi: 10.1021/ac000779v. [DOI] [PubMed] [Google Scholar]
- 19.Cheng J, et al. Preparation and hybridization analysis of DNA/RNA from E. coli on microfabricated bioelectronic chips. Nat Biotechnol. 1998;16:541–546. doi: 10.1038/nbt0698-541. [DOI] [PubMed] [Google Scholar]
- 20.Yager P, et al. Microfluidic diagnostic technologies for global public health. Nature. 2006;442:412–418. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- 21.Franke T, Abate AR, Weitz DA, Wixforth A. Surface acoustic wave (SAW) directed droplet flow in microfluidics for PDMS devices. Lab Chip. 2009;9:2625–2627. doi: 10.1039/b906819h. [DOI] [PubMed] [Google Scholar]
- 22.Franke T, Braunmüller S, Schmid L, Wixforth A, Weitz DA. Surface acoustic wave actuated cell sorting (SAWACS) Lab Chip. 2010;10:789–794. doi: 10.1039/b915522h. [DOI] [PubMed] [Google Scholar]
- 23.Kondoh J, Shimizu N, Matsui Y, Sugimoto M, Shiokawa S. Development of temperature-control system for liquid droplet using surface acoustic wave devices. Sens Actuators A Phys. 2009;149:292–297. [Google Scholar]
- 24.Neuzil P, Pipper J, Hsieh TM. Disposable real-time microPCR device: Lab-on-a-chip at a low cost. Mol Biosyst. 2006;2:292–298. doi: 10.1039/b605957k. [DOI] [PubMed] [Google Scholar]
- 25.Raghavan RV, Friend JR, Yeo LY. Particle concentration via acoustically driven microcentrifugation: MicroPIV flow visualization and numerical modelling studies. Microfluid Nanofluid. 2009;8:73–84. [Google Scholar]
- 26.Chisti Y. Hydrodynamic damage to animal cells. Crit Rev Biotechnol. 2001;21:67–110. doi: 10.1080/20013891081692. [DOI] [PubMed] [Google Scholar]
- 27.Paul R, et al. Shear stress related blood damage in laminar couette flow. Artif Organs. 2003;27:517–529. doi: 10.1046/j.1525-1594.2003.07103.x. [DOI] [PubMed] [Google Scholar]
- 28.Privorotskaya N, et al. Rapid thermal lysis of cells using silicon-diamond microcantilever heaters. Lab Chip. 2010;10:1135–1141. doi: 10.1039/b923791g. [DOI] [PubMed] [Google Scholar]
- 29.Moody A. Rapid diagnostic tests for malaria parasites. Clin Microbiol Rev. 2002;15:66–78. doi: 10.1128/CMR.15.1.66-78.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milne LM, Kyi MS, Chiodini PL, Warhurst DC. Accuracy of routine laboratory diagnosis of malaria in the United Kingdom. J Clin Pathol. 1994;47:740–742. doi: 10.1136/jcp.47.8.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson PE, Alker AP, Meshnick SR. Real-time PCR methods for monitoring antimalarial drug resistance. Trends Parasitol. 2005;21:278–283. doi: 10.1016/j.pt.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 32.Bourquin Y, Reboud J, Wilson R, Cooper JM. Tuneable surface acoustic waves for fluid and particle manipulations on disposable chips. Lab Chip. 2010;10:1898–1901. doi: 10.1039/c004506c. [DOI] [PubMed] [Google Scholar]
- 33.Eschbach E, Scharsack JP, John U, Medlin LK. Improved erythrocyte lysis assay in microtitre plates for sensitive detection and efficient measurement of haemolytic compounds from ichthyotoxic algae. J Appl Toxicol. 2001;21:513–519. doi: 10.1002/jat.797. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.