Abstract
A subclass of bacterial CLC anion-transporting proteins, phylogenetically distant from long-studied CLCs, was recently shown to be specifically up-regulated by F-. We establish here that a set of randomly selected representatives from this “CLCF” clade protect Escherichia coli from F- toxicity, and that the purified proteins catalyze transport of F- in liposomes. Sequence alignments and membrane transport experiments using 19F NMR, osmotic response assays, and planar lipid bilayer recordings reveal four mechanistic traits that set CLCF proteins apart from all other known CLCs. First, CLCFs lack conserved residues that form the anion binding site in canonical CLCs. Second, CLCFs exhibit high anion selectivity for F- over Cl-. Third, at a residue thought to distinguish CLC channels and transporters, CLCFs bear a channel-like valine rather than a transporter-like glutamate, and yet are F-/H+ antiporters. Finally, F-/H+ exchange occurs with 1∶1 stoichiometry, in contrast to the usual value of 2∶1.
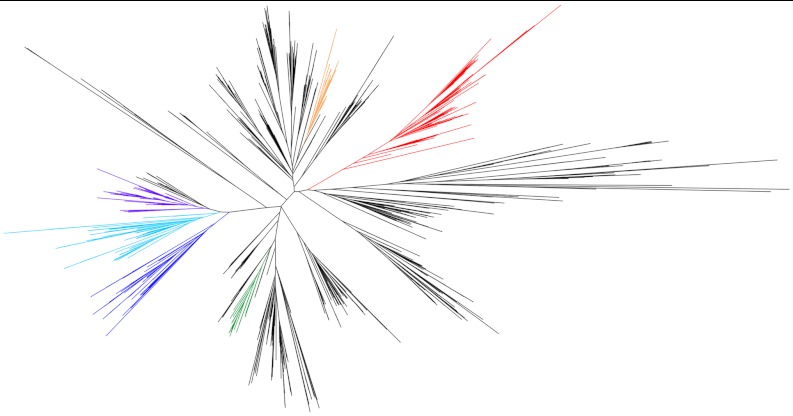
The CLC family of membrane proteins derives its name from its charter member, a double-barreled Cl- channel used by electric rays to generate high-power pulses to stun prey (1, 2). This family turned out to be split into proteins of two mechanistically disparate subtypes: anion channels and Cl-/H+ antiporters (3–6). CLCs participate in diverse biological tasks requiring transmembrane anion conductance, such as acidification of lysosomes, control of skeletal muscle excitability, renal regulation of blood pressure, and extreme acid resistance in enteric bacteria (7). Most CLCs thus far studied use Cl- for their physiological purposes, but 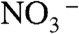 has been identified as the substrate anion in a plant vacuolar CLC (8). All of this known functional diversity resides in a remarkably narrow region of the family’s phylogeny (Fig. 1) that includes all of the eukaryotic CLCs and their closest bacterial counterparts; CLC-ec1, a Cl-/H+ antiporter from Escherichia coli that has been subject to extensive structural and mechanistic study (9–11) also resides in this region of the tree. But the CLC family is vast, with highly divergent sequences among its bacterial members. Many prokaryotic genomes encode CLCs, in some cases more than one, and some lack certain key residues found in every currently described CLC (10). What biological roles might these strange CLCs play?
has been identified as the substrate anion in a plant vacuolar CLC (8). All of this known functional diversity resides in a remarkably narrow region of the family’s phylogeny (Fig. 1) that includes all of the eukaryotic CLCs and their closest bacterial counterparts; CLC-ec1, a Cl-/H+ antiporter from Escherichia coli that has been subject to extensive structural and mechanistic study (9–11) also resides in this region of the tree. But the CLC family is vast, with highly divergent sequences among its bacterial members. Many prokaryotic genomes encode CLCs, in some cases more than one, and some lack certain key residues found in every currently described CLC (10). What biological roles might these strange CLCs play?
Fig. 1.
Phylogeny of the CLC superfamily. The clade that comprises F- riboswitch-controlled CLCs is highlighted in red. Other clades that include proteins for which functional or structural data is available are also highlighted: antiporters CLC-ec1 from E. coli, Synechocystis sp PCC6803, and Salmonella enterica in green; eukaryotic Cl- channels CLC-0, -1, and -2, and the Cl-/H+ antiporter from the thermophilic alga Cyanidioschyzon merolae in blue; the mammalian antiporters CLC-6 and -7, the yeast antiporter GEF1, and an Arabidopsis thaliana
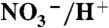 antiporter in cyan; the mammalian transporters CLC-3, -4, and -5 in purple; and a second CLC from E. coli, CLC-ec2, in orange.
antiporter in cyan; the mammalian transporters CLC-3, -4, and -5 in purple; and a second CLC from E. coli, CLC-ec2, in orange.
A first hint at an unusual CLC physiology in bacteria recently emerged from a small clade evolutionarily distant from the well studied members of the superfamily (Fig. 1). Baker and colleagues (12) described a conserved regulatory RNA motif that induces transcription of downstream genes upon binding F- ion; an X-ray crystal structure of a F--riboswitch was recently solved (13). The genes controlled by these riboswitches code for F--sensitive enzymes, including enolase and pyrophosphatase, as well as membrane proteins, including members of the CLC superfamily. The riboswitch-associated CLCs cluster within a single clade in the phylogenetic tree, although not all CLCs in this clade are regulated by riboswitches. Riboswitch-controlled CLCs differ substantially from the canonical members of the superfamily; for instance, CLC-psy, a homologue from the plant pathogen Pseudomonas syringae, shares only 22% identity with CLC-ec1 and lacks the serine and tyrosine residues that directly coordinate the central Cl- ion in all CLCs of known structure. Accordingly, these CLCs were suggested (12) to act as F- exporters that protect bacteria from environmental F- toxicity, an idea supported by the demonstration that CLC-psy rescues F--dependent growth-arrest in E. coli and catalyzes robust F- efflux in reconstituted liposomes.
Riboswitch-associated CLCs appear in a wide range of bacterial species: plant and human pathogens, soil and marine bacteria, and gram-negative and gram-positive organisms. Why would such bacteria have evolved dedicated F- export systems? Environmental levels of F-, though quite variable, are typically found in the 10–100 μM range (14), and fluoridation of public water supplies adds 50–100 μM to this. These levels are comparable to inhibition constants for enolase and pyrophosphatase; in acidic environments, bacteria would be particularly F--susceptible because HF is a weak acid (pKa = 3.4) highly permeable to biological membranes. Under such conditions, if a transport pathway for the anion were absent, F- would accumulate in the bacterial cytoplasm according to the pH gradient across the inner membrane. A F- pathway—either a channel or a H+-coupled antiporter—would provide a means of exporting the anion to well below its extracellular levels.
Because inorganic F- in biology is a largely unexplored topic, and as these F--controlled CLCs and their proposed F--exporting function are so unusual, we considered it worthwhile to survey this clade to gauge whether the F- resistance and transport reported for CLC-psy is a general property of these “CLCF” proteins. In addition, because CLCF proteins, like all known channel-type CLCs, have valine in the conserved position thought to distinguish channels from antiporters (15), we sought to experimentally confirm these CLCs as F- channels. We find that the CLCF proteins studied here protect E. coli against F- toxicity, and that in contrast to expectation, all are proton-coupled antiporters. One of these homologues, which allows detailed study by electrical recording, shows an unprecedented 1-to-1 F-/H+ exchange stoichiometry.
Results
Rescue of E. coli from F- Toxicity.
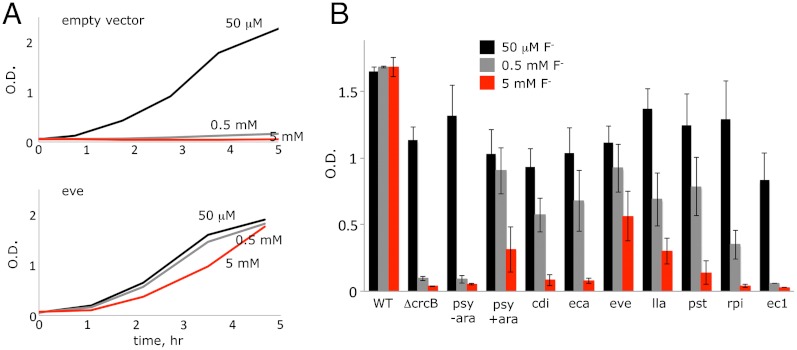
We have so far screened 29 members of the CLCF clade (12) for overexpression in E. coli (Table 1). Eight homologues produce protein of sufficient quality for studying F- transport in reconstituted proteoliposomes. We first examined these for their ability to protect E. coli against F- toxicity. E. coli does not carry any CLCF genes, but instead uses an exporter from the unrelated crcB family, deletion of which renders the bacterium highly sensitive to F- (12). We transformed this ΔcrcB strain with an arabinose-inducible rescue vector bearing CLCF genes to be tested, monitoring growth in LB supplemented with NaF. While wild-type cell growth is unaffected by 5 mM F-, the ΔcrcB strain is almost completely arrested by 0.5 mM F- (Fig. 2). Inhibition can be prevented by supplying the ΔcrcB cells with a CLCF homologue (Fig. 2A). The rescue vector successfully expressed protein for seven of the CLCF homologues, and all of these rescue ΔcrcB against F- toxicity (Fig. 2B, Table 1). It is notable that CLC-ec1, a Cl-/H+ antiporter, fails to rescue, consistent with its selectivity for Cl- over F- (9, 12).
Table 1.
Survey of CLCF homologues
| Homologue nickname | Source organism | NCBI reference sequence | t1/2 of F- efflux, sec | Protein yield, mg/L |
| CLC-cpi | Chitinophaga pinensis | YP_003123680.1 | 25 | 0.1 |
| CLC-cdi | Clostridium difficile | YP_001087131.1 | 7 | 0.2 |
| CLC-eca | Enterococcus casseliflavus | ZP_05647488.1 | 4 | 2.5 |
| CLC-eve | Eubacterium ventriosum | ZP_02027167.1 | 12 | 0.4 |
| CLC-lla | Lactococcus lactis | YP_001032764.1 | 8 | 0.5 |
| CLC-pst | Pirellula staleyi | YP_003370005.1 | 5 | 1.0 |
| CLC-psy | Pseudomonas syringae | ZP_07250577.1 | 30 | 1.5 |
| CLC-rpi | Ralstonia picketti | YP_001892513.1 | 28 | 0.6 |
CLCF homologues from Clostridium butyricum, Leeuwenhoekiella blandensis, Granulicatella elegans, Leptothrix cholodnii, Abiotrophia defectiva, Bacillus halodurans, Clostridium botulinum, and Agrobacterium radiobacter did not express detectable protein. Protein from Ruminococcus lactaris, Burkholderia pseudomallei, and Bacterioides coprocola could not be purified in stable, monodisperse dimer form. Protein from Clostridium botulinum E3 strain Alaska E43, Spirosoma linguale, Verrucomicrobium spinosum, Xanthomonas albilineans was inactive in anion transport assays. All CLC genes reported in this table are found downstream of a F- riboswitch except CLC-pst. All homologues mediate flux for F- (half-times indicated) but negligible flux for Cl-, and all show robust F--driven H+ antiport. F- efflux experiments were carried out with protein reconstituted at 10 μg/mg lipid in 300 mM K+ solutions, except for CLC-cpi and CLC-cdi, which used 300 mM Na+, which typically produces slightly higher rates than in K+. Values reported for t1/2 are means of duplicate determinations in this survey, except for CLC-eca and CLC-psy, which show < 5% variation over 3–5 measurements.
Fig. 2.
Rescue of E. coli from F- toxicity. (A) Representative growth curves for ΔcrcB strain transformed with an empty vector or with CLC-eve in LB supplemented with the indicated NaF. (B) Culture optical density after 3 h growth for wild-type E. coli or ΔcrcB with empty vector or indicated CLC genes. The label “psy-ara” refers to an uninduced culture of CLC-psy.
Fluoride Transport by CLCF Homologues.
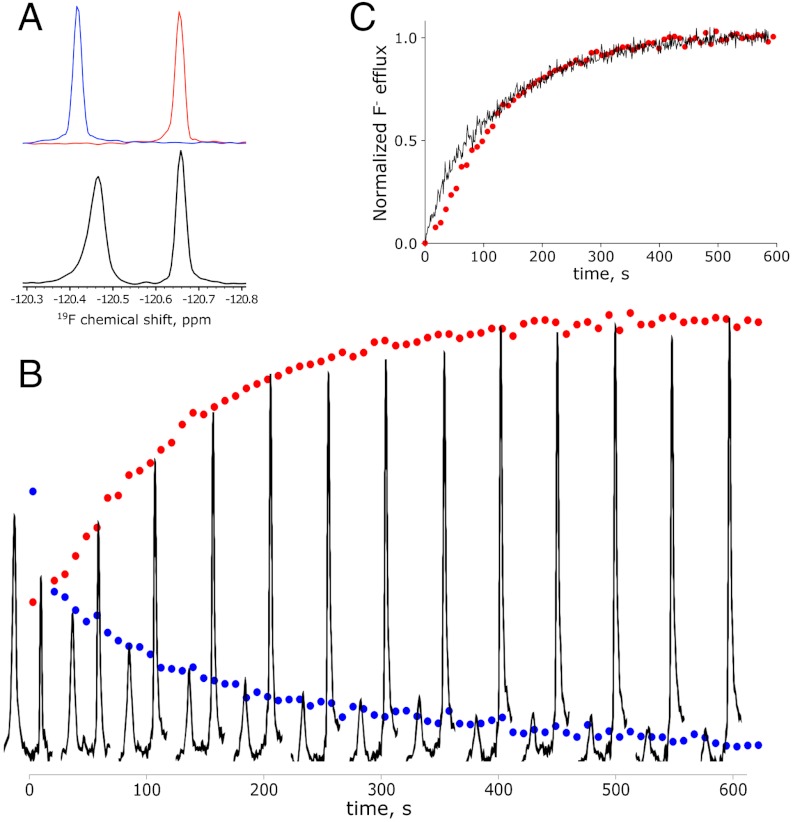
To establish whether protection by CLCFs reflects transmembrane export, we sought a direct measurement of F- transport. Radioactive, electrochemical, and fluorescent assays of F- are unsuitable for our purposes, so we turned to 19F NMR. With high natural abundance and sensitivity, the 19F nucleus offers especially felicitous characteristics. Most importantly, the 19F chemical shift is so sensitive to electronic environment that 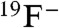 resonances in K+ vs. Na+ solutions are easily separable (16) (Fig. 3A). This difference may be exploited to monitor transport. CLC-psy proteoliposomes are loaded with 300 mM KF and diluted into isoosmotic Na2SO4 containing approximately 1 mM F-. Addition of valinomycin (Vln), a K+-specific ionophore, permits K+ to follow F- down its gradient out of the liposomes into the large external volume of Na+ solution. The ensuing transfer of
resonances in K+ vs. Na+ solutions are easily separable (16) (Fig. 3A). This difference may be exploited to monitor transport. CLC-psy proteoliposomes are loaded with 300 mM KF and diluted into isoosmotic Na2SO4 containing approximately 1 mM F-. Addition of valinomycin (Vln), a K+-specific ionophore, permits K+ to follow F- down its gradient out of the liposomes into the large external volume of Na+ solution. The ensuing transfer of 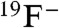 from a K+-rich internal solution to a Na+-rich external solution shows itself as a time-dependent decrease of the KF peak concomitant with an increase of the NaF peak (Fig. 3B). This time course yields a turnover rate of CLC-psy under these conditions of approximately 25 s-1, about 100-fold slower than CLC-ec1-mediated Cl- flux (17), about the same as Cl- flux in a cyanobacterial CLC (18), and far above the background F- leak in protein-free liposomes (12).
from a K+-rich internal solution to a Na+-rich external solution shows itself as a time-dependent decrease of the KF peak concomitant with an increase of the NaF peak (Fig. 3B). This time course yields a turnover rate of CLC-psy under these conditions of approximately 25 s-1, about 100-fold slower than CLC-ec1-mediated Cl- flux (17), about the same as Cl- flux in a cyanobacterial CLC (18), and far above the background F- leak in protein-free liposomes (12).
Fig. 3.
19F transport kinetics. (A) Upper box: overlain 19F NMR spectra of 3 mM KF (blue) or NaF (red) solution. Lower box: liposome sample with internal 300 mM KF and external 200 mM Na2SO4 spiked with 1 mM F-. (B) Kinetics of F- export from liposomes containing CLC-psy reconstituted at low protein density (approximately 1–2 transporters per liposome). Datapoints (blue for internal KF, red for external NaF) were collected every 9 s. Every fifth NMR spectrum is shown. (C) Overlay of NMR data for NaF appearance shown in (B) with F- efflux data obtained with light scattering measurements for the same batch of liposomes. Both datasets are normalized to the final value averaged between 500 and 600 s.
This 19F-NMR transport technique is satisfyingly direct and quantitative, but it is impractical for routine use. Beyond demanding daily untrammeled access to a high-resolution NMR facility, it requires large sample volumes and lacks the time resolution necessary for homologues with high transport rates. For these reasons, we employed an alternative assay based on light-scattering changes accompanying KF efflux (19). Though indirect, the light-scattering time course obtained for CLC-psy tracks the 19F NMR data well (Fig. 3C), while conferring the additional benefits of > 10-fold higher time resolution and 100-fold lower sample (and experimenter time) consumption. Further validation of this indirect method as a quantitative flux metric was obtained by comparing the light-scattering signal with direct electrochemical measurement of Cl- efflux in CLC-ec1-reconstituted liposomes (17). Henceforth, we employ this method for the broader survey of CLCF transport to be presented.
F- transport behavior was tested for the eight CLCF homologues that express biochemically stable protein and run as apparent homodimers in size-exclusion chromatography. We adopted a standard “zero-trans, zero voltage” condition for anion efflux, with 300 mM KF or KCl inside the liposomes and 300 mM K+ isethionate, an impermeant anion, in the external solution. All eight homologues show remarkable specificity for F-, with no discernable Cl- flux (Fig. 4, Table 1). CLC-psy is among the slowest of these F- transporters, and several homologues are roughly 10-fold faster. The only CLCF homologue that we could express from a gene without a riboswitch (CLC-pst) shows functional characteristics similar to the riboswitch-controlled CLCFs. We surmise, then, that F- export is general within the CLCF clade, regardless of the gene’s regulatory mechanism.
Fig. 4.
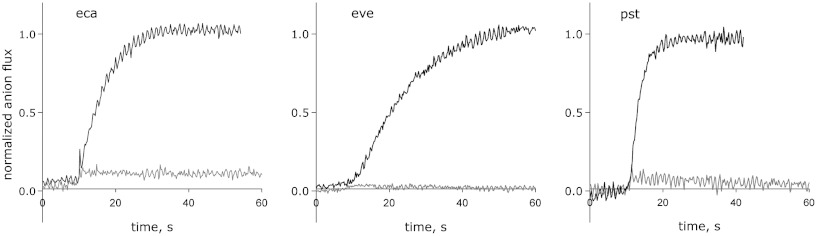
Anion transport in three CLCF homologues. Representative efflux data from indicated homologues are presented for F- (black traces) or Cl- (grey traces) monitored by light scattering. Transport was initiated by Vln 10 s after the beginning of the traces.
CLCF Proteins Are F-/H+ Antiporters.
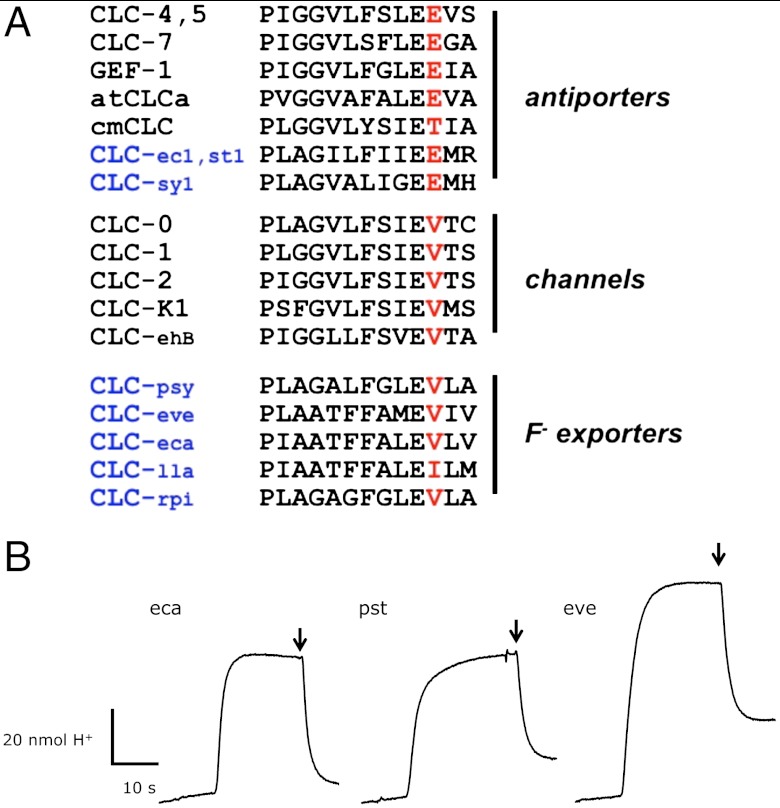
A fundamental question arising about any newly identified CLC protein concerns its mechanism of anion transport. Are these new CLCs F--selective channels or F-/H+ antiporters? Sequence perusal would suggest the former possibility, because the CLCFs have valine (or, rarely, isoleucine) at the position that in all known channels is valine and in nearly all known antiporters is glutamate (Fig. 5A). Either mechanistic incarnation could in principle meet the physiological requirements for F- export; the negative membrane potential sustained by most bacteria would expel anions through a channel mechanism, while coupling F- efflux to a proton gradient could provide an additional thermodynamic kick in acidic environments. We set up conditions for detecting H+ uptake coupled to anion efflux (3, 20): CLCF-reconstituted liposomes loaded with high KF and substantial buffer capacity suspended in a high-K+, low F-, weakly buffered solution. Addition of Vln allows F- efflux to proceed through the CLCF, and if this is coupled to proton uptake, the pH of the suspension should increase as H+ enters the liposome “uphill” against its thermodynamic gradient. A channel mechanism could not bring about such H+ uptake, as this would violate the second law of thermodynamics (3). As shown in pH recordings (Fig. 5B) all CLCF homologues tested catalyze F--driven H+ uptake. These experiments unambiguously demonstrate that the CLCFs examined here are F-/H+ antiporters, and not, as anticipated from sequence, F- channels.
Fig. 5.
Proton-coupled antiport in CLCF homologues. (A) Aligment of the PIGG-pen sequence of functionally verified CLC Cl- channels or Cl-/H+ antiporters. Black or blue lettering indicate eukaryotic or bacterial homologues, respectively. Eukaryotic homologues are from mammals (CLC-1,-2,-7, -K1), elasmobranch (CLC-0), plants (atCLCa, cmCLC), yeast (GEF1), and amoeba (CLC-ehB). (B) F- gradient-driven H+ uptake traces shown are representative of the full set of homologues studied. H+ uptake was monitored by time-dependent pH increase of the suspension of liposomes reconstituted with the CLCF homologues indicated. Transport was started by Vln, and FCCP was added (arrows) after steady-state uptake had been reached.
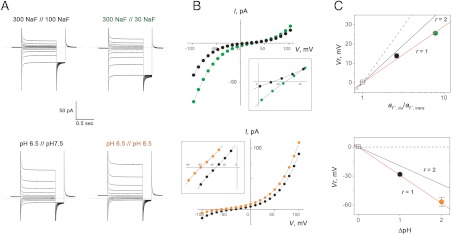
The magnitudes of H+ uptake observed here are substantial—within a factor of 2 of F- transport rates above. But these experiments do not permit quantitative determination of F-/H+ stoichiometry as F- uptake rates cannot be precisely enough measured from light-scattering traces. However, antiporter stoichiometry may be quantified by an electrophysiological method that records CLC-mediated current-voltage (I–V) relations in planar lipid bilayers separating solutions with pH or F- gradients (3, 21). In particular, the reversal potential Vr , the voltage at which current is zero, reflects the equilibrium point of the antiport reaction:
| [1] |
where r is the F-/H+ transport stoichiometry, and Ex is the Nernst equilibrium potential for each ion:
| [2] |
where [X] represents ion activity on the cis or trans side of the bilayer, and zx is the ion’s valence.
We selected the most highly expressed CLCF homologue, CLC-eca, for quantifying anion/H+ antiport stoichiometry by this method, expecting a value of 2, as with all previously studied CLC antiporters. Liposomes containing high density of CLC-eca protein were fused into planar bilayers, and macroscopic I–V curves from hundreds to thousands of antiporters were recorded in the presence of varying F- or pH gradients (Fig. 6). The result is clear and unexpected: CLC-eca exchanges F- for H+ with 1-to-1 stoichiometry.
Fig. 6.
F-/H+ antiport stoichiometry. CLC-eca proteoliposomes were incorporated into planar lipid bilayers, and I–V relations were determined in gradients of NaF (upper boxes) or pH (lower boxes). For F- gradients, pH was 6.5 on both sides; for pH gradients, F- was 300 mM. Gradients are indicated as cis side // trans side. (A) Raw current responses to families of 500-ms command voltage pulses from a holding potential of V = 0, followed by a 100-ms tail pulse to V = -100 mV. Zero level of current is indicated by dashed line. (B) I–V curves at end of command-voltage pulses. Colors of points match colors of labels in (A). Insets: blowups of I–V curves near reversal potential. (C) Reversal potential (Vr) as a function of gradients. Each point represents mean ± s.e. of 3–24 determinations, each in a separate bilayer. Solid lines represent predictions for F-/H+ antiporter stoichiometry indicated, Eq. 1, and dashed lines for F--specific channel mechanism. F- activity, aF, was calculated from tabulated mean ionic activity coefficients of NaF solutions.
Discussion
These experiments establish that the CLCF proteins, a minor clade within the CLC superfamily, export F- ions to protect bacteria against environmental F- challenge, and that they operate as F-/H+ antiporters rather than as electrodiffusive channels. Because these properties are seen with all of the homologues examined here, we suppose that they apply to the entire clade. Even with the small handful of homologues in this initial survey, mechanistic surprises emerge that set CLCFs apart from their Cl--transporting cousins. The most obvious of these concerns the key binding site for Cl- in the canonical CLCs, to which a pair of residues, the central serine and tyrosine (S107, Y445 in CLC-ec1), contribute hydroxyl dipoles for direct coordination (10). In plants, whose CLCs transport 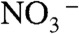 rather than Cl-, proline substitutes for the central serine (8, 22), and this substitution in other CLCs also switches ion selectivity towards
rather than Cl-, proline substitutes for the central serine (8, 22), and this substitution in other CLCs also switches ion selectivity towards 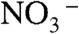 (23). These residues are strongly conserved in the familiar CLCs but are absent in alignments against the CLCFs. A conserved GNNLI/GMGLI sequence specific to the CLCF clade, however, is found in the same region as S107 in CLC-ec1, and homology models—admittedly suspect from the low sequence similarity to CLCs of known structure—place a conserved tyrosine near a conjectural central anion-binding site, in possible analogy to Y445 in CLC-ec1.
(23). These residues are strongly conserved in the familiar CLCs but are absent in alignments against the CLCFs. A conserved GNNLI/GMGLI sequence specific to the CLCF clade, however, is found in the same region as S107 in CLC-ec1, and homology models—admittedly suspect from the low sequence similarity to CLCs of known structure—place a conserved tyrosine near a conjectural central anion-binding site, in possible analogy to Y445 in CLC-ec1.
A second unexpected aspect of these transporters relates to ion selectivity. Canonical CLCs are rather weakly selective among Cl- and other monovalent anions like Br-, I-, 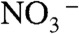 , or SCN-. Permeation of F- has been little studied, but the CLC-ec1 antiporter, the mammalian Cl- channel CLC-1, and a CLC channel from Entamoeba histolytica catalyze low but nonnegligible F- conduction (12, 24, 25). In contrast, CLCF antiporters greatly prefer F- over Cl- under the conditions used here, an ion selectivity especially intriguing from a bioinorganic chemical perspective. F-, strongly hydrated in aqueous solution (ΔH ∼ -125 kcal/mol, 35 kcal/mol stronger than for Cl-) is so loath to shed its water shell that it has been difficult to develop host-guest compounds selective for F- over Cl- (26, 27). The F--specific riboswitch achieves high selectivity by enveloping F- in a complex shell composed of Mg2+ and RNA phosphate (13), but CLCFs must extract and presumably desolvate F- ion from solution via protein moieties alone, without ionic cofactors, while shunning the more easily dehydrated Cl- ion.
, or SCN-. Permeation of F- has been little studied, but the CLC-ec1 antiporter, the mammalian Cl- channel CLC-1, and a CLC channel from Entamoeba histolytica catalyze low but nonnegligible F- conduction (12, 24, 25). In contrast, CLCF antiporters greatly prefer F- over Cl- under the conditions used here, an ion selectivity especially intriguing from a bioinorganic chemical perspective. F-, strongly hydrated in aqueous solution (ΔH ∼ -125 kcal/mol, 35 kcal/mol stronger than for Cl-) is so loath to shed its water shell that it has been difficult to develop host-guest compounds selective for F- over Cl- (26, 27). The F--specific riboswitch achieves high selectivity by enveloping F- in a complex shell composed of Mg2+ and RNA phosphate (13), but CLCFs must extract and presumably desolvate F- ion from solution via protein moieties alone, without ionic cofactors, while shunning the more easily dehydrated Cl- ion.
Third, we note that CLCF proteins present a glaring counterexample to a cherished regularity among other CLCs, namely the conservation of valine (in the channels) or glutamate (in most of the antiporters) at a specific site in the “PIGG-pen signature sequence” proposed to distinguish channels from antiporters (15). Although a recently discovered plant CLC antiporter with threonine at this position rather than glutamate weakens this sequence’s utility (28), the CLCFs provide the first example of a PIGG-pen valine in proton-coupled antiporters. On this basis, we hasten to demote this signature sequence to a mere mechanistic indicator.
A final oddity of CLCF proteins is their unprecedented 1-to-1 F-/H+ stoichiometry. All transport mechanisms previously proposed for CLC antiporters incorporate the coordinated counter movements of two Cl- ions with one H+ as a fundamental characteristic of CLC exchange machinery (28–30). The striking violation of 2-to-1 stoichiometry in CLCF-eca undermines this generality. The demands of planar bilayer recording constrain us to subject only a single CLCF to electrophysiological analysis, but we suspect from the phylogenetic similarities among these proteins that 1-to-1 stoichiometry will turn out to be a common feature of F-/H+ antiporters.
In summary, the CLCF antiporters represent a dramatic departure in sequence, function, and mechanism from the familiar members of the CLC superfamily. This work provides an initial outline of their idiosyncratic physiological role, exquisite F- selectivity, and unprecedented mechanistic quirks. We are struck by the several surprises already emerging from this survey. These surprises foreshadow a deeper and wider understanding of the entire CLC family to follow from more detailed studies of transport mechanism and prokaryotic F- biology and eventually from high-resolution structures.
Materials and Methods
Phylogenetic Tree.
CLC protein sequences were obtained from the PFAM database and were trimmed to exclude soluble domains. Multiple sequence alignment was performed for the full CLC domains using MUSCLE (31). From this preliminary coarse alignment, the transmembrane regions, which are more conserved, were identified based on comparison to the CLC-ec1 structure. These regions were retained, and variable loops and the extramembrane termini were removed. The remaining transmembrane helical regions were realigned, and manually adjusted using JalView (32). Phylogenetic analysis was conducted using PhyML 3.0 (33). The output tree was analyzed using Seaview (34) and FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
F- Toxicity Rescue.
Synthetic gene constructs coding for CLC homologues (Table 1) with C-terminal hexahistidine tags were purchased from Genscript and inserted into the pBAD18 expression vector using XbaI/HindIII restriction sites. E. coli (BW2113 background strain or ΔcrcB) was transformed with pBAD containing the CLC insert. Overnight cultures from single colonies were diluted 100-fold in LB and grown for approximately 2 doubling times (45 min) before inducing protein expression with 0.2% arabinose. After an hour, NaF was added to the desired concentration, whereupon growth was monitored by optical density. Protein expression was confirmed by Western blotting against the hexahistidine tag.
Protein Purification and Liposome Reconstitution.
For overexpression and biochemical applications, the coding sequences were inserted into a pASK vector with an N- or C-terminal hexahistidine tag (9). E. coli (BL21-DE3) was induced with anhydrotetracycline at an optical density of 1.0, and protein was expressed for 3 h or until optical density fell below 0.8. Cells were lysed by sonication and extracted with 40 mM decylmaltoside (DM) for 2 h at room temperature. After pelleting the cell debris, the clarified extract was passed over cobalt affinity beads (1 mL/L culture), washed with 100 mM NaCl, 10 mM imidazole, 5 mM DM, and eluted with solution containing 400 mM imidazole. Protein was further purified on a Superdex200 gel-filtration column in 100 mM NaCl, 10 mM NaF, 10 mM (2-acetamido)iminodiacetic acid, pH 6.5, 5 mM DM. Proteins eluting as monodisperse peaks at positions expected for a CLC homodimer were carried forward for functional analysis. Proteoliposomes were formed by dialysis of the micellar mix of protein (20–100 μg/mL), E. coli polar lipid (10 mg/mL), 5 mM DM against the desired intraliposomal solution at room temperature for 36 h. Liposomes were stored in aliquots at -80 °C until the day of use. Prior to functional assays, liposomes were extruded 21 times through a 400 nm membrane filter.
For the ultra-pure CLC-eca preparation required for planar bilayer recording, which cannot tolerate more than one part in 105 contamination by outer membrane porins endogenous to E. coli (21), the purification procedure included additional cobalt-column washes with 0.1% Triton-X-114 and 1 M NaCl, and substituted tandem ion-exchange chromatography for the Superdex200 step. Protein eluted from the colbalt column was diluted 10-fold into IE buffer (10 mM NaCl, 5 mM DM, 10 mM Mes-NaOH pH 6.5) and applied to a 2-mL cation-exchange column (Poros 50 HS). This column was then washed with 10 volumes each of IE buffer adjusted to pH 5.5, IE buffer+100 mM NaCl, and IE buffer+200 mM NaCl. Protein was eluted with IE buffer+500 mM NaCl. The eluate was concentrated approximately10-fold, diluted into IE buffer and applied to a 2-mL anion exchange column (Poros HQ beads), to which CLC-eca does not bind. The run through was concentrated and used for reconstitution as above, using 7.5 mg/mL 1-palmitoyl, 2-oleoyl phosphatidylethanolamine + 2.5 mg/mL 1-palmitoyl, 2-oleoyl phosphatidylglycerol (POPE/POPG) at a high protein density, 50–70 μg/mg lipid.
19F NMR Transport Assay.
For NMR analysis, liposomes were formed at a concentration of 20 mg lipid/mL, 40 μg protein/mL and loaded with 300 mM KF, 10 mM Hepes, 10 mM EDTA, pH 7.5. Immediately prior to data collection, the extraliposomal solution was exchanged for 200 mM Na2SO4, 10 mM Hepes, pH 7.5 by spinning through a 1.5-mL Sephadex G50 column and was supplemented with 10% D2O and 1 mM trifluoroacetate as an internal reference standard. This procedure resulted in approximately 1 mM F- remaining in the external solution. One-dimensional 19F NMR experiments were performed at 20 °C on a Varian Inova 500 MHz spectrometer equipped with a broadband probe. Each data set was recorded with an evolution time of 100 ms, an interscan delay of 1 s along with eight scans per transient, giving rise to a net acquisition time of approximately 9 s per spectrum. After recording the first spectrum to quantify the initial inside/outside F- signals, F- transport was initiated by the adding 1 μM Vln to the NMR tube with brief agitation. After a dead time of 15–20 s necessary for reinserting the sample into the spectrometer, F- efflux was followed by recording 128 spectra over approximately 20 min under steady-state conditions. The NMRPipe/NMRDraw software package was used to process the NMR data and quantify the peak intensities.
Light Scattering Assay of F- Efflux.
Anion efflux from liposomes was monitored as a change in 90° light scattering at 600 nm. In a typical experiment, a liposome sample containing 300 mM KF, 10 mM Hepes-KOH pH 7 was diluted 200-fold into 2 mL of a degassed isotonic solution containing 300 mM K+ isethionate, 10 mM Hepes-KOH pH 7 in a stirred cuvette. Baseline was allowed to stabilize for 20 s before flux was initiated by 1 μM Vln, which sets the liposome membrane potential to zero. Water efflux maintaining osmotic balance leads to time-dependent shrinking and flattening of the initially spherical liposomes, accompanied by an approximately 10% increase in 90° light scattering (19).
F--Driven H+ Pumping.
Liposomes were prepared with an internal solution of 300 mM KF, 10–25 mM Hepes, pH 7. The extraliposomal solution was exchanged for 300 mM K+ or Na+ isethionate, 1 mM Hepes, pH 7.2 and diluted 20-fold into 1.9 mL of the same buffer. Upon Vln addition, proton uptake was monitored continuously in a stirred cell by a glass electrode as an increase in pH of the external solution (3). The proton gradient was dissipated by 1 μM carbonylcyanide p-trifluoromethoxyphenylhydrazone (FCCP) after uptake had reached steady-state, and H+ transport was calibrated with a HCl standard solution.
Planar Lipid Bilayer Recording.
Planar bilayer recording of CLC-mediated anion current under voltage clamp was as previously described (21), using a POPE/POPG phospholipid mixture to form the bilayer and 150 mM NaF-1.5% agar salt bridges to connect the recording chambers to the Ag/AgCl electrodes through 150 mM KCl wells. Liposomes, after three rounds of freeze-thaw followed by approximately 10 s sonication, were fused into the bilayer with 300 mM NaF, 15 mM Mes-NaOH/tris-H2SO4 mixtures, depending on the pH, in the cis chamber, and 30 mM NaF, 15 mM buffer in the trans chamber, which is defined as electrical ground. CLC incorporation was monitored at -100 mV as the development of anion-selective current in discrete steps. After F- conductance had stabilized, gradients of F- or pH were established by perfusing the trans side with the desired solution. I–V relations were determined with 500-ms pulses from a holding potential of zero to command-voltages from -100 to +100 mV in 10-mV increments. Current output from a 505B amplifier (Warner Instruments) was low-pass filtered at 500 Hz and sampled at 10 kHz in pCLAMP software. Voltages were corrected for junction potential, always < 3 mV, and antiport stoichiometry was calculated from Eq. 1.
ACKNOWLEDGMENTS.
We thank Ronald R. Breaker for alerting us before publication to his discovery of riboswitch-controlled CLCs and for providing the ΔcrcB E. coli strain, and Jorge Arreola for sharing unpublished results of F- currents in CLC-2. We are also grateful to Dorothee Kern for facilitating the use of the Brandeis NMR facility and to Douglas Theobald for help with constructing the phylogenetic tree. R.O. is a HHMI Fellow of the Damon Runyon Cancer Research Foundation, DRG-2114-12 Z.W. was supported through Howard Hughes Medical Institute funding to Ronald R. Breaker.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.White MM, Miller C. A voltage-gated anion channel from electric organ of Torpedo californica. J Biol Chem. 1979;254:10161–10166. [PubMed] [Google Scholar]
- 2.Miller C. Open-state substructure of single chloride channels from Torpedo electroplax. Philos Trans R Soc Lond B Biol Sci. 1982;299:401–411. doi: 10.1098/rstb.1982.0140. [DOI] [PubMed] [Google Scholar]
- 3.Accardi A, Miller C. Secondary active transport mediated by a prokaryotic homologue of ClC Cl- channels. Nature. 2004;427:803–807. doi: 10.1038/nature02314. [DOI] [PubMed] [Google Scholar]
- 4.Picollo A, Pusch M. Chloride/proton antiporter activity of mammalian CLC proteins ClC-4 and ClC-5. Nature. 2005;436:420–423. doi: 10.1038/nature03720. [DOI] [PubMed] [Google Scholar]
- 5.Scheel O, Zdebik AA, Lourdel S, Jentsch TJ. Voltage-dependent electrogenic chloride/proton exchange by endosomal CLC proteins. Nature. 2005;436:424–427. doi: 10.1038/nature03860. [DOI] [PubMed] [Google Scholar]
- 6.Graves AR, Curran PK, Mindell JA. The Cl-/H+ antiporter CLC-7 is the primary chloride permeation pathway in lysozomes. Nature. 2008;453:788–792. doi: 10.1038/nature06907. [DOI] [PubMed] [Google Scholar]
- 7.Accardi A, Picollo A. CLC channels and transporters: proteins with borderline personalities. Biochim Biophys Acta. 2010;1798:1457–1464. doi: 10.1016/j.bbamem.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeAngeli A, et al. AtCLCa, a proton/nitrate antiporter, mediates nitrate accumulation in plant vacuoles. Nature. 2006;442:939–942. doi: 10.1038/nature05013. [DOI] [PubMed] [Google Scholar]
- 9.Maduke M, Pheasant DJ, Miller C. High-level expression, functional reconstitution, and quaternary structure of a prokaryotic ClC-type chloride channel. J Gen Physiol. 1999;114:713–722. doi: 10.1085/jgp.114.5.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dutzler R, Campbell EB, MacKinnon R. Gating the selectivity filter in ClC chloride channels. Science. 2003;300:108–112. doi: 10.1126/science.1082708. [DOI] [PubMed] [Google Scholar]
- 11.Miller C. ClC chloride channels viewed through a transporter lens. Nature. 2006;440:484–489. doi: 10.1038/nature04713. [DOI] [PubMed] [Google Scholar]
- 12.Baker JL, et al. Widespread genetic switches and toxicity resistance proteins for fluoride. Science. 2012;335:233–235. doi: 10.1126/science.1215063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren A, Rajashankar KR, Patel DJ. Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch. Nature. 2012;486:85–89. doi: 10.1038/nature11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinstein LH, Davison A, editors. Fluorides in the Environment: Effects on Plants and Animals. Cambridge, Mass: CABI Publishing; 2004. [Google Scholar]
- 15.Accardi A, et al. Separate ion pathways in a Cl-/H+ exchanger. J Gen Physiol. 2005;126:563–570. doi: 10.1085/jgp.200509417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deverell C, Shaumberg K, Bernstein HJ. 19F nuclear magnetic resonance chemical shift of alkali fluorides in light- and heavy-water solutions. J Chem Phys. 1968;49:1276–1283. [Google Scholar]
- 17.Walden M, et al. Uncoupling and turnover in a Cl-/H+ exchange transporter. J Gen Physiol. 2007;129:317–329. doi: 10.1085/jgp.200709756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jayaram H, Robertson JL, Wu F, Williams C, Miller C. Structure of a slow CLC Cl/H+ antiporter from a cyanobacterium. Biochemistry. 2011;50:788–794. doi: 10.1021/bi1019258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin AJ, Huster D, Gawrisch K, Nossal R. Light scattering characterization of extruded lipid vesicles. Eur Biophys J. 1999;28:187–199. doi: 10.1007/s002490050199. [DOI] [PubMed] [Google Scholar]
- 20.Accardi A, Lobet S, Williams C, Miller C, Dutzler R. Synergism between halide binding and proton transport in a CLC-type exchanger. J Mol Biol. 2006;362:691–699. doi: 10.1016/j.jmb.2006.07.081. [DOI] [PubMed] [Google Scholar]
- 21.Accardi A, Kolmakova-Partensky L, Williams C, Miller C. Ionic currents mediated by a prokaryotic homologue of CLC Cl- channels. J Gen Physiol. 2004;123:109–119. doi: 10.1085/jgp.200308935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zifarelli G, Pusch M. CLC transport proteins in plants. FEBS Lett. 2010;584:2122–2127. doi: 10.1016/j.febslet.2009.12.042. [DOI] [PubMed] [Google Scholar]
- 23.Picollo A, Malvezzi M, Houtman JC, Accardi A. Basis of substrate binding and conservation of selectivity in the CLC family of channels and transporters. Nat Struct Mol Biol. 2009;16:1294–1301. doi: 10.1038/nsmb.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rychkov GY, Pusch M, Roberts ML, Jentsch TJ, Bretag AH. Permeation and block of the skeletal muscle chloride channel, ClC-1, by foreign anions. J Gen Physiol. 1998;111:653–665. doi: 10.1085/jgp.111.5.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salas-Casas A, et al. Identification and functional characterization of EhClC-A, an Entamoeba histolytica ClC chloride channel located at plasma membrane. Mol Microbiol. 2006;59:1249–1261. doi: 10.1111/j.1365-2958.2006.05023.x. [DOI] [PubMed] [Google Scholar]
- 26.Cametti M, Rissanen K. Recognition and sensing of fluoride anion. Chem Commun. 2009;20:2809–2829. doi: 10.1039/b902069a. [DOI] [PubMed] [Google Scholar]
- 27.Wade CR, Broomsgrove AEJ, Aldridge S, Gabbai FP. Fluoride ion complexation and sensing using organoboron compounds. Chem Rev. 2010;110:3958–3984. doi: 10.1021/cr900401a. [DOI] [PubMed] [Google Scholar]
- 28.Feng L, Campbell EB, Hsiung Y, MacKinnon R. Structure of a eukaryotic CLC transporter defines an intermediate state in the transport cycle. Science. 2010;330:635–641. doi: 10.1126/science.1195230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller C, Nguitragool W. A provisional transport mechanism for a CLC-type Cl-/H+ exchanger. Philos Trans R Soc Lond B Biol Sci. 2008;364:175–180. doi: 10.1098/rstb.2008.0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Picollo A, Xu Y, Johner N, Berneche S, Accardi A. Synergistic substrate binding determines a the stoichiometry of transport of a prokaryotic H+∶Cl- exchanger. Nat Struct Mol Biol. 2012;19:525–531. doi: 10.1038/nsmb.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. (Oxford, England) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 34.Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]