Abstract
Combining synthetic biology and materials science will enable more advanced studies of cellular regulatory processes, in addition to facilitating therapeutic applications of engineered gene networks. One approach is to couple genetic inducers into biomaterials, thereby generating 3D microenvironments that are capable of controlling intrinsic and extrinsic cellular events. Here, we have engineered biomaterials to present the genetic inducer, IPTG, with different modes of activating genetic circuits in vitro and in vivo. Gene circuits were activated in materials with IPTG embedded within the scaffold walls or chemically linked to the matrix. In addition, systemic applications of IPTG were used to induce genetic circuits in cells encapsulated into materials and implanted in vivo. The flexibility of modifying biomaterials with genetic inducers allows for patterned placement of these inducers that can be used to generate distinct patterns of gene expression. Together, these genetically interactive materials can be used to characterize genetic circuits in environments that more closely mimic cells’ natural 3D settings, to better explore complex cell–matrix and cell–cell interactions, and to facilitate therapeutic applications of synthetic biology.
The complexity of cell signaling can be simplified by considering genetic networks composed of subsets of simpler parts, or modules. This simplification is the foundation of synthetic biology, where engineering paradigms are applied in rational and systematic ways to produce predictable and robust systems for understanding mechanisms of cellular function (1–6). The majority of work in synthetic biology has been in simple organisms, such as yeast and bacteria. However, as synthetic biology starts to expand to mammalian systems, it becomes increasingly more important to consider the environment in which the cells are grown. Biomaterials will play an important role in advancing synthetic biology within mammalian systems, because they provide highly controllable and tunable microenvironments where cells can behave as they do in vivo, in addition to organizing and delivering therapeutic cells to locations of interest. Our results show that interfacing synthetic biology and biomaterials can catalyze synthetic biology applications through engineered biomaterials that actively control genetic circuits in 3D scaffolds to more closely mimic the cells’ natural settings, in addition to providing mechanisms for translating synthetic biology for clinical applications (Fig. S1).
Biomaterials provide 3D environments for cell growth and have rapidly advanced our ability to investigate the coordinated interactions of many cellular phenomena because biomaterials recapitulate the in vivo setting better than traditional 2D cultures, where cells are grown in monolayers (7, 8). Physiological development, homeostasis, and regeneration each require a complex interplay of multiple signals that originate from the extracellular matrix (ECM) and from the intrinsic cellular control of gene products (9). While biomaterials provide a 3D environment characteristic of in vivo settings, mimicking and controlling this interactive environment is challenging, due to the dynamic nature and timing of gene expression during complex cellular events. One way to improve our ability to recreate aspects of the cellular niche is by coupling synthetic biology to biomaterials, thereby endowing the materials with the ability to regulate genetic circuits and dynamic gene expression patterns. These biomaterials will facilitate more advanced studies of complex intracellular networks, in addition to advancing clinical applications of synthetic biology. The therapeutic potential of cells carrying engineered genetic circuits has been seen through strategies employed to remediate or control diseases, such as type II diabetes (10), to promote biofilm degradation (5), and to treat cancer (11, 12). These strategies are based upon the tenet that effective cell therapies will require precise temporal and spatial regulation of gene expression, which may be controlled using synthetic gene circuits. One significant clinical constraint to translating synthetic biology is the survival, maintenance, and delivery of these therapeutic cells in the body. Another challenge is synthetic circuit robustness because performance consistency and long-term stability of cellular function in vivo is required for successful outcomes (13). One potential avenue for translating synthetic gene circuits to practical applications is by integrating synthetic biology with biomaterials. In this approach, the material is engineered to present a genetic inducer to cells with the synthetic gene circuits grown and cultured within the material. This approach provides additional levels of gene control because various materials provide unique techniques to release inducers providing flexibility of inducer exposure.
In addition to providing a path for translating synthetic biology, coupling biomaterials to synthetic biology could provide a novel way to create defined cellular microenvironments for sophisticated in vitro studies of cell behavior. Biomaterials can be engineered to create highly defined cellular microenvironments with multiple physical forms. The type of material used has significant application-dependent experimental and clinical relevance because cells gain distinct morphological and lineage characteristics when grown in different materials (14, 15). Examples of such scaffolds include electrospun fibers comprised of PCL [poly(γ-caprolactone)] sponges made from PLGA [poly(lactic-co-glycolic acid)] and hydrogels made from PEG [poly(ethylene glycol)]. For example, electrospun PCL fibrous scaffolds contain thin rod-like fibers that provide nanoscale textures for cells that can be randomly oriented or aligned in an orderly fashion (16). These attributes are important in many experimental and therapeutic applications. For instance, studying the effects of cell polarity, or in engineered tissues where alignment of cells is essential, such as the fibrous tissues of the musculoskeletal system (e.g., ligaments, tendons, and knee menisci) (17). By comparison, PLGA sponges are well-suited for bone tissue engineering because they provide a hard and highly porous microstructure with interconnected pores that is conducive to tissue in-growth (18, 19). In contrast, the soft porous hydrophilic environment of PEG hydrogels is advantageous for applications in soft tissue repair (20). Moreover, all of these materials can be chemically modified to further expand their applications for creating cellular niches that extend to other experimental and clinical approaches (21, 22), making them ideal for incorporating genetic inducers.
Results
IPTG Diffuses Through Various Materials and Activates Genetic Circuits in 3D.
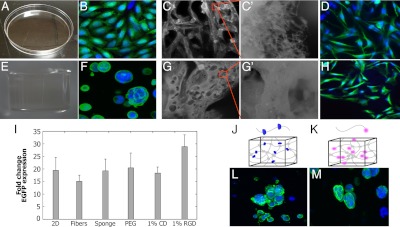
As an initial assessment of the applicability of coupling synthetic biology and biomaterials, the behavior of the mammalian genetic switch, LTRi_EGFP (23) was characterized in PCL fibers, PLGA sponges, and PEG hydrogels. LTRi encodes a combination of transcriptional repressors and RNAi inhibitors that enables dose dependent regulation of gene expression via the lactose analog, IPTG (isopropyl β-D-1-thiogalactopyranoside). Establishing IPTG efficacy in each class of material is important because each has its own application-specific attributes. Consistent with previous observations (23), we find that adding 500 uM of IPTG to the media of CHO cells stably transfected with LTRi_EGFP activates EGFP expression in 2D culture (Fig. 1A). More significantly, 3D cultures comprised of PCL electrospun fibers (Fig. 1C), PEG hydrogels (Fig. 1E), or PLGA sponges (Fig. 1G) allow for IPTG diffusion to occur resulting in activated EGFP expression. When quantified by Q-PCR, EGFP transcription is induced 20× over the control cells exposed to 250 pM IPTG (Fig. 1I). Fold induction was consistent between these different materials, despite the cells acquiring different morphologic characteristics on each material (Fig. 1 D, F, and H).
Fig. 1.
IPTG diffuses through biomaterials to activate genetic circuits in vitro. CHO cells stably transfected with LTRi_EGFP in the presence of 500 uM IPTG in the growth media. Green is EGFP immunofluorescence, and blue is DAPI nuclear staining. (A) Flat 2D tissue culture plate. (B) Confocal image of cells grown in 2D tissue culture plates. (C) Electrospun PCL fibers and (C′) their micro-scale texture. (D) Confocal image of cells grown on electrospun fibers. (E) PEG hydrogel. (F) Confocal image of cells growing in PEG hydrogels. (G) PLGA sponges and their (G′) porous architecture. (H) Confocal imaging of cells grown on sponges. (I) RT PCR of EGFP expression in the different materials with 500 uM IPTG in the media. Gene expression is compared to 250 pM IPTG induction in the media for each material. All expression levels were normalized to GAPDH as the reference gene. (J) Schematic of PEG functionalized with CD. (K) Schematic of PEG functionalized with RGD. (L) Confocal image of cells growing in 1% CD. (M) Confocal image of cells growing in 1% RGD. In all conditions, cells were analyzed 3 d after IPTG induction. Each error bar represents the mean of EGFP expression in at least four independent experiments. In this and all other figures, error bars represent standard deviations.
Modifying Biomaterials Can Increase Activation of Genetic Circuits.
An important attribute of these three classes of biomaterials is that they can be further modified with chemical or biological cues to create highly defined engineered niches for cells to grow. Examples of two such modifications include the incorporation of chemical functional groups and biological groups, such as peptides or proteins. To determine whether these modifications influence the performance of genetic circuits, CHO cells stably transfected with LTRi_EGFP were encapsulated in PEG hydrogels modified with α-cyclodextrin (CD) or RGD. α-CD is a six-membered sugar ring that complexes with the PEG chains (Fig. 1J) (24), adding hydrophobicity and functional moieties (25). PEG hydrogels containing 1% CD promote cell aggregation and clustering (Fig. 1L); however, these changes in cellular characteristics do not affect LTRi_EGFP function because EGFP activation is similar to cells grown in unmodified PEG (Fig. 1I). Another common modification is based upon the observation that cells require integrin receptors to bind ECM ligands for cell attachment, which influences and regulates cell migration, growth, differentiation, and apoptosis. One of these ligands is the RGD (R, arginine; G, glycine; and D, aspartic acid) peptide sequence that is an integrin-binding domain present in fibronectin and can be readily attached to PEG chains (Fig. 1K) (26, 27). In this case, LTRi_EGFP activation is enhanced by PEG functionalized with 1% RGD. EGFP expression in these cells is significantly higher than those grown in unmodified PEG (Fig. 1I), while morphological changes are minor (Fig. 1M). This may be a result of integrin activation, which has been shown to induce a vast number of structural and signaling changes within the cell, leading to changes in gene expression (28–32). Overall, we find that commonly used biomaterials provide a supportive 3D environment for cells harboring genetic circuits to grow, and that some modifications to the materials can be tuned to enhance sensitivity of the transcription response to IPTG. Together, these results suggest that materials can provide a 3D environment to characterize genetic circuits in settings that mimic cells’ natural milieu and facilitate cell–cell and cell–matrix interactions that can be engineered to increase gene products in the genetic circuits, in addition to providing a vehicle to deliver a significant number of cells with engineered genetic circuits for therapeutic modalities.
PLGA-IPTG Sponges.
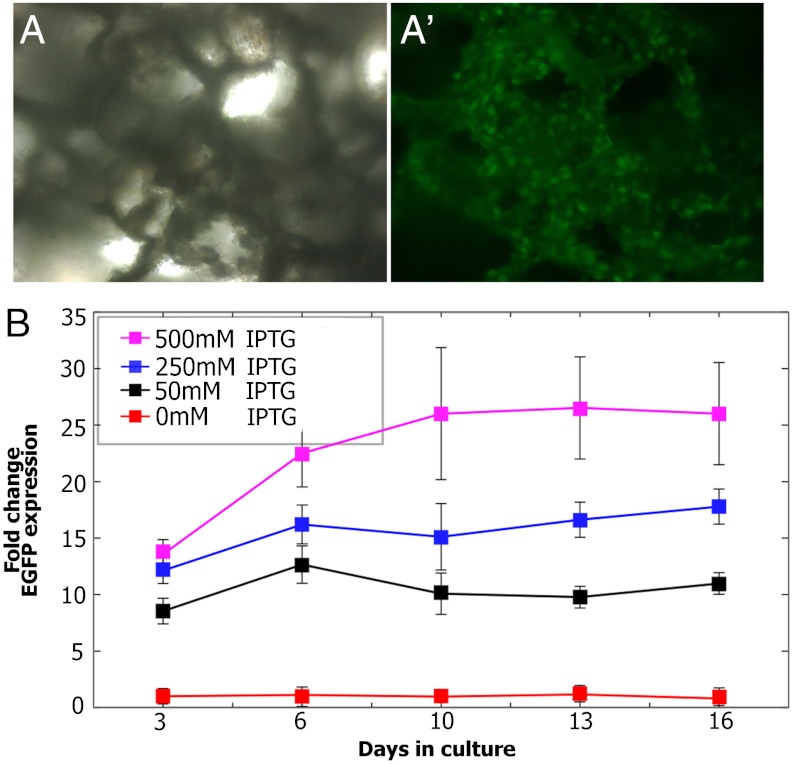
We propose that biomaterials may provide a venue to locally incorporate genetic inducers and, therefore, have the ability to dictate spatial and temporal control of gene expression. As an initial test of this hypothesis, IPTG was incorporated into porous PLGA sponges through a process that enables it to become trapped within the PLGA scaffolding. As the PLGA scaffold slowly degrades over time in culture, IPTG is released, allowing cellular uptake and activation of gene expression by the surrounding cells. The passive release of a genetic inducer from porous scaffolds has similarly been used to deliver the inducer, RSL1, from poly(ester urethane) urea (PEUU) films to overlying cells in culture (33). In contrast to RSL1, which is hydrophobic, IPTG is hydrophilic, soluble, and more amenable to delivery in culture and in vivo. HPLC analysis was used to measure the concentrations of IPTG loaded into sponges during processing and remained available to activate genetic circuits (Fig. S2). When tested after 6 d of culturing on PLGA-IPTG sponges, CHO cells stably transfected with LTRi_EGFP expressed robust levels of EGFP (Fig. 2A′). Long-term release of IPTG was assayed by quantifying EGFP expression using Q-PCR over 13 d of culture. Gene expression in response to IPTG was measured as fold induction over cells grown on PLGA sponges alone, with 250 pM IPTG added to the media. Results show that following an initial burst of IPTG release, there was a slower, sustained release of IPTG that was maintained for the subsequent days in culture (Fig. 2B). It is significant to note that the dose-dependent, tunable response of LTRi_EGFP is maintained in PLGA-IPTG sponges, with graded transcriptional responses to the different amount of IPTG loaded into the sponges. Furthermore, PLGA-IPTG is able to maintain regulation of this dose-dependent response throughout the entire course of the experiment.
Fig. 2.
PLGA-IPTG sponges. CHO cells stably transfected with LTRi_EGFP in the presence of 500 uM IPTG in the growth media. (A) Bright field image of PLGA-IPTG sponge seeded with CHO cells stably transfected with LTRi_EGFP. (A′) Fluorescent image of PLGA-IPTG sponge seeded with CHO cells stably transfected with LTRi_EGFP 6 d after seeding. (B) Q-PCR of EGFP expression in the PLGA-IPTG sponges. Gene expression of CHO cells stably transfected with LTRi_EGFP in PLGA-IPTG sponges is compared to 250 pM IPTG induction in the media. All expression levels were normalized to GAPDH as the reference gene.
PEG-IPTG Hydrogels Offer Spatial Control of Gene Expression.
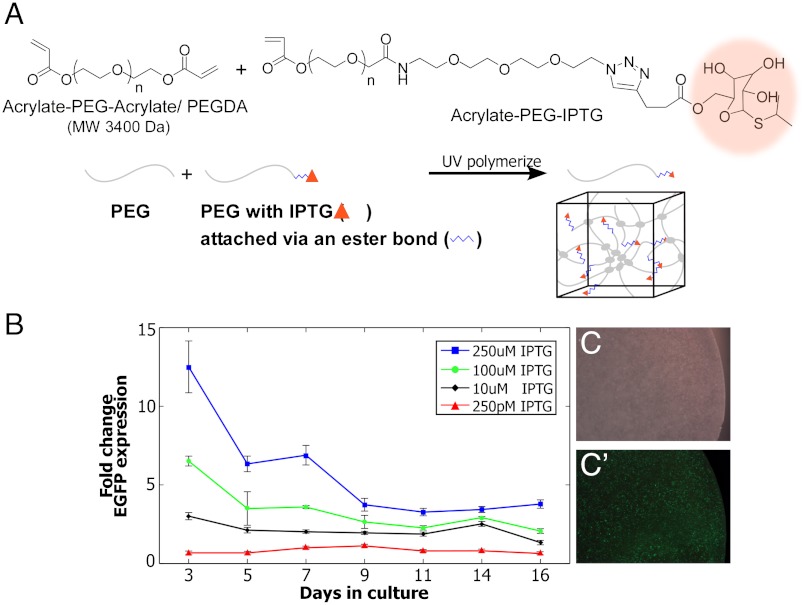
Coupling genetic inducers to biomaterials will advance the PLGA (Fig. 2) and PEUU (33) technologies and broaden the applications of synthetic biology by also enabling the spatial control of genetic circuits within a wide range of environments. To demonstrate this, we engineered PEG hydrogels to control the release of IPTG to cells with genetic circuits. Click chemistry, a versatile and compatible method to chemically functionalize materials, was employed to covalently link IPTG to the hydrogel network (34). IPTG was attached to the PEG chains via an ester bond to generate PEG-IPTG hydrogels (Fig. 3A and Fig. S3). An important aspect of the resulting ester bond linkage is that, over time, it undergoes hydrolysis, thereby locally releasing IPTG to the entrapped cells. Following 4 d of culture, EGFP expression was readily detected in CHO cells stably transfected with LTRi_EGFP (Fig. 3 C and C′). Long-term release of IPTG was assayed by quantifying EGFP expression using Q-PCR over 16 d of culture. Gene expression in response to IPTG was measured as fold induction over cells grown in PEG alone with 250 pM IPTG added to the media. Results are similar to the PLGA-IPTG results, showing an initial burst of IPTG release, followed by a slower sustained release of IPTG that was maintained for the subsequent days (Fig. 3B). Likewise, the dose-dependent, tunable response of LTRi_EGFP is maintained in PEG-IPTG hydrogels, with graded transcriptional responses to 10 uM, 100 uM, and 250 uM IPTG, and the PEG-IPTG material is able of regulating this dose-dependent response throughout the entire course of the experiment.
Fig. 3.
PEG-IPTG hydrogels. (A) IPTG (highlighted orange) was chemically attached to PEG chains via an ester bond using click chemistry. (B) Q-PCR of EGFP expression in the PEG-IPTG hydrogels. Gene expression of CHO cells stably transfected with LTRi_EGFP in PEG-IPTG hydrogels is compared to 250 pM IPTG induction in the media. All expression levels were normalized to GAPDH as the reference gene. (C) Bright field image of CHO cells stably transfected with LTRi_EGFP 3 d after encapsulation in PEG-IPTG (250 uM IPTG) gels. (C′) Fluorescent image of (C).
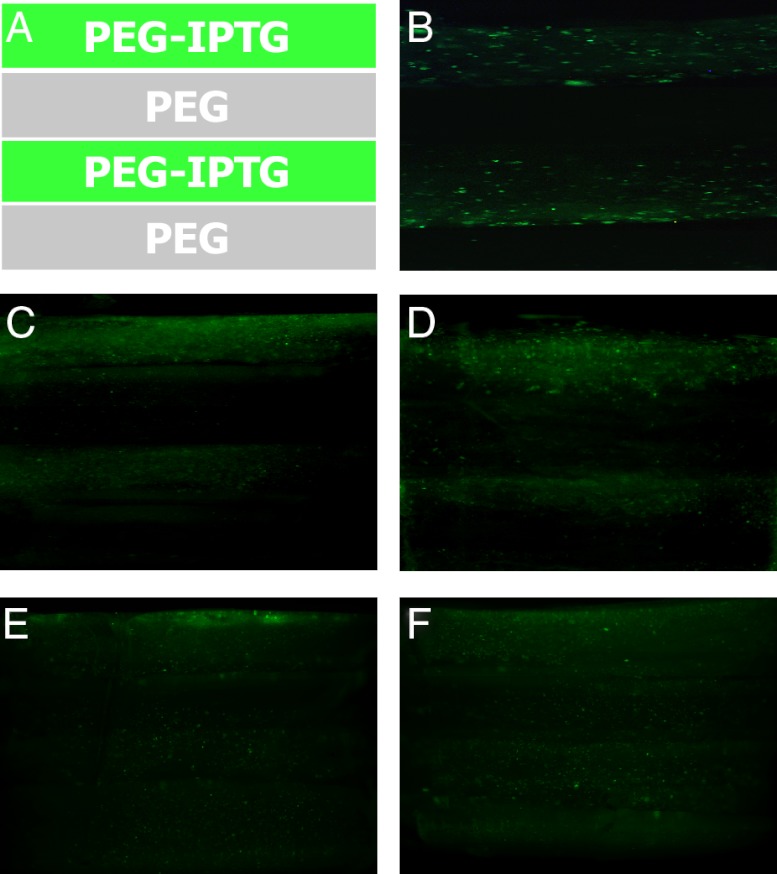
Using click chemistry to attach IPTG to PEG hydrogels allows for photo-patterning that enables distinct patterns of gene expression throughout the scaffold material. To demonstrate this, we layered PEG-IPTG (250 uM IPTG) and PEG hydrogels, each containing CHO cells stably transfected with LTRi_EGFP to spatially regulate EGFP expression (Fig. 4A). When viewed by fluorescent microscopy 3 d after encapsulation, endogenous EGFP fluorescence is restricted to layers containing PEG-IPTG and reveals the boundary between PEG and PEG-IPTG layers. Moreover, the spatial diffusion of IPTG following release from PEG is limited resulting in distinct borders, separating regions of fluorescent and nonfluorescent CHO cells. These fluorescent layers are maintained for approximately 10 d before decreasing in intensity and becoming less defined, albeit a region of nonactivated cells still remaining (Fig. 4 B–F). This reduction is likely due to IPTG diffusion between layers over time in culture.
Fig. 4.
Pattern formation using PEG-IPTG hydrogels. (A) Pattern formed using PEG-IPTG and PEG, all with CHO cells stably transfected with LTRi_EGFP. (B) 3 d after encapsulation. (C) 5 d after encapsulation. (D) 9 d after encapsulation. (E) 12 d after encapsulation. (F) 16 d after encapsulation.
In Vivo Induction of Genetic Circuits for Spatial and Temporal Control of Gene Expression.
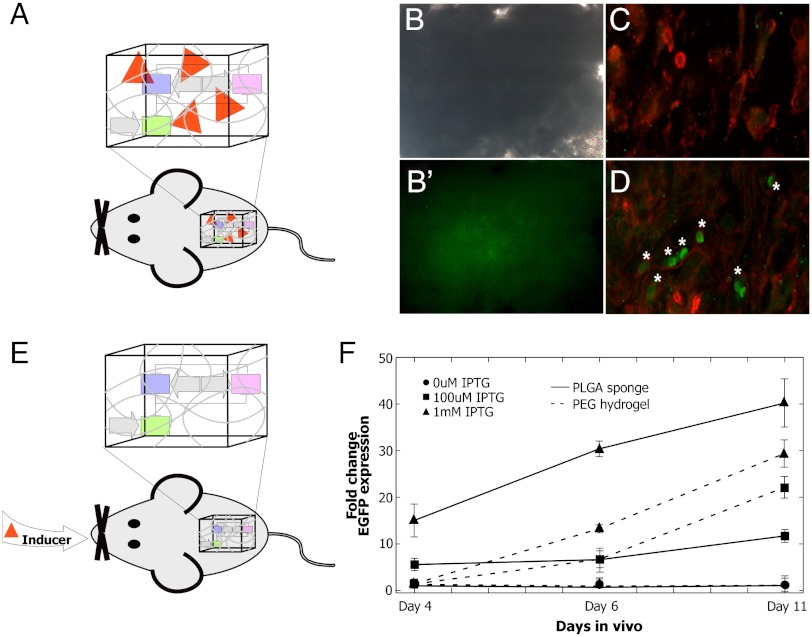
To translate our strategies of coupling synthetic biology and materials science, we investigated local (Fig. 5A) and systemic (Fig. 5E) induction of gene expression in vivo. For local spatial induction, PLGA-IPTG sponges containing CHO cells stably transfected with LTRi_EGFP were implanted subcutaneously into athymic mice (Fig. S4). The implants were removed 13 d after implantation, and EGFP expression was observed using confocal microscopy (Fig. 5D). Control implants consisted of PLGA sponges seeded with LTRi_EGFP CHO cells and showed no activation of EGFP expression (Fig. 5C). It has previously been reported that IPTG can be administered to animals to initiate gene activity in transgenic mice with no evidence of toxicity or lethality (35). More recently, Gitzinger et al. (36) show that injecting an analogous inducer, vanillic acid, in mice controls secreted alkaline phosphatase production in a synthetic circuit in vivo (36). For in vivo spatial and temporal induction, we sought to investigate whether we could activate and control EGFP expression by administering IPTG to the mice in their drinking water. IPTG is readily soluble in water so systemic administration is more practical experimentally and therapeutically than hydrophobic inducers, such as RSL1 or tamoxifen (33, 37). PLGA sponges and PEG hydrogels lacking IPTG but containing CHO cells stably transfected with LTRi_EGFP were implanted in the abdominal cavity of athymic mice. EGFP induction was initiated by the addition of IPTG to the drinking water. Following four, six, and 11 d of IPTG administration, EGFP expression was validated using fluorescent microscopy and quantitative PCR. Consistent with our previous in vitro results, the level of transgene expression was directly regulated by IPTG concentration and could be easily tuned by adding different amount of IPTG to the water (Fig. 5F). Importantly, control scaffolds (sponges and gels with CHO cells stably transfected with LTRi_EGFP and no IPTG in the water) demonstrated no EGFP induction during the course of the experiment (Fig. 5F).
Fig. 5.
Therapeutic applications of synthetic biology. (A) Local induction. PLGA sponges with and without IPTG containing CHO cells stably transfected with LTRi_EGFP were implanted subcutaneously of athymic mice. (B) Bright field image of sponges removed 13 d after implantation and (B′) EGFP expression was assessed using microscopy. (C) Confocal image of PLGA (no IPTG) sponge with CHO cells stably transfected with LTRi_EGFP immunolabeled with phalloidin and EGFP antibodies (D) Confocal image of PLGA-IPTG sponge with CHO cells stably transfected with LTRi_EGFP immunolabeled with phalloidin and EGFP antibodies. EGFP expressing cells (noted with *). (E) Systemic induction of genetic circuits in vivo. Sponges and hydrogels each containing CHO cells stably transfected with LTRi_EGFP were implanted subcutaneously or in the interperitoneum of athymic mice. Twenty-four h after the surgery, IPTG was added to their drinking water (0 mM, 100 uM, or 1 mM). The materials were removed four, six, and 11 d after initial introduction of IPTG to the drinking water. EGFP expression was assessed using RT PCR. All data was normalized to 250 pM IPTG induction in the media. All expression levels were normalized to the ribosomal protein, L19 as the reference gene.
Discussion
Altogether, these results demonstrate that integrating synthetic biology and materials science enable the production of biomaterials that are engineered to regulate the activity of genetic circuits carried by cells growing within the 3D matrix. As synthetic biology continues to expand into mammalian systems, the complex environment that mammalian cells naturally live in must be considered when designing and testing new circuits. Biomaterials provide a comprehensive toolbox for supporting cell and tissue growth that can be designed to deliver therapeutic cells that are also integrated into the environment in which they are placed in vivo. Employing these genetically interactive biomaterials will enable more comprehensive in vitro and in vivo models for understanding naturally occurring networks in higher organisms, studying cell–cell and cell–matrix interactions, and for the therapeutic control of genetic circuits in vivo (Fig. S1).
In this study, we introduce a versatile approach for activating genetic circuits by attaching the genetic inducer, IPTG, to several different classes of biomaterials. This versatility is important because different biomaterials promote distinct morphological and lineage characteristics that have significant application-dependent experimental and therapeutic relevance. Moreover, by including an inducible genetic circuit, these unique cellular differentiation outcomes can be further enhanced or tightly regulated by modulating the expression of downstream differentiation factors. In addition, the genetic inducer IPTG is already compatible with other sophisticated genetic circuits similar to LTRi that have been developed for reprogramming cells to perform desirable and predicable tasks (1, 38–40). Alternative genetic inducers have also been developed including vanillic acid and RSL1. Indeed, RSL1 has been delivered via passive diffusion from a material to initiate gene expression in cocultured cells. This raises the exciting potential that multiple genetic circuits activated by distinct chemical inducers could be combined within one material that could control various synthetic circuits with distinct dynamic patterns. One example is an AND-gate circuit in which gene expression would be turned on or off under specific well-defined biological circumstances. The result could be a genetically interactive matrix in which successive cellular differentiation events could be triggered in a spatial and/or temporal manner (33).
Conclusions
Several lines of evidence suggest that important developmental processes require dynamic control of gene expression (41–45). For example, during differentiation, it is unclear how long and how frequently a gene is actively transcribed; however, patterns of gene expression have been noted during various stages of development (46, 47). Coupling synthetic biology and materials science enables the engineering of biomaterials to generate niches that are capable of dynamic gene expression patterns, in addition to spatially and temporally controlling the expression of genes. Moreover, these engineered niches can provide an in vitro platform for understanding the mechanisms that regulate cellular phenomena, as well as the creation of relevant disease models that will enable the next generation of synthetic therapeutic technologies.
Materials and Methods
Cell Growth.
CHO cells stably transfected with LTRi_EGFP was previously reported (23). These cells were maintained in F12K medium containing 10% FBS and penicillin/streptomycin. For all experiments, the cells were grown in a 37 °C, humidified incubator with 5% CO2. Media changes were done every 2 to 3 d until harvesting. LTRi is commercially available from Origene (origene.com; cat #PS100060). See SI Materials and Methods and SI Text sections for details on the various biomaterials used, microscopy, and immunolabeling.
RNA Extraction and Quantitative Real-Time PCR (Q-PCR).
Total RNA was extracted and cDNA was synthesized (SI Text). Gene-specific primer sets are listed (Table S1).
Quantifying IPTG Loading Efficiency in PLGA-IPTG Sponges.
After the final wash of PLGA-IPTG sponges, sponges were redissolved in 100 uL Hexafluoro-2-propanol, 1 mL of chloroform was added, followed by 1.5 mL water. This solution was vortexed and sat at room temperature for 10 min, followed by a quick slow spin. The aqueous layer was removed and 1 mL water was readded to the organic mix. These steps were repeated two times. The final aqueous solution was lyophilized and prepared for HPLC analysis. Further details are available in SI Text.
Animal Experiments.
Mouse experiments were approved by the Johns Hopkins Animal Care and Use Committee. Animals were anesthetized with 3% isoflurane and received 0.05 mg/kg of intraperitoneal buprenorphine perioperatively. CHO cells stably transfected with LTRi_EGFP were either encapsulated in PEGDA hydrogels or seeded onto PLGA sponges the day before surgery. Hydrogels and sponges were implanted either subcutaneously into the dorsal region or in the intraperitoneum of six- to eight-wk-old male athymic nude mice for the specified amount of time. Harvested constructs were processed for imaging and/or RT-PCR assays. For the systemic induction experiments, the water bottles were changed in all cages and contained the specified amount of IPTG (0 uM IPTG, 100 uM IPTG, 1 mM IPTG) the day after surgery.
Supplementary Material
ACKNOWLEDGMENTS.
The authors thank M. Deans for comments on early versions of this manuscript and the members of the Elisseeff Laboratory for discussions. Funding was provided by the Maryland Technology Development Corporation (TEDCO) Stem Cell Research Fund for postdoctoral fellows (T.L.D. and A.S.) and the Jules Stein Professorship (J.H.E.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1204705109/-/DCSupplemental.
References
- 1.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 2.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 3.Becskei A, Serrano L. Engineering stability in gene networks by autoregulation. Nature. 2000;405:590–593. doi: 10.1038/35014651. [DOI] [PubMed] [Google Scholar]
- 4.O’Shaughnessy EC, Palani S, Collins JJ, Sarkar CA. Tunable signal processing in synthetic MAP kinase cascades. Cell. 2011;144:119–131. doi: 10.1016/j.cell.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu TK, Collins JJ. Dispersing biofilms with engineered enzymatic bacteriophage. Proc Natl Acad Sci USA. 2007;104:11197–11202. doi: 10.1073/pnas.0704624104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen YY, Jensen MC, Smolke CD. Genetic control of mammalian T-cell proliferation with synthetic RNA regulatory systems. Proc Natl Acad Sci USA. 2010;107:8531–8536. doi: 10.1073/pnas.1001721107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 8.Legant WR, et al. Measurement of mechanical tractions exerted by cells in 3D matrices. Nat Methods. 2010;7:969–971. doi: 10.1038/nmeth.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stevens MM, George JH. Exploring and engineering the cell surface interface. Science. 2005;310:1135–1138. doi: 10.1126/science.1106587. [DOI] [PubMed] [Google Scholar]
- 10.Ye H, Daoud-El Baba M, Peng RW, Fussenegger M. A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science. 2011;332:1565–1568. doi: 10.1126/science.1203535. [DOI] [PubMed] [Google Scholar]
- 11.Anderson JC, Clarke EJ, Arkin AP, Voigt CA. Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol. 2006;355:619–627. doi: 10.1016/j.jmb.2005.10.076. [DOI] [PubMed] [Google Scholar]
- 12.Xiang S, Fruehauf J, Li CJ. Short hairpin RNA-expressing bacteria elicit RNA interference in mammals. Nat Biotechnol. 2006;24:697–702. doi: 10.1038/nbt1211. [DOI] [PubMed] [Google Scholar]
- 13.Chen YY, Smolke CD. From DNA to targeted therapeutics: Bringing synthetic biology to the clinic. Sci Transl Med. 2011;3:106ps142. doi: 10.1126/scitranslmed.3002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 15.Fu J, et al. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat Methods. 2011;7:733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li WJ, Mauck RL, Cooper JA, Yuan X, Tuan RS. Engineering controllable anisotropy in electrospun biodegradable nanofibrous scaffolds for musculoskeletal tissue engineering. J Biomech. 2007;40:1686–1693. doi: 10.1016/j.jbiomech.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mauck RL, et al. Engineering on the straight and narrow: The mechanics of nanofibrous assemblies for fiber-reinforced tissue regeneration. Tissue Eng Part B Rev. 2009;15:171–193. doi: 10.1089/ten.teb.2008.0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karp JM, Shoichet MS, Davies JE. Bone formation on 2D poly(DL-lactide-co-glycolide) (PLGA) films and 3D PLGA tissue engineering scaffolds in vitro. J Biomed Mater Res A. 2003;64:388–396. doi: 10.1002/jbm.a.10420. [DOI] [PubMed] [Google Scholar]
- 19.Ochi K, et al. Use of isolated mature osteoblasts in abundance acts as desired-shaped bone regeneration in combination with a modified poly-DL-lactic-co-glycolic acid (PLGA)-collagen sponge. J Cell Physiol. 2003;194:45–53. doi: 10.1002/jcp.10185. [DOI] [PubMed] [Google Scholar]
- 20.Hillel AT, et al. Photo-activated composite biomaterial for soft tissue restoration in rodents and in humans. Sci Transl Med. 2011;3:93ra67. doi: 10.1126/scitranslmed.3002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deans TL, Elisseeff JH. Stem cells in musculoskeletal engineered tissue. Curr Opin Biotechnol. 2009;20:537–544. doi: 10.1016/j.copbio.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 22.Wang DA, et al. Multifunctional chondroitin sulphate for cartilage tissue-biomaterial integration. Nat Mater. 2007;6:385–392. doi: 10.1038/nmat1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deans TL, Cantor CR, Collins JJ. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell. 2007;130:363–372. doi: 10.1016/j.cell.2007.05.045. [DOI] [PubMed] [Google Scholar]
- 24.Miyake K, et al. Formation process of cyclodextrin necklace-analysis of hydrogen bonding on a molecular level. J Am Chem Soc. 2003;125:5080–5085. doi: 10.1021/ja026224u. [DOI] [PubMed] [Google Scholar]
- 25.Li J, et al. Self-assembled supramolecular hydrogels formed by biodegradable PEO-PHB-PEO triblock copolymers and alpha-cyclodextrin for controlled drug delivery. Biomaterials. 2006;27:4132–4140. doi: 10.1016/j.biomaterials.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 26.Liu JC, Heilshorn SC, Tirrell DA. Comparative cell response to artificial extracellular matrix proteins containing the RGD and CS5 cell-binding domains. Biomacromolecules. 2004;5:497–504. doi: 10.1021/bm034340z. [DOI] [PubMed] [Google Scholar]
- 27.Burdick JA, Anseth KS. Photo-encapsulation of osteoblasts in injectable RGD-modified PEG hydrogels for bone tissue engineering. Biomaterials. 2002;23:4315–4323. doi: 10.1016/s0142-9612(02)00176-x. [DOI] [PubMed] [Google Scholar]
- 28.Legate KR, Wickstrom SA, Fassler R. Genetic and cell biological analysis of integrin outside-in signaling. Genes Dev. 2009;23:397–418. doi: 10.1101/gad.1758709. [DOI] [PubMed] [Google Scholar]
- 29.Han J, et al. Reconstructing and deconstructing agonist-induced activation of integrin alphaIIbbeta3. Curr Biol. 2006;16:1796–1806. doi: 10.1016/j.cub.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 30.Clark EA, Brugge JS. Integrins and signal transduction pathways: The road taken. Science. 1995;268:233–239. doi: 10.1126/science.7716514. [DOI] [PubMed] [Google Scholar]
- 31.Liu Tsang V, et al. Fabrication of 3D hepatic tissues by additive photo-patterning of cellular hydrogels. FASEB J. 2007;21:790–801. doi: 10.1096/fj.06-7117com. [DOI] [PubMed] [Google Scholar]
- 32.Chen AA, et al. Humanized mice with ectopic artificial liver tissues. Proc Natl Acad Sci USA. 2011;108:11842–11847. doi: 10.1073/pnas.1101791108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baraniak PR, et al. Spatial control of gene expression within a scaffold by localized inducer release. Biomaterials. 2011;32:3062–3071. doi: 10.1016/j.biomaterials.2010.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Photo-degradable hydrogels for dynamic tuning of physical and chemical properties. Science. 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cronin CA, Gluba W, Scrable H. The lac operator-repressor system is functional in the mouse. Genes Dev. 2001;15:1506–1517. doi: 10.1101/gad.892001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gitzinger M, et al. The food additive vanillic acid controls transgene expression in mammalian cells and mice. Nucleic Acids Res. 2011;40:e37. doi: 10.1093/nar/gkr1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Imai T, Jiang M, Chambon P, Metzger D. Impaired adipogenesis and lipolysis in the mouse upon selective ablation of the retinoid X receptor alpha mediated by a tamoxifen-inducible chimeric Cre recombinase (Cre-ERT2) in adipocytes. Proc Natl Acad Sci USA. 2001;98:224–228. doi: 10.1073/pnas.011528898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stricker J, et al. A fast, robust and tunable synthetic gene oscillator. Nature. 2008;456:516–519. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ellis T, Wang X, Collins JJ. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat Biotechnol. 2009;27:465–471. doi: 10.1038/nbt.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedland AE, et al. Synthetic gene networks that count. Science. 2009;324:1199–1202. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Handwerger S, Aronow B. Dynamic changes in gene expression during human trophoblast differentiation. Recent Prog Horm Res. 2003;58:263–281. doi: 10.1210/rp.58.1.263. [DOI] [PubMed] [Google Scholar]
- 42.Yokoyama S, et al. Dynamic gene expression of Lin-28 during embryonic development in mouse and chicken. Gene Expr Patterns. 2008;8:155–160. doi: 10.1016/j.gep.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han F, et al. Dynamic changes in expression of heme oxygenases in mouse heart and liver during hypoxia. Biochem Biophys Res Commun. 2005;338:653–659. doi: 10.1016/j.bbrc.2005.08.100. [DOI] [PubMed] [Google Scholar]
- 44.Delgado I, et al. Dynamic gene expression during the onset of myoblast differentiation in vitro. Genomics. 2003;82:109–121. doi: 10.1016/s0888-7543(03)00104-6. [DOI] [PubMed] [Google Scholar]
- 45.Chubb JR, Trcek T, Shenoy SM, Singer RH. Transcriptional pulsing of a developmental gene. Curr Biol. 2006;16:1018–1025. doi: 10.1016/j.cub.2006.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aulehla A, Pourquie O. Oscillating signaling pathways during embryonic development. Curr Opin Cell Biol. 2008;20:632–637. doi: 10.1016/j.ceb.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki N, Furusawa C, Kaneko K. Oscillatory protein expression dynamics endows stem cells with robust differentiation potential. PLoS One. 2011;6:e27232. doi: 10.1371/journal.pone.0027232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.