Abstract
An essential aspect of innate immunity is recognition of molecular patterns on the surface of pathogens or altered self through the lectin and classical pathways, two of the three well-established activation pathways of the complement system. This recognition causes activation of the MASP-2 or the C1s serine proteases followed by cleavage of the protein C4. Here we present the crystal structures of the 203-kDa human C4 and the 245-kDa C4⋅MASP-2 substrate⋅enzyme complex. When C4 binds to MASP-2, substantial conformational changes in C4 are induced, and its scissile bond region becomes ordered and inserted into the protease catalytic site in a manner canonical to serine proteases. In MASP-2, an exosite located within the CCP domains recognizes the C4 C345C domain 60 Å from the scissile bond. Mutations in C4 and MASP-2 residues at the C345C–CCP interface inhibit the intermolecular interaction and C4 cleavage. The possible assembly of the huge in vivo enzyme–substrate complex consisting of glycan-bound mannan-binding lectin, MASP-2, and C4 is discussed. Our own and prior functional data suggest that C1s in the classical pathway of complement activated by, e.g., antigen–antibody complexes, also recognizes the C4 C345C domain through a CCP exosite. Our results provide a unified structural framework for understanding the early and essential step of C4 cleavage in the elimination of pathogens and altered self through two major pathways of complement activation.
Keywords: crystallography, pattern recognition, proteolysis, structural biology
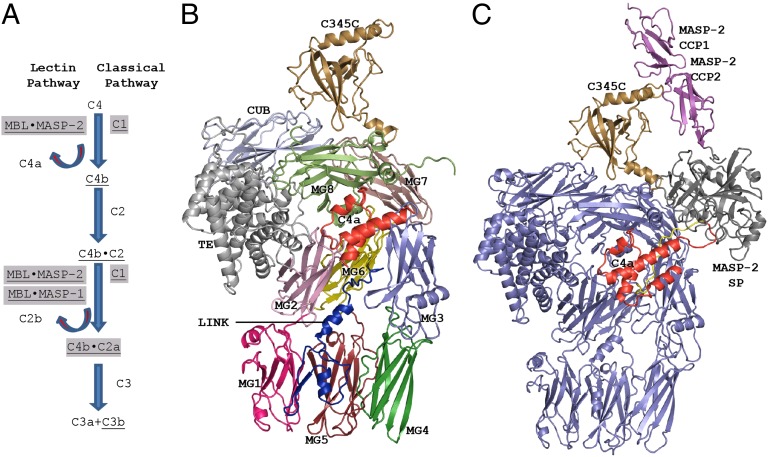
The ability of pattern-recognition molecules to bind foreign markers such as pathogen-associated molecular patterns is central to the innate immune defense. One such defense mechanism is complement, which is capable of recognizing molecular patterns associated with microbes and apoptotic or necrotic cells. Recognition causes activation of proteolytic enzyme cascades, resulting in cleavage of the complement proteins C3, C4, and C5. Fragments of these proteins have important effector functions through binding to host cell receptors and pathogen surfaces (1). In the lectin pathway of complement, four pattern recognition molecules, mannan-binding lectin (MBL) or H-, L-, or M-ficolin, may bind to surface-linked carbohydrates or acetyl groups on the surface of pathogens or damaged self-tissue (Fig. 1A) (2). The CL-11 protein is also a putative pattern recognition molecule acting in the lectin pathway (3). Pattern recognition leads to activation of the pattern recognition molecule-associated MASP-2 protease (4), and this enzyme then cleaves the 203-kDa protein C4 into the fragments C4a and C4b. The nascent C4b fragment becomes covalently linked through its reactive thioester (TE) to the surface bearing the pattern recognized (5). C4b recruits the zymogen C2, and subsequent C2 cleavage leads to formation of the surface-anchored and proteolytically active C3 convertase C4b⋅C2a, but C4b also contributes to immune clearance through interaction with the CR1 receptor (6). The classical pathway of complement, in which the MASP-2 paralogue C1s cleaves both C4 and C2, is initiated upon C1q recognition of, e.g., antigen–antibody complexes (7) and likewise results in C4b deposition and assembly of the C3 cleaving C4b⋅C2a complex on the surface recognized (Fig. 1A). Such C3 cleavage leads to alternative pathway amplification and, subsequently, cleavage of the C5 protein as well (8). The downstream outcome of complement activation through the lectin and classical pathways is therefore C3 and C5 fragments, which have important effector functions through binding to host cell receptors and pathogen surfaces. The activation of complement receptors ultimately elicits inflammatory responses directing immune cells and molecules to the point of infection, tagging of pathogens for phagocytosis, lysis of pathogens, and stimulation of the adaptive immune response (1).
Fig. 1.
Role of C4 in the complement system, the structure of C4, and its complex with MASP-2. (A) The proteolytic cascade starting upon pattern recognition by MBL or ficolins results in deposition of C4b and ultimately C3b. Active proteolytic enzymes are shown in gray boxes, and surface-associated proteins/complexes are underlined. Within the C1 complex, C1q recognizes the pattern leading to activation of the associated C1r and C1s SPs, but only C1s cleaves C4 and C2. (B) The structure of unbound C4 with the domains individually colored; see also Fig. S2A for a schematic representation. (C) Structure of the C4⋅MASP-2 complex. Except for C4a (red) and the C345C domain (brown), C4 is shown in blue. The MASP2 CCP domains (magenta) interact with the C4 C345C domain, whereas the catalytic SP domain (gray) recognizes C4 at the scissile bond region.
Results
Complete MBL⋅MASP-2 or ficolin⋅MASP-2 complexes bound to an activating carbohydrate structure and C4, to our knowledge, have never been reconstituted in a format suitable for crystallization. Instead, we used a MASP-2 fragment comprising the CCP1, CCP2, and the serine protease (SP) domains (9). By mutating the MASP-2 catalytic site serine 633 to alanine, we could crystallize the C4⋅MASP-2 complex, and in addition we crystallized C4 alone. We then determined the crystal structures of C4 and the enzyme–substrate complex at 3.6- and 3.75-Å resolution, respectively (Table S1, Fig. 1 B and C, and Fig. S1). Structure determination was promoted by the presence of two C4⋅MASP-2 complexes in the crystal, the known structure of the active MASP-2 fragment (10), and the ability to compare models of C4 either unbound or bound to MASP-2. As a result, the Rfree values of the structures are 0.27 and 0.24 for unbound C4 and C4⋅MASP-2, respectively (Table S1). C4 exists in two isoforms C4A and C4B, differing within six residues, and the vast majority of individuals express both isoforms (11). No attempt was made to separate the isoforms, and we assume that both structures contain a mixture of C4A and C4B (Figs. S1A and S2A). The structures revealed that C4 is structurally similar to its paralogues C3 and C5 (12–14), with six N-terminal MG domains (MG1–6) forming an irregular superhelical arrangement, the β-ring (Fig. 1B). The TE, MG8, and CUB domains form the tightly packed α-chain superdomain. The C4 TE bond formed between Cys-1010 and Gln-1,013 is buried between the MG8 domain and the α-helical TE domain as in C3 (12, 14). The C4a domain is wedged between the β-ring and the α-chain superdomain, whereas the MG7 and the C345C domains are located at the α-chain superdomain periphery (Fig. 1B).
MASP-2 Interactions with the Scissile C4 Region.
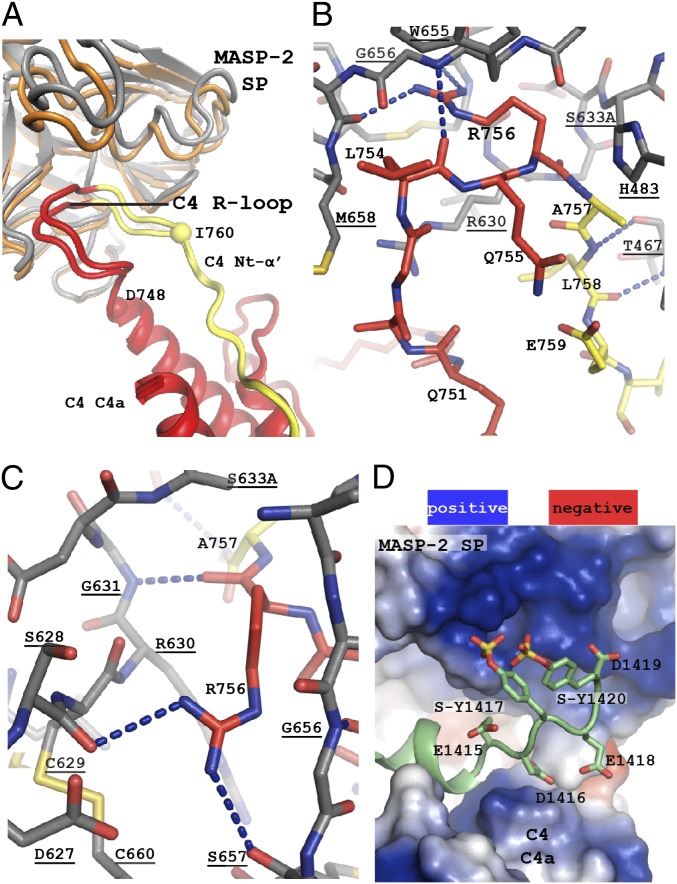
All three MASP-2 domains are in contact with C4, yielding an overall intermolecular interface of 1,800 Å2 (Fig. 1C and Fig. S2 C and D). The CCP domains contact the C4 C345C domain with an approximate interface area of 500 Å2. The MASP-2 SP domain interacts with an extended loop comprising C4 residues Asp-748–Ile-760 (Fig. 2 A–C). We shall refer to this loop as the R loop, which comprises the scissile bond region P2–P1–P1′–P2′ (Gln-755–Arg-756–Ala-757–Leu-758). In addition, the SP domain forms electrostatic interaction with the C4 sulfotyrosine region (see below) and a few contacts with the anchor region connecting the C4 MG8 and the C345C domains. The total interface area between the SP domain and C4 is ∼1,300 Å2. The SP domain loops A, B, E, D, 3, and 2, named according to ref. 10, interact with C4 (Fig. S2C). Whereas the R loop is bound identically to the SP domain in the two copies of the C4⋅MASP-2 complex in our crystal, there is a slight difference of the orientation of the SP domain relative to C4 residues outside the R loop (Fig. 2A). This flexibility is made possible by the extended R-loop conformation and makes one complex tighter and with better electron density for the SP domain; we describe this complex below. Because of the resolution of the C4⋅MASP-2 structure, hydrogen bonds and salt bridges mentioned below should be considered putative, although the presence of two copies of the C4⋅MASP-2 complex result in a better effective resolution than the nominal 3.75 Å.
Fig. 2.
The MASP-2 SP domain recognizes the C4 R loop and the SO3–Tyr region. Blue dotted lines indicate putative hydrogen bonds, and ionic interactions and MASP-2 labels are underlined. (A) Superposition of C4a (red) from the two complexes reveals a variation in the orientation of the SP domain (gray, tight; orange, open) made possible by the flexible R loop (red until Arg-756; yellow after Arg-756). Spheres at C4 Asp-748 and Ile-760 mark the R-loop boundaries. The Nt-α′ region comprises Ala-757–Arg-775. (B) Details of the interaction between the R loop (red and yellow carbon atoms) and the MASP-2 SP domain (gray carbon atoms). (C) The P1 Arg-756 side chain in the S1 pocket of MASP-2 and the Arg-756 carbonyl in the oxyanion hole. (D) The negatively charged SO3–Tyr region in C4 held between positively charged (blue) surface patches on the MASP-2 SP and the C4a domains.
The C4 R loop has the conformation of a twisted U, with a bend at C4 Gly-750–Gln-751 caused by van der Waals interactions between C4 Ala-752–Leu-754 and Met-658 in loop 2 of MASP-2 (Fig. 2B and Figs. S1B and S3A). The P1 arginine side chain is inserted into the MASP-2 S1 pocket and is held by hydrogen bonds to MASP-2 Ser-628 and -657 and an electrostatic interaction with Asp-627. The main chain oxygen of Arg-756 is placed in the oxyanion hole formed by the main-chain N–H of MASP-2 Gly-631 and Ser-633–Ala (Fig. 2C). In silico mutation of MASP-2 Ala-633 to serine in our structure suggests that the serine side chain from the catalytic triad can come within 2.8 Å of the P1 carbonyl C atom. P1′–P4′ residues Ala-757–Ile-760 adopt an extended conformation stabilized by putative main chain hydrogen bonds with MASP-2 Thr-467 in loop A and Gly-656. In a number of serpin–protease Michaelis complexes (15), likewise with their P1 side chain accommodated into the S1 site, the conformation of their P2–P2′ residues is rather similar to that of P2–P2′ residues in C4⋅MASP-2 (Fig. S3D). Furthermore, the main-chain conformations of C4 Gln-755–Glu-759 and residues Thr-29–Cys-33 in the MASP-2 inhibitor SGMI-2 bound to MASP-2 (16) strongly resemble each other (Fig. S3E), most likely due to the P1 residue carbonyl interaction with the oxyanion hole and main-chain hydrogen bonds between the P2′ residue and MASP-2 Thr-467 in both cases.
MASP-2 Exosites.
Within residues 1,405–1,427 located at the C terminus of the C4 α-chain are three sulfotyrosines (Fig. S2D), and together with seven glutamates/aspartates and the terminal carboxyl group, these provide 11 negative charges to this region. Residues 1,415–1,420 fold into an extended region sandwiched between a large, well-conserved, positively charged surface patch on the MASP-2 SP domain and the likewise positively charged C4a domain (Fig. 2D). The SO3 group of C4 Tyr-1,417 interacts electrostatically with MASP-2 Lys-503 (Fig. S1C), whereas C4 Asp-1419 faces MASP-2 Lys-450, Arg-578, and Arg-583. In the opposite direction, C4 Asp-1,416 is directed toward Lys-744 in C4a (Fig. 2D). These long-range electrostatic interactions suggest that the C4 sulfotyrosine region acts as flexible electrostatic “Velcro” between MASP-2 and the C4a domain.
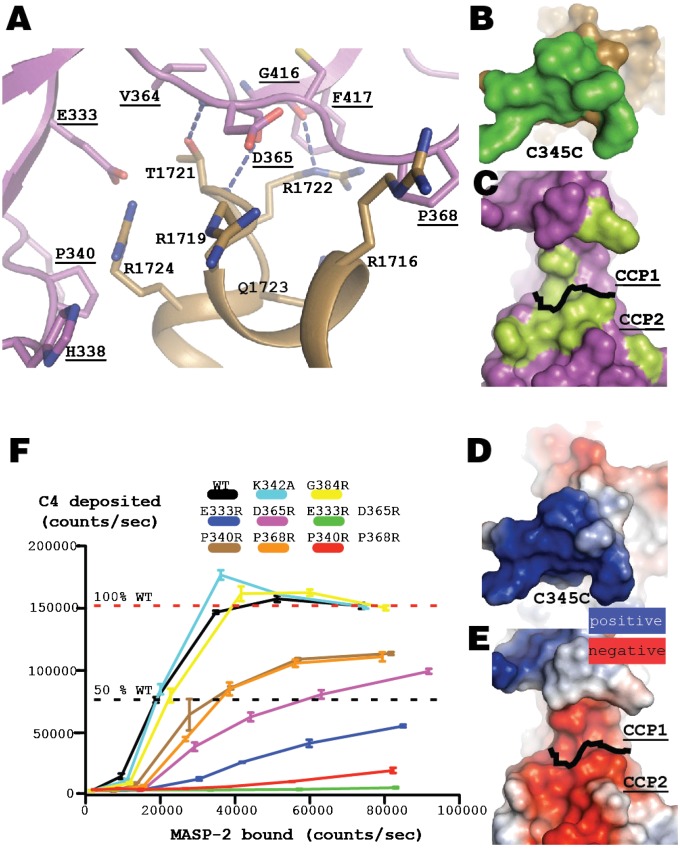
The CCP domains in both MASP-2 (9, 17, 18) and C1s (19, 20) have earlier been suggested to be important for C4 cleavage, but the regions of C4 possibly interacting with the CCP domains have never been identified. Our C4⋅MASP-2 structure reveals the MASP-2 regions forming what we will refer to as the CCP exosite. Conserved residues from both CCP domains provide an open negatively charged binding patch for four arginines within the C4 region 1,716–1,725 (Fig. 3 A–E and Fig. S2D) located immediately before the large C-terminal helix of the C345C domain. MASP-2 CCP1 Glu-333—strictly conserved in C1s and MASP-2—forms long-range electrostatic interactions with C4 Arg-1724. The main chain of MASP-2 Asp-365 interacts with Thr-1,721, and the aspartate side chain is facing C4 residues Arg-1,716, -1,719, and -1,724 (Fig. 3A). Overall, the intermolecular interactions formed by MASP-2 with both the C4 C345C domain and the sulfotyrosine region appear to be dominated by electrostatic forces and hydrogen bonds. In the C4 paralogue C3, the region equivalent to the MASP-2 interacting C4 region 1,716–1,725 is negatively charged, suggesting that electrostatic repulsion would prevent CCP exosite recognition of C3. Together with the C4-specific sulfotyrosine region, this feature could explain the strong preference for C4 over C3 as MASP-2 substrate (9).
Fig. 3.
Electrostatic interactions are important for C4–CCP exosite interactions. (A) Residues in the CCP–C4 interface. Dotted blue lines indicate putative hydrogen bonds or electrostatic interactions, and MASP-2 labels are underlined. (B) C4 C345C residues contacting MASP-2 (within 3.8 Å; green) mapped on the surface of C4. (C) MASP-2 CCP domain residues interacting with C4 (green residues). The view is related to that in B by a 180° rotation around a vertical axis. (D and E) Same view as in B and C, with the C345C domain and the CCP domains colored according to the surface electrostatic potential. (F) Deposition of C4 fragments is affected by CCP exosite mutations. Dilutions of wild-type or MASP-2 variants with CCP exosite mutations were added to microtiter wells coated with MBL bound to mannan. C4 was subsequently offered, and C4b was deposited on the surface upon cleavage by MASP-2. Counts represent fluorescence from europium bound to antibodies specific for C4 fragments or MASP-2.
Mutations in the CCP Exosite Affect C4 Recognition.
To validate the physiological relevance of the MASP-2 CCP exosite, we mutated residues in the CCP1 and CCP2 domains with surface-exposed side chains facing C4 in the complex to arginines aiming at introducing steric hindrance and electrostatic repulsion. These MASP-2 variants were associated with MBL bound to the yeast carbohydrate mannan, causing MASP-2 activation and cleavage of C4 and resulting in C4b deposition on the mannan surface (Fig. 3F). Mutating CCP1 Glu-333 or CCP2 Asp-365 severely reduced MASP-2 cleavage of C4, and the double mutant was unable to mediate cleavage. Mutation of CCP1 Pro-340 or CCP2 Pro-368 resulted in ∼60% cleavage activity, whereas the double mutant had a poor cleavage activity. The recently reported CCP1 Lys-342–Ala mutation (17) had no effect in accordance with the absence of Lys-342 from the C4 interface. All our mutants bound MBL and autoactivated as well as the wild type (Fig. S4 A and B), suggesting that impaired C4 cleavage activity was caused solely by weakened C4⋅MASP-2 interactions. We confirmed this hierarchy of activity for the MASP-2 variants in an even more physiological setting, where we measured C4 deposition onto mannan in MASP-2– deficient plasma reconstituted with the various forms of MASP-2 (Fig. S4C). In addition, surface plasmon resonance (SPR) measurements showed that the recombinant C4 C345C domain interacted with immobilized MASP-2 (Fig. S4D), with an apparent equilibrium dissociation constant KD of 21 nM and mutations in C4 residues interacting with MASP-2 in our structure affected this interaction. In summary, our functional experiments and biophysical measurements confirmed the importance of the C4⋅MASP-2 interactions at the CCP exosite observed in our crystal structure.
C1s Recognition of C4 and MASP-1 Discrimination.
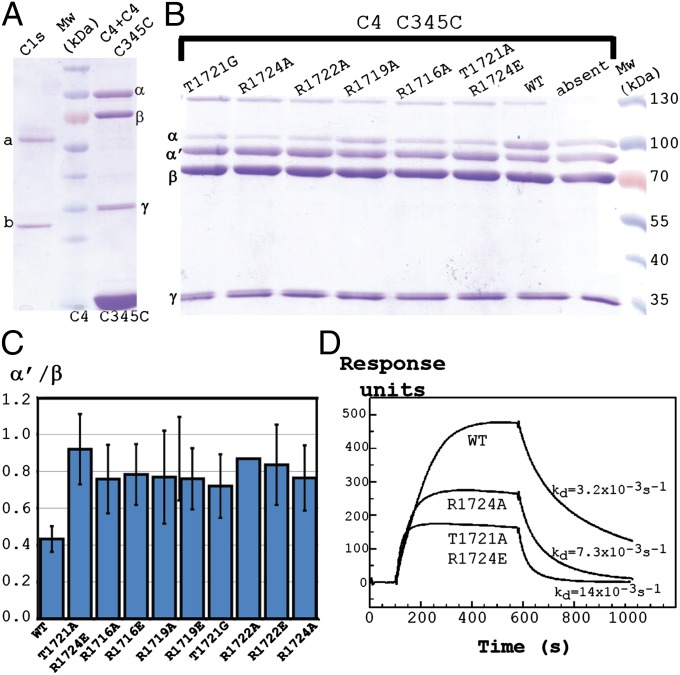
Based on prior data concerning the importance of the C1s CCP domains for C4 cleavage (19, 20), we hypothesized that the MASP-2 CCP exosite is conserved in C1s and interacts with the C4 C345C domain. In support of this hypothesis, we were able to inhibit C1s-mediated C4 cleavage by the addition of recombinant C345C domain in 30-fold molar excess relative to C4. Single mutations at five different C345C residues located in the MASP-2 interface all decreased the ability of the mutated C345C domain to inhibit C4 cleavage by C1s (Fig. 4 A–C). Similar to the C4⋅MASP-2 interaction, SPR measurements showed that the C1s interaction with the C4 C345C domain had an apparent dissociation constant KD of 37 nM (Fig. 4D), and mutations in C4 Arg-1,724 and Thr-1,721 affected this interaction. C1s (9) qualitatively shares the electrostatic properties at the CCP exosite, with MASP-2 supporting an interaction between C1s and C4 C345C similar to what we observed for C4⋅MASP-2 (Fig. S5A). Because a positively charged surface on the MASP-2 SP domain located next to the C4 sulfotyrosine region is also present on C1s (Fig. S5B), an interaction between the C1s SP domain and the C4 sulfotyrosine region may also occur. This surface area is identical to a very recently identified exosite in the C1s SP domain interacting with the C4 sulfotyrosine region, and mutations in this exosite decrease the efficiency of C4 cleavage (21). C4 sulfotyrosine interaction with C1s would also offer an explanation for the 10-fold higher concentration of C1s required to cleave nonsulforylated C4 compared with sulforylated C4 (22). Together, our experiments, prior functional data, and structural comparisons strongly support that the C1s SP domain interacts with the C4 SO3–Tyr region and that C1s contacts residues 1,716–1,725 in the C4 C345C domain through a CCP exosite. The inability of MASP-1 to cleave C4 may be due to the electrostatic properties of MASP-1 being different from those of MASP-2 and C1s, especially at the CCP site. Furthermore, a large MASP-1 insert in the SP domain loop B appeared to prevent insertion of the C4 R loop into the catalytic site (Fig. S5 C–E). MASP-3, the third protease of the lectin pathway, is an alternative splice product of the MASP-1 gene and contains a different SP domain but the same CCP modules. Their electrostatic properties probably explain why MASP-3, like MASP-1, is unable to cleave C4 and consequently cannot take over the function of MASP-2 in MASP-2–deficient or –depleted plasma.
Fig. 4.
C1s interacts with the C4 C345C domain. (A) SDS/PAGE analysis of input proteins used for the cleavage experiments. (B) SDS/PAGE analysis of C4 digestion by C1s at 37 °C for 30 min. The band at 130 kDa occurs only in the presence of the recombinant C345C domain, and mass spectrometry indicates that it contains a TE-mediated adduct of the C4 α′-chain and the C345C domain. (C) Comparison of cleavage inhibition by the C4 C345C variants measured as the ratio between the C4 α′-and β-chain after reaction starting from uncleaved C4. The α′/β ratios for the experiments containing wild-type (WT) or mutant C345C domain were normalized to the experiment conducted in the absence of the C345C domain. The cleavage series were repeated five times. The presence of the adduct at 130 kDa led to an underestimation of the α′/β ratio because a fraction of the α′-chain would be present in the 130-kDa band. (D) Assessment by SPR of the interaction between WT or mutated C4 C345C domain with immobilized C1s. An overlay of sensorgrams representing 40 nM (close to the calculated KD) WT C4 C345C and two mutants is shown. Dissociation constants (KD) are 36.9, 35.4, and 44.2 nM for WT C345C, C345C R1724A, and C345C T1721A,R1724E, respectively.
Conformational Changes in Enzyme and Substrate.
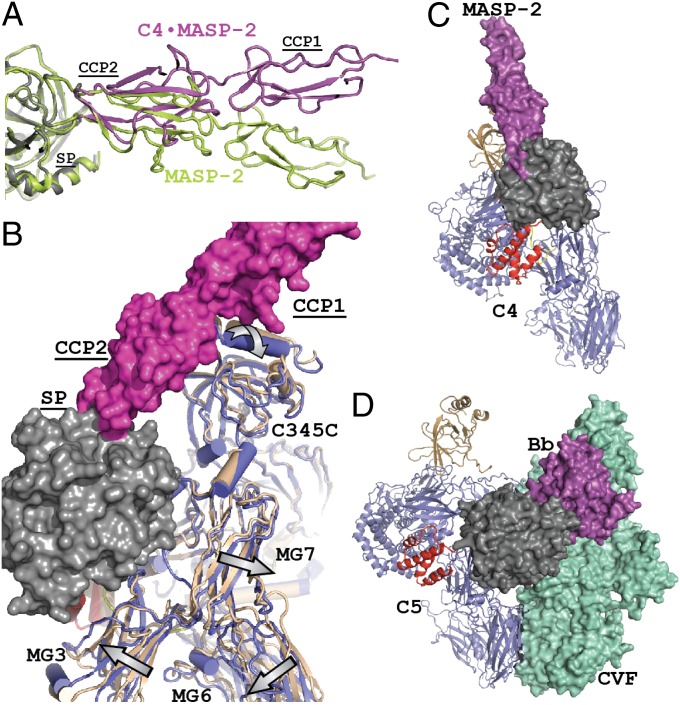
The conformation of the CCP1–CCP2 domain tandem is identical in C4-bound and zymogen MASP-2, whereas the SP domain has rotated by 24–29° (Fig. 5A) relative to the CCP2 domain of zymogen MASP-2 (23) or active unbound MASP-2 (10). The ability of MASP-2 to undergo this hinge movement at the CCP2–SP linker allows simultaneous substrate recognition by the CCP exosite and the SP domain. C4 binding induces only minor structural reorganization within the SP domain, suggesting that the exosite–C4 interactions do not further activate the SP domain. In accordance with this finding, the recombinant MASP-2 SP domain cleaves C2 efficiently, whereas at least the CCP2 domain is required for fast C4 cleavage (9). C4 also undergoes significant conformational changes upon binding to MASP-2 in two regions around the scissile bond. In unbound C4, the C4a α1-helix, the R loop, and downstream Nt-α′ residues are disordered. In the MASP-2 complex, C4a formed a four-helix bundle, and residues downstream of the R loop were ordered and held between the C4a α1-helix and the MG3 and MG8 domains (Fig. S6). MASP-2 recognition of the R loop most likely stabilized the conformation of downstream residues in the Nt-α′ region, and the CCP exosite interaction induces a rotation of the C4 C345C domain, which resulted in a 10° rotation of the MG1–5 domains (Fig. 5B and Movie S1). Hereby the MG3 domain could capture the C4a α1-helix between itself and the rest of the C4a domain, which presumably also locks the conformation of the Nt-α′ region located downstream of the scissile bond. Hence, not only does the MASP-2 CCP exosite interaction help to correctly orient the protease toward C4, but through a relay of domain movements it probably also stabilizes the R-loop conformation, which feasibly facilitates cleavage.
Fig. 5.
Conformational changes in MASP-2 and C4, the MBL⋅MASP-2⋅C4 complex, and comparison with convertases. (A) Comparison of zymogen MASP-2 (10) with C4-bound MASP-2 demonstrating the rotation of the CCP domains relative to the SP domain upon C4 binding. The two conformations were superimposed on their SP domains. (B) MASP-2 (magenta and gray surface) binding to C4 (blue cartoon, bound; light brown cartoon, unbound) induces rotations of 5–10° (directions indicated by gray arrows) of several C4 domains. The two C4 conformations were superimposed on their α-chain superdomains. (C and D) Comparison of the structure of the C4⋅MASP-2 complex (C) and a model of the C5⋅CVF⋅Bb complex (D) approximating complexes of substrates (C3 or C5) with their convertases (24). C4 and C5 are shown in cartoon; MASP-2 and Bb are shown in surface representation with their SP domain colored gray and the CCP/VWA domains colored magenta. CVF is shown as a green surface.
Discussion
Our structures of C4 and especially the C4⋅MASP-2 complex represent a crucial step forward in reaching a structure-based understanding of the series of events starting with pattern recognition in the lectin or classical pathway. The crystal structures presented here also allow us to compare how the structurally similar C3, C4, and C5 are cleaved by two distinct types of proteolytic enzymes, the C1s and MASP-2 based enzymes cleaving C4 in the lectin and classical pathway vs. the factor B and C2-based C3 and C5 convertases in the alternative and terminal pathway. The substrate recognition mechanism revealed by the C4⋅MASP-2 complex is fundamentally different from that identified in the C3 and C5 convertases (Fig. 5 C and D and Fig. S7). When C4 is cleaved by MASP-2 or C1s bound to a pattern recognition molecule, exosites in the MASP-2 or C1s CCP domains are required (9, 17, 18), and our results imply that their function is in the recognition of the C4 C345C domain. In the C3 and C5 convertases, large exosites are present in the noncatalytic C3b or C4b subunits recognizing the MG4, MG5, and probably also the MG7 domains, which are all located far from the C345C domain (24). In addition, the catalytic subunit of the convertases approaches much more horizontally relative to the scissile loop region in the substrate compared with the C4⋅MASP-2 complex (Fig. 5 C and D).
C4 is a remarkable substrate because it is cleavable by two types of enzymatically active complexes—depending on either MASP-2 or C1s as the catalytic subunit—becoming activated upon pattern recognition taking place within structurally very variable and unpredictable environments. In contrast, when C3 and C5 are substrates for the convertases, the primary substrate-binding noncatalytic C3b or C4b subunit is likely to be rather rigid (24). Substrate recognition by the convertases is therefore likely to be only slightly influenced by the environment in which the convertase is present. In C3 and C5, the scissile bond is presented in or close to an exposed loop with five to nine disordered residues (Fig. S7B), and in C5 the P1 residue Arg-751 is not even exposed, suggesting that this residue is not involved in the initial recognition between the convertase and the substrate. However, in C4 the scissile bond is located in a region with 22 disordered residues. The much higher flexibility of the scissile bond region in C4 offers an elegant solution to the problem of simultaneously being a substrate for two different proteolytic enzymes deposited in extremely variable environments. This high degree of flexibility may allow C4 to form initial contacts with the SP domain of C1s or MASP-2, approaching the protease from quite different orientations, and in combination with the flexibility of the MASP-2 and C1s containing proteolytic complexes discussed below, the flexibility of the scissile bond region may significantly promote formation of productive enzyme–C4 complexes.
Within a full enzyme–substrate MBL⋅MASP-2⋅C4 complex bound to a carbohydrate layer through the MBL carbohydrate recognition domains (CRDs), MASP-2 is bound to the MBL collagen stem through its CUB domains and firmly holds the substrate through contacts with primarily the C4 C345C domain and the C4 scissile bond region. This finding implies that the MG1–MG4–MG5 domains at the opposite end of C4 (Fig. 1 B and C) are expected to be oriented approximately in the same direction as the MBL CRDs toward the carbohydrate layer. In such an orientation, the TE would be directed into the carbohydrate layer during a C3b-like conformational change of nascent C4b, as previously suggested for convertase-bound C3b (24). The MBL⋅MASP-2⋅C4 complex is anticipated to be structurally flexible for several reasons. Considerable flexibility is present in MBL at the predicted kink of the collagen stem, and likewise the orientation of the CRDs relative to the collagen stem is variable. MBL is also heterogeneous by stoichiometry, with the smallest form being dimer of polypeptide trimers formed through the collagen stem, but such dimeric MBL–MASP complexes do not bind carbohydrate patterns with a high enough avidity to allow for efficient activation of complement. MBL trimers and tetramers are the dominating form occurring naturally in humans, but higher oligomers are also present, and their intersubunit orientation is quite variable (25). In MASP-2, flexibility is presumably present within the CUB2 domain (26), at the CUB2–CCP1 linkage (27), and at the CCP2–SP linkage, as shown here. Our C4⋅MASP-2 structure surprisingly revealed that the orientation between MASP-2 and C4 is also variable, and our structure of unbound C4 suggests high mobility of the scissile bond region and regions surrounding it. Obviously, all of these sources of flexibility increase the likelihood of forming productive MBL⋅MASP-2⋅C4 complexes within the highly variable glycan environment, and similar levels of flexibility can be expected for the ficolin⋅MASP-2⋅C4 and the C1⋅C4 complexes in the classical pathway, because ficolins and C1q share their basic architecture with MBL, as is also the case for MASP-2 and C1s.
Activation of complement triggers an aggressive proteolytic cascade, creating potent inflammatory effector molecules, with the risk of host tissue damage if not kept under tight control by an array of soluble and membrane-bound regulators (1, 28). Uncontrolled complement activation is seen in association with, e.g., sepsis, myocardial infarction, ischemic stroke, rheumatoid arthritis, glomerulonephritis, myasthenia gravis, lupus, age-related macular degeneration, and atypical hemolytic uremic syndrome (28, 29). Of particular relevance for the present study, MASP-2 inhibition has proven efficient in protection against ischemia/reperfusion injury (30).
Our C4⋅MASP-2 structure reveals in details the intermolecular interactions of a key event in the lectin pathway and should therefore facilitate further development of inhibitors of this pathway. Moreover, the exosite part of the C4 recognition by MASP-2 identified by our structure is likely to be common to both lectin and classical pathways. Thus, in conclusion, our results provide an essential structural framework for future studies of the lectin and classical pathway of complement activation and rationalize many prior functional studies of both pathways.
Materials and Methods
Human C4 was purified from human plasma by anion exchange chromatography, and recombinant MASP-2 CCP1–CCP2–SP S633A was prepared by refolding and activated with MASP-1 (9). Crystals of C4 and C4⋅MASP-2 were obtained by vapor diffusion and cryoprotected before data collection. The structures were determined by molecular replacement aided by single-wavelength anomalous diffraction phases for a Ta6Br12 derivative for the C4 structure. Rebuilding was done in O (31), and refinement was performed in PHENIX.REFINE (32). Full-length MASP-2 variants were expressed in HEK293F cells (33) and activated by their association with recombinant MBL bound to mannan-coated plates. C4 was then added and deposited upon MASP-2–mediated cleavage, and cleaved C4 was quantitated with a biotinylated monoclonal anti-C4 antibody through detection with europium-labeled streptavidin and fluorometry (34). The ability of the MASP-2 variants to mediate C4 deposition was also investigated in the presence of MASP-2–deficient plasma (33). Recombinant wild-type and mutated C4 C345C was expressed in Escherichia coli, purified by Ni2+–chelate chromatography and gel filtration, and tested in cleavage experiments with C1s at a molar ratio of 30:1 for (C4 C345C):C4. For SPR measurements, MASP-2 or C1s were immobilized on CM5 sensor chips, and the interaction with recombinant C4 C345C was measured at a flow rate of 5 μL/min. Data were analyzed by global fitting to a 1:1 binding model. A detailed description of experimental methods may be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the staff members at the European Synchrotron Radiation Facility and SOLEIL beamlines for help with data collection; L. Kristensen and J. Balczer for protein purification; S. Degn for help with expression; and A. M. Bundsgaard for help with SPR. G.R.A. was supported by Danscatt, the Lundbeck Foundation, the Lundbeck Foundation Nanomedicine Centre for Individualized Management of Tissue Damage and Regeneration, and a Hallas-Møller stipend from the Novo-Nordisk Foundation. P.G. was supported by Hungarian Scientific Research Fund (OTKA) Grant NK77978 and National Development Agency Grant KMOP-1.1.2-07/1-2008-0003. J.D. was supported by the János Bolyai Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. W.J.S. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4FXK and 4FXG).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208031109/-/DCSupplemental.
References
- 1.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: A key system for immune surveillance and homeostasis. Nat Immunol. 2010;11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thiel S. Complement activating soluble pattern recognition molecules with collagen-like regions, mannan-binding lectin, ficolins and associated proteins. Mol Immunol. 2007;44:3875–3888. doi: 10.1016/j.molimm.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Hansen S, et al. Collectin 11 (CL-11, CL-K1) is a MASP-1/3-associated plasma collectin with microbial-binding activity. J Immunol. 2010;185:6096–6104. doi: 10.4049/jimmunol.1002185. [DOI] [PubMed] [Google Scholar]
- 4.Thiel S, et al. A second serine protease associated with mannan-binding lectin that activates complement. Nature. 1997;386:506–510. doi: 10.1038/386506a0. [DOI] [PubMed] [Google Scholar]
- 5.Law SK, Dodds AW. The internal thioester and the covalent binding properties of the complement proteins C3 and C4. Protein Sci. 1997;6:263–274. doi: 10.1002/pro.5560060201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krych-Goldberg M, Atkinson JP. Structure-function relationships of complement receptor type 1. Immunol Rev. 2001;180:112–122. doi: 10.1034/j.1600-065x.2001.1800110.x. [DOI] [PubMed] [Google Scholar]
- 7.Kojouharova M, Reid K, Gadjeva M. New insights into the molecular mechanisms of classical complement activation. Mol Immunol. 2010;47:2154–2160. doi: 10.1016/j.molimm.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 8.Pangburn MK, Rawal N. Structure and function of complement C5 convertase enzymes. Biochem Soc Trans. 2002;30:1006–1010. doi: 10.1042/bst0301006. [DOI] [PubMed] [Google Scholar]
- 9.Ambrus G, et al. Natural substrates and inhibitors of mannan-binding lectin-associated serine protease-1 and -2: A study on recombinant catalytic fragments. J Immunol. 2003;170:1374–1382. doi: 10.4049/jimmunol.170.3.1374. [DOI] [PubMed] [Google Scholar]
- 10.Harmat V, et al. The structure of MBL-associated serine protease-2 reveals that identical substrate specificities of C1s and MASP-2 are realized through different sets of enzyme-substrate interactions. J Mol Biol. 2004;342:1533–1546. doi: 10.1016/j.jmb.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 11.Blanchong CA, et al. Genetic, structural and functional diversities of human complement components C4A and C4B and their mouse homologues, Slp and C4. Int Immunopharmacol. 2001;1:365–392. doi: 10.1016/s1567-5769(01)00019-4. [DOI] [PubMed] [Google Scholar]
- 12.Fredslund F, et al. The structure of bovine complement component 3 reveals the basis for thioester function. J Mol Biol. 2006;361:115–127. doi: 10.1016/j.jmb.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Fredslund F, et al. Structure of and influence of a tick complement inhibitor on human complement component 5. Nat Immunol. 2008;9:753–760. doi: 10.1038/ni.1625. [DOI] [PubMed] [Google Scholar]
- 14.Janssen BJ, et al. Structures of complement component C3 provide insights into the function and evolution of immunity. Nature. 2005;437:505–511. doi: 10.1038/nature04005. [DOI] [PubMed] [Google Scholar]
- 15.Huntington JA. Shape-shifting serpins—advantages of a mobile mechanism. Trends Biochem Sci. 2006;31:427–435. doi: 10.1016/j.tibs.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 16.Héja D, et al. Monospecific inhibitors show that both mannan-binding lectin-associated serine protease-1 (MASP-1) and -2 are essential for lectin pathway activation and reveal structural plasticity of MASP-2. J Biol Chem. 2012;287:20290–20300. doi: 10.1074/jbc.M112.354332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duncan RC, et al. Multiple domains of MASP-2, an initiating complement protease, are required for interaction with its substrate C4. Mol Immunol. 2012;49:593–600. doi: 10.1016/j.molimm.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 18.Rossi V, Teillet F, Thielens NM, Bally I, Arlaud GJ. Functional characterization of complement proteases C1s/mannan-binding lectin-associated serine protease-2 (MASP-2) chimeras reveals the higher C4 recognition efficacy of the MASP-2 complement control protein modules. J Biol Chem. 2005;280:41811–41818. doi: 10.1074/jbc.M503813200. [DOI] [PubMed] [Google Scholar]
- 19.Rossi V, Bally I, Thielens NM, Esser AF, Arlaud GJ. Baculovirus-mediated expression of truncated modular fragments from the catalytic region of human complement serine protease C1s. Evidence for the involvement of both complement control protein modules in the recognition of the C4 protein substrate. J Biol Chem. 1998;273:1232–1239. doi: 10.1074/jbc.273.2.1232. [DOI] [PubMed] [Google Scholar]
- 20.Bally I, Rossi V, Thielens NM, Gaboriaud C, Arlaud GJ. Functional role of the linker between the complement control protein modules of complement protease C1s. J Immunol. 2005;175:4536–4542. doi: 10.4049/jimmunol.175.7.4536. [DOI] [PubMed] [Google Scholar]
- 21.Duncan RC, et al. Identification of a catalytic exosite for complement component C4 on the serine protease domain of C1s. J Immunol. 2012;189:2365–2373. doi: 10.4049/jimmunol.1201085. [DOI] [PubMed] [Google Scholar]
- 22.Hortin GL, Farries TC, Graham JP, Atkinson JP. Sulfation of tyrosine residues increases activity of the fourth component of complement. Proc Natl Acad Sci USA. 1989;86:1338–1342. doi: 10.1073/pnas.86.4.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gál P, et al. A true autoactivating enzyme. Structural insight into mannose-binding lectin-associated serine protease-2 activations. J Biol Chem. 2005;280:33435–33444. doi: 10.1074/jbc.M506051200. [DOI] [PubMed] [Google Scholar]
- 24.Laursen NS, et al. Substrate recognition by complement convertases revealed in the C5-cobra venom factor complex. EMBO J. 2011;30:606–616. doi: 10.1038/emboj.2010.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensenius H, et al. Mannan-binding lectin: Structure, oligomerization, and flexibility studied by atomic force microscopy. J Mol Biol. 2009;391:246–259. doi: 10.1016/j.jmb.2009.05.083. [DOI] [PubMed] [Google Scholar]
- 26.Major B, et al. Calcium-dependent conformational flexibility of a CUB domain controls activation of the complement serine protease C1r. J Biol Chem. 2010;285:11863–11869. doi: 10.1074/jbc.M109.098541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teillet F, et al. Crystal structure of the CUB1-EGF-CUB2 domain of human MASP-1/3 and identification of its interaction sites with mannan-binding lectin and ficolins. J Biol Chem. 2008;283:25715–25724. doi: 10.1074/jbc.M803551200. [DOI] [PubMed] [Google Scholar]
- 28.Wagner E, Frank MM. Therapeutic potential of complement modulation. Nat Rev Drug Discov. 2010;9:43–56. doi: 10.1038/nrd3011. [DOI] [PubMed] [Google Scholar]
- 29.Degn SE, Jensenius JC, Thiel S. Disease-causing mutations in genes of the complement system. Am J Hum Genet. 2011;88:689–705. doi: 10.1016/j.ajhg.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwaeble WJ, et al. Targeting of mannan-binding lectin-associated serine protease-2 confers protection from myocardial and gastrointestinal ischemia/reperfusion injury. Proc Natl Acad Sci USA. 2011;108:7523–7528. doi: 10.1073/pnas.1101748108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 32.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stengaard-Pedersen K, et al. Inherited deficiency of mannan-binding lectin-associated serine protease 2. N Engl J Med. 2003;349:554–560. doi: 10.1056/NEJMoa022836. [DOI] [PubMed] [Google Scholar]
- 34.Thiel S, et al. Polymorphisms in mannan-binding lectin (MBL)-associated serine protease 2 affect stability, binding to MBL, and enzymatic activity. J Immunol. 2009;182:2939–2947. doi: 10.4049/jimmunol.0802053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.