Abstract
Despite decades of study, electron flow and energy conservation in methanogenic Archaea are still not thoroughly understood. For methanogens without cytochromes, flavin-based electron bifurcation has been proposed as an essential energy-conserving mechanism that couples exergonic and endergonic reactions of methanogenesis. However, an alternative hypothesis posits that the energy-converting hydrogenase Eha provides a chemiosmosis-driven electron input to the endergonic reaction. In vivo evidence for both hypotheses is incomplete. By genetically eliminating all nonessential pathways of H2 metabolism in the model methanogen Methanococcus maripaludis and using formate as an additional electron donor, we isolate electron flow for methanogenesis from flux through Eha. We find that Eha does not function stoichiometrically for methanogenesis, implying that electron bifurcation must operate in vivo. We show that Eha is nevertheless essential, and a substoichiometric requirement for H2 suggests that its role is anaplerotic. Indeed, H2 via Eha stimulates methanogenesis from formate when intermediates are not otherwise replenished. These results fit the model for electron bifurcation, which renders the methanogenic pathway cyclic, and as such requires the replenishment of intermediates. Defining a role for Eha and verifying electron bifurcation provide a complete model of methanogenesis where all necessary electron inputs are accounted for.
Keywords: hydrogenotrophs, H2:F420 oxidoreductase, ferredoxin, formate dehydrogenase
Methanogenesis is an anaerobic respiration carried out by a phylogenetically related group of Archaea within the phylum Euryarchaeota. Methanogens are divided into two metabolic types, those without and those with cytochromes (1). Methanogens without cytochromes use H2 as an electron donor and are termed hydrogenotrophic. Some species can substitute H2 with formate, and a few can use secondary alcohols. CO2 is the electron acceptor and is reduced to methane. Methanogens with cytochromes reduce certain methyl compounds or the methyl carbon of acetate to methane and are called methylotrophic. Many can also use H2 and CO2, as can hydrogenotrophic methanogens.
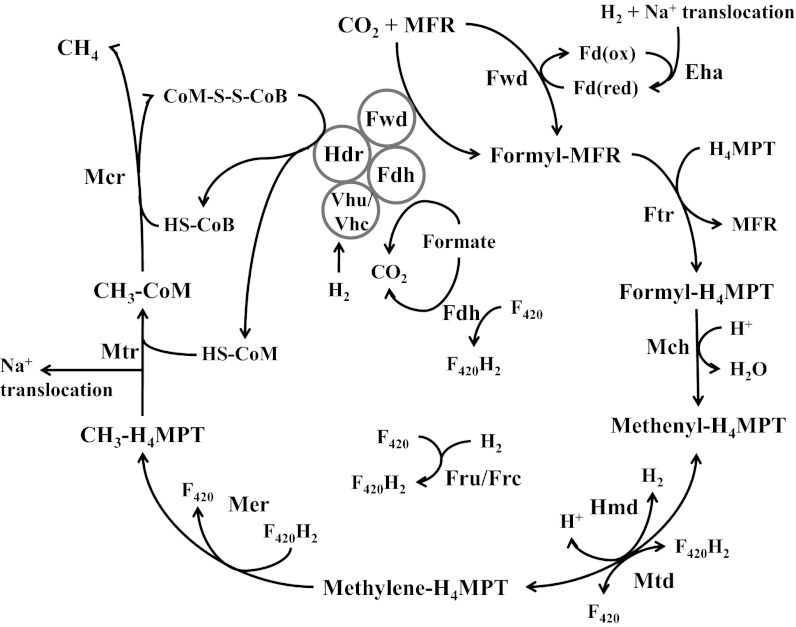
Although the pathways of methanogenesis have long been known, an understanding of energy conservation has been slower to emerge. Methanogens with and without cytochromes both export Na+ when a methyl group is transferred from the carrier tetrahydromethanopterin (H4MPT) to coenzyme M (CoM) (Fig. 1). The Na+ gradient across the membrane is used directly for ATP synthesis or is converted by an antiporter to a proton gradient. However, for methanogenesis from CO2, the initial reduction of CO2 to a formyl group attached to methanofuran (MFR) is endergonic. How energy is provided to drive this reaction is not well understood. Methanogens with and without cytochromes have membrane-associated energy-converting hydrogenases that couple the reduction of low-potential ferredoxins (Fd) to a chemiosmotic membrane gradient (2). If such a Fd donates electrons for CO2 reduction, an energy-converting hydrogenase is the conduit of energy for this reaction. Indeed, for methanogens with cytochromes, an energy-converting hydrogenase is required for CO2 reduction (3). However, the energy requirement for the first step in the pathway results in a need for additional energy conservation. This could be provided by the final step of methanogenesis, which involves an exergonic reduction of a heterodisulfide of two methanogenic cofactors (CoM-S-S-CoB) by heterodisulfide reductase (Hdr). Methanogens with cytochromes harvest the energy yielded in heterodisulfide reduction with a proton-exporting electron transport chain. However, methanogens without cytochromes lack this electron transport chain and an alternative explanation is required.
Fig. 1.
The methanogenic pathway. Eha, energy-converting hydrogenase A; Fdh, formate dehydrogenase; Fru and Frc, F420-reducing hydrogenases; Ftr, formyl-MFR:H4MPT formyltransferase; Fwd, formyl-MFR dehydrogenase; Hdr, heterodisulfide reductase; Hmd, H2-dependent methylene-H4MPT dehydrogenase; Mch, methenyl-H4MPT cyclohydrolase; Mcr, methyl-CoM reductase; Mer, methylene-H4MPT reductase; Mtd, F420-dependent methylene-H4MPT dehydrogenase; Mtr, methyl-H4MPT-CoM methyltransferase; Vhu and Vhc, F420-nonreducing (Hdr-associated) hydrogenases.
Here we present results supporting an emerging view of methanogenesis without cytochromes. The emerging model diverges from the conventional picture of a linear pathway of CO2 reduction to methane. Instead, a cyclical pathway involving electron bifurcation has been proposed (1) (Fig. 1). The reductions of the heterodisulfide and CO2 are coupled in the flavin-containing enzyme complex centered around Hdr. For each pair of electrons accepted, one electron is used for the exergonic reduction of CoM-S-S-CoB, and one is used to reduce a low-potential ferredoxin that in turn donates electrons for the reduction of CO2 to formyl-MFR. Hence, electron bifurcation, a nonchemiosmotic form of energy conservation, couples the exergonic and endergonic steps of methanogenesis and allows for the net availability of chemiosmotic energy for ATP synthesis. The electron bifurcation model renders methanogenesis a cyclic process, in which late steps are coupled by electron flow to the initial step, and explains why in cell extracts, CH4 production from CO2 requires an input of C-1 intermediates (4). Electron bifurcation is supported by experiments with whole cells (5), with purified enzymes (6), and by the characterization of an enzyme complex in which it could take place (7). However, these studies do not explain the presence in most methanogens without cytochromes of the energy-converting hydrogenase Eha that is apparently linked to the first step (2). Electron flux from this hydrogenase would appear to compete with flux from electron bifurcation as well as to consume chemiosmotic energy, leaving a deficit for ATP synthesis.
Whatever the correct model for energy conservation, it likely centers around reactions that reduce low-potential ferredoxins. Three such reactions are proposed to occur in methanogens without cytochromes. Two of these reactions are those mentioned above, the concomitant reduction of Fd and CoM-S-S-CoB that occurs in electron bifurcation, and the H2-dependent reduction of Fd by the energy-converting hydrogenase Eha, both of which are proposed to lead to the endergonic reduction of CO2 to formyl-MFR. A third such reaction, which reduces a Fd with another energy-converting hydrogenase, Ehb, functions in anabolic CO2 fixation reactions and does not appear to be involved in methanogenesis (8, 9).
Here we present an analysis of electron flow in methanogens without cytochromes, focusing on the role of H2 when formate is the electron donor for methanogenesis. We show that there are two pools of electrons that are distinguished by their substrate origins, their carriers, and their functions. One pool of electrons feeds into methanogenesis via coenzyme F420 as well as directly to Hdr from electron-donating growth substrates. Surprisingly, these electrons need not come from H2, even in hydrogenotrophic methanogens, but instead can come directly from formate. Another pool of electrons supports critical biosynthetic or anaplerotic steps, are carried by low-potential ferredoxins, and come only from H2. We show that only one hydrogenase, Eha, is the essential conduit of electrons from H2 and that Eha supports methanogenesis, but it does so in an anaplerotic and not a stoichiometric manner. Eha is needed only to replenish intermediates that are removed from the methanogenesis cycle by diversion to biosynthetic pathways, dilution of intermediates due to growth, or imperfect coupling in electron bifurcation as proposed previously (1). Electron bifurcation still accounts for the stoichiometric flow of electrons for methanogenesis. Our results therefore support the electron bifurcation model in vivo as well as demonstrating the function of Eha.
Results
Identification of an Additional H2:F420 Oxidoreductase Activity and Demonstration of a H2 Requirement for Growth.
Our initial question was whether H2 is a necessary substrate or intermediate for growth of hydrogenotrophic methanogens. Methanococcus maripaludis was an ideal species for addressing this question because it can substitute formate for H2 and mutations are easily generated (10). In the conventional view, during growth on formate, H2 generated from formate serves as the electron donor. Indeed, H2 is generated from formate and recycled in a poorly understood manner (5, 11). However, if there is no direct requirement for H2 in methanogenesis (5, 7), most of the hydrogenases encoded in the genome ought to be dispensable during growth on formate. The Hdr-associated hydrogenases (Vhu and Vhc) (Fig. 1), which provide electrons to the last reductive step of methanogenesis and potentially the first step via electron bifurcation (1, 6, 7), can be substituted by formate dehydrogenase (Fdh) during growth on formate (7). The F420-reducing hydrogenases (Fru and Frc) generate F420H2 for the second and third reductive steps of methanogenesis, but the Fdh is also F420 reducing (12, 13). The hydrogenase Hmd catalyzes the second reductive step directly with H2, but its function is redundant with Mtd, which uses reduced F420 for the same purpose (11). Finally, the anabolic energy-converting hydrogenase Ehb is nonessential in the presence of fixed carbon and is not required for methanogenesis (8, 9). Only Eha remains as possibly essential.
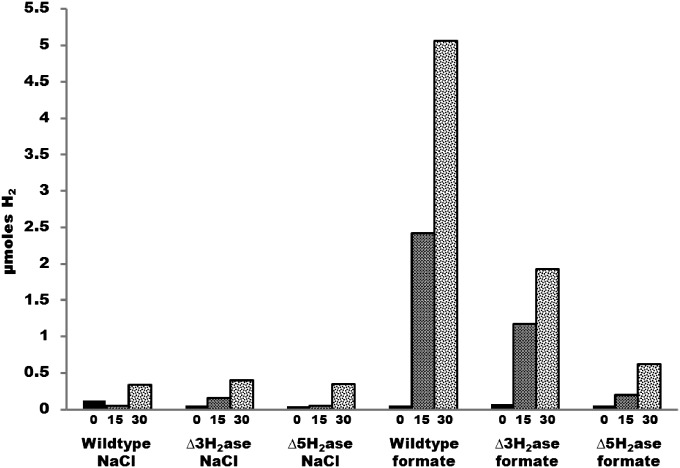
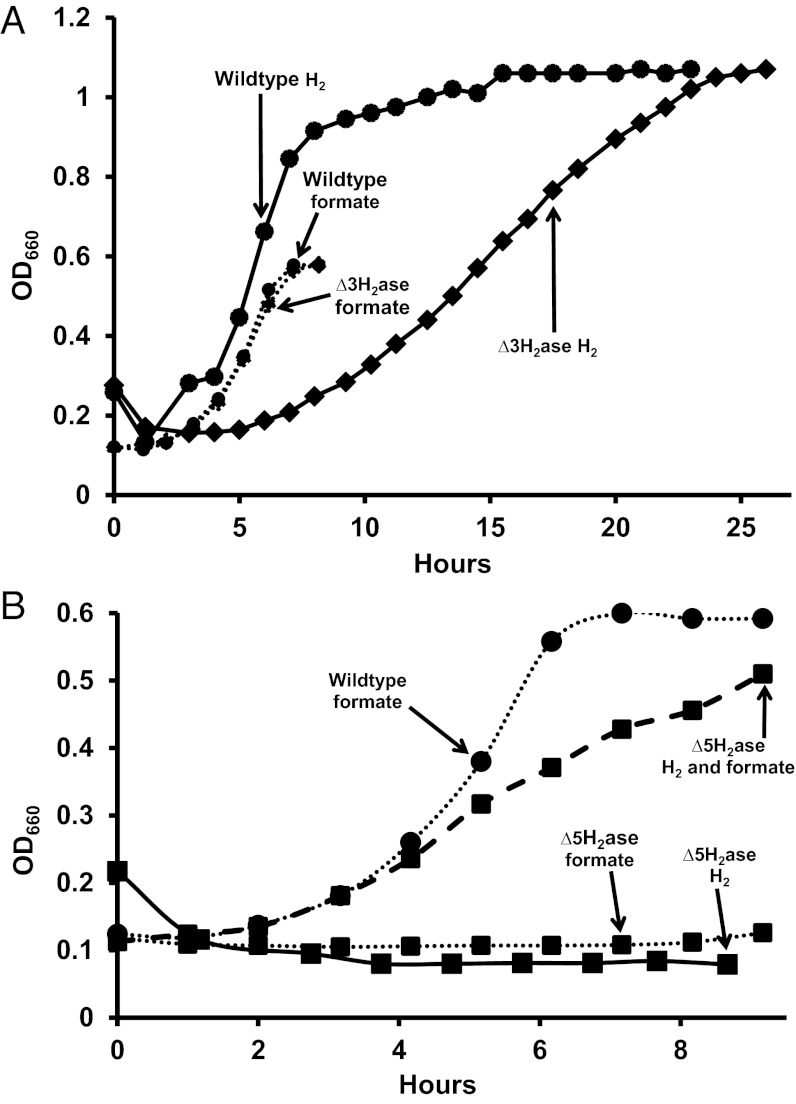
Based on the above considerations, we expected that H2 would not be needed as an intermediate for methanogenesis from formate. Indeed, experiments with cell suspensions have already shown that rates of methanogenesis can substantially exceed rates of H2 production from formate (5). As a further test, our approach here was to genetically remove formate-hydrogen lyase activity, so that H2 would not be produced from formate. If growth still occurred on formate without added H2, then H2 was not a required intermediate. Because Fdh is F420 reducing, removal of formate-hydrogen lyase activity amounts to the removal of F420:H2 oxidoreductase activity. Two such activities are known, the direct Fru or Frc activity and the Hmd-Mtd cycle (Fig. 1) (11). Therefore, deletion of fru, frc, and hmd should eliminate both modes of formate-hydrogen lyase activities. However, cell suspensions of a ∆fru∆frc∆hmd mutant (MM1290, henceforth designated ∆3H2ase) still produced substantial H2 from formate (Fig. 2). Furthermore, the mutant grew not only on formate as predicted, but also on H2, albeit poorly (Fig. 3A). Because F420H2 is essential for methanogenesis, a third pathway must exist for F420 reduction by H2.
Fig. 2.
H2 production by cell suspensions in the absence or presence of formate. Values in x axis are in minutes.
Fig. 3.
Requirements of the ∆3H2ase (A) and ∆5H2ase (B) mutants for H2 and formate for growth. For growth of the ∆5H2ase mutant on H2 and formate, 14.3 μmoles of H2 was added.
In a further attempt to remove F420:H2 oxidoreductase activity, vhu and vhc were deleted in the ∆3H2ase background, resulting in strain MM1289 containing deletions in five hydrogenases (∆fru∆frc∆hmd∆vhu∆vhc, ∆5H2ase). This strain required both formate and H2 for growth (Fig. 3B). This result suggested that F420:H2 oxidoreductase activity had been reduced to below the level needed to support growth on H2 alone. The low level of F420:H2 oxidoreductase activity in ∆5H2ase was verified by low H2 production from formate compared with wild type and ∆3H2ase in cell suspensions (Fig. 2). The third F420:H2 oxidoreductase activity is evidently Vhu/Vhc dependent and represents a previously uncharacterized electron flow pathway in methanogenic Archaea. Although further experiments are needed to characterize this pathway, it could involve Vhu/Vhc, Hdr, and Fdh, which is F420 reducing and like Vhu/Vhc is Hdr associated. The H2 requirement of the ∆5H2ase mutant demonstrates that, contrary to our initial expectation, H2 is a required intermediate during growth on formate and H2 is indeed required for growth of hydrogenotrophic methanogens.
H2 Requirement Is Quantitatively Low.
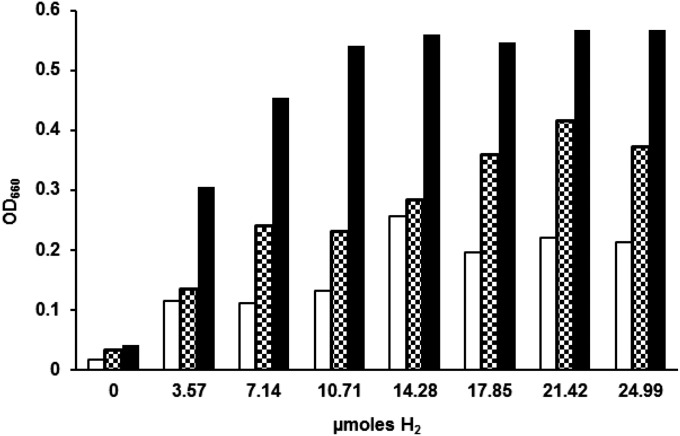
It was unclear whether the H2 that is required for growth of the ∆5H2ase mutant supports the catabolic process of methanogenesis or the anabolic process of CO2 fixation. Cultures of M. maripaludis grown on our formate medium use 1 mmole of formate (11). Therefore, if H2 were required for just one reductive step of methanogenesis, the amount of H2 needed would be ∼0.33 mmoles. However, we observed maximum growth with H2 as low as 10–15 μmoles (Fig. 4). At this level, H2 cannot be a substantial electron donor for methanogenesis. Instead, the H2 requirement may be anabolic and/or anaplerotic (see below). In fact, during autotrophic growth, about 35 μmol of H2 was required for each mg of cell dry weight formed in the ∆5H2ase mutant (Fig. 4) (for M. maripaludis, milligrams of dry weight/OD660/milliliter = 0.34) (5). Under these conditions, cells require 10.7 μmol of pyruvate and 3.9 μmol of acetate per milligram of cell dry weight for autotrophic growth (14). In methanogens, acetate biosynthesis requires two pairs of low potential electrons, one for formation of formyl-MFR and one for the acetyl-CoA synthase step. Pyruvate is formed from acetyl-CoA and requires one additional pair of low potential electrons. Thus, about 40 μmol of low potential electron pairs per milligram of cell dry weight are required for autotrophic growth, close to the value observed. If the H2 was required for generation of low potential electrons for anabolism, the addition of carbon sources to the medium should decrease the amount of H2 needed for growth. In fact, in the presence of acetate and acetate plus casamino acids, the amount of H2 required decreased to 17 and 8 μmol of H2 per milligram of cell dry weight, respectively (Fig. 4). Therefore, the H2 requirement appears at least partially anabolic.
Fig. 4.
H2 dose–response of ∆5H2ase mutant. Clear bars, mineral medium; checkered bars, 10 mM acetate added; solid bars, 10 mM acetate and casamino acids (0.2% wt/vol) added. All cultures contained 200 mM formate. Five-milliliter cultures were incubated until stationary phase and OD660 was measured.
H2 Is Not Required for Methanogenesis in Vitro but Stimulates Methanogenesis in Cell Suspensions.
We performed two additional experiments to further examine the nature of the H2 requirement. For both experiments, first Ehb was genetically eliminated from the ∆5H2ase mutant background to generate strain MM1284, which contained deletions in six hydrogenases (∆fruA∆frcA∆hmd∆vhuAU∆vhcA∆ehbN, ∆6H2ase). Eha was the sole remaining hydrogenase. Similar to the ∆5H2ase mutant, ∆6H2ase required H2 as well as formate for growth (Fig. S1). In the first experiment, in vitro CH4 production assays were performed (Fig. 5). These assays followed published reports (4, 15), which show that CH4 production from CO2 in vitro requires stimulation by the intermediate CH3-S-CoM and that the yield of CH4 is limited by the CH3-S-CoM added. In our assays, CH3-S-CoM was added and CH4 production continued presumably until CH3-S-CoM was depleted. CH4 production was measured in extracts of ∆6H2ase mutant or wild-type cells with either H2 or formate as electron donor. Extract of wild-type M. maripaludis produced substantial CH4 from CO2 with either electron donor. In contrast, the mutant extract produced substantial CH4 only from formate. H2 had no stimulatory effect on CH4 production from formate (Fig. S2).
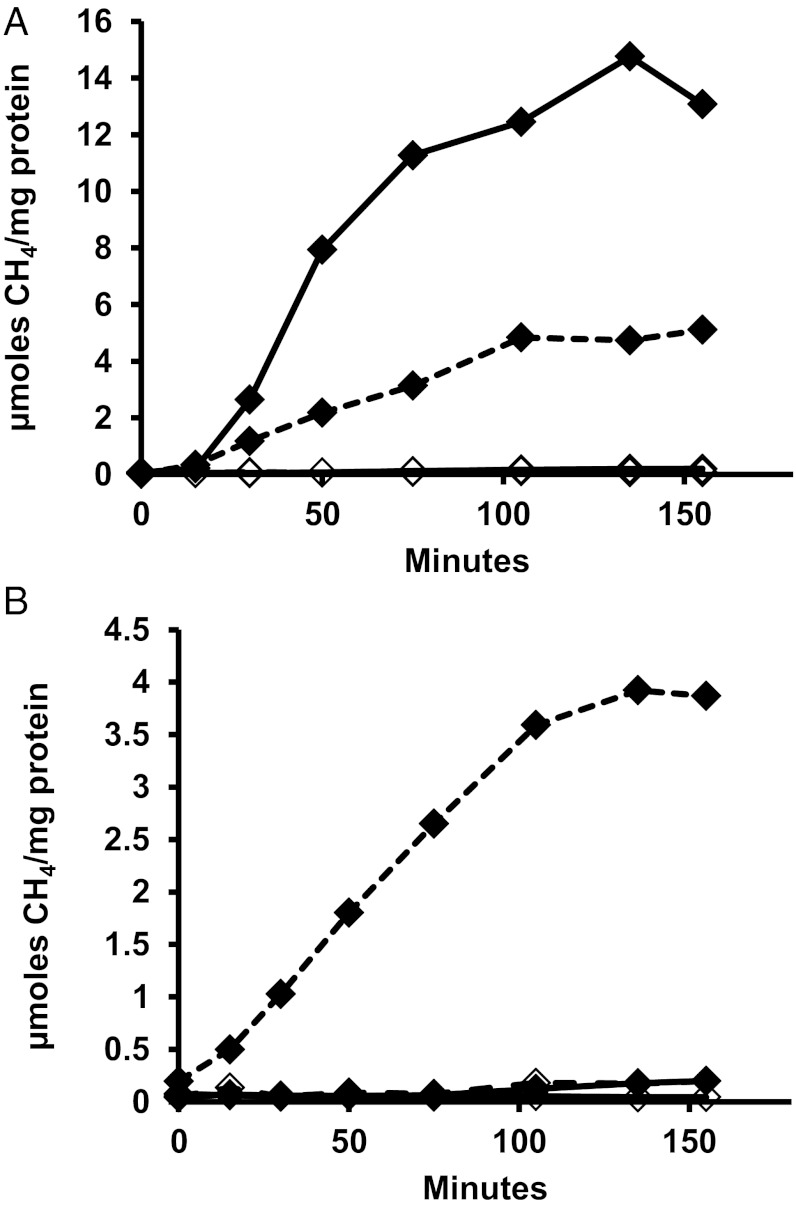
Fig. 5.
Methanogenesis in cell extracts from CH3-S-CoM and CO2 using H2 (solid line) or formate (dashed line) as the electron donor in cell extracts from (A) wild-type or (B) ∆6H2ase mutant. Each reaction contained 300 nmols of CH3-S-CoM and 200–350 μg of protein. Extracts with no CH3-S-CoM added are represented by open diamonds.
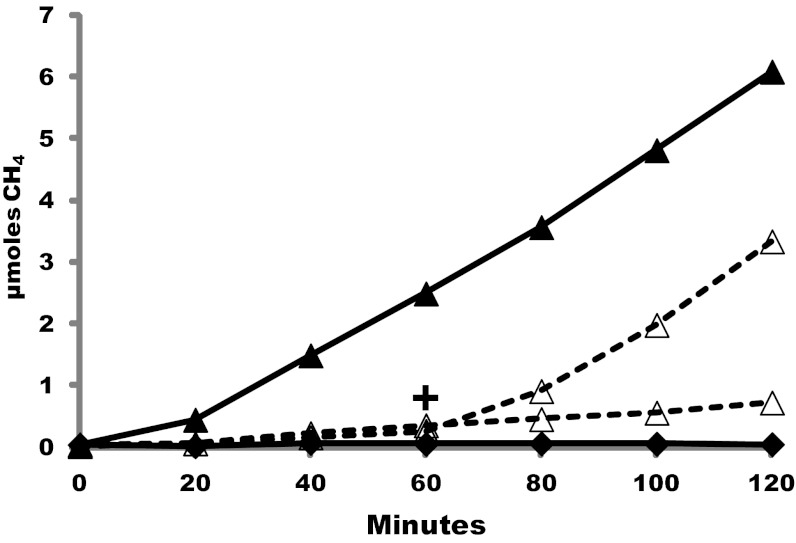
Next, we assayed CH4 production by cell suspensions of the ∆6H2ase mutant. With cell suspensions, stimulation by an intermediate in the pathway was not needed and CH3-S-CoM was not added. Methanogenesis occurred with formate but not with H2. Significantly, methanogenesis was greatly enhanced by H2, either present initially or added during the course of the assay (Fig. 6). Hence, H2 did not contribute to methanogenesis in vitro where the pathway intermediate CH3-S-CoM was added, but H2, presumably acting through Eha, stimulated methanogenesis in cell suspensions.
Fig. 6.
CH4 production by cell suspensions of the ∆6H2ase mutant. Black diamonds, H2 alone; open triangles, formate alone with (+) or without H2 addition [8% (vol/vol) final concentration] at 60 min; black triangles, H2 and formate.
Eha Is Essential for Growth of M. maripaludis.
To test whether Eha is essential, mutagenesis of ehaHIJ was attempted. The genes ehaH, I, and J encode the presumed cation translocator of the enzyme complex. Each is homologous to a portion of ehbF, for which a null allele has a strong phenotype (9). In preliminary experiments, the construction of an ehaHIJ allelic replacement with a puromycin resistance cassette was unsuccessful. An additional test of essentiality was sought. As before (11), our strategy was to determine whether negative selection to resolve a merodiploid would result in a deletion allele. All other things equal, if there is no growth disadvantage for a null allele, deletion mutants should arise with roughly the same frequency as wild-type alleles, and this occurred in a control experiment where a ehbF+-ΔehbF merodiploid was resolved (Table 1). However, resolution of the ehaHIJ+-ΔehaHIJ merodiploid in standard medium with H2 resulted in only wild-type clones. Similar results were obtained when formate rather than H2 was used. Eha could be involved in 2-ketoglutarate biosynthesis, because 2-ketoglutarate oxidoreductase depends on Fd (9), and high levels of glutamate in the medium (10 mM) could provide sufficient 2-ketoglutarate and remove the requirement for Eha. Alternatively, Eha might be involved in NAD+ reduction, and alanine dehydrogenase in methanococci generates NADH (16). However, when glutamate or alanine was added, still no mutations were obtained. In contrast, when the ehaHIJ mutagenesis experiment was performed in the presence of trans-complementation (Pnif-ehaHIJ), the majority of clones contained the deletion. These results strongly suggest that Eha is essential, consistent with the H2 requirement for growth of the ∆6H2ase mutant.
Table 1.
Resolution of merodiploids of ehb or eha
| No. of clones |
||
| Merodiploid | WT | Mutant |
| ΔehbF | 26 | 34 |
| ΔehaHIJ | 108 | 0 |
| ΔehaHIJ, formate | 24 | 0 |
| ΔehaHIJ, glu | 12 | 0 |
| ΔehaHIJ, ala | 38 | 0 |
| ΔehaHIJ + Pnif-ehaHIJ | 5 | 52 |
Number of clones with the wild-type (WT) or mutant allele following resolution of the merodiploids is given. Resolution was performed in complex medium with the indicated additions or trans-complementation by the plasmid expressing ehaHIJ from the Pnif promoter.
Discussion
Distinct Electron Pools Function in Hydrogenotrophic Methanogens.
Until recently, it was not known whether any of the hydrogenase activities in hydrogenotrophic methanogens could be eliminated. However, in past work we reported that some of these hydrogenases were unnecessary under some conditions. Thus, in separate strains we deleted genes encoding the F420-reducing hydrogenases (11), the Hdr-associated hydrogenases (7), and the hydrogen-using methylene-H4MPT dehydrogenase (11). Here we eliminated all three of these hydrogenase activities in a single strain (∆5H2ase) and found that both formate and H2 were required for growth, the former in quantities stoichiometrically sufficient for methanogenesis, and the latter in much smaller quantities. The mutant effectively separates two pools of electrons that ordinarily exchange via H2. One pool of electrons provides a stoichiometric supply of electrons for methanogenesis and flows through F420 and Hdr (Fig. 1). In the wild-type strain, either formate or H2 functions as electron donor for this pool. The ∆5H2ase and ∆6H2ase mutants, by eliminating H2:F420 oxidoreductase activities, disrupt electron flow from H2 and, as a result, formate is required as the stoichiometric electron donor for methanogenesis. The other pool of electrons supports biosynthesis, and as demonstrated here, anaplerotically replenishes methanogenesis (see below). This pool is carried by ferredoxins, and only H2 functions as electron donor. In the wild-type strain, H2 produced from formate allows the latter to function as sole electron donor. However, in the ∆5H2ase and ∆6H2ase mutants where electron flow between the two pools is blocked, H2 must be provided.
Function of Eha Is Essential, Anaplerotic, and Ancillary to Electron Bifurcation.
In the ∆5H2ase mutant, two hydrogenases remain, Eha and Ehb. Previous work has suggested that the role of Ehb is the reduction of Fd for anabolic CO2 fixation via acetyl-CoA synthase, pyruvate oxidoreductase, 2-ketoglutarate oxidoreductase, indole-pyruvate oxidoreductase, and 2-oxoisovalerate oxidoreductase (8, 9). Eha may play a role analogous to a related energy converting hydrogenase, Ech, which in methanogens with cytochromes generates reduced Fd for CO2 reduction to formyl-MFR (2). Because Eha and Ehb have different functions, they may reduce different ferredoxins. Although not yet proven biochemically, Eha could have specificity for a polyferredoxin associated with formylmethanofuran dehydrogenase (Fwd), the enzyme that catalyzes CO2 reduction to formyl-MFR (17), whereas Ehb could reduce a Fd associated with anabolic CO2 fixation reactions.
In methanogens without cytochromes, if the role of Eha is to reduce CO2 to formyl-MFR, how is this reconciled with electron bifurcation as the main pathway for delivery of electrons for the same step? Our data show that Eha needs to provide only a small portion of the electrons for this reaction. In the ∆5H2ase mutant, formate provided nearly all electrons for methanogenesis and the H2 requirement accounted for only up to 4% of the electrons for growth, much of this apparently for anabolic purposes. We propose that Eha functions in the reduction of CO2 to formyl-MFR, but does so anaplerotically. In the electron bifurcation model, hydrogenotrophic methanogenesis is a cyclic pathway where the first step is dependent on the last step (1). However, this model presents a dilemma. A constant pool of CoM-S-S-CoB is required, yet intermediates in methanogenesis will inevitably be diluted by growth and cell division, or lost due to a leaky electron bifurcating Hdr complex (1, 6). In addition, intermediates will diminish when methyl-H4MPT is diverted from methanogenesis to generate acetyl-CoA for autotrophic CO2 fixation (18). Our results show that Eha solves this dilemma by priming or recharging the cycle: it anaplerotically restores intermediates to the methanogenic pathway at the level of formyl-MFR. This model accounts for all of the following observations. First, only small amounts of H2 are required for growth. Second, Eha is essential even though electron bifurcation can account for a stoichiometric supply of electrons for methanogenesis. Third, even though H2 and Eha are essential, there is no need for H2 for methanogenesis in an in vitro assay where the intermediate CH3-S-CoM is added. Finally, for methanogenesis in cell suspensions where no intermediate is provided, H2 is stimulatory.
Why do most hydrogenotrophic methanogens, including M. maripaludis, maintain two ion-translocating energy-converting hydrogenases that reduce Fd and provide electrons to fix CO2? One such hydrogenase could suffice, and indeed, Eha can apparently recognize the Ehb-type ferredoxin and substitute for Ehb, albeit inefficiently (8, 9). Having both hydrogenases separates the recharging of methanogenesis from other anabolic activities and may optimize control over the separate processes. When conditions limit growth, anabolic CO2 fixation is unimportant but CO2 reduction to formyl-MFR for methanogenesis and ATP synthesis are still necessary for survival. Under these conditions, functional Eha is essential, and a functional Ehb could be detrimental.
Electron Flow and Energy Conservation in Hydrogenotrophic Methanogens.
Electron bifurcation at Hdr explains a decades-old dilemma regarding methanogenesis: How is net energy conservation achieved in hydrogenotrophic methanogens (1)? The results presented here verify that electron bifurcation must function in vivo and elaborate on the mechanisms that allow this to be the case, filling in the known gaps of a pathway that has been incomplete since methanogens were first discovered and grown in culture (19). A complete model for electron flow in methanogens without cytochromes can now be described: Methanogenesis is dependent upon F420-reducing enzymes and enzymes that feed electrons to Hdr for electron bifurcation. During growth on H2, these enzymes are the F420-reducing hydrogenases Fru and Frc and the Hdr-associated hydrogenases Vhu and Vhc. During growth on formate, both kinds of hydrogenases are unnecessary and Fdh performs both functions. Biochemical experiments have demonstrated CoM-S-S-CoB-dependent reduction of a clostridial Fd with H2 (6), but are still needed to prove that electrons flow from H2 or formate to Fwd concurrent with flow to Hdr. Nevertheless, in previous work, we showed that in M. maripaludis, Fdh as well as Vhu exist in a complex with Hdr, and that Fwd is in this complex as well (7). Hence, an enzyme complex exists that is suited for electron bifurcation with either H2 or formate. Fdh, either complexed or existing in an isolated form, also generates F420H2. Through electron bifurcation at Hdr and F420 reduction, formate provides the reducing equivalents to all four reductive steps of methanogenesis. A separate electron pool supports anabolism, which depends on electrons from H2 entering through Ehb and its associated Fd (8, 9). These isolated inputs keep the electron pools for catabolism and anabolism separated and under different regulatory control. Overlap between the two electron pools occurs when electrons from H2 enter methanogenesis through Eha. When low CoM-S-S-CoB concentrations limit the reduction of CO2 to formyl-MFR by electron bifurcation, Eha recharges methanogenesis.
Materials and Methods
Construction and Growth of Strains.
Unless otherwise stated, strains were grown in medium containing casamino acids and acetate, with H2 or formate as the electron donor (11). Strains and plasmids are shown in Table S1. Strain Mm901 was used as the wild-type strain unless otherwise stated. To construct plasmids pCRupt∆fruneo, pCRupt∆frcneo, and pCRupt∆hmdneo, insert DNA from plasmids pCRprt∆fruneo, pCRprt∆frcneo, and pCRprt∆hmdneo (11) was recloned into pCRuptneo (7). To construct plasmids pCRupt∆fruGBneo, pCRupt∆frcGBneo, and pCRupt∆ehbNneo, ∼0.5 kb of DNA upstream and downstream of the designated loci was obtained by PCR from genomic DNA and cloned into pCRuptneo. Primers are shown in Table S2. In each case, the resulting plasmid contained an in-frame deletion consisting of a start codon and a stop codon with intervening codons contained within an AscI site. Plasmid DNA was used to make deletions using a markerless mutagenesis method with neomycin for positive selection and 6-azauracil for negative selection as described (7, 20). When formate was present as electron donor, neomycin was increased to 5 mg/mL on liquid or solid medium (7). MM1290 (Mm901ΔfruAGBΔfrcAGBΔhmd, Δ3H2ase) was constructed by sequential deletion of frcA, hmd, fruA, fruG and B, and frcG and B. The first deletion was constructed using H2 as the growth substrate, and the remaining deletions used formate. MM1313 (MM901∆fruA∆frcA∆vhuAU∆vhcA, Δ4H2ase) was constructed from Mm1272 (MM901∆vhuAU∆vhcA) (7) by deletion of fruA and frcA using formate. MM1289 (MM901∆fruA∆frcA∆hmd∆vhuAU∆vhcA, Δ5H2ase) was then constructed by deleting hmd. Finally, MM1284 (MM901∆fruA∆frcA∆hmd∆vhuAU∆vhcA∆ehbN, Δ6H2ase) was constructed by deleting ehbN. During construction of the Δ5H2ase and Δ6H2ase mutants, cultures were grown with formate and H2. All deletions were confirmed by PCR.
Essentiality of Eha.
Merodiploids were constructed containing deletions of the ehaHIJ genes and ehbF (Fig. S3 and Table S1), and the generation of deletion mutants by recombination-based resolution of the merodiploids was attempted using wild-type strain Mm900 as described (20). Among the resulting clones, those containing deletion mutations and wild-type alleles were distinguished by PCR. A trans-complementing plasmid (pMEV1nif::ehaHIJ) (Table S1) was constructed on a replicative vector with ehaHIJ under control of the nitrogen-regulated nif promoter (21, 22). To test the effect of trans-complementation, merodiploids containing this plasmid were plated with alanine or ammonia, and the merodiploids were resolved. In preliminary experiments, ehaHIJ deletion strains could be obtained with either nitrogen source, and ammonia was used henceforth.
H2 and CH4 Production.
To measure H2 production by cell suspensions, cultures (5 mL) were grown on formate to OD660 between 0.5 and 0.6, and cells were pelleted, washed, and resuspended anaerobically in the same volume of assay buffer (modified from ref. 5, 50 mM Mops pH 7.0, 400 mM NaCl, 20 mM KCl, 20 mM MgCl2, 1 mM CaCl2, 5 mM DTT, and 1 mM bromoethanesulfonate). Tubes were then flushed with N2 for 10–30 min to remove residual H2. At time = 0 min, an initial gas sample was taken, then the assay was initiated by addition of 40 mM (final concentration) sodium formate (pH 7) or NaCl and incubated at 37 °C with shaking. At 15-min and 30-min time points, the headspace was sampled and transferred to butyl rubber stoppered 5-mL vials, mouth ID 13 mm × OD 20 mm (Wheaton; catalog no. 223685) preflushed with N2. H2 was analyzed with a SRI Instruments gas chromatography (GC) model 8610C equipped with a 6 foot × one-eighth inch stainless steel Molecular Sieve 5A packed column and a reduced gas detector. The carrier gas was He (20 psi), oven temperature was 130 °C, and detector temperature was 290 °C. CH4 production by cell suspensions was measured as above except cells were grown to mid-log phase (OD660 ∼0.25), bromoethanesulfonate was omitted from the assay buffer, and the headspace was directly analyzed on a Buck Scientific model 910 GC equipped with a flame ionization detector provided with air (16 psi) and H2 (26 psi). The carrier gas was He (24 psi). To measure CH4 production by cell-free extracts, cells were washed and suspended in buffer (modified from ref. 4) 100 mM Trizma base, 15 mM MgCl2, 5 mM ATP, 2 mM 2-mercaptoethanol, 500 μM FAD+, with or without 50 mM formic acid, and pH adjusted to 7.1 with 1 M HCl). The suspension was sonicated with a Misonix XL-2000 series sonicator to disrupt the cells. Debris was removed by centrifugation at 16,000 × g, and 200 μL supernatant was placed in a 5-mL serum vial and preincubated for 10 min at room temperature. To start the assay, the headspace was flushed with N2:CO2 (80:20, for formate) or H2:CO2 (80:20), and 1.5 mM CH3-S-CoM was added (4). Methanogenesis was monitored as described above.
Supplementary Material
Acknowledgments
We thank Dave Stahl for use of the gas chromatography in hydrogen measurements and Birte Meyer for assistance. This work was supported by Grant DE-FG02-05ER15709 from the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, US Department of Energy. A portion of the work on the essentiality of Eha was supported by Grant R24 GM074783 from the National Institute of General Medical Sciences. K.C.C. was supported in part by Public Health Service, National Research Service Award T32 GM07270, from the National Institute of General Medical Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 15084.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208779109/-/DCSupplemental.
References
- 1.Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. Methanogenic archaea: Ecologically relevant differences in energy conservation. Nat Rev Microbiol. 2008;6:579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- 2.Hedderich R. Energy-converting [NiFe] hydrogenases from archaea and extremophiles: Ancestors of complex I. J Bioenerg Biomembr. 2004;36:65–75. doi: 10.1023/b:jobb.0000019599.43969.33. [DOI] [PubMed] [Google Scholar]
- 3.Meuer J, Kuettner HC, Zhang JK, Hedderich R, Metcalf WW. Genetic analysis of the archaeon Methanosarcina barkeri Fusaro reveals a central role for Ech hydrogenase and ferredoxin in methanogenesis and carbon fixation. Proc Natl Acad Sci USA. 2002;99:5632–5637. doi: 10.1073/pnas.072615499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gunsalus RP, Wolfe RS. Stimulation of CO2 reduction to methane by methylcoenzyme M in extracts Methanobacterium. Biochem Biophys Res Commun. 1977;76:790–795. doi: 10.1016/0006-291x(77)91570-4. [DOI] [PubMed] [Google Scholar]
- 5.Lupa B, Hendrickson EL, Leigh JA, Whitman WB. Formate-dependent H2 production by the mesophilic methanogen Methanococcus maripaludis. Appl Environ Microbiol. 2008;74:6584–6590. doi: 10.1128/AEM.01455-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaster AK, Moll J, Parey K, Thauer RK. Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea. Proc Natl Acad Sci USA. 2011;108:2981–2986. doi: 10.1073/pnas.1016761108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costa KC, et al. Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase. Proc Natl Acad Sci USA. 2010;107:11050–11055. doi: 10.1073/pnas.1003653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Major TA, Liu Y, Whitman WB. Characterization of energy-conserving hydrogenase B in Methanococcus maripaludis. J Bacteriol. 2010;192:4022–4030. doi: 10.1128/JB.01446-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Porat I, et al. Disruption of the operon encoding Ehb hydrogenase limits anabolic CO2 assimilation in the archaeon Methanococcus maripaludis. J Bacteriol. 2006;188:1373–1380. doi: 10.1128/JB.188.4.1373-1380.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leigh JA, Albers SV, Atomi H, Allers T. Model organisms for genetics in the domain Archaea: Methanogens, halophiles, Thermococcales and Sulfolobales. FEMS Microbiol Rev. 2011;35:577–608. doi: 10.1111/j.1574-6976.2011.00265.x. [DOI] [PubMed] [Google Scholar]
- 11.Hendrickson EL, Leigh JA. Roles of coenzyme F420-reducing hydrogenases and hydrogen- and F420-dependent methylenetetrahydromethanopterin dehydrogenases in reduction of F420 and production of hydrogen during methanogenesis. J Bacteriol. 2008;190:4818–4821. doi: 10.1128/JB.00255-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Widdel F, Wolfe R. Expression of secondary alcohol dehydrogenase in methanogenic bacteria and purifcation of the F420-specific enzyme from Methanogenium thermophilum strain TC1. Arch Microbiol. 1989;152:322–328. [Google Scholar]
- 13.Schauer NL, Ferry JG, Honek JF, Orme-Johnson WH, Walsh C. Mechanistic studies of the coenzyme F420 reducing formate dehydrogenase from Methanobacterium formicicum. Biochemistry. 1986;25:7163–7168. doi: 10.1021/bi00370a059. [DOI] [PubMed] [Google Scholar]
- 14.Yang YL, Glushka JN, Whitman WB. Intracellular pyruvate flux in the methane-producing archaeon Methanococcus maripaludis. Arch Microbiol. 2002;178:493–498. doi: 10.1007/s00203-002-0480-9. [DOI] [PubMed] [Google Scholar]
- 15.Bobik TA, Wolfe RS. Physiological importance of the heterodisulfide of coenzyme M and 7-mercaptoheptanoylthreonine phosphate in the reduction of carbon dioxide to methane in Methanobacterium. Proc Natl Acad Sci USA. 1988;85:60–63. doi: 10.1073/pnas.85.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu JP, Ladapo J, Whitman WB. Pathway of glycogen metabolism in Methanococcus maripaludis. J Bacteriol. 1994;176:325–332. doi: 10.1128/jb.176.2.325-332.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hochheimer A, Schmitz RA, Thauer RK, Hedderich R. The tungsten formylmethanofuran dehydrogenase from Methanobacterium thermoautotrophicum contains sequence motifs characteristic for enzymes containing molybdopterin dinucleotide. Eur J Biochem. 1995;234:910–920. doi: 10.1111/j.1432-1033.1995.910_a.x. [DOI] [PubMed] [Google Scholar]
- 18.Berg IA, et al. Autotrophic carbon fixation in archaea. Nat Rev Microbiol. 2010;8:447–460. doi: 10.1038/nrmicro2365. [DOI] [PubMed] [Google Scholar]
- 19.Barker H. Studies upon the methane fermentation. IV. The isolation and culture of Methanobacterium omelianskii. Antonie van Leeuwenhoek J. Microbiol. Serol. 1940;6:201–220. [Google Scholar]
- 20.Moore BC, Leigh JA. Markerless mutagenesis in Methanococcus maripaludis demonstrates roles for alanine dehydrogenase, alanine racemase, and alanine permease. J Bacteriol. 2005;187:972–979. doi: 10.1128/JB.187.3.972-979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dodsworth JA, Cady NC, Leigh JA. 2-Oxoglutarate and the PII homologues NifI1 and NifI2 regulate nitrogenase activity in cell extracts of Methanococcus maripaludis. Mol Microbiol. 2005;56:1527–1538. doi: 10.1111/j.1365-2958.2005.04621.x. [DOI] [PubMed] [Google Scholar]
- 22.Lie TJ, Wood GE, Leigh JA. Regulation of nif expression in Methanococcus maripaludis: roles of the euryarchaeal repressor NrpR, 2-oxoglutarate, and two operators. J Biol Chem. 2005;280:5236–5241. doi: 10.1074/jbc.M411778200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.