Abstract
Alzheimer’s disease (AD) is characterized by the presence of toxic protein aggregates or plaques composed of the amyloid β (Aβ) peptide. Various lengths of Aβ peptide are generated by proteolytic cleavages of the amyloid precursor protein (APP). Mutations in many familial AD-associated genes affect the production of the longer Aβ42 variant that preferentially accumulates in plaques. In the case of sporadic or late-onset AD, which accounts for greater than 95% of cases, several genes are implicated in increasing the risk, but whether they also cause the disease by altering amyloid levels is currently unknown. Through loss of function studies in a model cell line, here RNAi-mediated silencing of several late onset AD genes affected Aβ levels is shown. However, unlike the genes underlying familial AD, late onset AD-susceptibility genes do not specifically alter the Aβ42/40 ratios and suggest that these genes probably contribute to AD through distinct mechanisms.
Keywords: cystatin, genome-wide association studies, late-onset, epistasis
Alzheimer’s disease (AD) is the most common form of dementia, with a predicted incidence in the global population of 1 in 85 by 2050 (1). The disease is degenerative, eventually leading to death, and there is currently no cure; therefore insights into the molecular causes of the disease are urgently required (2, 3). AD is pathologically characterized by the presence of amyloid plaques and tangles in the brain (4, 5). These plaques are composed of insoluble aggregates of Aβ peptides, which are derived from the amyloid precursor protein (APP) via cleavage by the proteases, β- and γ-secretases (6, 7). However, whether amyloid plaques are causative or a consequential feature of AD is still unclear (8, 9).
AD can be divided into different types based on age of onset and genetic predisposition. Sporadic or late onset AD (LOAD) accounts for over 95% of cases and begins after the age of 65 years. Early onset or familial AD (FAD) is rare and usually manifests by age 60. Although FAD is far less common, its associated familial mutations underlie the predominant molecular model of the disease on which many of the drugs currently under clinical development are based (5). These FAD-associated mutations are mainly found in components involved in the production of Aβ peptides such as APP and presenilins. Cleavage of APP by β- and γ -secretases generates Aβ. Whereas β-secretase cleavage of APP releases the soluble ectodomain of APP (sAPPβ) and the β-cleaved c-terminal fragment (β-CTF), γ-cleavage of the latter releases the APP intracellular domain (AICD) and Aβ peptides. Interestingly, γ-cleavage generates Aβ of different lengths, such as Aβ38, Aβ40, Aβ42, through a mechanism that is still poorly understood. β-Secretase activity is conferred by the transmembrane type I protein, β-site APP cleaving enzyme 1 (BACE1) (10), whereas cleavage by γ secretase is mediated by an intramembrane protease complex composed of Nicastrin, Aph1, Pen-2, and the catalytic components presenilins (11, 12). FAD-associated mutations have been found in the APP and presenilin genes, and have been shown to affect the production or aggregation of Aβ peptides (13). Mutations in the presenilin-1 (PS1) and presenilin-2 (PS2) genes largely affect the production specifically of the Aβ42 peptide form, which preferentially accumulates as amyloid plaques. These discoveries lent strong support to the amyloid hypothesis put forward 20 y ago, which assigned a causative role for amyloid plaques in AD (14). However, although FAD and LOAD are clinically indistinguishable, it remains unclear whether they share a common underlying molecular cause.
The molecular basis for the more common late onset AD is currently not fully understood, although genome-wide association studies (GWASs) have identified associated risk genes and loci (15–17). Because mutations associated with FAD change the ratio of Aβ42/40 by increasing the production of Aβ42, which is linked to amyloid plaque formation (13, 18), we therefore investigated whether the genes linked to LOAD would have the same effect.
Results and Discussion
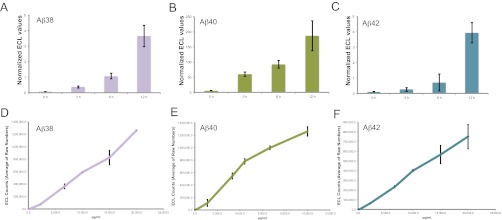
Because Aβ levels in an organism are determined by various factors including clearance, metabolism, aggregation, and vascular deposition, we took advantage of a simple assay using human cells that robustly produce Aβ38, Aβ40, and Aβ42 peptides similar to neurons (Fig. 1 A–C) to quantitatively monitor the effects of individual genes on amyloid production and changes in Aβ42/40 ratios (19, 20). The associated multiplexed electrochemiluminescence assay enables the quantification of Aβ38, 40, and 42 levels from the same sample in the same well, avoiding intermeasurement variations. All measurements were performed so that the values lay well within the linear range (Fig. 1 D–F). These cells are also amenable to gene knockdown using RNA interference (RNAi) (Figs. 2A and 3 A and B, and Fig. S1 A–C).
Fig. 1.
(A–C) Quantification of cell culture derived and synthetic Aβ peptides. Aβ38 (A), 40 (B), and 42 (C) levels in supernatant of HeLa swAPP cells after 0 h, 3 h, 6 h, and 12 h were analyzed using the triplex ECL assay (MSD). The values are given as ECL counts normalized to cell viability. (A–C): Linearity of the detection for each of the analytes in the multiplex ECL assay platform was determined for different concentrations of synthetic Aβ38 (D), Aβ40 (E), and Aβ42 (F) peptides.
Fig. 2.
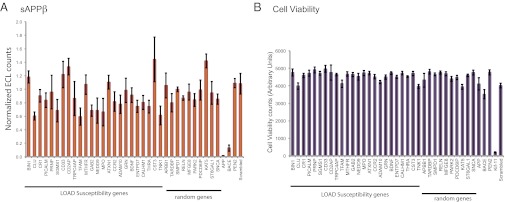
β-Cleavage of APP is not specifically affected by genes involved in LOAD susceptibility. (A) HeLa swAPP cells were transfected with siRNAs to the corresponding genes, and the supernatant was analyzed for sAPPβ 72 h after transfection (with 3 h medium exchange) using duplex ECL array (MSD). The values are given as ECL counts normalized to cell viability and relative to that of a scrambled (MedGC) control. The genes (BIN1, CLU, CR1, PICALM, CD33, and CD2AP) are among the top 10 AlzGene meta-analysis results (ranking is based on P value, and top 10 have P values <0.00001) listed in the AlzGene database. APP, BACE1, Pen-2, and scrambled (MedGC) serve as both transfection and assay controls. (B) Cell viability: HeLa swAPP cells were incubated for 72 h after siRNA transfection and subjected to a cell viability assay using AlamarBlue. Results are shown as fluorescence counts using a microplate absorbance/fluorometer plate reader (Molecular Devices Spectramax Gemini XS). Effects of positive (KIF11) and negative (scrambled) controls are shown among the other corresponding silenced genes. Note that only Kif11 silencing produces dramatic effects in cell viability.
Fig. 3.
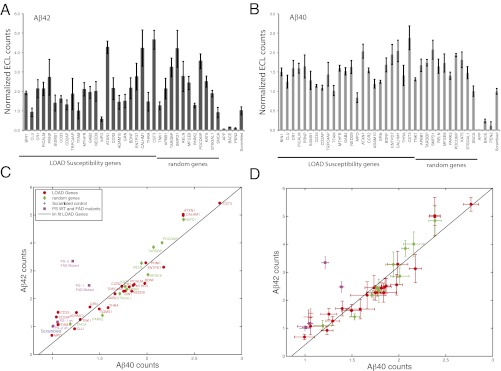
Genes involved in LOAD susceptibility do not affect Aβ42/40 levels. HeLa- swAPP cells were transfected with the corresponding siRNAs, and the medium was analyzed for (A) Aβ42 and (B) Aβ40. The values are given as ECL counts normalized to cell viability and relative to that of medium GC scrambled control. Aβ42 vs. Aβ40 plot without (C) and with (D) error bars. Aβ42 ECL counts, normalized to Aβ42 counts of the scrambled control (Scrambled), plotted vs. normalized Aβ40 counts for AD risk genes [LOAD Genes (red ●)], random control genes (green ◆). Aβ42 and Aβ40 counts of FAD genes, PS1, and PS2 mutations along with the wild type controls (PS1 WT, PS2 WT) are indicated by a pink ■, and normalized to those of mock transfected control. A linear fit was added as a guide. Error bars in A and B are SD and in (D) are SEM. Note that only the FAD mutants of PS1 and PS2 alter the Aβ42 to Aβ40 ratio (C and D).
RNAi was used to systematically silence 24 genes linked to LOAD (the top genes in the AlzGene database, www.alzgene.org) that were expressed in this cellular system (19). As random controls, we chose 10 genes that were implicated in other neurodegenerative diseases including Parkinson’s disease and frontotemporal lobar degeneration but for which no polymorphisms have been associated with AD (Table S1). In addition, as controls for Aβ production, we silenced APP, BACE1 (the rate-limiting β-secretase enzyme), and Pen-2 (a component of γ-secretase). Knockdown of Kif11, which is a kinesin involved in cell division, was used as a transfection control (Fig. 2B and Fig. S1A). Quantitative RT-PCR analyses before and after silencing showed that effective knockdown of the genes was achieved (Table S2). First we examined the levels of sAPPβ, which is an indicator of β-secretase mediated cleavage of APP (Fig. 2A). As expected, silencing APP and BACE1 dramatically decreased sAPPβ levels, whereas Pen-2 knockdown did not, validating the silencing protocol and also the assay (Fig. 2A). Interestingly, silencing of cystatin C (CST3), a potent inhibitor of lysosomal and extracellular proteases, led to an increase in sAPPβ levels. However, silencing of most of the LOAD-susceptibility genes did not significantly alter the levels of sAPPβ, suggesting that, unlike some of the FAD-linked mutations, they do not affect β-secretase activity. The exceptions were silencing of CLU, CD2AP, GAB2, CD2AP, TFAM, ENTPD7, THRA, and TNK1, as well as the random control genes KAT5 and SNCA (Fig. 2 and Table S3).
Next, by measuring the levels of Aβ42 and Aβ40, we examined whether the LOAD-susceptibility genes would affect the γ-secretase–mediated cleavage of APP. Although both Aβ40 and Aβ42 are products of APP cleavage by β- and γ-secretases, Aβ42 has been shown to be the primary indicator of AD disease pathology as the FAD mutations in presenilins specifically increase Aβ42 levels (21). Therefore, we analyzed whether the LOAD-risk genes also affected Aβ42 levels. As expected, knocking down Pen-2, which is a key component of γ-secretase, dramatically reduced both Aβ42 and Aβ40 levels (Fig. 3 A and B) without affecting sAPPβ levels (Fig. 2A). In contrast, silencing 17 of the 24 LOAD-susceptibility genes increased the levels of Aβ42 (Fig. 3A and Table S3). These genes included CST3, which encodes for the cysteine protease inhibitor cystatin C. CST3 is a LOAD risk gene and has been previously shown to negatively regulate amyloid deposition in animals (22). We show that silencing of CST3 slightly increased sAPPβ levels and significantly increased Aβ levels. Our results lend a unique molecular explanation as to how CST3 could regulate amyloid levels. In addition, we found that most of the risk genes except for CLU, CD2AP, and TFAM upon silencing, had significant effect on Aβ42 levels. Interestingly, however, we also found a similar increase in Aβ42 levels upon silencing the 10 random control genes (except for SNCA), which are not linked to AD but are involved in other neurodegenerative diseases (Fig. 3A). In addition, silencing of many of the LOAD-risk and the random genes also increased Aβ40 levels (Fig. 3B and Table S3). These effects on Aβ42 and Aβ40 were not due to differences in cell viability (Fig. 2B). When the values of Aβ42 were plotted against Aβ40, a linear correlation was observed for all of the genes, showing that there was no specific effect on Aβ42 levels or the Aβ42/40 ratios (Fig. 3 C and D). Similar effects were also observed for Aβ38, another variant of Aβ that has been suggested to have protective effects in AD (Fig. S2 A–C and Table S3). In contrast, when the same cell line was transfected with plasmids expressing FAD mutations in PS1 or PS2, as expected, we observed a specific increase in Aβ42 levels but not of Aβ40, compared with WT PS1, WT PS2, and control transfected cells (Fig. 3 C and D and Fig. S3). In the Aβ42/40 2D plot, presenilin mutants patently deviated from the trend toward Aβ42 (Fig. 3 C and D), confirming that these early onset FAD mutations indeed change the Aβ42/40 ratio. These results reveal that, unlike the FAD-linked mutations, most of the LOAD-susceptibility genes, and the random genes that are associated with other neurodegenerative diseases, alter the levels of both Aβ42 and Aβ40 (Fig. 3 A and B) but not the ratios of Aβ42/40 values.
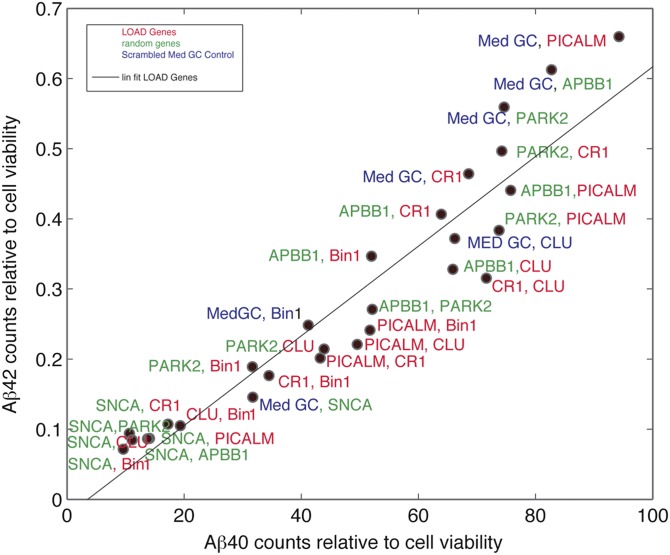
In the absence of effects of the LOAD-risk genes alone, we hypothesized that epistatic interactions between these genes may alter Aβ42/40 ratios, given that certain genes have been speculated to interact with each other in an epistatic manner and thus influence the disease risk (23, 24). To test this, we performed a combinatorial RNAi screen silencing seven genes (four LOAD risk, three random genes) against one another in a 7 × 7 mini array format. Analysis of Aβ40 and Aβ42 levels from the supernatants of the single and double knockdowns showed no evidence for epistatic interactions between genes affecting Aβ42/40 ratios (Fig. 4). Taken together, our results conclusively show that, unlike early onset mutations, susceptibility genes associated with the risk for sporadic AD, in a loss of function manner, do not specifically alter Aβ42/40 ratios.
Fig. 4.
Epistatic interactions among the genes involved in the risk of AD do not affect Aβ42/40 levels. HeLa-swAPP cells were transfected with siRNAs targeting the corresponding genes and the medium was analyzed for Aβ42 and Aβ40. The values are given as ECL counts normalized to cell viability. MedGC represents scrambled siRNA control. Aβ42 vs. Aβ40 plot in which normalized Aβ42 counts were plotted vs. normalized Aβ40 counts for AD risk genes (gene symbols are represented in red), random control genes (gene symbols are represented in green), and MedGC control (represented in blue). A linear fit was added as a guide.
The question of how late onset genes and gene loci confer risk for sporadic AD and how relevant the amyloid hypothesis is to the sporadic form of the disease are important for a better understanding of the disease and also for devising effective therapies (16). Although the amyloid cascade hypothesis explains how APP and PS mutations contribute to FAD, how LOAD genes affect the risk is still not understood (25). Our study addressed this question by capitalizing on loss of function RNAi studies in an Aβ-producing human cell line model. Because Aβ levels in an organism are determined by various factors, including clearance, metabolism, aggregation into smaller aggregates, plaques, and vascular deposition, and because genes could contribute to amyloid levels at multiple levels, it has proved difficult to address whether specific genes directly influence amyloid production in a model organism. In support of this, risk genes such as ApoE, Clusterin, and CCR2 have been shown to modulate amyloid deposition; but whether they influenced the production of the peptide or affected the clearance, thereby leading to amyloid deposition, is not well understood, highlighting the problems associated with studying the role of risk genes at the organismal level (26–28). Moreover, there are substantial technical limitations of neurons and cortical/hippocampal slice cultures, such as poor transfection and silencing efficiency, making it currently unfeasible to perform such a quantitative study using a neuronal set-up. We therefore took advantage of a simple cellular assay to monitor whether certain genes, via loss of function, could affect amyloid production or change the Aβ42/40 ratios (19). Here we showed that the LOAD risk genes do not specifically affect β-cleavage of APP, production of Aβ40 or Aβ42, or the Aβ42/40 ratios, unlike the early onset FAD mutations. Surprisingly, both the LOAD and the genes that are linked to other neurodegenerative diseases that we used in our study perturbed the levels of Aβ40 and Aβ42. This indicates that the genes involved in other neurodegenerative diseases that were used as random controls might be involved in regulating Aβ levels. We also showed that epistatic interactions between the risk genes did not change the Aβ42/40 ratios, making it unlikely that combinatorial effects of these genes would contribute to the increased risk via altering the Aβ42/40 ratios (17, 23). Our results also suggest that the reduced Aβ42/40 levels in the CSF of LOAD patients probably stem from clearance defects rather than from altered Aβ42 production (29, 30). It is tempting to speculate that unlike FAD, amyloid might be a consequence in LOAD pathology (8, 31, 32). Indeed, the failure of drugs aimed at reducing amyloids, either by immunization or via γ-secretase inhibitors, which are currently in clinical trials, supports this speculation (32–35), as does accumulating evidence that amyloid-independent mechanisms, such as neuroinflammation, mitochondrial dysfunction, or synaptic dysfunction, contribute to the disease (3, 8, 36). Nonetheless, we cannot completely rule out the possibility that the LOAD genes could play a role in the vulnerability to Aβ-mediated toxicity in late-onset AD (37). Because we used RNAi silencing to study the effect of these genes on amyloid production, our results reflect only their effect on Aβ, in a loss-of-function manner. Hence, we cannot entirely rule out the possibility that the polymorphisms of these genes or certain epistatic interactions of these genes with environmental factors could cause the pathology through altering amyloid levels. Also we do not know to which extent these polymorphisms affect the expression of these genes in the affected individuals. One of the major limitations of our study is that we used the 2010–2011 Alzgene database based on the meta-analysis of the GWASs conducted on LOAD, which is updated constantly, and we studied only the effect of the genes in the list. As few of the top genes were not expressed in the model cell line studied (e.g., APOE, MS4A6A, MS4A4E), we could not study their effect on amyloid and hence do not know whether these genes affect AD by altering the levels of Aβ42/40. However, with the gene set that we studied, we conclusively show that the LOAD genes, when silenced using RNAi in this model cell line, do not specifically alter Aβ42/40 ratios.
Methods
Cell Culture.
HeLa cells expressing the Swedish mutant of APP (HeLa swAPP) cells were cultured in DMEM (Invitrogen) at 37 °C and 5% (vol/vol) CO2 in a humidified incubator. Media were supplemented with 10% (vol/vol) FCS (Invitrogen), 1% (vol/vol) penicillin/streptomycin (Gibco) 0.1% (vol/vol) G418 antibiotic (Carl Roth), and 0.1%(vol/vol) selective antibiotic Zeocin (Invitrogen).
siRNAs.
All siRNAs are chemically synthesized stealth siRNAs from Invitrogen. A pool of four different siRNA against 21 AlzGenes (three for CLU), 10 random genes (negative controls), three positive controls for transfection, silencing, and assay (APP, BACE1, and PEN2), Med-GC (transfection/silencing negative control) and KIF-11 (transfection positive control) were transfected into HeLa swAPP cells (Table S1).
siRNA Reverse Transfection.
Transfection complexes in quadruplicate were prepared in Opti-mem serum-free medium (Invitrogen) by mixing 0.3 μL of Oligofectamine (Invitrogen) and 5 nM of siRNA. HeLa sw APP cells at a density of 3,500 cells per well were seeded in 96-well format after the addition of transfection complexes.
Cell Proliferation Assay.
Sixty-nine hours (for sAPPβ) or 60 h (for Aβ38, Aβ40, or Aβ42) after siRNA transfection and subsequently analyzed with an alamar blue cell proliferation assay (AbD Serotec BUF012B) using a Plate reader with excitation at 544 nm and emission at 590 nm (Molecular Devices Spectramax Gemini XS) according to the manufacturer’s recommended protocol.
SDS/PAGE and Immunoblotting.
For detection of intracellular proteins, whole cell extracts were prepared using a lysis buffer (1% Nonidet P-40 and 0.1% SDS) supplemented with proteinase inhibitors. Extracts were subjected to SDS/PAGE using precast gels (Invitrogen). In all cases, gel loading was normalized to total protein content in the cell extract (using BCA assay). Proteins were transferred onto nitrocellulose membranes, which were then blocked with PBS containing 5% (wt/vol) dry skim milk for at least 1 h at RT. The membranes were then incubated with primary antibody 6E10 (Covance, 1:5,000), followed by the appropriate HRP-conjugated secondary antibody for at least 1 h at RT. Both antibodies were diluted in 5% milk/PBS 0.05% Tween-20. Immunoblotted proteins were detected using an enhanced chemiluminescence kit (Pierce).
Electrochemiluminescence Assay.
MSD 96-well MULTI-ARRAY Human Multiplex Kits were used (Meso Scale Discovery) to measure the level of sAPPβ, Aβ38, Aβ40, and Aβ42. To obtain standards for the quantifications and to determination of assay detection limits, dilution series of the analytes (synthetic sAPPβ and Aβ1–38/40/42 peptides) were prepared and measured in triplicate. For sAPPβ and Aβ–38/40/42 determination, supernatants of KD samples were collected, cleared by centrifugation, and further processed according to the manufacturer's instructions. sAPPβ, Aβ38, Aβ40, and Aβ42 peptides were detected with a monoclonal antibody and quantified by electrochemiluminescence assay using a SECTOR Imager 6000 reader (Meso Scale Discovery). Electrochemiluminescence (ECL) readings were normalized to viability assay readings and relative to Med-GC (scrambled) values.
Plasmid Transfection.
HeLa sw APP cells were transfected with mock plasmid (pCDNA), or plasmids expressing WT Presenilin 1 (PS1), PS1L166P mutant, WT Presenilin2 (PS2), or PS2 N141I using Lipofectamine reagent (Invitrogen), according to the manufacturer’s protocol. Twenty-four hours after transfection, the medium was exchanged, and conditioned medium was collected for 12 h; this was analyzed for Aβ40 and Aβ42 using the Meso Scale Discovery ECL platform, as described in the previous section.
RT-PCR Analysis.
Seventy-two hours after transfection, RNA was prepared using the RNeasy Plus Mini kit (Qiagen catalog no. 74136). Purity of RNAs (A260/A280 and A260/A230) and concentration were measured using Nanodrop spectrophotometer. A 2-μg quantity of total RNA was used for reverse transcription with oligo-dT primer using SuperScript first-strand synthesis system for RT-PCR (Invitrogen) according to the manufacturer’s recommended protocol. Real-time primers were designed in a way that all spanned exon/exon boundaries on the cDNA. PCR was performed using validated primers (Microsynth) for the corresponding human genes (Table S1), iTaq SYBR Green Supermix with ROX (Bio-Rad) and 50 ng/μL cDNA by a 7900HT Fast Real-Time PCR system (Applied Biosystems).
Assays were performed in quadruplicate, and expression levels of genes were normalized against GAPDH controls. Levels of Med-GC cDNA as an internal control were normalized to GAPDH cDNA according to the ΔΔCt method.
siRNA Reverse Transfections for Epistasis Analysis.
Transfection complex containing the desired siRNAs mix were prepared in Opti-mem medium (Invitrogen) by mixing 0.3 μL Oligofectamine (Invitrogen) and 5 nM of each siRNA. HeLa-swAPP cells at a density of 3,500 cells/well were seeded in a 96-well plate after addition of transfection complexes.
Supplementary Material
Acknowledgments
The authors thank Jonas Ries for help with data analysis; Martin Schwab, Christoph Hock, Hanns Mohler, Jonas Ries, and Roger Nitsch for their valuable comments on the work; Harald Steiner and Alice Suelzen for providing the WT, PS1, and PS2 constructs; and G. Minakaki for help with Western blotting. This study was funded by the Velux Stiftung, a Swiss National Science Foundation grant (Project: Role of beta-secretase cleavage, 31003A_130755), and the National Center of Competence in Research program of the Swiss National Science Foundation. The authors acknowledge Alzforum (www.alzforum.org) and the AlzGene database as the sources for the risk genes tested here.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201632109/-/DCSupplemental.
References
- 1.Brookmeyer R, Johnson E, Ziegler-Graham K, Arrighi HM. Forecasting the global burden of Alzheimer's disease. Alzheimer's Dementia. 2007;3:186–191. doi: 10.1016/j.jalz.2007.04.381. [DOI] [PubMed] [Google Scholar]
- 2.Frisoni GB, Hampel H, O’Brien JT, Ritchie K, Winblad B. Revised criteria for Alzheimer’s disease: What are the lessons for clinicians? Lancet Neurol. 2011;10:598–601. doi: 10.1016/S1474-4422(11)70126-0. [DOI] [PubMed] [Google Scholar]
- 3.Ankarcrona M, Mangialasche F, Winblad B. Rethinking Alzheimer’s disease therapy: Are mitochondria the key? J Alzheimers Dis. 2010;20(Suppl 2):S579–S590. doi: 10.3233/JAD-2010-100327. [DOI] [PubMed] [Google Scholar]
- 4.Small SA, Gandy S. Sorting through the cell biology of Alzheimer’s disease: Intracellular pathways to pathogenesis. Neuron. 2006;52:15–31. doi: 10.1016/j.neuron.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mangialasche F, Solomon A, Winblad B, Mecocci P, Kivipelto M. Alzheimer’s disease: Clinical trials and drug development. Lancet Neurol. 2010;9:702–716. doi: 10.1016/S1474-4422(10)70119-8. [DOI] [PubMed] [Google Scholar]
- 6.De Strooper B. Proteases and proteolysis in Alzheimer disease: A multifactorial view on the disease process. Physiol Rev. 2010;90:465–494. doi: 10.1152/physrev.00023.2009. [DOI] [PubMed] [Google Scholar]
- 7.Rajendran L, Annaert W. Membrane trafficking pathways in Alzheimer’s disease. Traffic. 2012;13:759–770. doi: 10.1111/j.1600-0854.2012.01332.x. [DOI] [PubMed] [Google Scholar]
- 8.Pimplikar SW, Nixon RA, Robakis NK, Shen J, Tsai LH. Amyloid-independent mechanisms in Alzheimer’s disease pathogenesis. J Neurosci. 2010;30:14946–14954. doi: 10.1523/JNEUROSCI.4305-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herrup K. Reimagining Alzheimer’s disease—an age-based hypothesis. J Neurosci. 2010;30:16755–16762. doi: 10.1523/JNEUROSCI.4521-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vassar R, Kovacs DM, Yan R, Wong PC. The beta-secretase enzyme BACE in health and Alzheimer’s disease: Regulation, cell biology, function, and therapeutic potential. J Neurosci. 2009;29:12787–12794. doi: 10.1523/JNEUROSCI.3657-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thinakaran G, Koo EH. Amyloid precursor protein trafficking, processing, and function. J Biol Chem. 2008;283:29615–29619. doi: 10.1074/jbc.R800019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spasic D, Annaert W. Building gamma-secretase: The bits and pieces. J Cell Sci. 2008;121:413–420. doi: 10.1242/jcs.015255. [DOI] [PubMed] [Google Scholar]
- 13.Borchelt DR, et al. Familial Alzheimer’s disease-linked presenilin 1 variants elevate Abeta1-42/1-40 ratio in vitro and in vivo. Neuron. 1996;17:1005–1013. doi: 10.1016/s0896-6273(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 14.Hardy JA, Higgins GA. Alzheimer’s disease: The amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 15.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: The AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 16.Harold D, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hollingworth P, et al. Alzheimer’s Disease Neuroimaging Initiative CHARGE Consortium EADI1 Consortium Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duff K, et al. Increased amyloid-beta42(43) in brains of mice expressing mutant presenilin 1. Nature. 1996;383:710–713. doi: 10.1038/383710a0. [DOI] [PubMed] [Google Scholar]
- 19.Rajendran L, et al. Efficient inhibition of the Alzheimer’s disease beta-secretase by membrane targeting. Science. 2008;320:520–523. doi: 10.1126/science.1156609. [DOI] [PubMed] [Google Scholar]
- 20.Rajendran L, et al. Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc Natl Acad Sci USA. 2006;103:11172–11177. doi: 10.1073/pnas.0603838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Strooper B, Vassar R, Golde T. The secretases: Enzymes with therapeutic potential in Alzheimer disease. Nat Rev Neurol. 2010;6:99–107. doi: 10.1038/nrneurol.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mi W, et al. Cystatin C inhibits amyloid-beta deposition in Alzheimer’s disease mouse models. Nat Genet. 2007;39:1440–1442. doi: 10.1038/ng.2007.29. [DOI] [PubMed] [Google Scholar]
- 23.Combarros O, Cortina-Borja M, Smith AD, Lehmann DJ. Epistasis in sporadic Alzheimer’s disease. Neurobiol Aging. 2009;30:1333–1349. doi: 10.1016/j.neurobiolaging.2007.11.027. [DOI] [PubMed] [Google Scholar]
- 24.Belbin O, et al. Investigation of 15 of the top candidate genes for late-onset Alzheimer’s disease. Hum Genet. 2011;129:273–282. doi: 10.1007/s00439-010-0924-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunter S, Friedland RP, Brayne C. Time for a change in the research paradigm for Alzheimer’s disease: The value of a chaotic matrix modeling approach. CNS Neurosci Ther. 2010;16:254–262. doi: 10.1111/j.1755-5949.2009.00117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Irizarry MC, et al. Modulation of A beta deposition in APP transgenic mice by an apolipoprotein E null background. Ann N Y Acad Sci. 2000;920:171–178. doi: 10.1111/j.1749-6632.2000.tb06919.x. [DOI] [PubMed] [Google Scholar]
- 27.DeMattos RB, et al. Clusterin promotes amyloid plaque formation and is critical for neuritic toxicity in a mouse model of Alzheimer’s disease. Proc Natl Acad Sci USA. 2002;99:10843–10848. doi: 10.1073/pnas.162228299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El Khoury J, et al. Ccr2 deficiency impairs microglial accumulation and accelerates progression of Alzheimer-like disease. Nat Med. 2007;13:432–438. doi: 10.1038/nm1555. [DOI] [PubMed] [Google Scholar]
- 29.Blennow K, Hampel H. CSF markers for incipient Alzheimer’s disease. Lancet Neurol. 2003;2:605–613. doi: 10.1016/s1474-4422(03)00530-1. [DOI] [PubMed] [Google Scholar]
- 30.Mawuenyega KG, et al. Decreased clearance of CNS beta-amyloid in Alzheimer’s disease. Science. 2010;330:1774. doi: 10.1126/science.1197623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selkoe DJ. Resolving controversies on the path to Alzheimer’s therapeutics. Nat Med. 2011;17:1060–1065. doi: 10.1038/nm.2460. [DOI] [PubMed] [Google Scholar]
- 32.Seabrook GR, Ray WJ, Shearman M, Hutton M. Beyond amyloid: The next generation of Alzheimer’s disease therapeutics. Mol Interv. 2007;7:261–270. doi: 10.1124/mi.7.5.8. [DOI] [PubMed] [Google Scholar]
- 33.Sperling RA, Jack CR, Jr, Aisen PS. Testing the right target and right drug at the right stage. Sci Trans Med. 2011;3:111–133. doi: 10.1126/scitranslmed.3002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider LS, Lahiri DK. The perils of Alzheimer’s drug development. Curr Alzheimer Res. 2009;6:77–78. doi: 10.2174/156720509787313871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambamurti K, et al. Targets for AD treatment: Conflicting messages from γ-secretase inhibitors. J Neurochem. 2011;117:359–374. doi: 10.1111/j.1471-4159.2011.07213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreira PI, Carvalho C, Zhu X, Smith MA, Perry G. Mitochondrial dysfunction is a trigger of Alzheimer’s disease pathophysiology. Biochim Biophys Acta. 2010;1802:2–10. doi: 10.1016/j.bbadis.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 37.Treusch S, et al. Functional links between Aβ toxicity, endocytic trafficking, and Alzheimer’s disease risk factors in yeast. Science. 2011;334:1241–1245. doi: 10.1126/science.1213210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.