Abstract
The prototypical prokaryotic potassium channel KcsA alters its pore depending on the ambient potassium; at high potassium, it exists in a conductive form, and at low potassium, it collapses into a nonconductive structure with reduced ion occupancy. We present solid-state NMR studies of KcsA in which we test the hypothesis that an important channel-inactivation process, known as C-type inactivation, proceeds via a state similar to this collapsed state. We test this using an inactivation-resistant mutant E71A, and show that E71A is unable to collapse its pore at both low potassium and low pH, suggesting that the collapsed state is structurally similar to the inactivated state. We also show that E71A has a disordered selectivity filter. Using site-specific K+ titrations, we detect a local change at E71 that is coupled to channel collapse at low K+. To gain more insight into this change, we site specifically measure the chemical shift tensors of the side-chain carboxyls of E71 and its hydrogen bond partner D80, and use the tensors to assign protonation states to E71 and D80 at high K+ and neutral pH. Our measurements show that E71 is protonated at pH 7.5 and must have an unusually perturbed pKa (> 7.5) suggesting that the change at E71 is a structural rearrangement rather than a protonation event. The results offer new mechanistic insights into why the widely used mutant KcsA–E71A does not inactivate and establish the ambient K+ level as a means to populate the inactivated state of KcsA in a controlled way.
Keywords: membrane proteins, ion channel, chemical-shift anisotropy
Potassium channels constitute a highly conserved family of intrinsic membrane proteins that are responsible for the passage of K+ ions across membranes, and are involved in many important physiological functions. The molecular mechanisms by which these channels are regulated, specifically the control mechanisms by which they can be closed or inactivated, are of significant interest.
One such control mechanism is C-type inactivation, which is a K+- and voltage-dependent closure of the outer mouth of the channel after channel opening. C-type inactivation was first described as a millisecond timescale process in Shaker channels (1), and a similar process occurs in a wide range of bacterial potassium channels, including KcsA and KirBac, and in the mammalian channel hERG, albeit at different rates. Understanding the molecular basis for C-type inactivation in mammalian potassium channels is of significant clinical interest because of its direct relation to cardiac malfunction and the development of a variety of pharmaceuticals (2, 3). The inactivated state is a transiently populated kinetic intermediate in the K+ transport cycle and stabilizing it on the timescale of structural studies is a challenge. Identifying the molecular structure of this transient intermediate could yield mechanistic insight into the molecular details of inactivation. In this paper, we test the idea that the C-type inactivated state of the channel can be studied by lowering the ambient K+ concentration.
We conduct our studies on the prokaryotic K+ channel, KcsA, which is native to the soil bacteria Streptomyces lividans and bears striking similarities to mammalian potassium channels in terms of its ion selectivity and gating properties. Because of its experimental convenience and homology to key regions of mammalian channels, it has become a model system for mechanistic studies of potassium channels. KcsA alters the structure of its pore depending on the ambient potassium concentration; at high potassium, it exists in a conductive form, and at low potassium, it collapses into a nonconductive structure with lowered ion occupancy (4). The channel is gated by pH, and after pH-dependent activation, it inactivates in a K+- and voltage-dependent manner on the timescale of milliseconds to seconds (5).
The different conformations accessed as a function of K+ and pH illustrate the importance of conformational plasticity for channel function. NMR is a great tool to study conformational dynamics. Solid-state NMR offers the unique opportunity to study conformational dynamics of membrane proteins in a native bilayer environment and has recently been used to investigate mechanistic questions in systems like proton channels (6), members of the rhodopsin family (7), and potassium channels (8, 9). We began our studies of KcsA by reporting the structural and qualitative dynamic characterization of the two limiting states of the selectivity filter at high and low potassium, and measuring site-specific potassium affinities in the pore (9). In this paper, we focus on the low K+-induced “collapsed” state of KcsA.
Several experiments have suggested that the C-type inactivated state of KcsA may resemble the low K+ collapsed state. Our own measurements of the slow exchange rate (kf + kr < 500/s) of the collapsed state with respect to the conductive state was one indication of a possible equivalence because C-type inactivation is also slow and K+-dependent (9). Additionally, recent crystallographic studies of various KcsA mutants show that when the pH gate of the KcsA is artificially opened, the structure and ion occupancy of the filter resemble the low K+ collapsed state (10). We use a mutation of KcsA, E71A, which is known to be inactivation-resistant and has been extensively characterized by electrophysiology (11), together with our knowledge of the spectroscopic signatures of the wild-type collapsed state at low K+ to ask the question: Can the inactivation-resistant mutant E71A collapse its filter at low K+? If it can, then it is unlikely that the inactivated state resembles the low K+ state; but if it cannot, then this result, together with the slow exchange rate and the crystallographic results, makes a very good case for structural equivalence of the low K+ state and the inactivated state.
E71A is an important mutant of KcsA. E71 is part of an intricate hydrogen bond network involving the side chains of E71, D80, and W67, a water molecule, and also backbone atoms of G77 and Y78 (Fig. 1 and Table S1). Mutating residues in this network affects ion selectivity (12), gating (13), and tetramer stability (14). Because of suppressed inactivation, E71A is a “constitutively open” K+ channel, widely used for kinetic studies (15, 16). Despite its wide use, there is no clear mechanism in the literature explaining why E71A does not inactivate. Previous work using mutations and electrophysiology have suggested that the ionized E71 and D80 side chains in close proximity destabilize the conductive state of the pore (5). Crystallography, however, shows that the local structure and hydrogen bond environment are fairly similar between the conductive and collapsed/inactivated structures. At the resolution of the available crystal structures, the protonation states of E71 and D80 are tentative (Table S1). NMR studies of a KcsA–Kv1.3 chimera suggest that a proton is loaded onto the side chain E71 as the K+is lowered at neutral pH. (8). Electrostatic pKa calculations indicate that the side chain of E71 has an unusually high pKa, and at pH 7.5, the side chain of E71 must be protonated (17–19). Thus, there is an inconsistency in the literature regarding the protonation state of E71. In this paper, we use site-specific chemical shift tensor measurements to make a much less ambiguous measurement of the protonation state of E71 and its hydrogen bond partner, D80, which is a key piece of evidence to understand the mechanism of inactivation. We also characterize the structural dynamics of the E71A mutant and show how it is different from the wild-type channel.
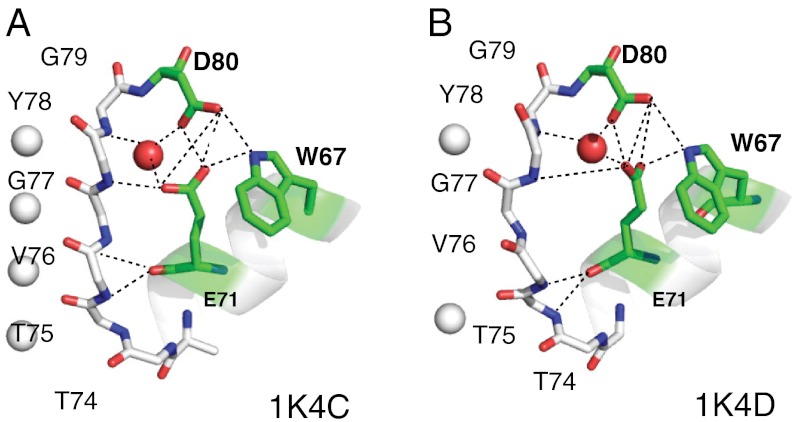
Fig. 1.
The network of hydrogen bonds that tether the selectivity filter to the pore helix and involve residues E71, D80, Y78, G77, T75, and W67 in KcsA are shown. (A) Shows the high K+, conductive state (PDB 1K4C). (B) Shows the collapsed, low K+ state (PDB 1K4D). Notice that although the E71–D80–W76 hydrogen bond network is generally maintained between the two states, the geometry of the network between E71 and D80 becomes more compact and the hydrogen bond Y78 N–E71 CO2 becomes much weaker at low potassium. A water molecule between E71 and D80 is included in the majority (approximately 80%) of all crystal structures of WT-KcsA with a resolution lower than 2.5 Å. Heavy-atom distances corresponding to the hydrogen bonds are listed in Table S1.
Our studies of wild-type and E71A KcsA lead to unique insights into the molecular control of C-type inactivation in KcsA. The results also illustrate the use of solid-state NMR methods to answer important mechanistic questions about membrane proteins while studying them in a native bilayer environment.
Results
E71A Does Not Collapse Its Filter at Low [K+] or Low pH.
We measured NMR chemical shifts of both wild-type KcsA and the mutant KcsA–E71A at two K+ levels, 25 mM K+ and 0.2 μM K+, at a neutral pH of 7.5. The channel was embedded in a native-like lipid bilayer composed of 9∶1 DOPE and DOPS lipids. Our results show that the inactivation-resistant mutant E71A is indeed unable to access the collapsed state of the channel, even at an ambient potassium level of as low as 0.2 μM, which is about an order of magnitude lower than the titration point for the WT filter collapse (9). This conclusion is supported by several previously characterized markers of filter collapse at various other sites in the filter including T74 and V76 (Fig. 2A). We also measured both the wild-type channel and the mutant E71A at high K+ (25 mM) and two pH conditions: pH 7.5 and pH 3.5. Because of the coupling of C-type inactivation with pH-dependent activation, some fraction of the selectivity filter in WT-KcsA is expected to exist in the inactivated state at low pH. This phenomenon has been described in recent NMR studies (20). Our spectra suggest that although the wild-type channel shows some signs of entering a collapsed-like state at T74 and V76, the mutant E71A remains completely in the conductive state even at pH 3.5 (Fig. 2B). The mutant channel’s inability to access a collapsed state at both low potassium and low pH conditions, together with our previously measured slow exchange rate (9), are strong evidence for the structural similarity between the collapsed state of the filter and the C-type inactivated state.
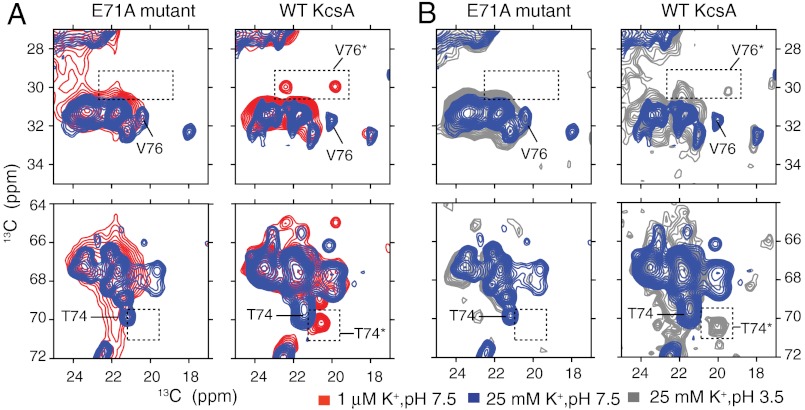
Fig. 2.
Sections of 13C–13C 2D spectra of both WT-KcsA and the inactivation-resistant mutant E71A are shown with key markers highlighted. Samples are in 9∶1 DOPE∶DOPS lipid bilayers and measured at 0–10 °C. (A) Spectra at pH 7.5 and two K+ concentrations, approximately 1 μM (red) and 25 mM (blue), show that the characteristic collapsed-state chemical shifts (V76* and T74*) are detected for the WT protein at low K+, but not detected for the mutant E71A at low ambient [K+], which instead remains conductive. The absence of the collapsed state for E71A and the presence for the WT channel is reproduced at several markers, including V76 Cβ-Cγ (30 ppm, 19.5 ppm) and T74 Cβ-Cγ (70.3 ppm, 20.5 ppm). (B) Spectra at high [K+] and two pH conditions, 3.5 (gray) and 7.5 (blue), are shown for both WT-KcsA and the mutant E71A. The WT low-pH spectra show some signature collapsed-state shifts suggesting that at low pH, a state similar to the collapsed state is sampled. These data show that, unlike the WT, the E71A filter does not collapse at low K+ or at low pH.
Disorder in Selectivity Filter of E71A–KcsA.
The mutant E71A exhibits structural heterogeneity in some parts of the filter at both high and low potassium. We have qualitatively characterized this heterogeneity by comparing spectra of wild-type KcsA at high potassium (25 mM K+, pH 7.5) with E71A–KcsA under the same conditions. As shown in Fig. 3, E71A exhibits more than one conformation at several sites, including W67, Y78, T74, and D80. In some cases, the alternative states of the amino acids can be observed as a second set of shifts through the entire spin system. Based on our assignments and chemical shift analysis (Fig. S1 and S2), these shifts most likely correspond to an alternate conformation of the filter that exists simultaneously in the sample and is in very slow exchange on the NMR timescale. The relatively narrow linewidth of the peaks indicates that the heterogeneity consists of at least two distinct conformations of the filter, rather than a broad continuum of states. The alternate conformation could potentially be the “flipped” structure of the filter that was observed in a small fraction of E71A crystals (21) (Fig. S2).
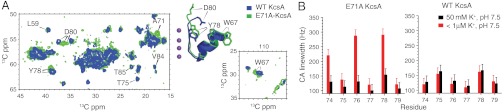
Fig. 3.
A summary of the linewidth and spectral quality of the KcsA mutant E71A at high and low potassium is shown. Samples are in 9∶1 DOPE∶DOPS lipid bilayers and measured at 0–10 °C. (A) The structural perturbation and heterogeneity observed at high potassium in a 2D correlation spectrum of the mutant (green) at sites W67, Y78, and D80 are shown and overlaid onto a WT-KcsA spectrum (blue) under the same conditions. The disordered and perturbed residues are mapped onto the structure of the KcsA filter with the conductive state (1K4C, blue) and the minor flipped conformation seen in some X-ray structures (like 2ATK, green). The narrow linewidth of marker peaks and other nearby residues indicates that at high K+, the mutant is correctly folded and the disorder likely results from two structurally distinct populations in a local region, and not a broad continuum of states. (B) An analysis of the Cα linewidths in the selectivity filter of wild-type KcsA and mutant E71A is shown. When [K+] is < 1 μM, residues in the wild-type filter shift but maintain a narrow NMR linewidth, whereas particular peaks in the mutant filter (Y78, V76, and T74) appear to be significantly broader. The data suggest that at low potassium, the selectivity filter in E71A exhibits significantly more disorder or dynamics than the wild-type filter.
The behavior of E71A at low potassium is very different from the wild-type channel, showing much broader Cα linewidths. Curiously, at high ambient potassium, both the mutant and the WT show comparably narrow linewidths (Fig. 3B), suggesting that the presence of potassium as a ligand is exceptionally important for the stability of E71A. Of course, the NMR linewidth in a uniformly labeled protein sample can be affected by a variety of technical issues like the static field strength, homonuclear decoupling efficiency, sample temperature and temperature gradients, sample hydration level, and shimming. These effects would be expected to affect all the residues and marker peaks, although the decoupling efficiency may be differential. In order to eliminate systematically some of these effects, all comparative linewidth spectra were acquired at the same sample temperature of approximately 0–10 °C. The datasets were acquired and processed identically so that parameters like the digital resolution and decoupling efficiency remained constant. The data were reproduced at two different static field strengths and the hydration level of the samples was maintained at 85% relative humidity using a constant humidity chamber.
The linewidth effects we report are associated specifically with some lines and not obviously related to spin system type. The affected sites are also not all in the selectivity filter. Therefore, these effects at low K+ are more likely caused by conformational disorder, either dynamic or static. In contrast to the disorder described for the high K+ state of E71A, which appears to be slow exchange based on lineshapes, the disorder in the low K+ state is compatible with millisecond to microsecond motions in that for each line observed, there is not a resolved pair of lines but rather a general broadening (Fig. 3 and Fig. S2). In both cases, the data suggest that the interactions between the E71 side chain and the selectivity filter provide a set of constraints on the conformation of the selectivity filter that are lost when E71 is mutated to an alanine.
Changes in E71 and D80 as a Function of [K+] in WT-KcsA.
Our results on the mutant E71A described above suggested that E71 plays an important role in controlling the channel’s ability to access the collapsed or inactivated state. Because this state is typically accessed at low K+, we measured the isotropic chemical shifts of E71 and D80 as a function of ambient potassium ranging from 0.2 μM to 50 mM at pH 7.5 (see Fig. S1 and Table S2 for the assignment of E71 and D80). The data, summarized in Fig. 4, show a significant upfield change from 182.5 ppm to 180.2 ppm in chemical shift for the E71 side-chain carboxyl atom as the potassium concentration is decreased from 10 μM to < 1 μM. The backbone carbon and nitrogen chemical shifts of E71 do not show significant change, and none of the D80 chemical shifts show significant changes. The NMR chemical shift is a sensitive indicator of local electronic environment and can report on both structural changes and changes in the electrostatic environments of atoms. Because the change in E71 is restricted to the terminal carboxyl group, it is very likely that it reports on a local change in the electronic environment of the E71 side-chain carboxyl.
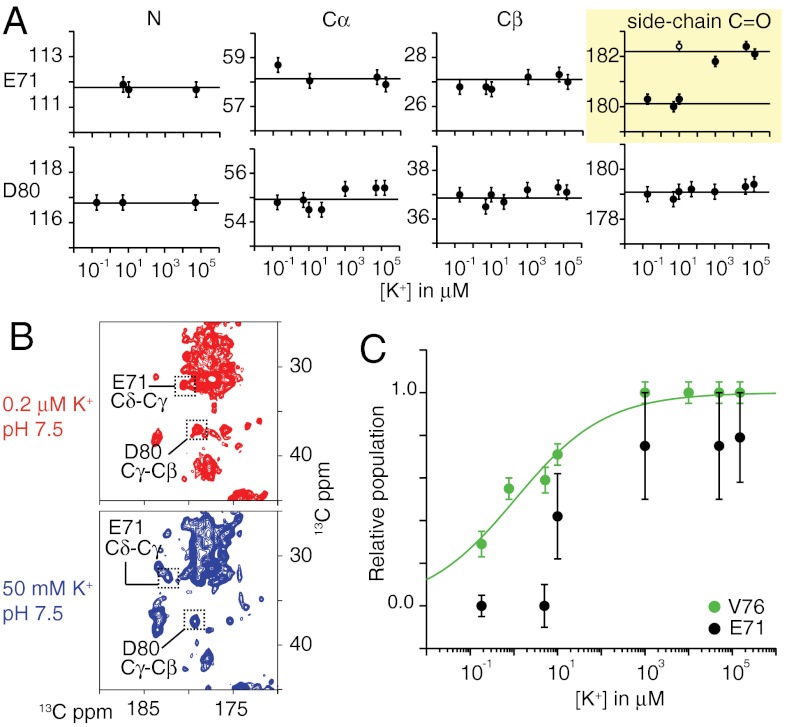
Fig. 4.
The potassium-dependent isotropic chemical shifts of E71 and D80 are shown. (A) The 13C chemical shifts of E71 (Top) and D80 (Bottom) in WT-KcsA are plotted as a function of ambient [K+] at pH 7.5. The data show a large downfield movement in the side-chain carboxyl chemical shift of E71 (in yellow) as [K+] is increased, but no change in the backbone shifts. At [K+] = 10 μM, both states of E71 are observed, indicated by the filled and hollow markers. The D80 isotropic shifts remain unchanged by potassium. (B) Shows 13C–13C correlation spectra highlighting the E71 and D80 carboxyl chemical shifts at the endpoints of the titration. (C) The relative population of the deprotonated state of E71 based on its NMR signal intensity is overlaid with populations from another selectivity filter marker, V76. The V76 curve (fit to a two-state binding model) shows that the transition from the conductive state to the collapsed state occurs between [K+] = 1 μM and 10 μM. The data show that change in the E71 side chain occurs in the same potassium-concentration regime as the structural collapse of the filter based on Kd measurements of selectivity filter markers. Therefore these two processes are very likely coupled.
The observed upfield shift in E71 also occurs when the ambient K+ level is changed from 1 μM to 10 μM, which is the same range in which several other marker peaks in the selectivity filter indicate collapse. Fig. 4B plots the relative population of the high K+ form of E71 observed at 182.5 ppm as a function of the K+ level, and compares it to our previously published curve documenting the structural collapse of the filter using the marker V76 (9). It is clear that the transitions in both of these curves occur in the same potassium-concentration regime, suggesting that the change in E71 is coupled to the slow, potassium-dependent structural collapse of the selectivity filter. The data also show that D80 remains unaffected through the collapsing process. The ionization states of E71 and D80 have been of some interest in computational studies (see above); therefore we pursued this question further.
The Protonation State of E71 and D80 in WT-KcsA.
As discussed in the Introduction, previous NMR studies have documented a similar upfield change in the chemical shift of the E71 side chain during conversion into the low K+ form, and interpreted it as the protonation of E71 (8). Their interpretation is based on typical isotropic chemical shifts of protonated and deprotonated carboxyls. However, detailed studies of hydrogen bonding in amino acid model compounds suggest that the 13C carboxyl isotropic shift can sometimes be a poor indicator of the actual protonation state of a carboxyl group, and that the anisotropic components of the chemical shift tensor are often much more sensitive to changes in the protonation state (22, 23). Because solid-state NMR offers us the unique ability to measure site-specific chemical shift tensors, we used slow magic-angle-spinning (MAS) experiments to extract site specifically the anisotropic components of chemical shift tensors of the E71 and D80 side-chain carboxyls.
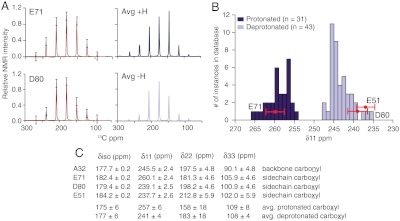
Fig. 5A shows the observed spinning sideband pattern for the E71 and the D80 side-chain carboxyl carbons at 25 mM K+ and a pH of 7.5. The extracted principal components of the chemical shift tensors are listed in Fig. 5C along with two control residues: the E51 carboxyl, which has a surface-exposed side chain that is definitely deprotonated at pH 7.5, and a standard backbone carbonyl of the α-helical residue A32 (Fig. S3). The tensor lineshapes of E71 and D80 are very different, which is already a clue that they are likely not in the same protonation state. To establish quantitatively the protonation state of E71 and D80, we analyzed the magnitude of the δ11 tensor component, which is typically oriented in the plane of carboxyl oxygens and is known to be one of the clearest indicators of protonation. A comparison of our values of the δ11 tensor component for E71, D80, and E51, to a distribution of values from a known database (22) of 43 deprotonated and 31 protonated carboxyls is shown in Fig. 5B. It is very clear that at high potassium and neutral pH, the E71 δ11 belongs to the protonated distribution, and the D80 and E51 δ11s belong to the deprotonated distribution. These distributions are clearly separated. Data collected at neutral pH and low potassium indicate that E71 and D80 retain the same protonation states at high and low potassium. As a control we also measured the tensor of the backbone carbonyl of the α-helical residue A32 and compared it to previously measured α-helical tensors (24). The agreement is good. In order to confirm that the shape of the E71 tensor was not a result of fast proton exchange in a low-barrier hydrogen bond, we simulated the effects of a fast-limit proton exchange on the sideband patterns of a typical carboxyl carbon. The fast exchange results are inconsistent with sideband patterns that we observe at E71 (Fig. S4) and with marked differences between the E71 and the D80 tensors.
Fig. 5.
A summary of the chemical-shift anisotropy measurements at E71 and D80 is shown. Measurements were made at 25 mM K+, pH 7.5, and temperature of 0–10 °C. (A) The experimentally derived sideband intensities from slow magic-angle spinning spectra (black) are shown together with the best fit from SPINEVOLUTION (red). The error bars represent the average signal:noise of the spectrum. (A, Right) The simulated tensors based on the average values of a protonated (dark) and deprotonated (light) carboxyl are shown. The principle components of the chemical shift tensors of E71, D80, and two other control residues (A32 and E51) were derived from these fits and are listed in C. The errors reflect the standard deviation in the values obtained from the fit. (B) Plots the distribution of the δ11 component from a previously published database (22) of protonated (dark) and deprotonated (light) carboxyl tensors of known crystal structure. Our measured δ11 values for E71, D80, and E51 are overlaid in red onto this distribution. (The y-offset of E51 is for clarity.) From the data it is very clear that E71 is protonated and D80 and E51 are both deprotonated at neutral pH.
Through solid-state NMR methods, we were able to adjudicate between prior NMR and theoretical assessments of the state and role of ionization in the selectivity filter. The chemical shift tensors of E71 and D80 clearly show that at high K+ and a pH of 7.5, E71 is protonated and D80 is deprotonated. Thus, E71 must have an extremely elevated pKa (> 7.5, instead of the expected value of approximately 4.5), which is consistent with predictions from various electrostatic and molecular dynamics simulations.
Discussion
Our data on the inactivation-resistant mutant E71A show that the collapsed state accessed at low K+ is indeed structurally similar to the C-type inactivated state of the channel. The C-type inactivated state is typically populated transiently for a few milliseconds to seconds after the pH gate is opened and is therefore difficult to study by equilibrium methods. We show that lowering the ambient K+ level is an experimentally feasible way of accessing the inactivated state. Controlling the K+ level can be used to modulate the relative populations of the inactivated state with respect to the conductive state, which facilitates future biophysical studies of this clinically important process. To gain more mechanistic insight, we determined the protonation state of E71 and its hydrogen bond partner, D80, and showed that E71 is protonated at pH 7.5 and therefore has an unexpectedly high pKa, whereas D80 is deprotonated at pH 7.5. The results are significant for both scientific and technical reasons. Since description of the first crystal structure of KcsA (25), there has been a lot of interest in the protonation state of E71 because of its proximity to the selectivity filter. A perturbed pKa for E71 had been predicted by multiple electrostatic simulations; specifically, Ranatunga et al. predicted a pKa of > 14.5 for E71 (17) and Bernache et al. also predicted a perturbation of approximately +10 pKa units for E71 (18). Our own simulations using the X-ray coordinates 1K4C and 1K4D and two different publicly available simulation programs—H ++ (26) and MCCE (27)—over a range of different protein dielectric constants also qualitatively support an elevated pKa for E71 (Fig. S5 and Table S3). The data in this paper are previously undescribed unambiguous experimental measurements that validate these predictions. We show that E71 is protonated at neutral pH and high K+.
The 2.3-ppm K+-dependent upfield change in the carboxyl chemical shift of E71, which occurs simultaneously with the structural collapse of the channel and had previously been interpreted as a protonation event (8), is therefore a result of a local structural change in the side chain of E71 upon K+ depletion and not protonation. Crystallography (Fig. 1 and Table S1) suggests that the E71–D80 interaction is generally conserved between the conductive and collapsed states: The carboxyl groups of E71 and D80 are engaged in a syn–syn interaction with each other and with a conserved water molecule in both states. However, the E71 carboxyl looses a key hydrogen bond with the Y78 backbone amide as the channel collapses. We also observe a change in the isotropic shift of the Y78 amide 15N at low K+, we suspect that the 2.3-ppm shift in E71 is reporting on the weakening/loss of a hydrogen bond to Y78 and on a minor structural rearrangement to compensate for the loss.
Our results help us understand why the mutant E71A fails to inactivate. Functional studies have shown that replacing E71 with a histidine accelerates C-type inactivation, but most other substitutions (including serine, valine, isoleucine, threonine, cysteine, and glutamine) reduce inactivation in KcsA (28). Thus, the suppression of C-type inactivation in E71A is unlikely to be an effect of smaller side-chain volume or a specific chemical effect of the alanine. Instead, our data show a significantly more disordered filter in the E71A mutant. Molecular dynamics simulations have also suggested that mutations at E71 can lead to increased dynamics in the filter (28). Crystallography on the E71A mutant has indicated disorder in the form of an alternative flipped conformation of the mutant filter (12). It is intriguing to consider whether the conductive state of the E71A mutant is preferentially stabilized by this disorder—i.e., the population of additional states entropically stabilizes the conductive state in E71A, and shifts the K+-dependent equilibrium between the conductive and inactivated/collapsed states into a regime that is beyond our detection (Kd ≪ 1 μM). Such a hypothesis would also explain how a variety of different mutations at E71 could all lead to suppression of C-type inactivation (to various extents), as long as they also lead to increased disorder.
Sequence alignment shows that a glutamic acid in position 71 is not conserved across all potassium channels. In other channels, it is often substituted by large nonpolar amino acids, like valine and isoleucine, and smaller polar amino acids, like serine (Fig. S6). Mutations at the position equivalent to E71 affect C-type inactivation in both hERG and Kv channels (29). We suspect that the manipulation of the contacts and hydrogen bonds at this position using different amino acids is a mechanism to tune the relative stability of the conductive state of the channel, and thereby allow for diversity in the kinetics of C-type inactivation across various channels.
From a technical standpoint, our results demonstrate the use of chemical-shift anisotropy to measure functionally important protonation states of side chains in a native membrane environment—a measurement that is unique to solid-state NMR. The magnitude of the chemical-shift anisotropy tensor is typically described by three parameters—δ11, δ22, and δ33—which measure the extent of the shielding in three orthogonal directions around the carboxyl carbon of interest, and are averaged to give the isotropic shift. The isotropic shift is the most commonly measured NMR observable, but in the case of the E71 side-chain carboxyl, an analysis based purely on its isotropic chemical shift leads to the wrong protonation state for this residue (Fig. S7). The unusually high isotropic chemical shift of the protonated E71 (approximately 182.5 ppm versus an average of approximately 179 ppm) is caused by an unusually high value for the δ22 component, which suggests a strong hydrogen bond acceptor positioned near the side-chain carbonyl, and is consistent with the positioning of the conserved crystallographic water near the E71 carbonyl. The strong hydrogen bond indicates that the protons of this water molecule are oriented towards the carbonyl of E71. Our data and simulations preclude the existence of a low-barrier hydrogen bond between E71 and D80, and suggest that the proton primarily resides near the carboxyl of E71 (Fig. S4). It has long been known from model compounds that the δ11 and δ22 elements of the chemical shift tensor are more reliable indicators of protonation than the isotropic shift. Here, we illustrate how such anisotropic measurements can make a big difference in the context of mechanistic studies of an important membrane protein.
Conclusion
In this paper, we use a two-pronged approach to investigate the role of a key residue, E71, for KcsA function. First, by using the mutant E71A we showed that the low K+-induced collapsed state of KcsA is structurally similar to the C-type inactivated state of the channel and that removing the E71 side chain leads to a disordered selectivity filter. Second, we use anisotropic chemical shifts to make a uniquely direct, conclusive measurement of the side-chain protonation states of E71 and D80 in KcsA and show that the pKa of E71 is indeed highly perturbed (> 7.5) compared to solution, as was predicted by electrostatic calculations. These results together provide mechanistic insights into the control of channel inactivation, which is an important physiological process.
Materials and Methods
NMR Sample Preparation.
The WT-KcsA and KcsA–E71A gene cloned into a PASK90 vector were overexpressed in Escherichia coli JM83 cells as described previously (9). The WT-KcsA tetramer is detected as a clean band at approximately 70 kDa in SDS gels at 25 °C. E71A–KcsA is denatured by SDS and runs as a band at approximately 18 kDa. The mutant E71A is unstable when stored in detergent at 4 °C and was therefore stored in liposomes at -80 °C and used as quickly as possible. The protein was reconstituted into 9∶1 wt/wt DOPE/DOPS liposomes as described previously (9). The ionic strength of the dialysis buffer was kept constant by replacing K+ with Na+. Low K+ concentrations were determined by atomic absorption spectroscopy. The hydration level of samples was maintained at 85% using a home-built hydration chamber. Samples were analyzed by cryo-EM to check for the bilayer structure and packed in Bruker 4-mm or 3.2-mm rotors.
NMR Spectroscopy.
NMR spectra were collected on two spectrometers: a Bruker Avance DRX-750 MHz spectrometer (with a 4-mm HXY probe spinning at 14 kHz and a 3.2-mm HXY probe spinning at 14 KHz) and a Bruker Avance 2 900 MHz spectrometer (with a 3.2-mm standard-bore E-free probe). Typical 90° pulse lengths for 1H were approximately 2.5 μs on the standard HXY probes and approximately 3 μs on the E-free probe. Spectra were collected at a spectrometer-set temperature of 240 K. At 14-KHz spinning, our calibration show that the actual sample temperature was 0–10 °C during signal acquisition. Chemical shift tensors were measured at 5.555-KHz magic-angle spinning and fit using SPINEVOLUTION (SI Text). All 13C chemical shifts were referenced externally to the adamantane line at 40.48 ppm. Using the recommended gyromagnetic ratio from the Biological Magnetic Resonance Data Bank, 15N shifts were referenced internally to the carbon shifts.
Supplementary Material
ACKNOWLEDGMENTS.
The authors thank the McDermott group, especially Wenbo Li, for help with troubleshooting SPINEVOLUTION; Ivan Sergeyev for help with installing MCCE; and Benjamin Wylie and Segolene Laage for helpful discussions. We also thank the laboratory of Crina Nimigean for the E71A plasmid, Marilyn Gunner for helpful advice about electrostatic simulations, and Dr. Boris Itin at the New York Structural Biology Center for support with high-field instrumentation. Professor McDermott is a member of the New York Structural Biology Center, a Strategically Targeted Academic Research (STAR) center supported by the New York State Office of Science, Technology, and Academic Research. This work was supported by grants from the National Institutes of Health: National Institutes of Health Grant P41 GM66354 supported NMR resources, and this work was supported by National Institutes of Health Grant R01 GM 88724.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211900109/-/DCSupplemental.
References
- 1.Starkus JG, Kuschel L, Rayner MD, Heinemann SH. Ion conduction through C-type inactivated Shaker channels. J Gen Physiol. 1997;110:539–550. doi: 10.1085/jgp.110.5.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanguinetti MC, Tristani-Firouzi M. hERG potassium channels and cardiac arrhythmia. Nature. 2006;440:463–469. doi: 10.1038/nature04710. [DOI] [PubMed] [Google Scholar]
- 3.Bowlby M, Peri R, Zhang H, Dunlop J. hERG (KCNH2 or Kv11.1) K+ channels: Screening for cardiac arrhythmia risk. Curr Drug Metab. 2008;9:965–970. doi: 10.2174/138920008786485083. [DOI] [PubMed] [Google Scholar]
- 4.Zhou Y, Morais-Cabral JH, Kaufman A, Mackinnon R. Chemistry of ion coordination and hydration revealed by a K channel Fab complex at 2.0 A resolution. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 5.Cordero-Morales JF, Cuello LG, Perozo E. Voltage-dependent gating at the KcsA selectivity filter. Nat Struct Mol Biol. 2006;13:319–322. doi: 10.1038/nsmb1070. [DOI] [PubMed] [Google Scholar]
- 6.Cady SD, et al. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature. 2010;463:689–692. doi: 10.1038/nature08722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Struts AV, Salgado GFJ, Brown MF. Solid-state 2H NMR relaxation illuminates functional dynamics of retinal cofactor in membrane activation of rhodopsin. Proc Natl Acad Sci USA. 2011;108:8263–8268. doi: 10.1073/pnas.1014692108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ader C, et al. Coupling of activation and inactivation gate in a K+ channel: Potassium and ligand sensitivity. EMBO J. 2009;28:2825–2834. doi: 10.1038/emboj.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhate MP, Wylie BJ, Tian L, McDermott AE. Conformational dynamics in the selectivity filter of KcsA monitored by solid-state NMR. J Mol Biol. 2010;401:155–166. doi: 10.1016/j.jmb.2010.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuello LG, Jogini V, Cortes DM, Perozo E. Structural mechanism of C-type inactivation in K+ channels. Nature. 2010;466:203–208. doi: 10.1038/nature09153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cordero-Morales JF, et al. Molecular driving forces determining potassium channel slow inactivation. Nat Struct Mol Biol. 2007;14:1062–1069. doi: 10.1038/nsmb1309. [DOI] [PubMed] [Google Scholar]
- 12.Cheng WWL, McCoy JG, Thompson AN, Nichols CG, Nimigean CM. Mechanism for selectivity-inactivation coupling in KcsA potassium channels. Proc Natl Acad Sci USA. 2011;108:5272–5277. doi: 10.1073/pnas.1014186108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cordero-Morales JF, Jogini V, Chakrapani S, Perozo E. A multipoint hydrogen-bond network underlying KcsA C-type inactivation. Biophys J. 2011;100:2387–2393. doi: 10.1016/j.bpj.2011.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi H, Heginbotham L. Functional influence of the pore helix glutamate in the KcsA K+ channel. Biophys J. 2004;86:2137–2144. doi: 10.1016/S0006-3495(04)74273-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson AN, Posson DJ, Parsa PV, Nimigean CM. Molecular mechanism of pH sensing in KcsA potassium channels. Proc Natl Acad Sci USA. 2008;105:6900–6905. doi: 10.1073/pnas.0800873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piasta KN, Theobald DL, Miller C. Potassium-selective block of barium permeation through single KcsA channels. J Gen Physiol. 2011;138:421–436. doi: 10.1085/jgp.201110684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ranatunga KM, Shrivastava IH, Smith GR, Sansom MS. Side-chain ionization states in a potassium channel. Biophys J. 2001;80:1210–1219. doi: 10.1016/S0006-3495(01)76097-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernèche S, Roux B. The ionization state and the conformation of Glu-71 in the KcsA K+ channel. Biophys J. 2002;82:772–80. doi: 10.1016/S0006-3495(02)75439-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bucher D, Guidoni L, Rothlisberger U. The protonation state of the Glu-71/Asp-80 residues in the KcsA potassium channel: A first-principles QM/MM molecular dynamics study. Biophys J. 2007;93:2315–2324. doi: 10.1529/biophysj.106.102509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imai S, Osawa M, Takeuchi K, Shimada I. Structural basis underlying the dual gate properties of KcsA. Proc Natl Acad Sci USA. 107:6216–6221. doi: 10.1073/pnas.0911270107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cordero-Morales JF, et al. Molecular determinants of gating at the potassium-channel selectivity filter. Nat Struct Mol Biol. 2006;13:311–318. doi: 10.1038/nsmb1069. [DOI] [PubMed] [Google Scholar]
- 22.Gu Z, Zambrano R, McDermott A. Hydrogen bonding of carboxyl groups in solid-state amino acids and peptides: Comparison of carbon chemical shielding, infrared frequencies, and structures. J Am Chem Soc. 1994;116:6368–6372. [Google Scholar]
- 23.Gu Z, McDermott A. Chemical shielding anisotropy of protonated and deprotonated carboxylates in amino acids. J Am Chem Soc. 1993;115:4282–4285. [Google Scholar]
- 24.Wylie BJ, et al. Chemical-shift anisotropy measurements of amide and carbonyl resonances in a microcrystalline protein with slow magic-angle spinning NMR spectroscopy. J Am Chem Soc. 2007;129:5318–5319. doi: 10.1021/ja0701199. [DOI] [PubMed] [Google Scholar]
- 25.Doyle D, Cabral J, Pfuetzner R, Kuo A, Gulbis J. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 26.Gordon J, et al. H ++: A server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 2005;33:W368–W371. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Georgescu RE, Alexov E, Gunner MR. Combining conformational flexibility and continuum electrostatics for calculating pKas in proteins. Biophys J. 2002;83:1731–1748. doi: 10.1016/S0006-3495(02)73940-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chakrapani S, et al. On the structural basis of modal gating behavior in K+ channels. Nat Struct Mol Biol. 2010;18:67–74. doi: 10.1038/nsmb.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferrer T, et al. Molecular coupling in the human ether-a-go–go-related gene-1 (hERG1) K+ channel inactivation pathway. J Biol Chem. 2011;286:39091–39099. doi: 10.1074/jbc.M111.292060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.