Abstract
To investigate the role of histone H3K27 demethylase UTX in embryonic stem (ES) cell differentiation, we have generated UTX knockout (KO) and enzyme-dead knock-in male ES cells. Deletion of the X-chromosome-encoded UTX gene in male ES cells markedly decreases expression of the paralogous UTY gene encoded by Y chromosome, but has no effect on global H3K27me3 level, Hox gene expression, or ES cell self-renewal. However, UTX KO cells show severe defects in mesoderm differentiation and induction of Brachyury, a transcription factor essential for mesoderm development. Surprisingly, UTX regulates mesoderm differentiation and Brachyury expression independent of its enzymatic activity. UTY, which lacks detectable demethylase activity, compensates for the loss of UTX in regulating Brachyury expression. UTX and UTY bind directly to Brachyury promoter and are required for Wnt/β-catenin signaling-induced Brachyury expression in ES cells. Interestingly, male UTX KO embryos express normal levels of UTY and survive until birth. In contrast, female UTX KO mice, which lack the UTY gene, show embryonic lethality before embryonic day 11.5. Female UTX KO embryos show severe defects in both Brachyury expression and embryonic development of mesoderm-derived posterior notochord, cardiac, and hematopoietic tissues. These results indicate that UTX controls mesoderm differentiation and Brachyury expression independent of H3K27 demethylase activity, and suggest that UTX and UTY are functionally redundant in ES cell differentiation and early embryonic development.
In vitro, embryonic stem (ES) cells are capable of differentiating into three germ layers, ectoderm, endoderm, and mesoderm, which mimics the early stage of embryonic development in vivo (1). Transcription factor Brachyury (T) is highly expressed in the primitive streak during gastrulation and is required for mesoderm formation (2). Mutation of Brachyury gene in mice causes defective formation of posterior mesoderm, failure of notochord morphogenesis, and embryonic death around 10 d of gestation (2). Brachyury expression is directly regulated by Wnt/β-catenin signaling in mesoderm and in ES cells. Wnt/β-catenin signaling promotes de-phosphorylation of β-catenin, which enters nucleus and binds transcription factor LEF1/TCF1 to activate Brachyury expression (3, 4). Brachyury also positively regulates Wnt/β-catenin signaling. Such a positive auto-regulatory loop between Brachyury and Wnt/β-catenin signaling maintains the mesodermal progenitor cells and is essential for posterior mesoderm development in vertebrates (5).
The Polycomb Repressive Complex 2 (PRC2) is critical for the proper differentiation of ES cells. PRC2 is localized on a large number of developmental regulator genes in ES cells. Disruption of PRC2 in ES cells markedly decreases the global levels of H3K27 di- and trimethylation (H3K27me2 and H3K27me3) and leads to up-regulation of many developmental regulator genes (6, 7).
UTX belongs to a subfamily of JmjC domain-containing proteins that also includes UTY and JMJD3. Previously, we and others identified UTX and Jmjd3 as histone H3K27 demethylases (8–12). UTX gene is located on the X chromosome, whereas UTY gene is on the Y chromosome and thus only expressed in male cells. UTY is a paralog of the X-linked UTX and shares 88% sequence homology with UTX protein. Unlike UTX, UTY lacks detectable histone demethlase activity in vitro (8, 12). The viability data from male and female UTX KO mice indicate a largely functional redundancy between UTX and UTY during male embryonic development (13).
UTX has been shown to regulate myocyte differentiation, heart development, and T-box transcription factor target gene expression (13–15). However, UTX functions in ES cell differentiation and early embryonic development are unclear. Using UTX KO, conditional KO, and enzyme-dead knock-in ES cells, here we show that UTX, but not its H3K27 demethylase activity, is required for ES cell differentiation into mesoderm. Mechanistically, UTX and UTY are redundant and they directly control the induction of Brachyury, a master regulator of mesoderm differentiation. Consistently, female UTX KO (UTX−/−) embryos, but not male UTX KO (UTX−/Y) ones that express UTY normally, show severe defects in Brachyury expression and mesoderm development.
Results
Generation of UTX KO, Conditional KO, and Enzyme-Dead Knock-In Male ES Cell Lines.
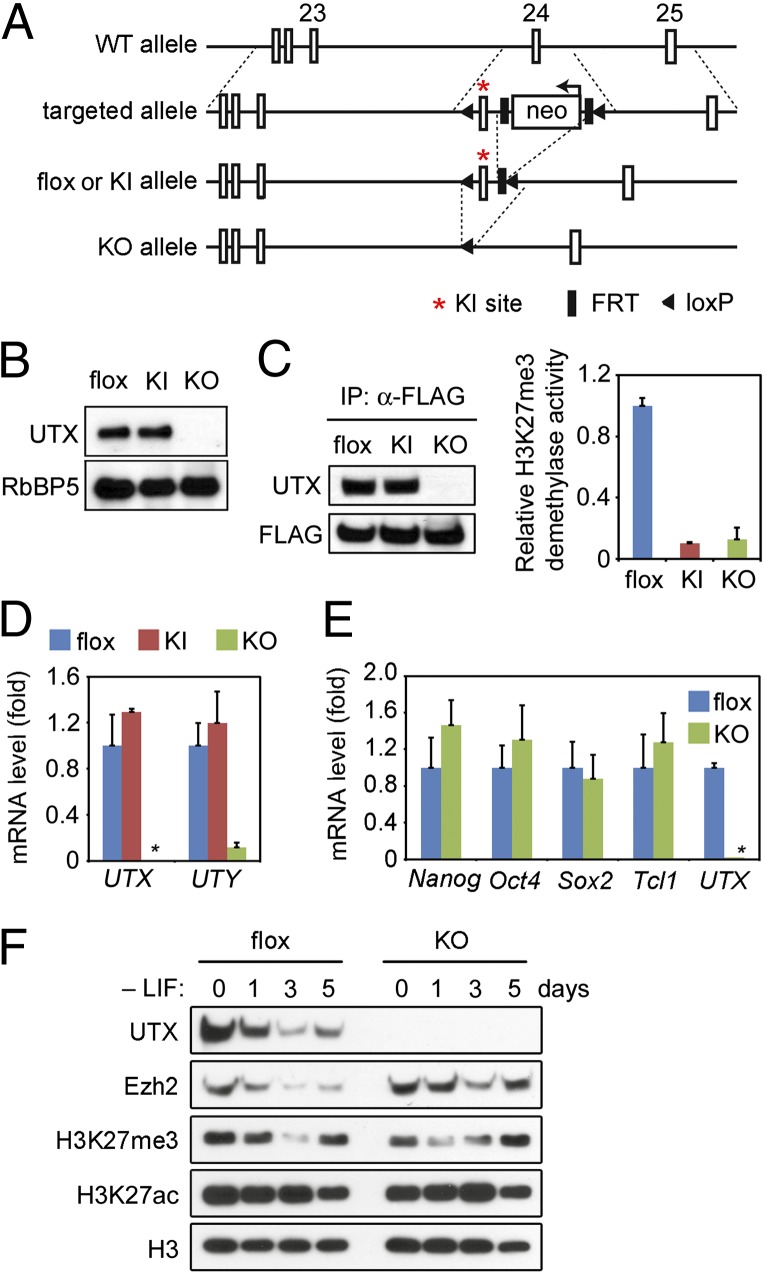
Most ES cell lines including the ones used here are male and carry one allele of UTX and one allele of UTY. To investigate the role of UTX in ES cell differentiation and animal development, we established male UTX conditional KO (flox) ES cell lines by gene targeting (Fig. 1A). Exon 24 was flanked by two loxP sites in the flox allele. Deletion of exon 24 from the flox allele by Cre recombinase resulted in the KO allele. His1146 (H1146) and Glu1148 (E1148) encoded by exon 24 are essential for UTX demethylase activity (11, 16). To identify UTX functions that depend on its demethylase activity, we also generated the enzyme-dead knock-in (KI) allele by mutating H1146 and E1148 of endogenous UTX to Ala (Fig. 1A). The genotypes of flox (UTXflox/Y; Y stands for UTY), KI (UTXKI/Y), and KO (UTX−/Y) cell lines were confirmed by genomic PCR (Fig. S1 A and B). The H1146A and E1148A mutations in the KI allele were confirmed by sequencing (Fig. S1C). UTX was expressed at similar levels in flox and KI cells but absent in KO cells (Fig. 1B). Histone demethylase assay in vitro confirmed that H1146 and E1148 were essential for UTX demethylase activity (Fig. S1D). Endogenous UTX from the KI ES cells was also deficient in H3K27 demethylase activity (Fig. 1C). Quantitative reverse-transcriptase PCR (qRT-PCR) confirmed deletion of exon 24 in KO cells. Interestingly, UTY mRNA levels were similar in flox and KI cells but decreased markedly in UTX KO cells, indicating that in male ES cells, UTX protein, but not its enzymatic activity, controls UTY expression (Fig. 1D).
Fig. 1.
Characterization of UTX KO, conditional KO, and knock-in male ES cells. (A) Schematic representation of mouse UTX wild-type (WT) allele, targeted allele, conditional KO (flox) allele, enzyme-dead knock-in (KI) allele, and KO allele. Deletion of neo selection cassette from the targeted allele by FLP recombinase generates the flox allele. Deletion of exon 24 from the flox allele by Cre generates the KO allele. The KI allele carries H1146A and E1148A mutations (represented by the red asterisk) in exon 24. (B) Immunoblotting of nuclear extracts prepared from flox (UTXflox/Y; Y stands for UTY), KI (UTXKI/Y) and KO (UTX−/Y) ES cells using indicated antibodies. (C) Endogenous UTX in KI ES cells is deficient for H3K27 demethylase activity. Nuclear extracts were prepared from flox, KI and KO cells stably expressing FLAG-tagged PA1, a unique subunit of the MLL3/4 histone methyltransferase complex that strongly associates with UTX (11, 32). Endogenous UTX was pulled down from the nuclear extracts by affinity purification of MLL3/4 complex using anti-FLAG antibody as described (32), followed by a demethylase assay using the JMJD3/UTX demethylase activity assay kit (Epigenase no. P-3084). (Left) Immunoblotting of immunoprecipitated endogenous UTX and FLAG-PA1. (Right) Demethylase assay. (D) UTX, but not its demethylase activity, controls UTY expression in male ES cells. UTX and UTY mRNA levels were analyzed by qRT-PCR. The absence of UTX expression in KO cells was indicated by an asterisk. (E) UTX is dispensable for self-renewal of ES cells. Expression of markers for undifferentiated ES cells was analyzed by qRT-PCR. (F) Deletion of UTX has little effect on the global H3K27me3 level during ES cell differentiation. flox and KO ES cells were cultured without LIF for 0, 1, 3, or 5 d. Nuclear extracts were analyzed by immunoblotting. Quantitative PCR data in all figures are presented as means ± SD.
Deletion of UTX and the subsequent loss of UTY did not affect expression of transcription factors critical for ES cell self-renewal, including Nanog, Oct4, Sox2, and Tcl1 (17) (Fig. 1E). The absence of UTX and UTY in ES cells had little effects on the population doubling time and colony-formation ability either (Fig. S2 A and B). Moreover, alkaline phosphatase, a marker for undifferentiated ES cells, was expressed at similar levels in flox and KO cells (Fig. S2C). Removing the cytokine LIF from medium would initiate spontaneous differentiation of ES cells cultured in the absence of fibroblast feeder cells (18). Alkaline phosphatase staining showed that, compared with flox cells, KO cells formed similar amount of differentiated colonies after LIF removal (Fig. S2C). These results indicate that UTX and UTY are dispensable for ES cell self-renewal and initial differentiation potential.
We also examined global H3K27me3 and H3K27ac levels during spontaneous differentiation of UTX KO cells. Consistent with a previous report (19), both Ezh2 and H3K27me3 levels decreased during spontaneous differentiation of ES cells. However, deletion of UTX did not lead to obvious changes of the global levels of H3K27me3 and H3K27ac (Fig. 1F). Interestingly, UTX protein level decreased along with that of Ezh2, suggesting that the reduced H3K27me3 level in differentiated cells is due to reduced Ezh2 level.
UTX has been reported to regulate Hox genes as well as genes encoding Rb-binding proteins (10–12, 20). However, we found that the loss of both UTX and UTY in ES cells did not affect retinoic acid (RA)-induced Hoxb1 expression, constitutive expression of Hoxb7, or expression of genes encoding Rb-binding proteins such as HBP1 and RbBP6 (Fig. S3 A–C and E). Similar results were obtained from mouse embryonic fibroblasts (MEFs) (Fig. S3 D and F). Thus, UTX and UTY are dispensable for expression of Hox genes and genes encoding Rb-binding proteins in ES cells and MEFs.
UTX Controls Mesoderm Differentiation of ES Cells Independent of H3K27 Demethylase Activity.
Next, we investigated whether UTX was required for ES cell differentiation using well established protocols (7, 19, 21–23). Cells were cultured on Petri dishes in the absence of LIF to form 3D multicellular aggregates called embryoid bodies (EBs). Differentiation of EBs recapitulates the generation of all three primary germ layers, ectoderm, mesoderm, and endoderm, during embryonic gastrulation (1). EBs were treated with RA to promote ES cells to differentiate along the neural ectoderm lineage and become neuron precursor (NP) cells expressing Nestin and Tubb3 (β-III Tubulin) (21). Immunostaining revealed robust expression of Tubb3 in both flox and KO cells (Fig. S4A). By examining gene expression before and after differentiation, we observed marked decreases of ES cell markers Nanog and Oct4 and marked increases of NP markers Nestin and Tubb3 in both flox and KO cells (Fig. S4B). These results indicate that UTX is dispensable for RA-induced ES cell differentiation toward NP cells.
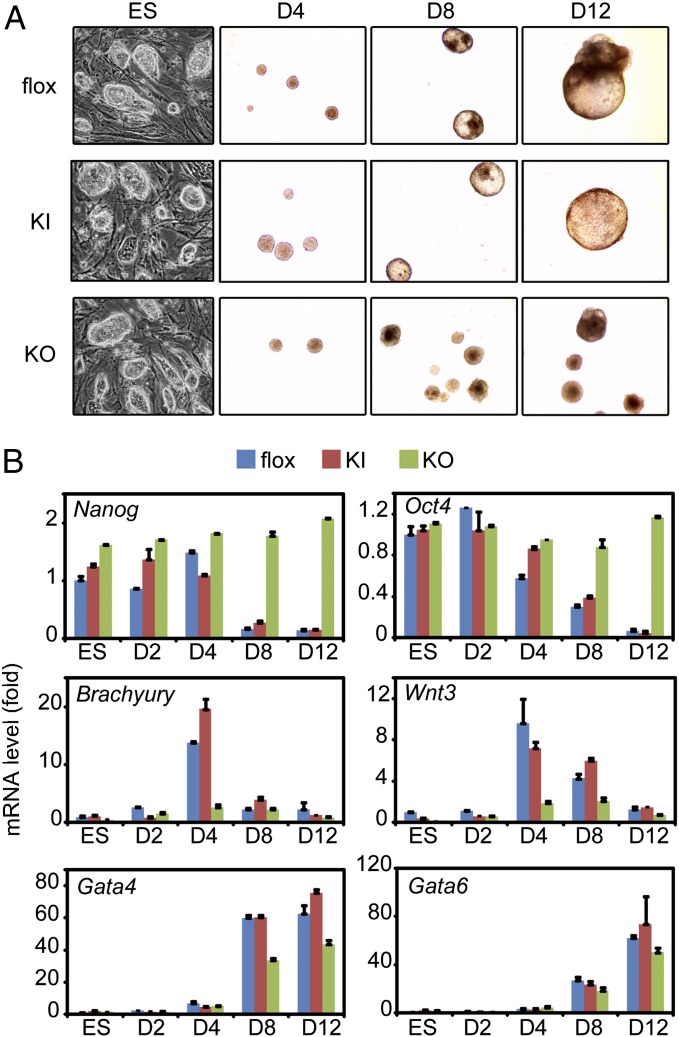
To investigate whether UTX was required for mesoderm and endoderm differentiation of ES cells, EBs were cultured for 12 d in the absence of RA (23), because RA strongly inhibits mesoderm differentiation (5). As expected, flox cells formed cystic EBs starting from day 8. In contrast, KO cells formed much smaller EBs with no obvious cystic morphology, indicating a severe differentiation defect (Fig. 2A). Consistent with the morphologies, during differentiation of the flox cells, expression of ES cell markers Nanog and Oct4 decreased markedly while expression of mesoderm markers Brachyury and Wnt3 increased markedly and reached the peak levels at day 4 and then dropped. In contrast, KO EBs retained Nanog and Oct4 expression and failed to induce Brachyury and Wnt3 during differentiation. The induction of endoderm markers Gata4 and Gata6 was largely unaffected by UTX deletion (Fig. 2B). The same sets of experiments were repeated using the KI cells carrying the enzyme-dead UTX allele. Surprisingly, the KI cells pheno-copied the flox ones, suggesting that the enzymatic activity of UTX is dispensable for mesoderm differentiation of ES cells (Fig. 2 A and B). Together, these results indicate that UTX controls mesodermal differentiation of ES cells independent of its H3K27 demethylase activity.
Fig. 2.
UTX controls mesodermal differentiation of ES cells independent of H3K27 demethylase activity. (A) Microscopic pictures of flox, KI, and KO ES cells before differentiation and EBs at day 4, day 8, and day 12 of differentiation. (B) qRT-PCR analysis of ES cell markers (Nanog, Oct4), mesoderm markers (Brachyury, Wnt3) and endoderm markers (Gata4, Gata6) at indicated time points of differentiation. D2, D4, D8, and D12 refer to day 2, 4, 8, and 12, respectively.
Brachyury Is a Direct UTX Target Gene in ES Cells.
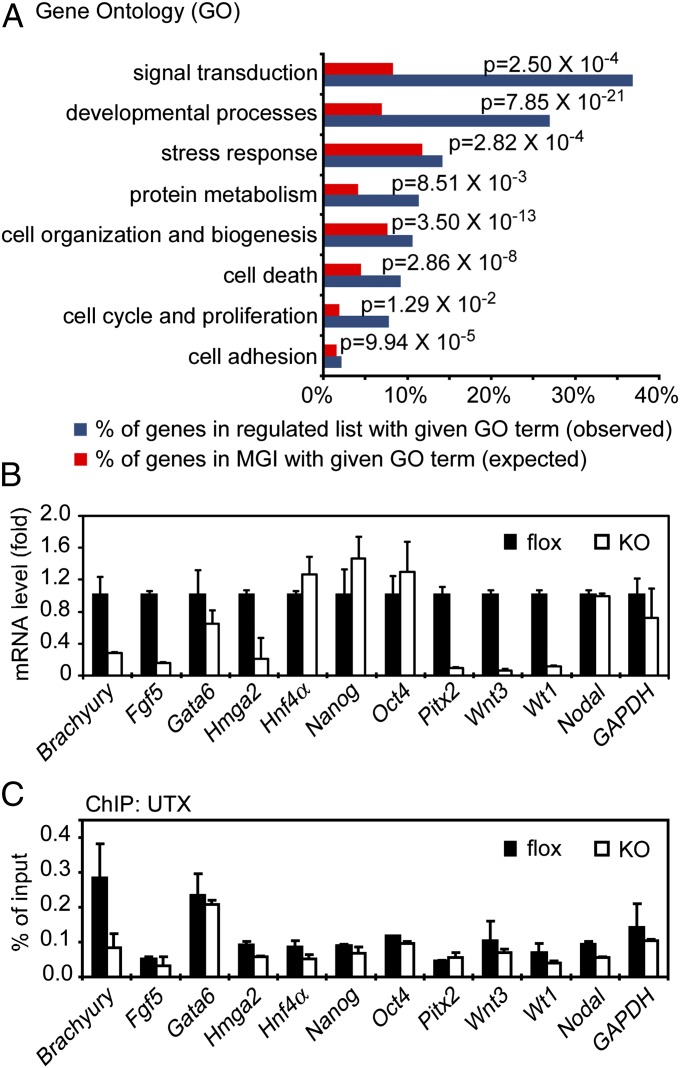
To understand the mechanism underlying the defective mesoderm differentiation in UTX KO cells, we compared gene expression profiles between flox and KO cells. Microarray analysis revealed that deletion of UTX in ES cells led to over 2.5-fold decreased expression of 141 genes (Table S1). Gene ontology analysis revealed that 52 genes were associated with developmental process, which was the most significant functional category (Fig. 3A). qRT-PCR confirmed that deletion of UTX decreased expression of developmental genes such as Brachyury, Fgf5, Hmga2, Pitx2, Wnt3, and Wt1 in undifferentiated ES cells. However, expression of ES cell markers Nanog and Oct4 and endoderm markers Gata6 and Hnf4a was largely unaffected (Fig. 3B). Although expression of Brachyury and Wnt3 decreased over threefold in the KO cells, expression of Nodal, another key regulator of mesoderm differentiation, was not affected by UTX deletion. By chromatin immunoprecipitation (ChIP) assays in flox and KO cells, we observed specific enrichment of endogenous UTX on the proximal promoter of Brachyury but not any other genes examined (Fig. 3C). Thus, UTX directly and selectively regulates Brachyury gene in ES cells. Because Brachyury is essential for mesoderm formation, the identification of Brachyury as a direct target gene of UTX provides a possible mechanism by which UTX controls mesoderm differentiation of ES cells.
Fig. 3.
Brachyury is a direct UTX target gene in ES cells. (A) Gene ontology (GO) analysis of down-regulated genes identified in microarray. Genes with over 2.5-fold decrease in UTX KO cells compared with flox cells were subjected to GO analysis. Dark blue bars represent the observed percentage of down-regulated genes in a particular GO category. Red bars represent the expected percentage in all GO-annotated genes among the entire mouse genome recorded in the MGI database. P value of each term was determined using Fisher’s exact test. (B) Confirmation of microarray data by qRT-PCR. (C) ChIP assays of UTX on proximal promoters of the indicated genes in flox and KO ES cells.
UTX and UTY Are Required for Wnt/β-Catenin Signaling-Induced Brachyury Expression.
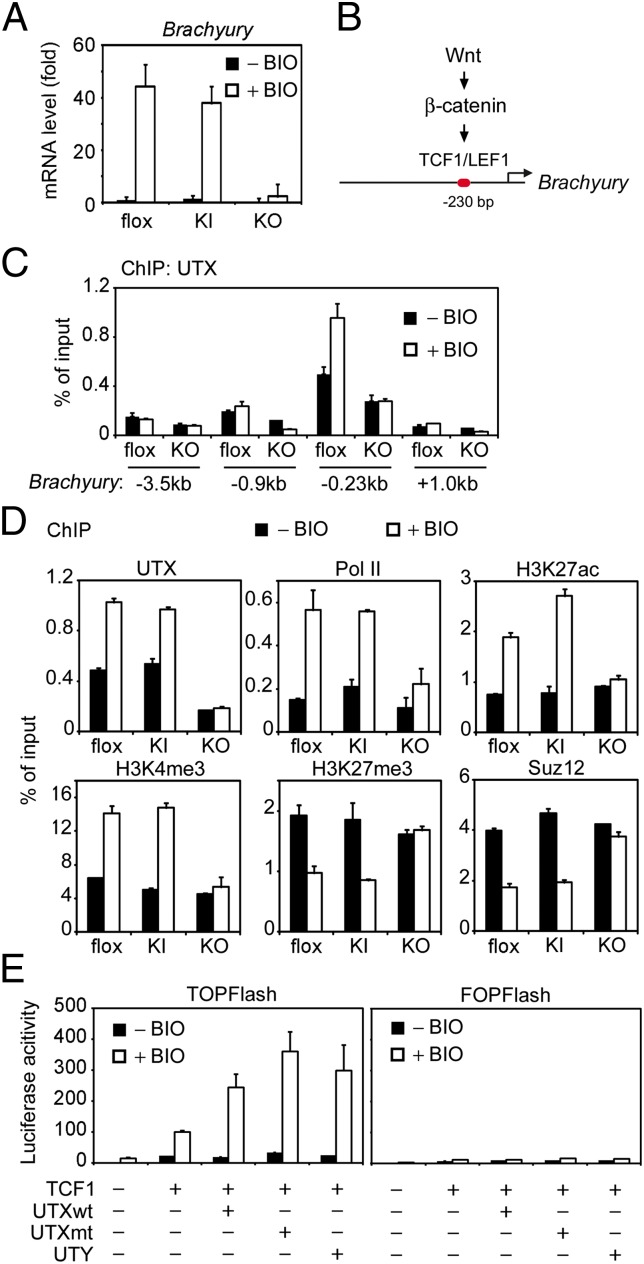
Because Brachyury is a direct target gene of Wnt/β-catenin signaling (3, 4), we treated ES cells with BIO, a small molecule inhibitor of GSK3β kinase and a robust activator of Wnt/β-catenin signaling, to induce Brachyury expression (24). Deletion of UTX in ES cells severely impaired BIO-induced Brachyury expression. In contrast, BIO-induced Brachyury expression was normal in the KI cells (Fig. 4A), indicating that UTX protein, but not its demethylase activity, is required for Wnt/β-catenin signaling-induced Brachyury expression in ES cells.
Fig. 4.
UTX is required for Wnt/β-catenin signaling-induced Brachyury expression. (A–D) flox, KI, and KO ES cells were passaged twice on 0.1% gelatin-coated dishes to remove feeder cells and then cultured on gelatin-coated dishes. Cells were treated with 2 μM BIO for 48 h in the absence of LIF. (A) UTX, but not its H3K27 demethylase activity, is required for BIO-induced Brachyury expression in ES cells. Brachyury expression was determined by qRT-PCR. (B) Schematic representation of Brachyury proximal promoter containing the Wnt/β-catenin signaling-responsive TCF1/LEF1 binding site. (C) ChIP assays of UTX on the indicated regions of Brachyury gene. (D) ChIP assays of levels of UTX, RNA polymerase II (Pol II), indicated histone modifications, and the Suz12 subunit of PRC2, around the TCF1/LEF1 binding site on Brachyury proximal promoter. (E) UTX and UTY function as TCF1 coactivators independent of demethylase activity. HeLa cells were transfected with 30 ng of TCF1 plasmid, 30 ng of TOPFlash or FOPFlash, and 150 ng of pCS2 plasmids expressing wild-type (UTXwt) or enzyme-dead (H1146A and E1148A) mutant UTX (UTXmt) or UTY. Twenty-four hours later, cells were treated with 2 μM BIO for 24 h, followed by luciferase assays using dual-luciferase assay kit (Promega).
ChIP assays revealed UTX enrichment in the region surrounding the Wnt/β-catenin-responsive TCF1/LEF1 binding site on Brachyury promoter (Fig. 4 B and C). BIO treatment increased UTX level on Brachyury promoter in flox and KI cells but not in KO cells (Fig. 4D). Consistent with the BIO-induced Brachyury expression in Fig. 4A, BIO induced enrichment of RNA polymerase II (Pol II) as well as positive histone marks H3K27ac and H3K4me3 on the Brachyury promoter in flox and KI cells but not in KO cells. Interestingly, BIO attenuated H3K27me3 level on Brachyury promoter in flox and KI cells but not in KO cells, which was likely due to the loss of PRC2 (represented by the Suz12 subunit) on the same promoter (Fig. 4D). These results suggest that UTX directly regulates Wnt/β-catenin signaling-induced Brachyury expression in ES cells independent of H3K27 demethylase activity.
Next, we asked whether the enzymatically inactive UTY could compensate for the loss of UTX in ES cells. Because endogenous UTY level is dramatically reduced in UTX KO ES cells, we stably expressed human UTY in the KO cells (Fig. S5A). Interestingly, ectopic expression of UTY in UTX KO cells rescued not only BIO-induced Brachyury expression (Fig. S5B), but also BIO-induced increases of Pol II, H3K27ac, and H3K4me3 and decreases of H3K27me3 and Suz12, on Brachyury promoter (Fig. S5C). Furthermore, BIO induced UTY enrichment to the same region that UTX bound on Brachyury promoter (Fig. S5C).
We next tested whether UTX and UTY could regulate TCF1 transcription activity using β-catenin responsive luciferase reporter TOPFlash containing seven repeats of TCF/LEF binding sites, with FOPFlash as the negative control. We found that not only wild-type and enzyme-dead UTX, but also UTY, were able to enhance BIO-induced TCF1 transcription activity (Fig. 4E). Collectively, these results suggest that UTX and UTY function as TCF1 coactivators to promote Wnt/β-catenin signaling-induced Brachyury expression in ES cells independent of H3K27 demethylase activity.
UTX Is Required for Brachyury Expression and Mesoderm Development in Mice.
To verify the ES cell results, we knocked out UTX gene in mice. Because of the early embryonic lethality in female UTX KO mice (see below), we first generated male KO mice (UTX−/Y) by crossing male wild-type (UTX+/Y) with female UTX+/− mice (Fig. S6A). Male UTX KO mice could survive until the end of embryonic development but died around birth, due to neuron tube closure defect and inability to breathe (Fig. S6 B and C). Thus, deletion of the single UTX allele in male mice did not affect mesoderm development. Unlike in the male ES cells, deletion of UTX in male embryos did not affect UTY expression (Fig. S6D), suggesting a functional redundancy between UTX and UTY in regulating mesoderm development in male embryos.
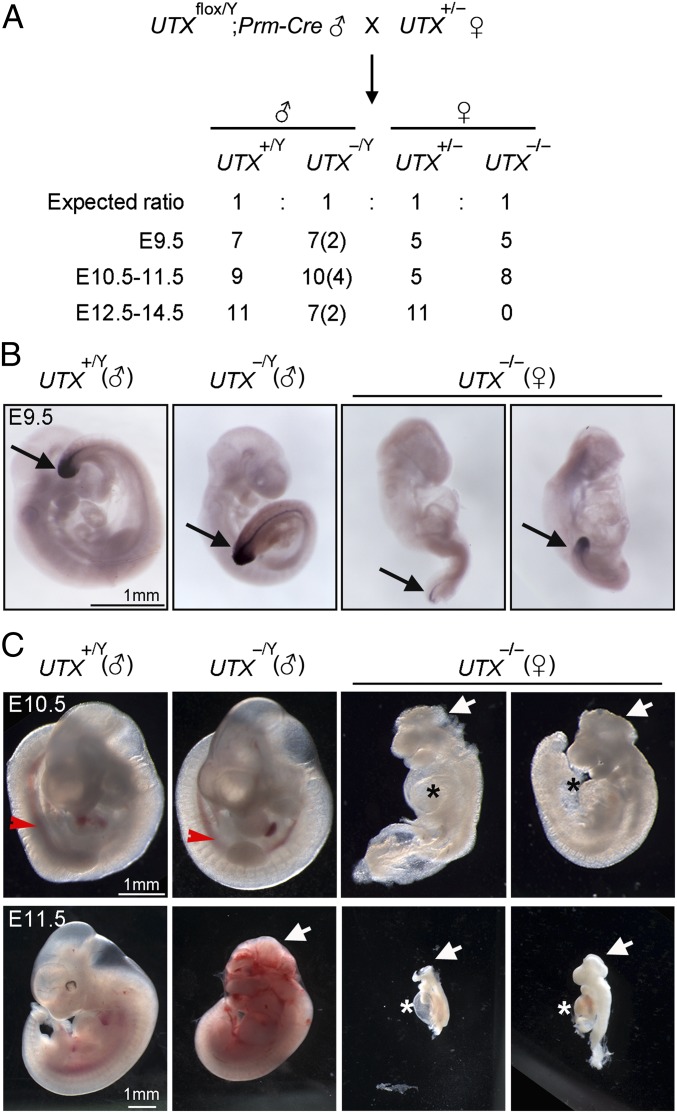
Next, we deleted both alleles of UTX in female mice where UTY gene is absent. Because of the perinatal lethality of male UTX KO mice, female UTXflox/flox mice were crossed with male Prm1-Cre mice expressing Cre under the control of sperm-specific Prm1 promoter. The resulting male UTXflox/Y;Prm1-Cre mice, which had UTX deletion in the sperms, were crossed with female UTX+/− to generate female UTX KO (UTX−/−) embryos (Fig. 5A). Consistent with a recent report (13), UTX−/− female embryos were not viable after embryonic day E10.5, although degenerated dead UTX−/− embryos were found at E11.5 (Fig. 5). The vast majority of E 9.5–10.5 UTX−/− embryos displayed abnormal or truncated posterior bodies (Fig. 5 B and C and Fig. S6F), which pheno-copied homozygous Brachyury mutant mice (25). In the remaining two E9.5 UTX−/− embryos with relatively intact posterior body formation, we observed markedly reduced Brachyury expression (Fig. 5B and Fig. S6E).
Fig. 5.
UTX is required for Brachyury expression and mesoderm development in mice. (A) The mating strategy for generating female UTX KO embryos (UTX−/−) and the genotyping result. The numbers of embryos with execenphaly are indicated in the parentheses. (B) Decreased Brachyury expression in female but not male UTX KO E9.5 embryos. Brachyury was detected by WISH. Black arrows indicate the tail bud and posterior notochord where Brachyury is expected to express. (C) UTX−/− female embryos show defects in neural tube closure, cardiac development and hematopoiesis. Representative images of E10.5 and E11.5 embryos with indicated genotypes are shown. Abnormal or truncated posterior notochords were observed in UTX−/− embryos. Red arrowheads indicate red blood cells in Aorta-Gonad-Mesonephros, which were absent in UTX−/− embryos. Asterisks indicate cardiac effusion in UTX−/− embryos, represented by swollen pericardial sacs. White arrows point to the defects in neural tube closure.
In E10.5 wild-type embryos, red blood cells were visible in Aorta-Gonad-Mesonephros, where early hematopoietic stem cells arise (26). In contrast, anemia was observed in UTX−/− embryos, suggesting a critical role of UTX in hematopoiesis (Fig. 5C). The majority of UTX−/− embryos also displayed severe defects in heart development and neural tube closure (Fig. 5C). In contrast, male UTX KO littermates (UTX−/Y) at E 9.5–10.5 were morphologically normal except for the neural tube closure defect in some of them (Fig. 5C), suggesting that UTX and UTY are largely redundant in regulating mesoderm development. Taken together, these results indicate that UTX is required for Brachyury expression and mesoderm development in mice.
Discussion
In this paper, we show UTX controls UTY expression in ES cells independent of its enzymatic activity. Deletion of UTX and the subsequent loss of UTY do not affect global H3K27me3 level, Hox gene expression, or ES cell self-renewal. However, UTX, but not its enzymatic activity, is essential for mesoderm differentiation and Brachyury expression in ES cells. Unlike in the ES cells, UTX is dispensable for UTY expression in mice. UTX and the enzymatically inactive UTY are functionally redundant in early embryonic development. Female UTX KO mice, which lack UTY gene, show severe defects in Brachyury expression and mesoderm development. Together, these results indicate an essential role of UTX protein, but not its H3K27 demethylase activity, in regulating mesoderm development and Brachyury expression.
Regulation of ES Cell Differentiation by UTX.
Recently, Lee et al. reported that UTX is dispensable for ES cell self-renewal and that UTX is required for UTY expression in ES cells but not in embryos (13), which are consistent with our results. We further demonstrate that UTX controls UTY expression independent of its enzymatic activity. However, the essential role of UTX in mesoderm differentiation and Brachyury expression in ES cells was not observed by Lee et al. (13). The reason of this discrepancy is that we induce EB differentiation toward mesoderm in the absence of RA (21, 22), but Lee et al. included RA. RA has been shown to strongly inhibit mesoderm differentiation by down-regulating Brachyury expression (5). Consistently, we found that RA strongly inhibits Brachyury expression during ES cell differentiation (Fig. S7). Our ES cell data are highly consistent with our observation on the essential role of UTX in mesoderm development and Brachyury expression in mice.
Regulation of Brachyury Expression by UTX.
Our findings that Brachyury is a direct UTX target gene and that UTX is required for Wnt/β-catenin signaling-induced Brachyury expression in ES cells provide a mechanism to explain the essential role of UTX for ES cell differentiation toward mesoderm. These results are also consistent with recent reports that Wnt/β-catenin signaling is dispensable for the self-renewal of ES cells cultured under appropriate conditions (i.e., on feeder cells in the presence of LIF) and that Wnt/β-catenin signaling appears to be selectively required for mesoderm formation during ES cell differentiation (27).
We confirmed a previous report that BIO-induced Brachyury expression is accompanied by decreases of PRC2 and PRC2-mediated H3K27me3 on Brachyury promoter (24). However, BIO treatment decreases H3K27me3 on Brachyury promoter in both flox and KI cells but not in KO cells, indicating that UTX regulates Wnt/β-catenin signaling-induced Brachyury expression independent of its H3K27 demethylase activity. Because PRC2 and H3K27me3 are dispensable for BIO-induced Brachyury expression in ES cells (24), there is no need to get the H3K27 demethylase activity involved to activate Brachyury expression. The BIO-induced decrease of H3K27me3 on Brachyury promoter could be due to another H3K27 demethylase, Jmjd3, which is recruited to the Brachyury promoter upon BIO treatment (24). However, it remains unclear whether Jmjd3 is required for BIO-induced Brachyury expression.
UTX has been shown to function as a transcription coactivator and recruit Brg1 chromatin remodeling complex to facilitate promoter accessibility of target genes in T cells and cardiac myocytes (13, 14). Whether UTX recruits Brg1 complex to Brachyury promoter remains to be investigated. Consistent with the coactivator role of UTX, UTX is required for Pol II recruitment to Brachyury promoter.
Regulation of Early Embryonic Development by UTX and UTY.
Mutation of Brachyury in mice leads to abnormal posterior notochord organization and posterior truncation (25). Consistent with the requirement of UTX for Wnt/β-catenin signaling-induced Brachyury expression in ES cells, UTX−/− embryos show posterior truncation and decreased Brachyury expression. Interestingly, knockdown of UTX in zebrafish also leads to truncated posterior notochord and abnormal posterior structure, indicating defective dorsal mesoderm formation (12). Together, these results suggest an evolutionally conserved role of UTX in mesoderm development.
In addition to previously reported defects in heart development in female UTX KO mice and neural tube closure defects in both female and male UTX KO mice (13, 28), we observed anemia in female UTX KO mice. Because both hematopoietic cells and cardiac myocytes are derived from mesoderm (1), the defective mesoderm development likely contributes to anemia and impaired heart development in UTX−/− embryos. Whether UTX plays direct, tissue-specific roles in regulating hematopoiesis and heart development will require deletion of UTX in a tissue-specific manner. It is possible that UTX regulates expression of other Wnt target genes, which may also contribute to the regulation of mesoderm development. However, Brachyury is the master regulator of mesoderm development. Regulation of Brachyury expression by UTX likely represents the major mechanism for the role of UTX in mesoderm development.
Our data suggest that UTX and UTY are redundant and regulate early embryonic development independent of demethylase activities. Highly consistent with our result, a very recent paper reports that Caenorhabditis elegans UTX is essential for normal development in a demethylase activity independent manner (29). However, the fact that female UTX+/− mice appear normal whereas male UTX−/Y mice show perinatal lethality suggests distinct functions of UTX and UTY in peri-/postnatal development. Given UTY lacks detectable demethylase activity (8, 12), it is possible that the H3K27 demethylase activity of UTX may be involved in late embryonic and/or postnatal development. Generating the enzyme-dead UTX KI mice will help understand the physiological roles of UTX demethylase activity.
Materials and Methods
Plasmids, Antibodies, Chemicals, Cell Culture, and Differentiation.
The details are described in SI Materials and Methods. UTXflox/flox primary MEFs were isolated from E12.5 UTXflox/flox female embryos. 3T3 immortalization and retroviral infection of MEFs were done as described (30).
Generation of UTX KO, flox, and KI ES Cell Lines.
The design of gene targeting constructs and the generation and genotyping of UTX KO, flox, and KI ES cell lines are described in SI Materials and Methods (also see Fig. 1A and Fig. S1A). For each of the flox, KO, and KI ES cell lines, three individual colonies were identified by PCR screen and used for characterization and differentiation experiments.
Generation of UTX KO and Conditional KO Mice.
Generation of UTX conditional KO male (UTXflox/Y) and female (UTXflox/flox) mice, as well as the mating strategies to generate UTX KO male (UTX−/Y) and female (UTX−/−) mice, are described in SI Materials and Methods (also see Fig. S6A and Fig. 5A). All mouse work was approved by the Animal Care and Use Committee of National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK).
Microarray, qRT-PCR, ChIP, Immunoblotting, Immunofluorescence, and WISH.
For microarray analysis, flox and KO ES cells were passaged twice on 0.1% gelatin-coated dishes to remove feeder cells and then cultured in ES medium on gelatin-coated dishes. Total RNAs were purified and analyzed on Mouse Genome 430 2.0 Array (Affymetrix) as described (31). Data are deposited in National Center for Biotechnology Information GEO database (accession no. GSE35415). Gene ontology (GO) analysis of genes that show decreased expression in KO cells was done using the MGI Slim Chart Tool (http://www.informatics.jax.org/gotools/MGI_GO_Slim_Chart.html). qRT-PCR with 18S rRNA as control and ChIP with PCR quantitation using SYBR Green kit were performed as described (30). Data are presented as means ± SD. The sequences of SYBR Green primers are listed in Table S2. Immunoblotting of nonhistone proteins and immunofluorescence staining were done as described (8). Immunoblotting of histone modifications was done using acid extracted histones (30). Whole-mount in situ hybridization (WISH) of E9.5 mouse embryos was done using a Brachyury probe from Chuxia Deng (NIDDK, NIH, Bethesda, MD).
Supplementary Material
Acknowledgments
We thank R. Moon, D. Forrest, C. Deng, and C. Cepko for plasmids and reagents and C. Li for technical help on WISH. This work was supported by the Intramural Research Program of the NIDDK, National Institutes of Health (to K.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data reported in this paper has been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE35415).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1204166109/-/DCSupplemental.
References
- 1.Keller G. Embryonic stem cell differentiation: Emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- 2.Wilkinson DG, Bhatt S, Herrmann BG. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature. 1990;343:657–659. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- 3.Arnold SJ, et al. Brachyury is a target gene of the Wnt/beta-catenin signaling pathway. Mech Dev. 2000;91:249–258. doi: 10.1016/s0925-4773(99)00309-3. [DOI] [PubMed] [Google Scholar]
- 4.Yamaguchi TP, Takada S, Yoshikawa Y, Wu N, McMahon AP. T (Brachyury) is a direct target of Wnt3a during paraxial mesoderm specification. Genes Dev. 1999;13:3185–3190. doi: 10.1101/gad.13.24.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin BL, Kimelman D. Brachyury establishes the embryonic mesodermal progenitor niche. Genes Dev. 2010;24:2778–2783. doi: 10.1101/gad.1962910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 7.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong S, et al. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci USA. 2007;104:18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swigut T, Wysocka J. H3K27 demethylases, at long last. Cell. 2007;131:29–32. doi: 10.1016/j.cell.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 10.Agger K, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 11.Lee MG, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 12.Lan F, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 13.Lee S, Lee JW, Lee S-K. UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev Cell. 2012;22:25–37. doi: 10.1016/j.devcel.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell. 2010;40:594–605. doi: 10.1016/j.molcel.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seenundun S, et al. UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. EMBO J. 2010;29:1401–1411. doi: 10.1038/emboj.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith ER, et al. Drosophila UTX is a histone H3 Lys27 demethylase that colocalizes with the elongating form of RNA polymerase II. Mol Cell Biol. 2008;28:1041–1046. doi: 10.1128/MCB.01504-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macarthur BD, Ma’ayan A, Lemischka IR. Systems biology of stem cell fate and cellular reprogramming. Nat Rev Mol Cell Biol. 2009;10:672–681. doi: 10.1038/nrm2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams RL, et al. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 19.Walker E, et al. Polycomb-like 2 associates with PRC2 and regulates transcriptional networks during mouse embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2010;6:153–166. doi: 10.1016/j.stem.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang JK, et al. The histone demethylase UTX enables RB-dependent cell fate control. Genes Dev. 2010;24:327–332. doi: 10.1101/gad.1882610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bibel M, et al. Differentiation of mouse embryonic stem cells into a defined neuronal lineage. Nat Neurosci. 2004;7:1003–1009. doi: 10.1038/nn1301. [DOI] [PubMed] [Google Scholar]
- 22.Keller GM. In vitro differentiation of embryonic stem cells. Curr Opin Cell Biol. 1995;7:862–869. doi: 10.1016/0955-0674(95)80071-9. [DOI] [PubMed] [Google Scholar]
- 23.Dovey OM, Foster CT, Cowley SM. Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proc Natl Acad Sci USA. 2010;107:8242–8247. doi: 10.1073/pnas.1000478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dahle O, Kumar A, Kuehn MR. Nodal signaling recruits the histone demethylase Jmjd3 to counteract polycomb-mediated repression at target genes. Sci Signal. 2010;3:ra48. doi: 10.1126/scisignal.2000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herrmann BG. Expression pattern of the Brachyury gene in whole-mount TWis/TWis mutant embryos. Development. 1991;113:913–917. doi: 10.1242/dev.113.3.913. [DOI] [PubMed] [Google Scholar]
- 26.Dzierzak E, Medvinsky A. The discovery of a source of adult hematopoietic cells in the embryo. Development. 2008;135:2343–2346. doi: 10.1242/dev.021279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe K, Dai X. A WNTer revisit: New faces of β-catenin and TCFs in pluripotency. Sci Signal. 2011;4:pe41. doi: 10.1126/scisignal.2002436. [DOI] [PubMed] [Google Scholar]
- 28.Cox BJ, et al. Phenotypic annotation of the mouse X chromosome. Genome Res. 2010;20:1154–1164. doi: 10.1101/gr.105106.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vandamme J, et al. The C. elegans H3K27 demethylase UTX-1 is essential for normal development, independent of its enzymatic activity. PLoS Genet. 2012;8:e1002647. doi: 10.1371/journal.pgen.1002647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin Q, et al. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho YW, et al. Histone methylation regulator PTIP is required for PPARgamma and C/EBPalpha expression and adipogenesis. Cell Metab. 2009;10:27–39. doi: 10.1016/j.cmet.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho Y-W, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem. 2007;282:20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.