Abstract
The p53 protein plays a central role in the prevention of tumorigenesis. Cellular stresses, such as DNA damage and aberrant oncogene activation, trigger induction of p53 that halts cellular proliferation and allows cells to be repaired. If cellular damage is beyond the capability of the repair mechanisms, p53 induces apoptosis or cell cycle arrest, preventing damaged cells from becoming cancerous. However, emerging evidence suggests that the function of p53 needs to be considered as isoform-specific. Here, we report that the expression profile of p53 can be shifted toward inhibitory p53 isoforms by the pathogenic bacterium Helicobacter pylori, which is known for its strong association with gastric cancer and gastric mucosa-associated lymphoid tissue lymphoma. We found that interaction of H. pylori with gastric epithelial cells, mediated via the cag pathogenicity island, induces N-terminally truncated ∆133p53 and ∆160p53 isoforms in human cells. Induction of an orthologous p53 isoform, ∆153p53, was also found in H. pylori-infected Mongolian gerbils. The p53 isoforms inhibit p53 and p73 activities, induce NF-κB, and increase survival of infected cells. Expression of ∆133p53, in response to H. pylori infection, is regulated by phosphorylation of c-Jun and activation of activator protein-1–dependent transcription. Together, these results provide unique insights into the regulation of p53 protein and may contribute to the understanding of tumorigenesis associated with H. pylori.
Keywords: gastric tumor, AP-1, c-Jun
The gram-negative bacterium Heicobacter pylori infects approximately half of the world’s population and is a strong risk factor for gastric cancer and gastric mucosa-associated lymphoid tissue (MALT) lymphoma. H. pylori pathogenesis is determined largely by interactions between bacterial factors and host cells. The most well-characterized virulence determinants are the vacuolating cytotoxin A (vacA) and cag pathogenicity island, a 40-kb region of DNA that encodes a type IV secretion system (T4SS). The T4SS forms a syringe-like pilus structure used for the injection of bacterial components directly into host cells, triggering complex alterations of the host signaling pathways. Aberrant activation of multiple oncogenic cascades (PI3K/Akt, Wnt/β-catenin, Ras/Erk, and others) and DNA damage induced by H. pylori directly or indirectly, through chronic inflammatory responses, are thought to result in premalignant lesions and subsequent cancer development (1).
Oncogenic stress and DNA damage normally activate the p53 protein. Depending on the nature of the stress signal and cellular context, activated p53 induces apoptosis, cell cycle arrest, or cellular senescence that, in turn, inhibits proliferation of damaged cells, maintains the genome integrity, and prevents tumorigenesis (2, 3). In gastric cancer, p53 is inactivated by mutations in ∼40% of tumor cases. H. pylori infection has been shown to enhance mutagenesis in the p53 gene (4). However, emerging evidence suggests that during precancerous stages, H. pylori compromises the p53 function by mutation-independent mechanisms. We have previously demonstrated that proteasomal degradation of p53 is significantly induced by CagA bacterial protein in gastric epithelial cells infected with H. pylori (5). This process is regulated by cellular apoptosis-stimulating protein of p53 2 (ASPP2) protein (6). Inhibition of p53 transcription activity and p53-dependent apoptosis by H. pylori has also been reported in other cell types, including B cells, the cell of origin of MALT lymphoma (7).
The tumor suppressor function of p53 is well characterized, yet much less is known regarding the function of recently discovered p53 isoforms. Based on the mechanisms of transcriptional regulation, p53 isoforms can generally be divided into two groups, those produced from the main P1 promoter and those produced from the alternative intragenic P2 promoters (8). The latter group, represented by a human ∆133p53 isoform, has been suggested to promote tumorigenesis. This is consistent with a recent finding of strong spontaneous tumorigenesis in mice expressing the ∆122p53 protein that mimics the human ∆133p53 isoform (9). In human colon adenomas, the ∆133p53 isoform has been found to inhibit p53-mediated replicative senescence, and its increase may signal an escape from the senescence barrier during the progression from adenoma to carcinoma (10). Elevated levels of ∆133p53 have also been reported in a number of human tumors (11).
Here, we report that the interaction of gastric epithelial cells with H. pylori leads to up-regulation of truncated p53 isoforms in a T4SS-dependent manner. These isoforms increase transcriptional activity of NF-κB and inhibit p53 and p73. Together, our studies provide a previously undescribed link between p53 isoforms and H. pylori that may be an important component of tumorigenesis associated with this bacterium.
Results
H. pylori Induces p53 Isoforms in Vitro and in Vivo.
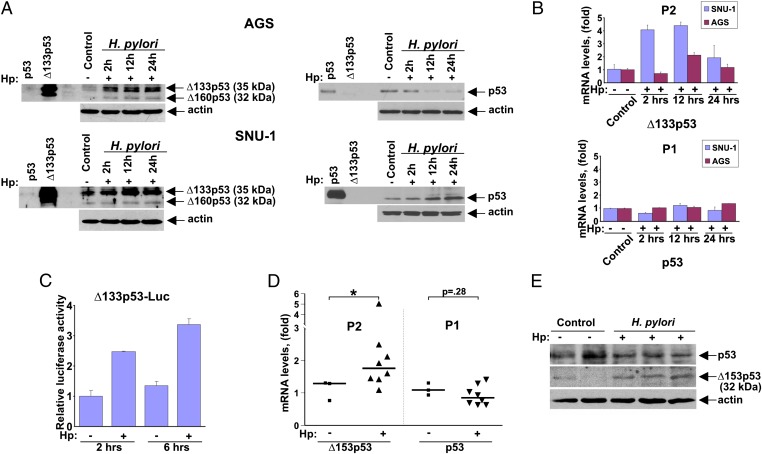
To investigate regulation of the p53 protein, AGS (and SNU-1) gastric epithelial cells harboring WT p53 were co-cultured with H. pylori (strain J166) and analyzed for protein expression of p53 isoforms. Surprisingly, we found strong up-regulation of truncated p53 isoforms, ∆133p53 and ∆160p53, in both cell lines following co-culture with H. pylori (Fig. 1A). Isoform induction was further confirmed using a panel of p53 antibodies and isoform-specific shRNA (full blots are shown in Figs. S1 and S2). Notably, regulation of full-length p53 protein was varied between cell lines, with a significant down-regulation of p53 in AGS cells and an increase in SNU-1 cells. This is consistent with previous observations showing different dynamics of p53 inhibition in cell lines co-cultured with H. pylori (5).
Fig. 1.
Truncated p53 isoforms are up-regulated by H. pylori. (A) Protein lysates were prepared from control (−) AGS cells (Upper) and SNU-1 cells (Lower) or from those co-cultured (+) with H. pylori strain J166 for the indicated time and analyzed for expression of p53 and Δ133p53 (Δ160p53) isoforms by Western blotting using DO-1 (Right) and DO-11 (Left) antibodies, respectively. (B) Bar graph represents qRT-PCR analyses of p53 transcripts produced from the P2 (Upper) and P1 (Lower) promoters in SNU-1 and AGS cells co-cultured with H. pylori for the indicated time. Data are normalized to HPRT1 mRNA expression. The mRNA expression of p53 isoforms in uninfected cells is arbitrarily set at 1. (C) SNU-1 cells transfected with a luciferase reporter (Δ133p53-Luc) were co-cultured with H. pylori (Hp) for the indicated time and analyzed using a dual-luciferase reporter assay. (D) Expression of p53 transcripts derived from the P1 and P2 promoters in the gastric mucosa harvested from gerbils infected with H. pylori strain 7.13 for 3 d. H. pylori infection leads to increased expression of Δ153p53 mRNA. *P < 0.05 vs. uninfected control. (E) Expression analysis of p53 and Δ153p53 proteins in gerbils infected with H. pylori strain 7.13 for 3 d using Western blotting with DO-1 and CM1 antibodies, respectively. p53 and Δ153p53 protein migrated as bands with molecular masses of 52 kDa and 32 kDa.
We next examined the transcription of the p53 gene using primers that specifically amplify transcripts derived from either the P1 or P2 promoter in SNU-1 and AGS cells. The analysis revealed a striking difference between these two groups of transcripts. We found that whereas levels of p53 mRNA produced by the P1 promoter did not change, the P2 transcripts were induced by H. pylori. The greatest difference was seen in SNU-1 cells cocultured with H. pylori J166 (Fig. 1B). To confirm activation of the P2 promoter, we next performed a luciferase reporter analysis in SNU-1 cells. Fig. 1C shows that the reporter, controlled by a 1.5-kb fragment of the P2 p53 gene promoter, was significantly induced following co-culture with H. pylori.
To investigate regulation of the p53 gene promoters in vivo, we used the Mongolian gerbil (Meriones unguiculatus) model, because gastric pathology in this system more closely recapitulates human infection than in murine models (12). Because gerbil p53 isoforms were unknown, we used a transcript walking approach to identify the corresponding p53 isoforms, which we then sequenced. We found that, similar to human and murine counterparts, the gerbil TP53 gene contains two promoters and produces a ∆153p53 transcript that is highly homologous to previously described human ∆133p53 (∆160p53) and murine ∆157p53 isoforms (Fig. S3). To define the gerbil isoforms more robustly, Mongolian gerbils (n = 8) were inoculated with the cag+ rodent-adapted H. pylori strain 7.13, whereas a control group of animals (n = 3) received sterile broth alone. Three days after infection, gastric tissues were collected and examined for expression of p53 isoforms using quantitative PCR (qPCR) and Western blotting. Similar to our analyses in vitro, we found significant up-regulation of ∆153p53 transcripts, whereas p53 mRNA levels remained unchanged or slightly decreased (Fig. 1D). Taking advantage of significant protein similarities between human and gerbil isoforms, we identified a human antibody (CM1; Novocastra) that recognizes gerbil ∆153p53 protein. We confirmed a significant increase of ∆153p53 protein in the stomachs of infected gerbils using Western blotting (Fig. 1E). Another p53 antibody (DO-11; AbDseroTec) also detected increased levels of ∆153p53 protein (Fig. S3). In contrast to ∆153p53 isoform, levels of full-length p53 were not significantly altered (Fig. 1E). Together, these results demonstrate that H. pylori infection results in increased expression of truncated p53 isoforms in vitro and in vivo.
∆133p53 Isoform Suppresses p53 and p73 Transcriptional Activities and Increases Cell Survival of H. pylori-Infected Cells.
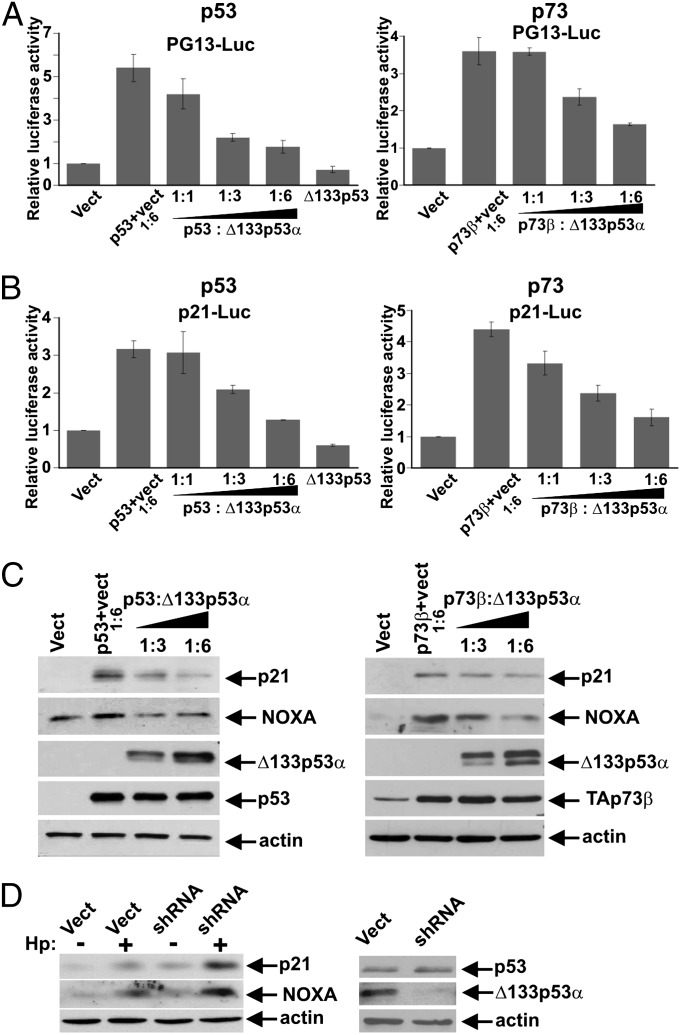
We next assessed the role of ∆133p53 in our cell culture model. SNU-1 cells were cotransfected with p53 and an increasing amount of ∆133p53 expression plasmid and were used for reporter assays. As an initial approach, we used the PG13-Luc reporter that contains multiple repeats of the p53 consensus binding site. Given that another member of the p53 family, p73, is activated in response to H. pylori (13), p73 activity was also analyzed using the same approach. We found that the transcriptional activities of both p53 and p73 proteins were strongly inhibited by ∆133p53 in a dose-dependent manner (Fig. 2A). A similar pattern of inhibition was observed using another p53/p73 reporter, p21-pWWP-Luc, which is controlled by a 2.4-kb promoter fragment of the CDKN1A gene that encodes the CDK inhibitor, p21 (Fig. 2B). These results were confirmed by Western blotting, where expression of p21/WAF1 and the proapoptotic p53 target protein, NOXA, was strongly inhibited by ectopic expression of ∆133p53 (Fig. 2C). We next determined whether the endogenous ∆133p53 protein affects the expression of p53/p73 targets. SNU-1 cells were stably transfected with a shRNA plasmid that selectively targets the ∆133p53 isoform but not p53 (Fig. 2D) and were analyzed for expression levels of p21 and NOXA proteins following co-culture with H. pylori. Our analyses revealed that down-regulation of ∆133p53 increases expression of p21 and NOXA proteins, further supporting the role of ∆133p53 in regulation of p53 target genes (Fig. 2D).
Fig. 2.
Δ133p53 suppresses p53 and p73 transcriptional activities. (A) Transcriptional activities of p53 (Left) and p73 (Right) were determined using PG13-Luc reporter in SNU-1 cells following cotransfection with Δ133p53 at the indicated molar ratios. Luciferase activity in cells transfected with an empty vector (Vect) is arbitrarily set at 1. (B) Same as in A, but p21-pWWP-Luc reporter was used. (C) p53-null cells, Kato III, were cotransfected with Δ133p53 and p53 (Left) or p73 (Right) at the indicated ratios and analyzed for expression of p21 and NOXA proteins by Western blotting. (D) Expression of p21 and NOXA proteins was analyzed in SNU-1 cells, in which levels of endogenous Δ133p53 isoform were suppressed with shRNA. (Right) shRNA plasmid selectively targets the ∆133p53 isoform but not p53.
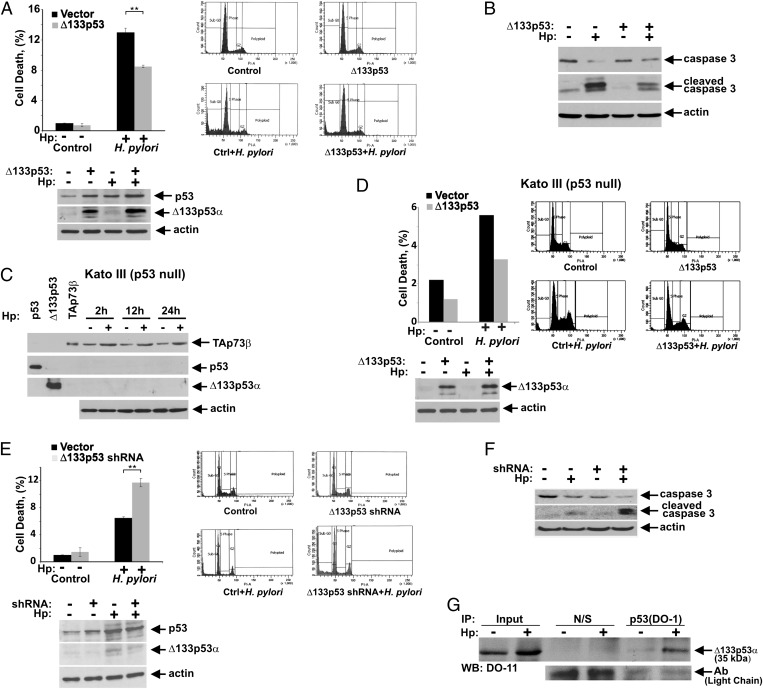
In view of the fact that H. pylori damages epithelial cells and potentiates apoptosis (14), we next examined the effect of ∆133p53 on the viability of gastric epithelial cells. Because SNU-1 cells express endogenous p53 and ∆133p53 proteins (Fig. 1A), we first assessed whether a further increase in ∆133p53 protein levels affects cell death induced by H. pylori. Cell survival was measured by flow cytometry in SNU-1 cells stably transfected with either empty vector or ∆133p53α expression plasmid and co-cultured with H. pylori strain J166 for 24 h. Transfection of ∆133p53 significantly increased survival of infected cells, although its effect may be diminished by the presence of the endogenous protein (Fig. 3A). Caspase 3 cleavage was also reduced, suggesting that ectopic expression of ∆133p53 inhibits H. pylori-induced apoptosis (Fig. 3B).
Fig. 3.
Δ133p53 protein increases cell survival of H. pylori-infected cells. (A) SNU-1 cells stably transfected with Δ133p53 or empty vector were co-cultured with H. pylori (Hp) strain J166 for 24 h and analyzed for cell death by flow cytometry using propidium iodide (PI) staining. The percentage of cells in sub-G1 is shown. The Δ133p53 transfection significantly increases survival of H. pylori-infected cells (**P = 0.002; n = 3). (Lower) Expression of Δ133p53 protein in analyzed cells is shown. (B) Same as in A, but apoptosis was analyzed by Western blotting with antibodies recognizing uncleaved and cleaved caspase 3. (C) p73 is strongly up-regulated by H. pylori in p53-null Kato III cells. Cells were co-cultured with H. pylori strain J166 and analyzed for expression of TAp73β, p53, and Δ133p53 proteins at the indicated time using p73, DO-1, and DO-11 antibodies. (D) Same as in A, but Kato III cells were analyzed. (Lower) Expression of exogenous Δ133p53 protein. (E) SNU-1 cells, stably transfected with either shRNA against Δ133p53 or scrambled control vector, were co-cultured with H. pylori strain J166 for 24 h and analyzed as described in A. Down-regulation of Δ133p53 significantly increases cell death induced by H. pylori (**P = 0.009; n = 3). (Lower) Expression of endogenous Δ133p53 protein is shown. (F) Same as in E, but apoptosis was analyzed by Western blotting with antibodies recognizing uncleaved and cleaved caspase 3. (G) Δ133p53 protein physically interacts with p53. Protein lysates from SNU-1 cells co-cultured with H. pylori (+) for 12 h or left uninfected (−) were immunoprecipitated with either p53 (DO-1) antibody or nonspecific mouse IgG (N/S). The p53–Δ133p53 protein complexes were analyzed by Western blotting with the p53 (DO-11) antibody, which recognizes the Δ133p53 isoform. Equal antibody loading was verified by detecting light chains of antibodies.
Previously, it has been reported that p73 activation contributes to apoptosis induced by H. pylori (13). To assess whether ∆133p53 affects p73-dependent apoptosis, we studied apoptosis in the gastric cancer cell line Kato III. These cells lack p53 expression and strongly up-regulate TAp73β following co-culture with H. pylori (Fig. 3C). Similar to WT p53 SNU-1 cells, ∆133p53 transfection strongly increased survival of infected cells, implying that ∆133p53 inhibits p73-induced cell death (Fig. 3D, cell cycle analysis is illustrated in Fig. S3). To confirm the role of ∆133p53 further, we next evaluated endogenous ∆133p53 function. Cell death was analyzed in SNU-1 cells in which ∆133p53 was down-regulated via the use of specific shRNA, which not only decreased steady-state levels of endogenous ∆133p53 protein but diminished induction of ∆133p53 by H. pylori (Fig. 3E, Lower). This resulted in a significant increase in caspase 3 cleavage and cell death, supporting a model in which relative expression levels of ∆133p53 and full-length p53 (and TAp73) isoforms regulate activities of p53 and p73 in H. pylori-infected cells (Fig. 3 E and F).
We next analyzed the mechanism of ∆133p53 inhibition in more detail. Because p53 functions as a tetramer and ∆133p53 shares the oligomerization domain with p53, one explanation for the inhibitory function of ∆133p53 is that ∆133p53 forms transcriptionally defective oligomeric complexes with p53. To assess this possibility, protein interactions between p53 and ∆133p53 isoforms were examined in cells co-cultured with H. pylori. Using an antibody, DO-1, that recognizes p53 but not ∆133p53 (8), we were able to coimmunoprecipitate p53 and ∆133p53 complexes. Importantly, we found that H. pylori strongly enhances the binding of ∆133p53 to p53 (Fig. 3G). In a separate series of experiments, we also investigated binding of ∆133p53 to p73. Our results indicated that ∆133p53 inhibits p73 through distinct mechanism(s), because we were unable to co-immunoprecipitate endogenous ∆133p53 with TAp73 in SNU-1 cells (Fig. S3). Combined, our results show that the ∆133p53 isoform enhances survival of cells infected by H. pylori.
∆133p53 Isoform Induces NF-κB Activity.
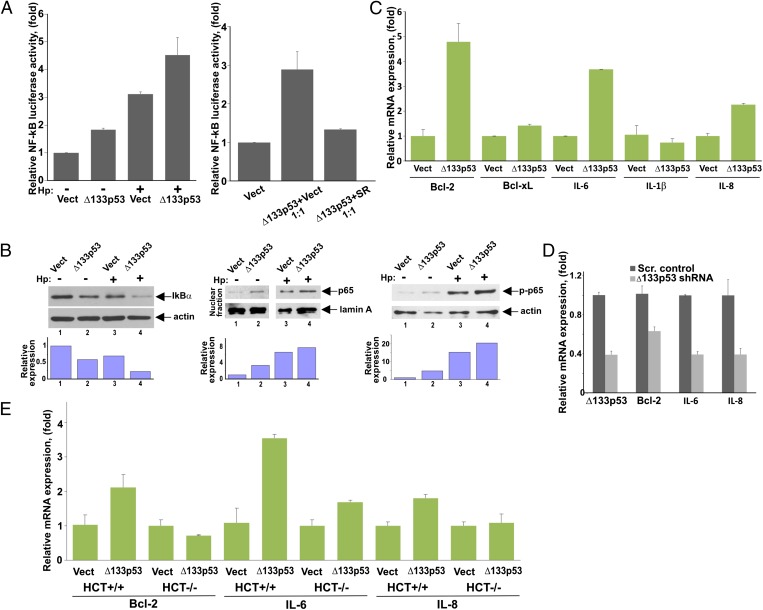
Based on observations that NF-κB is strongly induced by H. pylori and on previous data demonstrating cross-talk between NF-κB and p53 signaling pathways, we further defined the role of ∆133p53 in cells infected with H. pylori. To determine whether ∆133p53 affects NF-κB activity, SNU-1 cells were transfected with ∆133p53 and NF-κB activity was then measured using a luciferase reporter analysis in the presence or absence of H. pylori (Fig. 4A, Left). We found that cells transfected with ∆133p53 exhibit significantly higher activation of the pNFκB-Luc reporter than control cells, indicating that ∆133p53 induces NF-κB activity. To confirm the specificity of our analysis, the reporter analysis was repeated in the presence of the NF-κB inhibitor, IκB super repressor (SR). We found that SR blocks ∆133p53 activity, implying that activation of the reporter is specifically mediated by NF-κB but not by other transcription factors (Fig. 4A, Right). Similar results were obtained using another cell line, AGS (Fig. S4).
Fig. 4.
Δ133p53 protein regulates NF-κB activity. (A) NF-κB activity was assessed in SNU-1 cells transfected with either Δ133p53 expression vector (Δ133p53) or empty control vector (Vect) and co-cultured with H. pylori (Hp) (+) for 8 h or left uninfected (−). NF-κB activity was assessed using the pNFκB-Luc reporter. (Right) Reporter analysis was repeated in the presence of the NF-κB inhibitor, IκB SR. SNU-1 cells were cotransfected with Δ133p53 vector and SR (or empty vector) at a 1:1 ratio. (B) SNU-1 cells stably transfected with Δ133p53 or empty control vector were co-cultured with H. pylori for 3 h and analyzed for IκBα protein (Left) as well as nuclear localization of p65 (RelA) protein (Center) and its phosphorylation at serine 536 (Right) by Western blotting. The graphs represent the results of densitometric analysis of immunoblots and depict actin (or lamin A)-normalized protein expression. (C) SNU-1 cells were transfected with either Δ133p53 or empty control vector for 48 h and analyzed for mRNA expression of the indicated NF-κB target genes by qPCR. (D) AGS cells were stably transfected with a specific shRNA against Δ133p53 or scrambled (Scr.) control vector and were analyzed for mRNA expression of Bcl-2, IL-6, and IL-8 by qPCR. (E) Same as in C, but isogenic cell lines HCT116+/+ (WT p53) and HCT116−/− (p53-null) were used.
To examine NF-κB activity directly, we next analyzed IκBα protein as well as the subcellular location of p65 (RelA) protein and its phosphorylation at Ser536, which are required for NF-κB activation. Consistent with the NF-κB reporter analyses, transfection of ∆133p53 decreased protein levels of IκBα and induced phosphorylation and nuclear translocation of endogenous RelA protein (Fig. 4B). We then analyzed transcription of NF-κB target genes that are known to be induced by H. pylori, including IL-6, IL-1β, Bcl-xL, IL-8, and Bcl-2. We found that ectopic expression of ∆133p53 induces a subset of these NF-κB targets (Fig. 4C). Of note, induction levels were varied between targets, with the highest induction levels seen with IL-6, Bcl-2, and IL-8. To confirm the role of ∆133p53 in these signaling events further, endogenous ∆133p53 was down-regulated using a specific shRNA and transcript levels of IL-6, Bcl-2, and IL-8 were analyzed in AGS cells. As shown in Fig. 4D, ∆133p53 shRNA significantly inhibited the expression of IL-6, Bcl-2, and IL-8. These results raised a question regarding the role of full-length p53. To address it, the aforementioned experiments were repeated in p53-null Kato III cells. We found that transfection of ∆133p53 failed to induce NF-κB activity and NF-κB target genes in these cells, pointing out the important role of p53 (Fig. S4). To confirm these findings further, we took advantage of isogenic HCT116 cell lines that differ only in their p53 status. Expression of NF-κB targets (IL-6, Bcl-2, and IL-8) was compared in p53+/+ and p53−/− HCT116 cells after transfection with ∆133p53. Consistent with our findings in Kato III cells, induction of the analyzed NF-κB targets was significantly decreased in p53−/− HCT116 cells, revealing that ∆133p53 is involved in the regulation of NF-κB activity in a p53-dependent manner (Fig. 4E).
Activator Protein-1 Regulates Transcription of ∆133p53 in H. pylori-Infected Cells.
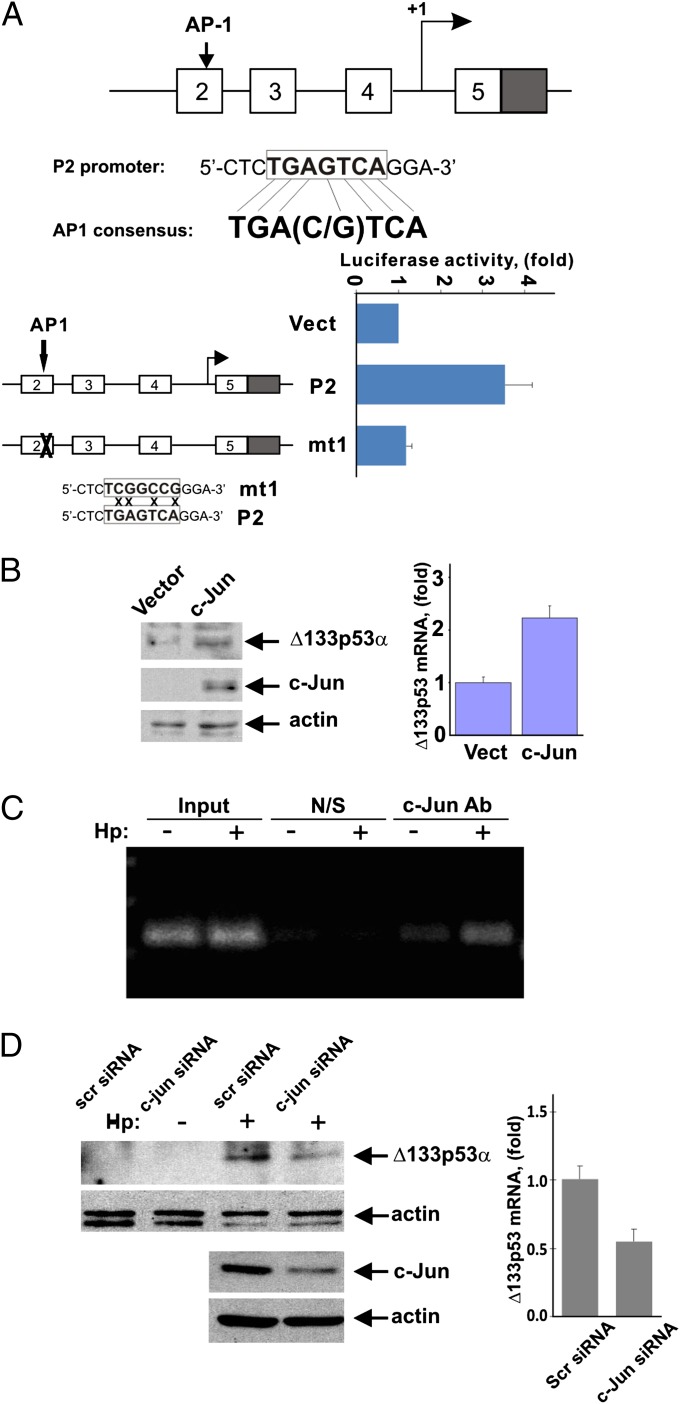
Having demonstrated an increase in ∆133p53 mRNA following infection with H. pylori, we next analyzed the regulation of ∆133p53 transcription. Based on qPCR and the reporter analyses shown in Fig. 1C, we concluded that H. pylori may activate transcription from the P2 promoter. To investigate this possibility, we performed a P2 promoter analysis for potential cis-regulatory elements in silico using Genomatix MatInspector software. Among potential regulators, activator protein-1 (AP-1) transcription factor, a heterodimeric or homodimeric transcription complex that comprises members of the Jun and Fos protein families, had one of the highest matrix similarities. The identified response element located at position -1061/-1055 of the P2 promoter was found to be identical to the AP-1 consensus binding site [TGA(C/G)TCA] (Fig. 5A). To evaluate the contribution of AP-1 to ∆133p53 transcription, SNU-1 cells were cotransfected with a c-Jun expression plasmid and ∆133p53 luciferase reporters controlled by either a 1.5-kb fragment of the P2 promoter or its AP-1 binding site mutant (Fig. 5A). We found that c-Jun significantly increases reporter activity, whereas mutation of the AP-1 binding site causes its suppression, indicating that AP-1 is involved in regulation of the P2 promoter. To assess whether c-Jun affects endogenous ∆133p53, SNU-1 cells transfected with a c-Jun expression plasmid were analyzed for ∆133p53 protein. Consistent with the reporter analysis, c-Jun transfection significantly up-regulated ∆133p53 mRNA and protein (Fig. 5B).
Fig. 5.
AP-1 transcription factor is involved in the regulation of p53 isoforms. (A) Location of AP-1 binding site in the p53 P2 promoter are shown. The activity of luciferase reporters controlled by the 1.5-kb P2 promoter or its AP-1 binding site mutant was analyzed 24 h after transfection of c-Jun. Luciferase activity in cells transfected with empty vector (Vect) is arbitrarily set at 1. (Lower) Four nucleotides were mutated to inactivate the AP-1 binding site. (B) Protein and mRNA levels of Δ133p53 were determined in SNU-1 cells transfected with c-Jun or empty control vector. (C) DNA binding of c-Jun to the native P2 p53 gene promoter was analyzed by ChIP in SNU-1 cells co-cultured with H. pylori strain J166 for 12 h (+) or left uninfected (−). An antibody recognizing phospho-c-Jun (Ser63) was used for detection of the c-Jun binding. (D) Protein and mRNA levels of Δ133p53 were analyzed in SNU-1 cells transfected with either c-Jun siRNA or scrambled control siRNA for 48 h and co-cultured with H. pylori strain J166 for an additional 12 h.
To exclude the possibility of indirect regulation of ∆133p53 transcription by c-Jun, we co-cultured SNU-1 cells with H. pylori strain J166 and analyzed c-Jun promoter binding using ChIP with the phospho-Jun (Ser63) antibody, which recognizes the activated form of c-Jun. ChIP analysis revealed direct binding of phosphorylated c-Jun to the P2 promoter, which was significantly increased by H. pylori (Fig. 5C). We next assessed expression of ∆133p53 protein in SNU-1 cells in which c-Jun was down-regulated using a specific siRNA. As shown in Fig. 5D, c-Jun siRNA inhibits the induction of ∆133p53 mRNA and protein. Thus, our results show that ∆133p53 expression is mediated by the AP-1 transcription factor.
H. pylori cag-Pathogenicity Island Mediates Induction of p53 Isoforms.
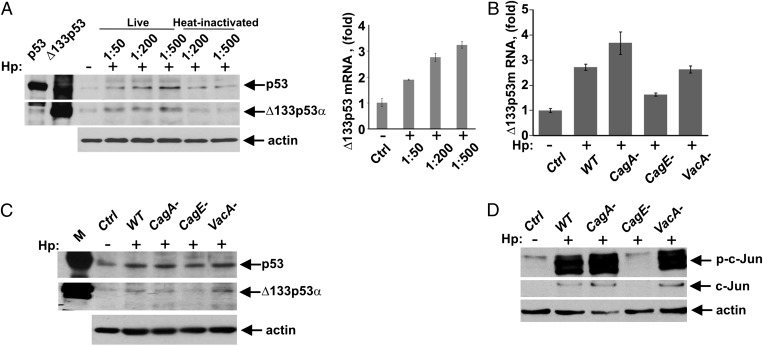
To define the role of H. pylori virulence factors in this process, we co-cultured SNU-1 cells with the cag+ H. pylori strain J166 at different multiplicities of infection (MOI: 50–500) and analyzed expression of ∆133p53. Expression of ∆133p53 mRNA and protein was augmented in a dose-dependent manner (Fig. 6A, representative full blots are shown in Fig. S4). However, even at an MOI of 500, heat-inactivated bacteria failed to elicit induction of ∆133p53, suggesting that viable bacteria are needed for the induction of ∆133p53. To elucidate the specific bacterial factors regulating ∆133p53 expression, isogenic cagA-, vacA-, and cagE-null mutants were generated in H. pylori strain J166. The cagE-null mutant has a defective secretion system, whereas cagA- and vacA-null mutants lack the expression of VacA and CagA virulence factors, respectively. SNU-1 cells were co-cultured with WT or isogenic H. pylori mutants for 12 h and analyzed for ∆133p53 mRNA and protein expression (Fig. 6 B and C), as well as for phosphorylation of c-Jun at serine 63, indicative of c-Jun activation (Fig. 6D). We found that deletion of the vacA-null mutant did not have an effect on either ∆133p53 or c-Jun phosphorylation (Fig. 6 C and D). The deletion of cagA-null mutant was also minimally effective. In contrast, the cagE-null mutant, which does not form a T4SS, lost the ability to induce AP-1 and ∆133p53 proteins, showing that a functional T4SS plays a primary role in the regulation of ∆133p53 protein by H. pylori. Notably, specific siRNA against c-Jun suppressed induction of ∆133p53 by WT, cagA-null, and vacA-null bacteria (Fig. S4).
Fig. 6.
Induction of p53 isoforms is mediated by the T4SS. (A) Protein lysates were prepared from control SNU-1 cells (−) or those co-cultured with H. pylori (Hp) (+) strain J166 at the indicated cell/bacteria ratios or with heat-inactivated bacteria and analyzed for expression of Δ133p53 protein by Western blotting. (Right) qPCR analysis of Δ133p53 transcripts in SNU-1 cells co-cultured with H. pylori is shown. Data are normalized to HPRT1 mRNA expression. The Δ133p53 mRNA level in uninfected cells is arbitrarily set at 1. (B) SNU-1 cells were co-cultured with WT H. pylori strain J166 or its isogenic cagA-, cagE-, or vacA-null mutants and analyzed for Δ133p53 mRNA by qPCR. SNU-1 cells were co-cultured with H. pylori as described in B for 12 h and analyzed for expression of p53 and Δ133p53 proteins (C), as well as c-Jun protein levels and its phosphorylation at position Ser63 (D), by Western blotting.
Discussion
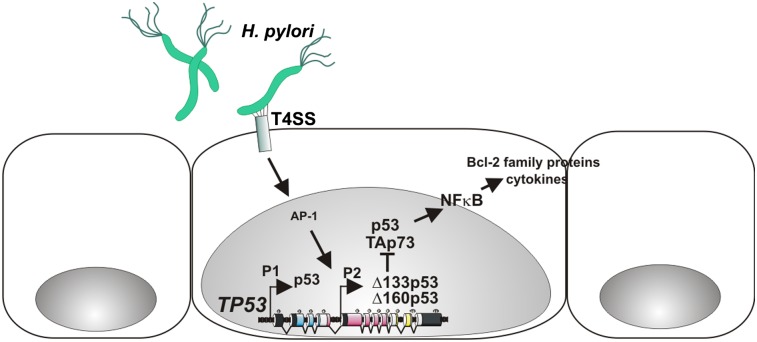
Here, we report that H. pylori alters the expression profile of p53 isoforms, resulting in up-regulation of N-terminally truncated isoforms of p53. These isoforms inhibit the transcriptional activity of p53 and increase survival of epithelial cells infected by H. pylori. Mechanistically, H. pylori increases the binding between ∆133p53 and p53 proteins that, in turn, inhibits p53-dependent transcription. Our studies also demonstrated that ∆133p53 inhibits the transcriptional activity of the TAp73 protein that has been shown to mediate H. pylori-induced apoptosis (13). Therefore, ∆133p53 has an effect through the inhibition of both members of the p53 family. Interestingly, we were unable to coimmunoprecipitate TAp73 and ∆133p53 proteins, suggesting that ∆133p53 inhibits p73 through distinct mechanism(s). Because ∆133p53 retains a significant portion of the DNA binding domain, one possibility is that this isoform may interfere with the binding of p73 to target gene promoters as has been reported for p53 (15). Further studies will be needed to elucidate these mechanisms fully. The ∆133p53 transcript encodes two isoforms, ∆133p53 and ∆160p53 (16). Both proteins were found to be induced by H. pylori, although ∆133p53 is the predominant molecular species. In Mongolian gerbils, we were able to identify only one transcript produced from the P2 promoter, ∆153p53. This isoform is orthologous to human ∆133p53 (∆160p53) and murine ∆157p53 isoforms. In this respect, the expression profile of p53 isoforms in gerbils is similar that in mice, where a single ∆157p53 isoform produced from the P2 promoter is currently known (17).
Our studies also show that ∆133p53 is engaged in the regulation of the NF-κB pathway. Increased expression of ∆133p53 was found to lead to the induction of NF-κB transcription activity and NF-κB target genes, such as proinflammatory cytokines and antiapoptotic Bcl-2 family proteins. Therefore, ∆133p53 inhibits apoptosis not only by directly inhibiting p53 and p73 but through prosurvival signals fostered by the NF-κB pathway. A relative contribution of each mechanism remains unclear, although our analysis suggests that ∆133p53 retains the ability to inhibit cell death without a significant induction of NF-κB in p53-deficient cells. Mechanistically, activation of NF-κB by ∆133p53 is linked to inhibition of p53, given that NF-κB activation is hampered in p53-null cells. These findings are in line with the general concept of “functional antagonism” between p53 and NF-κB pathways (18).
Another important finding directly related to the regulation of ∆133p53 is the role of AP-1–dependent transcription. Our studies identified the AP-1 binding site in the ∆133p53 promoter that is actively engaged in ∆133p53 regulation in H. pylori-infected cells. Activation of AP-1 is mediated by host–bacterial interactions through the cag-T4SS, which leads to phosphorylation of c-Jun and its binding to the ∆133p53 promoter. These findings are consistent with previous observations that ERK/MAPK and stress-activated protein kinase/c-Jun NH2-terminal kinase (SAPK/JNK) pathways as well as upstream signaling regulators, such as Ras and Rho-GTPases that regulate AP-1, are induced by H. pylori (19–21). However, our data show that AP-1 specifically activates transcription from the P2 promoter despite the presence of the AP-1 binding site within the main P1 promoter (22). The basis for this selectivity remains unclear. One possibility is that additional transcriptional regulators are involved. It is also possible that H. pylori not only induces ∆133p53 transcription but stabilizes the ∆133p53 protein by posttranslational mechanisms.
Together, our studies suggest that in the process of evolutionary adaptation to the host environment, H. pylori has developed molecular mechanisms to counteract p53/p73 function. Inhibition of p53 may provide advantages to H. pylori and allow bacteria to alter the cellular homeostasis without triggering cell cycle arrest or apoptosis. As a consequence, however, this alteration may increase the risk for tumor development. Further definition of these mechanisms will contribute to a deeper understanding of the role of p53 in H. pylori infection and gastric tumorigenesis.
Materials and Methods
Cell Culture and H. pylori Strains.
The human gastric cancer cell lines AGS and SNU-1 harboring WT p53 and p53-null Kato III cells were grown in F-12 medium (Invitrogen) supplemented with 10% (vol/vol) FBS. The p53 WT and null isogenic colorectal cancer cell lines (HCT116+/+ and HCT116−/−) were maintained in DMEM (Invitrogen). The cagA+ H. pylori clinical strain J166 and rodent-adapted strain 7.13 were grown in Brucella broth with 5% FBS for 18 h, harvested by centrifugation, and added to gastric cells at an MOI of 100 or as indicated. Isogenic cagA-, cagE-, and vacA-null mutants were constructed by insertional mutagenesis using aphA and selected with 25 μg/mL kanamycin. Heat-inactivated H. pylori bacteria were generated by heating the bacteria to 80 °C for 10 min.
Plasmids, siRNA, and Antibodies.
Plasmids expressing human p53; TAp73β proteins; and luciferase reporters PG13-Luc, p21-pWWP-Luc, and pNFκB-Luc were described previously (23–25). shRNA vector (pSuper-NEO-Δ133p53-shRNA) against human p53 isoforms was constructed by inserting 5′-GCCGTCTTCCAGTTGCTTT-3′ sequence into pSuper-Neo-GFP vector (Oligoengine). The Δ133p53-pcDNA3 expression vector and Δ133p53-Luc (pGL3-P2-Luc) luciferase reporter were kindly provided by J. C. Bourdon (University of Dundee, Dundee, United Kingdom). c-Jun-pcDNA3 expression plasmid was a kind gift from P. Datta (Vanderbilt University, Nashville, TN). siRNA against human c-Jun protein and corresponding negative controls were from Ambion.
The following antibodies were used: p53 (DO-1), p21WAF (Ab-1) from Calbiochem; p53 (DO-11) from AbDseroTec; p53 (CM1) from Novocastra; p73 from Bethyl; p-c-Jun (KM-1), NF-κB p65(A), and nonspecific mouse IgG, p63 (4A4) from Santa Cruz Biotechnology; caspase-3 (8G10), cleaved caspase 3 (Asp175), c-Jun (L70B11), p-NF-κB (Ser536), β-Actin, and IκBα (L35A5) from Cell Signaling; lamin A from Chemicon; and NOXA from Imgenex.
Western blot data were quantified by densitometry using ImageJ software (National Institutes of Health).
Gerbil and H. pylori Infection.
Four- to eight-week-old pathogen-free Mongolian gerbils purchased from Harlan Laboratories were orogastrically challenged with Brucella broth containing 5 × 109 H. pylori strain 7.13 or broth alone and euthanized at 3 d postchallenge. mRNA and proteins were extracted from the stomachs and analyzed for expression of Δ153p53 isoform using RT-PCR and Western blotting. All animal studies were performed according to protocols approved by Vanderbilt University Institutional Animal Care and Use Committee.
Luciferase Reporter Assay, RNA Extraction, PCR, and Mutagenesis.
Luciferase activity was measured using the Dual Luciferase Reporter Assay Kit (Promega) according to the manufacturer’s protocol. RNA was isolated and reverse-transcribed as described previously (5). qPCR was performed using the iCycler thermal cycler (Bio-Rad). Primers sequences are shown in Table S1.
Site-directed mutagenesis was carried out using the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene) and confirmed by direct sequencing. The AP-1 binding motif within the p53 P2 promoter was mutagenized from TGAGTCA to TCGGCCG, as described previously (26).
Immunoprecipitation, ChIP, and Subcellular Fractionation.
To analyze protein interactions between p53 and Δ133p53 isoforms, AGS or SNU-1 cells were co-cultured with H. pylori strain J166 for 8 h and lysed using a standard protocol (5). The prepared cell lysates from infected and control (uninfected) cells were immunoprecipitated with the p53 (DO-1) antibody and analyzed by Western blotting using the p53 (DO-11) antibody. The latter antibody detects both p53 isoforms, whereas the former DO-1 antibody only recognizes p53.
ChIP analyses were carried out as described previously (27). Binding of the c-Jun protein to the P2 promoter was analyzed with the following ChIP primers: 5′-GCTCCTGAGGTGTAGACGCCAA-3′ and 5′-CCTGCAACCCACTAGCGAGC-3′.
Subcellular fractionation was carried out using the NE-PER Nuclear and Cytoplasmic Extraction Kit (Pierce) according to the manufacturer’s protocol. β-Actin and lamin A proteins were used as cytoplasmic and nuclear fractionation markers, respectively.
Cell Survival Analysis.
Cell death was measured by flow cytometry (13). Briefly, cells were fixed in 70% ethanol overnight, stained with 50 μg/mL propidium iodide (PI) for 1 h, and analyzed for DNA content using the FACScan flow cytometer (Bio-Rad).
Statistical Analysis.
Statistical analysis was performed using the Student t test and the Wilcoxon rank-sum test depending on the dataset. Results were expressed as mean values (± SEM unless otherwise noted). Results were considered significant if P < 0.05.
Supplementary Material
Acknowledgments
This work was supported by National Cancer Institute Grant NIH CA138833, Vanderbilt Ingram Cancer Center Grant P30 CA68485, and Vanderbilt Digestive Disease Research Center Grant DK058404.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 15094 (volume 109, number 38).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205664109/-/DCSupplemental.
References
- 1. Polk DB, Peek RM, Jr. Helicobacter pylori: Gastric cancer and beyond. Nat Rev Cancer 10:403–414. [DOI] [PMC free article] [PubMed]
- 2.Levine AJ, Hu W, Feng Z. The P53 pathway: What questions remain to be explored? Cell Death Differ. 2006;13:1027–1036. doi: 10.1038/sj.cdd.4401910. [DOI] [PubMed] [Google Scholar]
- 3.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 4.Matsumoto Y, et al. Helicobacter pylori infection triggers aberrant expression of activation-induced cytidine deaminase in gastric epithelium. Nat Med. 2007;13:470–476. doi: 10.1038/nm1566. [DOI] [PubMed] [Google Scholar]
- 5.Wei J, et al. Regulation of p53 tumor suppressor by Helicobacter pylori in gastric epithelial cells. Gastroenterology. 2010;139:1333–1343. doi: 10.1053/j.gastro.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buti L, et al. Helicobacter pylori cytotoxin-associated gene A (CagA) subverts the apoptosis-stimulating protein of p53 (ASPP2) tumor suppressor pathway of the host. Proc Natl Acad Sci USA. 2011;108:9238–9243. doi: 10.1073/pnas.1106200108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Umehara S, Higashi H, Ohnishi N, Asaka M, Hatakeyama M. Effects of Helicobacter pylori CagA protein on the growth and survival of B lymphocytes, the origin of MALT lymphoma. Oncogene. 2003;22:8337–8342. doi: 10.1038/sj.onc.1207028. [DOI] [PubMed] [Google Scholar]
- 8.Khoury MP, Bourdon JC. The isoforms of the p53 protein. Cold Spring Harb Perspect Biol. 2010;2:a000927. doi: 10.1101/cshperspect.a000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slatter TL, et al. Hyperproliferation, cancer, and inflammation in mice expressing a Δ133p53-like isoform. Blood. 2011;117:5166–5177. doi: 10.1182/blood-2010-11-321851. [DOI] [PubMed] [Google Scholar]
- 10.Fujita K, et al. p53 isoforms Delta133p53 and p53beta are endogenous regulators of replicative cellular senescence. Nat Cell Biol. 2009;11:1135–1142. doi: 10.1038/ncb1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei J, Zaika E, Zaika A. p53 Family: Role of Protein Isoforms in Human Cancer. J Nucleic Acids. 2012;2012:687359. doi: 10.1155/2012/687359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kodama M, Murakami K, Nishizono A, Fujioka T. Animal models for the study of Helicobacter-induced gastric carcinoma. J Infect Chemother. 2004;10:316–325. doi: 10.1007/s10156-004-0353-z. [DOI] [PubMed] [Google Scholar]
- 13.Wei J, et al. Interaction of Helicobacter pylori with gastric epithelial cells is mediated by the p53 protein family. Gastroenterology. 2008;134:1412–1423. doi: 10.1053/j.gastro.2008.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia HH, Talley NJ. Apoptosis in gastric epithelium induced by Helicobacter pylori infection: Implications in gastric carcinogenesis. Am J Gastroenterol. 2001;96(1):16–26. doi: 10.1111/j.1572-0241.2001.03447.x. [DOI] [PubMed] [Google Scholar]
- 15.Marcel V, et al. p53 regulates the transcription of its Delta133p53 isoform through specific response elements contained within the TP53 P2 internal promoter. Oncogene. 2010;29:2691–2700. doi: 10.1038/onc.2010.26. [DOI] [PubMed] [Google Scholar]
- 16.Marcel V, et al. Δ160p53 is a novel N-terminal p53 isoform encoded by Δ133p53 transcript. FEBS Lett. 2010;584:4463–4468. doi: 10.1016/j.febslet.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 17.Marcel V, et al. Biological functions of p53 isoforms through evolution: Lessons from animal and cellular models. Cell Death Differ. 2011;18:1815–1824. doi: 10.1038/cdd.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ak P, Levine AJ. p53 and NF-κB: Different strategies for responding to stress lead to a functional antagonism. FASEB J. 2010;24:3643–3652. doi: 10.1096/fj.10-160549. [DOI] [PubMed] [Google Scholar]
- 19.Mitsuno Y, et al. Helicobacter pylori induced transactivation of SRE and AP-1 through the ERK signalling pathway in gastric cancer cells. Gut. 2001;49(1):18–22. doi: 10.1136/gut.49.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer-ter-Vehn T, Covacci A, Kist M, Pahl HL. Helicobacter pylori activates mitogen-activated protein kinase cascades and induces expression of the proto-oncogenes c-fos and c-jun. J Biol Chem. 2000;275:16064–16072. doi: 10.1074/jbc.M000959200. [DOI] [PubMed] [Google Scholar]
- 21.Naumann M, et al. Activation of activator protein 1 and stress response kinases in epithelial cells colonized by Helicobacter pylori encoding the cag pathogenicity island. J Biol Chem. 1999;274:31655–31662. doi: 10.1074/jbc.274.44.31655. [DOI] [PubMed] [Google Scholar]
- 22.Kirch HC, Flaswinkel S, Rumpf H, Brockmann D, Esche H. Expression of human p53 requires synergistic activation of transcription from the p53 promoter by AP-1, NF-kappaB and Myc/Max. Oncogene. 1999;18:2728–2738. doi: 10.1038/sj.onc.1202626. [DOI] [PubMed] [Google Scholar]
- 23.Soutto M, et al. Loss of TFF1 is associated with activation of NF-κB-mediated inflammation and gastric neoplasia in mice and humans. J Clin Invest. 2011;121:1753–1767. doi: 10.1172/JCI43922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.el-Deiry WS, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 25.Vilgelm AE, et al. Interactions of the p53 protein family in cellular stress response in gastrointestinal tumors. Mol Cancer Ther. 2010;9:693–705. doi: 10.1158/1535-7163.MCT-09-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geimonen E, et al. Activation of protein kinase C in human uterine smooth muscle induces connexin-43 gene transcription through an AP-1 site in the promoter sequence. J Biol Chem. 1996;271:23667–23674. doi: 10.1074/jbc.271.39.23667. [DOI] [PubMed] [Google Scholar]
- 27.Zaika E, et al. p73 protein regulates DNA damage repair. FASEB J. 2011;25:4406–4414. doi: 10.1096/fj.11-192815. [DOI] [PMC free article] [PubMed] [Google Scholar]