Abstract
Sensitivity is essential in CD8+ T-cell killing of virus-infected cells and tumor cells. Although the affinity of the T-cell receptor (TCR) for antigen is relatively low, the avidity of T cell-antigen–presenting cell interactions is greatly enhanced by increasing the valence of the interaction. It is known that TCRs cluster into protein islands after engaging their cognate antigen (peptides bound to MHC molecules). Here, we show that mouse Kb class I molecules segregate into preformed, long-lasting (hours) clusters on the antigen-presenting cell surface based on their bound viral peptide. Peptide-specific Kb clustering occurs when source antigens are expressed by vaccinia or vesicular stomatitis virus, either as proteasome-liberated precursors or free intracellular peptides. By contrast, Kb–peptide complexes generated by incubating cells with synthetic peptides are extensively intermingled on the cell surface. Peptide-specific complex sorting is first detected in the Golgi complex, and compromised by removing the Kb cytoplasmic tail. Peptide-specific clustering is associated with increased T-cell sensitivity: on a per-complex basis, endogenous SIINFEKL activates T cells more efficiently than synthetic SIINFEKL, and wild-type Kb presents endogenous SIINFEKL more efficiently than tailless Kb. We propose that endogenous processing generates peptide-specific clusters of class I molecules to maximize the sensitivity and speed of T-cell immunosurveillance.
Keywords: MHC class I clustering, CD8 T cell recognition, dual-color TIRF imaging, antigen processing/presentation, intracellular trafficking
CD8+ T cells recognize MHC class I molecules bearing oligopeptides derived largely from proteasome degraded proteins. Recognition is based on the activation of the T-cell receptor (TCR) signaling complex. Sensitivity, a key feature of T-cell immunosurveillance, enables detection of low copy number peptides and accelerates recognition of virus-infected cells, where speed is of the essence, because cells must be killed before the release of progeny virus, which can occur within hours of infection.
Although the intrinsic affinity of the TCR for peptide–MHC complexes (pMHC) is low (1, 2), the functional avidity of the T cell-antigen–presenting cell interaction is greatly enhanced by increasing the valence of the interaction. When exposed to cognate pMHC on the antigen presenting cell surface, TCRs cluster into protein islands (3, 4), increasing their sensitivity for activation. Class I molecules are delivered to the cell surface in clusters (5–8). T-cell sensitivity is increased by class I clustering (9), but the relevance of clustering to detecting viral and other endogenously generated peptides has yet to be established. Because clusters contain 50 or fewer class I molecules (5), it is difficult to see how this could enhance detecting viral peptides early after infection, when viral peptides are of low abundance and would be statistically unlikely to be present in the same cluster.
What if, however, there were a mechanism for enhancing T-cell immunosurveillance by delivering viral peptides to the cell surface in preformed clusters? Here, we use TCR-like reagents to explore the generation of peptide-specific clusters by virus-infected cells and cells exposed to synthetic cognate peptides.
Results
Peptide-Specific Clusters Predominate on the Cell Surface.
To localize peptide-specific MHC class I complexes by immunofluorescence, we used the 25-D1.16 mAb (10) or 2C m67 TCR (11), which demonstrate high specificity for mouse H-2 Kb class I molecule complexed respectively with model antigenic peptides SIINFEKL (SIIN) or SIYRYYGL (SIYR). To facilitate detection, we infected l-Kb cells (L929 mouse fibroblasts stably transfected with Kb) with recombinant vaccinia viruses (VVs) expressing SIIN or SIYR as ubiquitin (Ub) fusion proteins. These peptides are immediately liberated in saturating amounts from nascent Ub by highly active cellular ubiquitin hydrolases (12, 13) [this mechanism is used for natural Ub synthesis, which is liberated from Ub-Ub or Ub-ribosomal protein fusions (14)].
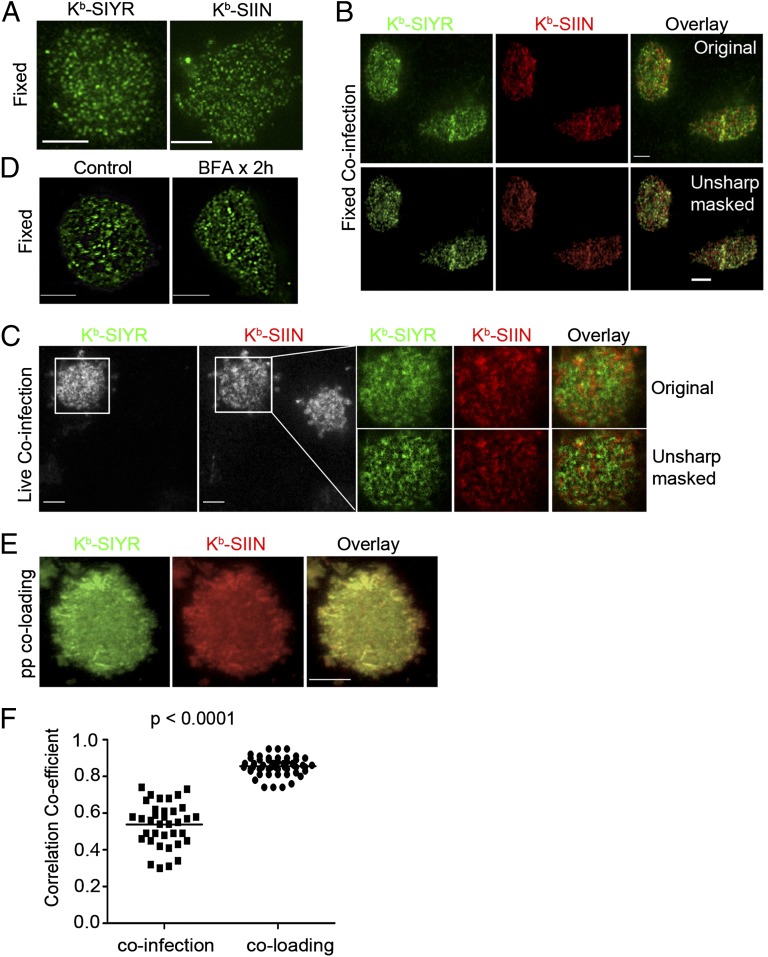
We used total internal reflection fluorescent (TIRF) microscopy to selectively image pMHC complexes at, or just below, the plasma membrane (15). Imaging fixed, nonpermeabilized cells at 4 h post infection (p.i.) with VV-Ub-SIIN or VV-Ub-SIYR revealed that both 25-D1.16 and 2C m67 detected (via indirect immunofluorescence) their cognate pMHCs in highly clustered structures of 200–900 nm diameter (Fig. 1A). Imaging coinfected cells clearly revealed a distinct spatial separation of Kb–SIIN and Kb–SIYR complexes (Fig. 1B). Fixation immobilizes pMHCs (confirmed by photobleaching following indirect staining), preventing antibody-induced redistribution, but potentially introduces artifactual clustering.
Fig. 1.
Endogenously generated SIYR– and SIIN–Kb complexes are expressed in distinct cell-surface clusters. (A) (Left) l-Kb cells 4 h p.i. with VV expressing Ub-liberated SIYR were stained with biotinylated 2Cm67, followed by secondary staining with Alexa 488-conjugated Streptavidin. After fixation, cells were imaged with TIRF microscopy; (Right) LKb cells infected with SIIN-expressing VV were fixed, surface stained with 25D1.16 and then stained with an Alexa647-conjugated goat anti-mouse IgG secondary antibody. TIRF was performed using a Leica AF6000× microscope equipped with a 100× objective. (B) Four hours post coinfection with VV expressing Ub-liberated SIYR and SIIN (MOI = 1 for each), l-Kb cells were costained with biotinylated 2Cm67 and Alexa647-conjugated 25D1.16, followed by secondary staining with Dylight488-conjugated Streptavidin. Unsharp mask processing was performed with Metamorph Imaging Series 7.1. (C) Four hours p.i. with VV expressing Ub-liberated SIYR or SIIN (MOI = 1 for each), l-Kb cells were stained live with a mixture of directly conjugated monovalent reagents, Alexa488-conjugated 2Cm67 and Alexa647-conjugated 25D1.16. Stained cells were visualized live with dual-color TIRF. Enlarged images showed the relative localization between Kb–SIYR and Kb–SIIN on the cell surface. Unsharp mask processing was done with Metamorph Imaging Series 7.1. Note that of two cells in field, only one is coinfected, providing an internal specificity control for staining. (D) Kb–SIIN still clustered after abrogation of surface class I MHC delivery by BFA. l-Kb cells were incubated with BFA at 10 μg/mL starting at 4 h p.i. with VV expressing Ub-liberated SIIN (MOI = 1). Two hours later, cells were fixed with paraformaldehyde, surface stained with 25D1.16 and Alexa488-conjugated goat anti-mouse IgG secondary antibody. (E) Exogenous Kb–SIYR and Kb–SIIN complexes colocalize. Human β2-microglobulin (hβ2m)-sensitized l-Kb cells loaded with a mixture of synthetic SIYR and SIIN peptides (5 μM each), stained live with a mixture of Alexa488-2Cm67 and Alexa647-25D1.16 Fab and imaged immediately by dual-color TIRF. (F) Statistical analysis of the colocalization coefficients between exogenous peptide coloading and endogenous coinfection conditions in l-Kb cells. More than 30 images were collected from each condition. After background subtraction, the correlation coefficient R was calculated with the Image Correlation 1o plugin of National Institutes of Health Image J software.
To avoid fixation or reagent-based cross-linking artifacts, we imaged live cells with monovalent preparations of directly conjugated 25-D1.16 (Fab) and 2C m67 (naturally monovalent), taking care to remove multivalent and aggregated species. Due to the lower affinity of monovalent agents and lack of amplification from secondary reagents, the signal-to-noise ratio suffers. Exploiting the minute focusing volume of TIRF, we enhanced the signal by imaging cells in the presence of direct conjugates. Images were collected simultaneously by illuminating with two wavelengths and collecting with dual aligned detectors (16). At 4 h post-co-infection with VV-Ub-SIIN and VV-Ub-SIYR, surface Kb–SIIN and Kb–SIYR complexes were each highly clustered and still predominantly nonoverlapping (Fig. 1C).
To determine the kinetic stability of clusters, we incubated infected cells 4 h p.i. with brefeldin A (BFA) to abrogate cell-surface delivery of new pMHC. This revealed that clusters remain spatially segregated on a peptide-specific basis for at least 2 h, and are therefore stable structures (Fig. 1D).
Quantitative flow cytometry indicates that 4 h p.i. ∼50,000 Kb–SIIN complexes are present at the surface of cells infected with VV-mCherry-Ub-SIIN (Fig. S1), similar to our previous findings (17, 18). Because we detect ∼25% of the cell surface in the TIRF imaging volume, which contains ∼250 clusters, we can estimate that each cluster contains ∼50 Kb–peptide complexes. This is similar to the class I clusters described by Edidin (5), and roughly matches the number of TCRs per cluster detected by Lillemeier et al. (3).
Exogenously Loaded Synthetic Peptides Do Not Generate Highly Clustered pMHC Complexes.
To examine the potential contribution of endogenous peptide loading per se to clustering, we generated pMHC at the cell surface by incubating cells with synthetic peptides. As a control, l-Kb cells exposed to a mixture of Alexa488-SIIN and Alexa647-SIIN synthetic peptides demonstrate the high degree of colocalization expected, and also little clustering by live-cell TIRF (Fig. S2A). A near-identical pattern was observed when SIIN-pulsed cells were incubated with a mixture of 25D1.16 Fab conjugated with either Alexa488 or Alexa647 (Fig. S2B). It is important to note that cells pulsed with equimolar SIIN and SIYR then incubated with Alexa647-conjugated 25D1.16 Fab and Alexa488-conjugated 2Cm67 demonstrated extensive colocalization and little clustering (Fig. 1 E and F) [similar results were obtained switching the fluorescent labels (Fig. S2C)].
Together, these findings show that TIRF is capable of detecting colocalization, and that clustering is not induced by 25D1.16 or 2C m67. Rather, the mutually exclusive clustering of Kb–SIIN and Kb–SIYR complexes requires endogenous antigen presentation.
Viral infection Generates Peptide-Selective Clusters in Distinct Intracellular Compartments.
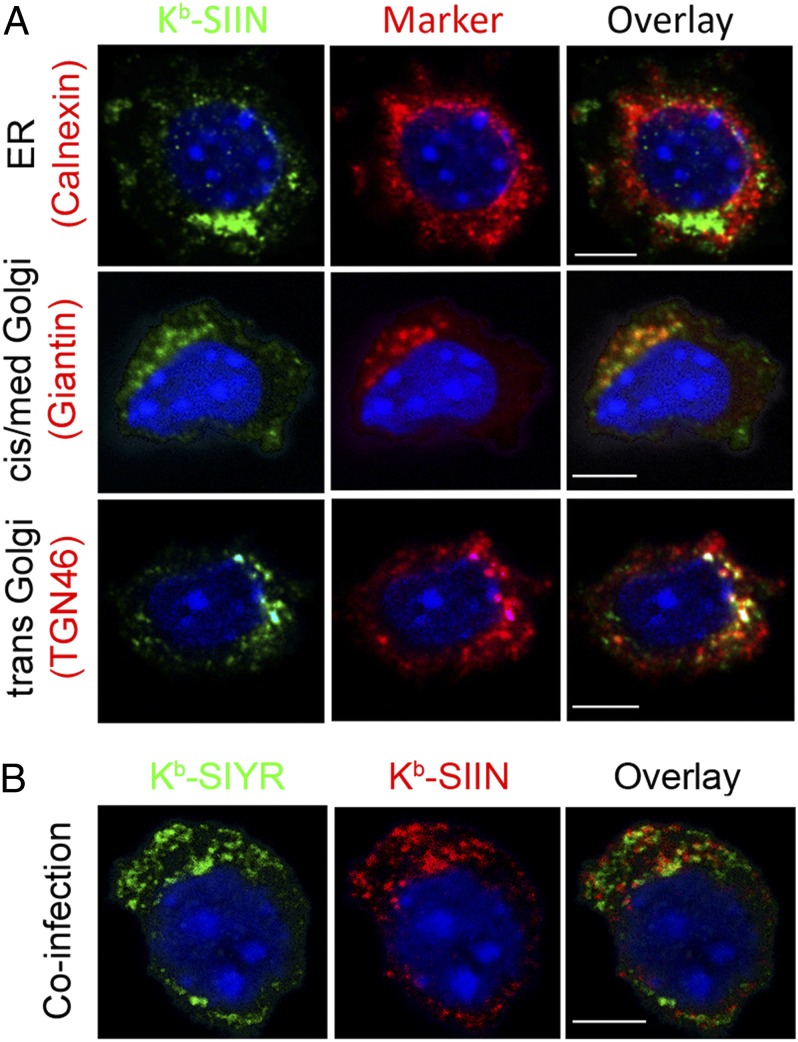
To gain mechanistic insight into peptide-specific clustering, we indirectly stained fixed and permeabilized cells with 25D1.16 4 h p.i. with VV-Ub-SIIN. Laser-scanning confocal microscope imaging with marker antibodies (Abs) revealed that Kb–SIIN complexes are detected in the distal-GC (Giantin, TGN 46 staining) and cis-GC (Giantin staining), but not the endoplasmic reticulum (ER) (calnexin staining) (Fig. 2A), ER exit sites (Sec 23 staining), or ER-GC intermediate compartment (ERGIC 53 staining) (Fig. S3A).
Fig. 2.
Kb–SIIN and Kb–SIYR complexes exist in distinct subcellular compartments. (A) Four hours p.i. with VV-expressing Ub-liberated SIIN (MOI = 1), cells were stained intracellularly with 25D1.16 and antibodies against the indicated intracellular markers, followed by the appropriate secondary antibody staining. (B) Four hours p.i. with VVs expressing Ub-liberated SIYR and SIIN (each at MOI = 1), cells were stained intracellularly using 2C m67 and 25-D1.16.
The absence of ER 25D1.16 staining is surprising, because the ER is well established as the principal site of class I assembly with transporter associated with antigen processing (TAP)-transported peptides (19). The absence of 25D1.16 ER staining is probably not due to rapid transport of peptide-loaded MHC complexes from the ER, because prolonged incubation of infected cells at 15 °C, which greatly retards ER export of nascent membrane proteins (20), failed to reveal Kb–SIIN complexes in the ER. As expected, we easily detected Kb molecules in the ER using pAbs specific for the Kb cytoplasmic tail (Fig. S3B), or Kb–SIIN complexes if we used BFA to fuse the ER and GC (21) (Fig. S3C). Could it be that TAP loads SIINFEKL in the cis-GC (22–25)? This possibility is unlikely, because we fail to detect TAP1-GFP in the GC of l-Kb (Fig. S3D). Based on this finding, we tentatively conclude that peptide loading occurs in the ER, but that other factors preclude detection of pMHC complexes by 25-D1.16 or 2C m67.
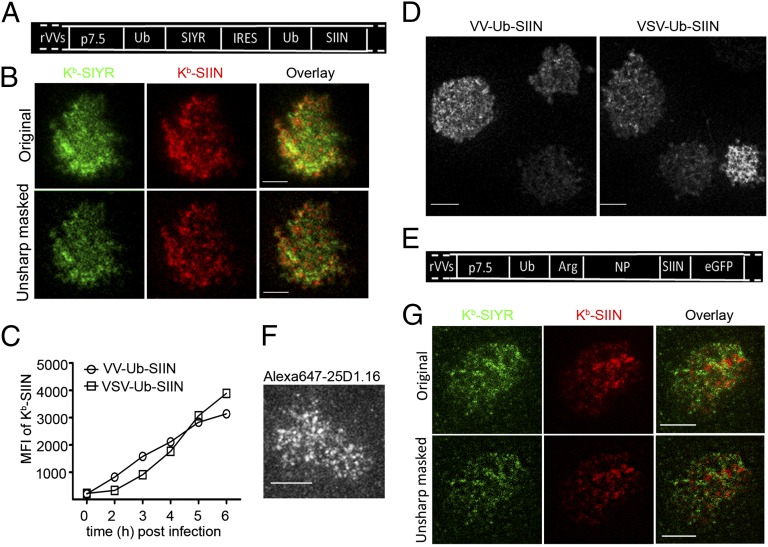
We next coinfected cells with VV-Ub-SIIN and VV-Ub-SIYR. Intracellular costaining with 25D1.16 and 2C m67 revealed surprising noncolocalization between 25-D1.16 and 2C m67 staining in the GC and other intracellular compartments (Fig. 2B). VV synthesizes its gene products in viral factories (26). Under the infection conditions used with high multiplicity coinfection, factories from individual viruses fuse at the resolution of light microscopy (26). Still, it was possible that compartmentalized loading was related to generating mRNAs from distinct input virions. To examine this possibility, we engineered a recombinant VV that expresses SIIN and SIYR from a single mRNA with an internal ribosomal entry site (IRES) sequence (Fig. 3A). The low expression of the IRES-driven peptide (SIIN) necessitated using a monovalent secondary anti-mouse IgG Ab to increase the signal. Still, we could clearly detect Kb–SIIN and Kb–SIYR as distinct clusters via live-cell TIRF (Fig. 3B), demonstrating that peptide-specific clusters can arise from a single viral factory.
Fig. 3.
Characterizing compartmentalized presentation. (A) Ub-liberated SIYR or SIIN peptides were expressed by VV from a single mRNA with an IRES under the p7.5 VV promoter as indicated. (B) Because of low levels of SIINFEKL expression from the IRES, it was necessary to treat l-Kb cells for 3 d with IFN-γ to increase antigen presentation. Six hours after infecting with the VV-Ub-SIYR-IRES-Ub-SIIN (MOI = 1) cells were stained with Alexa488-2Cm67 and Alexa568-25D1.16 Fab, followed by secondary staining of the Kb–SIIN complexes with Dylight549-conjugated goat anti-mouse IgG Fab to enhance the signal in the corresponding channel. (C) Expression of surface Kb–SIIN between VV-Ub-SIIN and VSV-Ub-SIIN infections. At indicated times p.i., we determined relative levels of surface Kb–SIIN complexes via flow cytometry after Alexa647-25D1.16 staining. (D) Kb–SIIN generated from recombinant vesicular stomatitis virus clusters on the cell surface. Four hours p.i. with either VV-Ub-SIIN or VSV-Ub-SIIN, we visualized surface Kb–SIIN complexes with Alexa647-25D1.16 Fab in live TIRF. (E) Construct used to visualize SIIN presentation from a full-length protein consisting of Ub-Arg-NP-SIIN-eGFP. Due to rapid N-end rule degradation, Kb–SIIN complexes are generated at 3× the rate relative to NP-SIIN-GFP or ovalbumin itself. (F) Six hours p.i. VV-Ub-Arg-NP-SIIN-eGFP, l-Kb cells were labeled live with Alexa647-conjugated 25D1.16 Fab and visualized live with TIRF. (G) Six hours after coinfection with VV-Ub-Arg-NP-SIIN and VV-Ub-Arg-NP-SIYR, l-Kb cells were stained live with a mixture of directly conjugated monovalent reagents Alexa488-conjugated 2Cm67 and Alexa647-conjugated 25D1.16. Stained cells were visualized live with dual-color TIRF.
Is cluster formation limited to peptides expressed by VV? We found that cells expressing SIIN from a recombinant vesicular stomatitis virus encoding GFP-Ub-SIIN express surface complexes with similar kinetics (Fig. 3C) to VV-infected cells, and generate clusters similar to those generated by VV-expressed SIIN (Fig. 3D). Thus, pMHC clusters are generated by two viruses with very different replication cycles.
Proteasome Liberated Viral Peptides Also Form Clusters.
At this point, we have studied viruses that generate peptides as products liberated from Ub-fusions proteins. We extended these findings to peptides generated by proteasomes from a rapidly degraded nucleoprotein (NP)-SIIN fusion protein (Fig. 3E). Although sensitivity precludes detection of intracellular Kb–SIIN complexes, live-cell TIRF 6 h following infection clearly revealed the presence of Kb–SIIN clusters (Fig. 3F) (27). Despite the lower levels of Kb–SIIN expression relative to cells expressing Ub-liberated SIIN, clusters were of similar intensity, implying that clusters are released from their loading sites after achieving a threshold number.
We used live-cell TIRF to examine cells coinfected with VVs expressing rapidly degraded NP fused to either SIIN or SIYR. Once again, distinct surface Kb clusters segregated based on their peptide cargo (Fig. 3G), extending this phenomenon to proteasome-dependent–antigen processing.
Peptide-Specific Cluster Segregation Requires the Kb Cytoplasmic Tail.
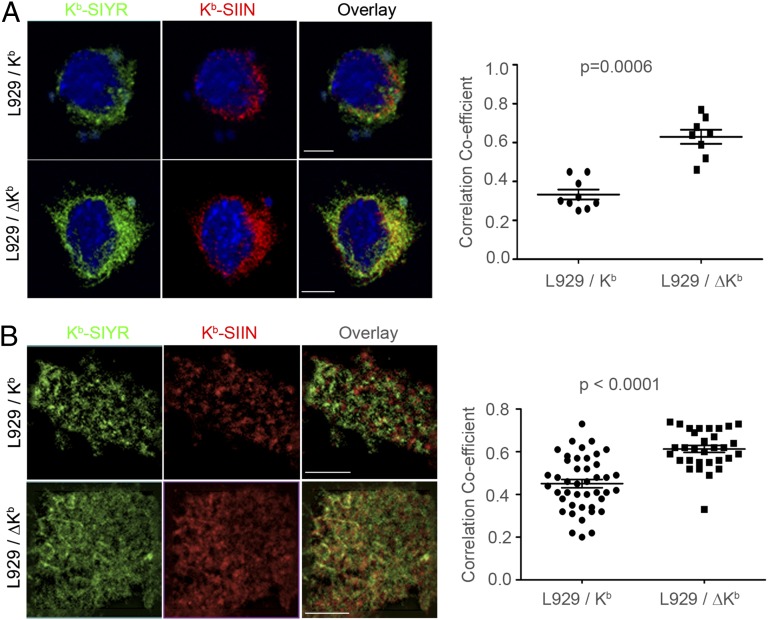
The class I cytoplasmic domain modulates class I intracellular trafficking (28–33). Removal of the Kb cytoplasmic domain (ΔKb) had little effect on class I cell surface expression, kinetics of Kb–SIIN expression, gross clustering of Kb–SIIN complexes, intracellular distribution, or efficiency of Kb–SIIN generation from cytosolic or defective ribosomal product (DRiP)-liberated peptides (Fig. S4 A–E). Kb tail deletion, however, significantly reduced the intracellular (Fig. 4A) and cell-surface segregation (Fig. 4B) of Kb–SIIN and Kb–SIYR clusters in coinfected cells. That maximal spatial segregation of SIIN and SIYR complexes depends on the cytoplasmic tail of Kb provides an important functional control that clustering is not an artifact associated with detection of the complexes with 25D-1.16/2Cm67.
Fig. 4.
Segregation of Kb–peptide clusters is impaired when Kb is truncated of cytoplasmic tail. (A) L929 cells stably transfected with wild-type (L929/Kb) or cytoplasmic tail-deleted Kb (L929/ΔKb) were coinfected with VV-Ub-SIIN and VV-Ub-SIYR (MOI = 1 for each). Four hours later, intracellular Kb–SIYR and Kb–SIIN complexes were costained with 2Cm67 and 25D1.16 as described in Fig. 1C. The colocalization between Kb–SIYR and Kb–SIIN complexes was analyzed with NIH Image J. The P value is 0.0006 for colocalization correlation coefficient analyzed with ∼10 images from each cell line by Mann-Whitney nonparametric two-tail analysis. (B) Cells infected as in A were stained with Alexa488-2Cm67 and Alexa647-25D1.16 Fab and imaged using dual-color live TIRF. Forty images for each condition were used for statistical analysis. The P value is less than 0.0001 using a Mann-Whitney nonparametric two-tail analysis.
This observation extends the function of the class I cytoplasmic domain, previously shown to be involved with class I plasma membrane internalization and endosomal trafficking (28–33). Because the efficiency of Kb loading with SIIN is not affected by the loss of the cytoplasmic tail, we infer that ΔKb properly associates with TAP and functions normally in the peptide-loading complex. This implies that the cytoplasmic tail affects peptide segregation only after Kb release from the loading complex, consistent with a role for the tail in maintaining Kb–peptide clusters in the GC and plasma membrane.
Peptide Clustering Enhances T-Cell Sensitivity.
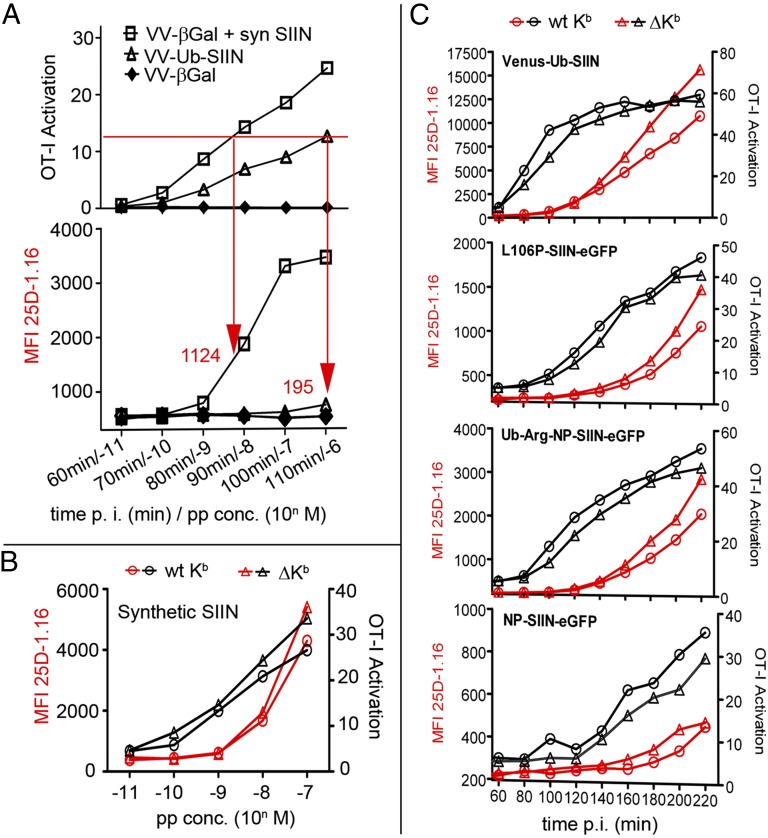
Do clusters enhance T-cell sensitivity? We compared activation of OT-I transgenic T cells (specific for Kb–SIIN) by virus-infected (cluster generating) vs. synthetic SIIN exposed l-Kb cells (no clusters). To compare a similar range of sublimiting numbers of Kb–SIIN complexes, we infected cells for increasing times with VV-Ub-SIIN, abrogating antigen presentation by exposing cells to BFA during the T-cell activation assay. In parallel, we exposed cells to increasing amounts of synthetic SIIN. Cells were then assessed for their ability to activate OT-I IFN-γ synthesis by intracellular cytokine staining and for Kb–SIIN expression by binding of Alexa 647 25D1.16. Because VV-induced changes in antigen-presenting cells that could potentially influence T-cell activation, we exposed cells infected with a non-SIIN-expressing VV-βGal to synthetic SIIN.
OT-I cells are a more sensitive measure of Kb–SIIN expression than 25D1.16 staining, as originally reported (10). We could, however, detect 25D1.16 binding to VV-Ub-SIIN infected cells at 110 min postinfection, a time when T-cell activation was not saturated. At this time point, infected cells gave a 25D1.16 signal 195 mean fluorescent intensity (MFI) units above background levels. To achieve the same level of T-cell activation (∼12%), nearly six times as many Kb–SIIN complexes, 1,124 MFI units, were present on peptide-sensitized cells (Fig. 5A).
Fig. 5.
Kb–SIIN clustering is associated with increased T-cell sensitivity. (A) L929/Kb cells infected with VV-Ub-SIIN (MOI = 2) or VV-βGal (control) were coincubated with OT1 CD8 T cells (E:T = 1:1) for 1.5 h in the presence of brefeldin A. IFN-γ expression was measured by flow cytometry for intracellular anti-IFN-γ expression, gating for CD8α+ cells. In parallel, L929/Kb cells were stained with 25D1.16 for surface Kb–SIIN expression. For peptide loading, hβ2m sensitized L929/Kb cells were infected with VV-βGal for 2 h followed by peptide incubation for 1 h at 4 °C. Arrows and numbers indicate the MFI of Kb–SIIN required in each condition to achieve equivalent levels of OT1 activation. (B) L929/Kb or tailless L929/ΔKb cells pretreated with human β2m were loaded with SIIN peptide and tested for their ability to activate OT-I cells as in A. (C) OT-I activation tested as in A with L929/Kb or tailless L929/ΔKb cells after infection with indicated recombinant VV expressing SIIN in the form of peptide (Venus-Ub-SIIN) or full-length rapidly degraded (L106P-SIIN-eGFP, Ub-Arg-NP-SIIN-eGFP) or stable (NP-SIIN-eGFP) proteins.
If clustering of endogenously generated peptides enhances OT-I activation, then ΔKb, which exhibits less clustering than WT Kb, should be less efficiently recognized per Kb–SIIN complex expressed (as determined by 25D1.16 binding). OT-I cells are triggered with equal efficiency per Kb–SIIN complex when synthetic SIIN is presented by ΔKb vs. WT Kb (Fig. 5B), establishing that removing the tail does not negatively impact Kb T-cell activation function. Next, we infected cells with VVs expressing SIIN in the context of an Ub-fusion protein (Venus-Ub-SIIN), rapidly degraded protein (L106P-SIIN-eGFP, Ub-R-NP-SIIN-eGFP), or stable protein (NP-SIIN-eGFP). In each case, for nearly all time points p.i., OT-I cells were better activated by WT vs. ΔKb for equivalent or even diminished levels of Kb–SIINFEKL expression (Fig. 5C).
Taken together, the efficiency of OT-I activation on a per complex basis parallels the degree of clustering. Synthetic peptides demonstrate a large decrease in efficiency vs. endogenous peptides in parallel with a near-complete decrease in clustering. Removing the cytoplasmic tail has a less marked effect on T-cell activation efficiency in parallel with a partial decrease in clustering. These findings support the conclusion that the peptide-specific clustering associated with endogenous antigen processing enhances T-cell recognition.
Discussion
Despite the availability of class I peptide–specific reagents for 15 y (10, 34), there is little published on their detection of intracellular complexes generated by endogenous antigen processing. Makler et al. (35) exclusively detected an abundant naturally processed CMV peptide complexed with HLA-A2 in the GC and plasma membrane. Also, we were unable to detect Kb complexed with either SIIN or SIYR in the ER, detecting each complex in the GC and plasma membrane. Our inability to detect TAP beyond the ER strongly supports ER peptide loading. Kb–peptide complexes could be rapidly transported from the ER, resulting in subdetectable steady-state levels. Arguing against this possibility, however, we did not detect complexes when class I export from the ER was retarded by 15 °C incubation. Further, Spiliotis et al. (36) reported that peptide-loaded class I molecules accumulate at ER export sites awaiting transport to the GC.
We therefore favor the idea that 25-D1.16 and 2C-m67 are unable to detect their epitopes in the ER possibly due to steric interference from proteins that participate in loading class I molecules. Or, as an alternative, class I molecules may undergo a conformational alteration upon release from the loading complex needed to create the respective epitopes. The clear staining of the ER by 25-D1.16 after BFA treatment, which merges the ER and early GC compartments, as reported (10), is consistent with either of these possibilities. We note that although we previously used 25-D1.16 to detect Kb–SIIN complexes in the ER of cells incubated with synthetic SIIN, we were unable to demonstrate the export of these complexes from the ER (37), which suggests that they were aberrantly loaded.
Our most important finding is that endogenous antigen processing generates intracellular clusters of class I molecules segregated on the basis of their peptide cargo that are maintained for hours after their delivery to the cell surface. Clusters could derive from compartmentalized translation, resulting in localized TAP-mediated transport and loading of peptides generated from the translation products of an individual mRNA (38). Although the generation of distinct clusters from the IRES construct argues against this interpretation, we are not certain that the two gene products are mediated by distinct ribosome populations. Alternatively, clusters might arise by peptide-based sorting. Because clusters are not generated when synthetic peptides are loaded exogenously, this implies that such sorting is limited to endogenously loaded complexes, perhaps during their export from the ER, because we detect clustering in the cis-GC. Based on biophysical imaging studies, Pentcheva and Edidin (39) concluded that class I molecules cluster before they exit from the ER, and further, that wild-type class I molecules and a mutated version unable to bind TAP formed separate clusters. Could it be that the clusters they observed also segregate based on their peptide cargo?
Cluster segregation is partially dependent on the Kb cytoplasmic tail. MHC class I tail truncations are known to increase the mobility of class I molecules on the cell surface (32, 40), suggesting that the Kb tail helps to maintain clusters once they are formed, rather than participating in cluster generation. Indeed, removing the tail did not detectably affect the efficiency of Kb–SIIN loading from various VV-encoded SIIN-containing gene products.
Edidin and colleagues have reported that class I clustering increases the sensitivity of T-cell recognition (6, 9, 39). Our findings imply that this is due to clustering at the level of individual peptides. Such clustering would greatly facilitate the 2D binding properties known to be important in T-cell activation (3, 41, 42), and synergize with the recognition events involving self-peptide/MHC and the TCR/CD8 complex (43, 44). We show that at equivalent levels of Kb–SIIN expression, virus-infected cells are recognized more efficiently than synthetic peptide-sensitized cells, and that Kb tail deletion reduces T-cell activation on a per-complex basis consistent with enhanced triggering by preformed clusters with endogenous peptides.
Clustering could be particularly important for recognition of low abundance peptides, and could contribute to the high sensitivity of tumor-specific T cells that recognize peptides from low abundance translation products. Peptide segregation might also mitigate the effects of naturally processed self-antagonist peptides on T-cell recognition. Clearly, much remains to be learned about the organization of MHC class I peptide complexes on the antigen presenting cell (APC) surface and the biological effects of peptide-specific clustering.
Materials and Methods
We infected LKb cells with rVVs expressing antigenic SIINFEKL (SIIN) or SIYRYYGL (SIYR) at the multiplicity of infection (MOI) indicated and detected Kb–peptide complexes with mAb 25D1.16 or TCR-like molecule 2C m67. Intracellular staining was visualized using Leica TCS-SP5 DMI6000, and surface staining was visualized using a custom-made dual-color TIRF microscope (16). Kb surface expression was measured using a BD LSR II flow cytometer. OT-I CD8 T-cell lines stimulated ex vivo were coincubated with infected wild-type L929/Kb cells or cytoplasmic tail-deleted L929/ΔKb cells, the activation was measured by intracellular cytokine IFN-γ expression in OT1 T cells. Exogenous peptides were loaded onto cells incubated overnight with human β2-microglobulin (5 μg/mL). Details of experimental conditions and analysis are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Phil Holler for original engineering of the 2C-m67 TCR, and Natalie Bowerman for development of the single-chain form of the TCR for detection. This work was generously supported by the National Institute of Allergy and Infectious Diseases Division of Intramural Research and National Institutes of Health Grant P01 CA097296 (to D.M.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208696109/-/DCSupplemental.
References
- 1.Matsui K, Boniface JJ, Steffner P, Reay PA, Davis MM. Kinetics of T-cell receptor binding to peptide/I-Ek complexes: Correlation of the dissociation rate with T-cell responsiveness. Proc Natl Acad Sci USA. 1994;91:12862–12866. doi: 10.1073/pnas.91.26.12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corr M, et al. T cell receptor-MHC class I peptide interactions: Affinity, kinetics, and specificity. Science. 1994;265:946–949. doi: 10.1126/science.8052850. [DOI] [PubMed] [Google Scholar]
- 3.Lillemeier BF, et al. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat Immunol. 2010;11:90–96. doi: 10.1038/ni.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varma R, Campi G, Yokosuka T, Saito T, Dustin ML. T cell receptor-proximal signals are sustained in peripheral microclusters and terminated in the central supramolecular activation cluster. Immunity. 2006;25:117–127. doi: 10.1016/j.immuni.2006.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edidin M. Class I MHC molecules as probes of membrane patchiness: From biophysical measurements to modulation of immune responses. Immunol Res. 2010;47:265–272. doi: 10.1007/s12026-009-8159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matko J, Bushkin Y, Wei T, Edidin M. Clustering of class I HLA molecules on the surfaces of activated and transformed human cells. J Immunol. 1994;152:3353–3360. [PubMed] [Google Scholar]
- 7.Lavi Y, Gov N, Edidin M, Gheber LA. Lifetime of major histocompatibility complex class-I membrane clusters is controlled by the actin cytoskeleton. Biophys J. 2012;102:1543–1550. doi: 10.1016/j.bpj.2012.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lavi Y, Edidin MA, Gheber LA. Dynamic patches of membrane proteins. Biophys J. 2007;93:L35–L37. doi: 10.1529/biophysj.107.111567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fooksman DR, Grönvall GK, Tang Q, Edidin M. Clustering class I MHC modulates sensitivity of T cell recognition. J Immunol. 2006;176:6673–6680. doi: 10.4049/jimmunol.176.11.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porgador A, Yewdell JW, Deng Y, Bennink JR, Germain RN. Localization, quantitation, and in situ detection of specific peptide-MHC class I complexes using a monoclonal antibody. Immunity. 1997;6:715–726. doi: 10.1016/s1074-7613(00)80447-1. [DOI] [PubMed] [Google Scholar]
- 11.Holler PD, Chlewicki LK, Kranz DM. TCRs with high affinity for foreign pMHC show self-reactivity. Nat Immunol. 2003;4:55–62. doi: 10.1038/ni863. [DOI] [PubMed] [Google Scholar]
- 12.Fruci D, et al. Quantifying recruitment of cytosolic peptides for HLA class I presentation: Impact of TAP transport. J Immunol. 2003;170:2977–2984. doi: 10.4049/jimmunol.170.6.2977. [DOI] [PubMed] [Google Scholar]
- 13.Lev A, et al. Compartmentalized MHC class I antigen processing enhances immunosurveillance by circumventing the law of mass action. Proc Natl Acad Sci USA. 2010;107:6964–6969. doi: 10.1073/pnas.0910997107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finley D, Bartel B, Varshavsky A. The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature. 1989;338:394–401. doi: 10.1038/338394a0. [DOI] [PubMed] [Google Scholar]
- 15.Mattheyses AL, Simon SM, Rappoport JZ. Imaging with total internal reflection fluorescence microscopy for the cell biologist. J Cell Sci. 2010;123:3621–3628. doi: 10.1242/jcs.056218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crites TJ, Chen L, Varma R. A TIRF microscopy technique for real-time, simultaneous imaging of the TCR and its associated signaling proteins. J Vis Exp. 2012;(61):e3892. doi: 10.3791/3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Princiotta MF, et al. Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity. 2003;18:343–354. doi: 10.1016/s1074-7613(03)00051-7. [DOI] [PubMed] [Google Scholar]
- 18.Anton LC, Yewdell JW, Bennink JR. MHC class I-associated peptides produced from endogenous gene products with vastly different efficiencies. J Immunol. 1997;158:2535–2542. [PubMed] [Google Scholar]
- 19.Wearsch PA, Cresswell P. The quality control of MHC class I peptide loading. Curr Opin Cell Biol. 2008;20:624–631. doi: 10.1016/j.ceb.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saraste J, Kuismanen E. Pre- and post-Golgi vacuoles operate in the transport of Semliki Forest virus membrane glycoproteins to the cell surface. Cell. 1984;38:535–549. doi: 10.1016/0092-8674(84)90508-7. [DOI] [PubMed] [Google Scholar]
- 21.Doms RW, Russ G, Yewdell JW. Brefeldin A redistributes resident and itinerant Golgi proteins to the endoplasmic reticulum. J Cell Biol. 1989;109:61–72. doi: 10.1083/jcb.109.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleijmeer MJ, et al. Location of MHC-encoded transporters in the endoplasmic reticulum and cis-Golgi. Nature. 1992;357:342–344. doi: 10.1038/357342a0. [DOI] [PubMed] [Google Scholar]
- 23.Baas EJ, et al. Peptide-induced stabilization and intracellular localization of empty HLA class I complexes. J Exp Med. 1992;176:147–156. doi: 10.1084/jem.176.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paulsson KM, et al. Association of tapasin and COPI provides a mechanism for the retrograde transport of major histocompatibility complex (MHC) class I molecules from the Golgi complex to the endoplasmic reticulum. J Biol Chem. 2002;277:18266–18271. doi: 10.1074/jbc.M201388200. [DOI] [PubMed] [Google Scholar]
- 25.Ghanem E, et al. The transporter associated with antigen processing (TAP) is active in a post-ER compartment. J Cell Sci. 2010;123:4271–4279. doi: 10.1242/jcs.060632. [DOI] [PubMed] [Google Scholar]
- 26.Katsafanas GC, Moss B. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe. 2007;2:221–228. doi: 10.1016/j.chom.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antón LC, et al. Intracellular localization of proteasomal degradation of a viral antigen. J Cell Biol. 1999;146:113–124. doi: 10.1083/jcb.146.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lizée G, et al. Control of dendritic cell cross-presentation by the major histocompatibility complex class I cytoplasmic domain. Nat Immunol. 2003;4:1065–1073. doi: 10.1038/ni989. [DOI] [PubMed] [Google Scholar]
- 29.Capps GG, Van Kampen M, Ward CL, Zúñiga MC. Endocytosis of the class I major histocompatibility antigen via a phorbol myristate acetate-inducible pathway is a cell-specific phenomenon and requires the cytoplasmic domain. J Cell Biol. 1989;108:1317–1329. doi: 10.1083/jcb.108.4.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basha G, et al. MHC class I endosomal and lysosomal trafficking coincides with exogenous antigen loading in dendritic cells. PLoS ONE. 2008;3:e3247. doi: 10.1371/journal.pone.0003247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodríguez-Cruz TG, et al. Natural splice variant of MHC class I cytoplasmic tail enhances dendritic cell-induced CD8+ T-cell responses and boosts anti-tumor immunity. PLoS ONE. 2011;6:e22939. doi: 10.1371/journal.pone.0022939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edidin M, Zúñiga MC, Sheetz MP. Truncation mutants define and locate cytoplasmic barriers to lateral mobility of membrane glycoproteins. Proc Natl Acad Sci USA. 1994;91:3378–3382. doi: 10.1073/pnas.91.8.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vega MA, Strominger JL. Constitutive endocytosis of HLA class I antigens requires a specific portion of the intracytoplasmic tail that shares structural features with other endocytosed molecules. Proc Natl Acad Sci USA. 1989;86:2688–2692. doi: 10.1073/pnas.86.8.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andersen PS, et al. A recombinant antibody with the antigen-specific, major histocompatibility complex-restricted specificity of T cells. Proc Natl Acad Sci USA. 1996;93:1820–1824. doi: 10.1073/pnas.93.5.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makler O, Oved K, Netzer N, Wolf D, Reiter Y. Direct visualization of the dynamics of antigen presentation in human cells infected with cytomegalovirus revealed by antibodies mimicking TCR specificity. Eur J Immunol. 2010;40:1552–1565. doi: 10.1002/eji.200939875. [DOI] [PubMed] [Google Scholar]
- 36.Spiliotis ET, Manley H, Osorio M, Zúñiga MC, Edidin M. Selective export of MHC class I molecules from the ER after their dissociation from TAP. Immunity. 2000;13:841–851. doi: 10.1016/s1074-7613(00)00081-9. [DOI] [PubMed] [Google Scholar]
- 37.Day PM, Yewdell JW, Porgador A, Germain RN, Bennink JR. Direct delivery of exogenous MHC class I molecule-binding oligopeptides to the endoplasmic reticulum of viable cells. Proc Natl Acad Sci USA. 1997;94:8064–8069. doi: 10.1073/pnas.94.15.8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yewdell JW. DRiPs solidify: Progress in understanding endogenous MHC class I antigen processing. Trends Immunol. 2011;32:548–558. doi: 10.1016/j.it.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pentcheva T, Edidin M. Clustering of peptide-loaded MHC class I molecules for endoplasmic reticulum export imaged by fluorescence resonance energy transfer. J Immunol. 2001;166:6625–6632. doi: 10.4049/jimmunol.166.11.6625. [DOI] [PubMed] [Google Scholar]
- 40.Capps GG, Pine S, Edidin M, Zúñiga MC. Short class I major histocompatibility complex cytoplasmic tails differing in charge detect arbiters of lateral diffusion in the plasma membrane. Biophys J. 2004;86:2896–2909. doi: 10.1016/S0006-3495(04)74341-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang J, et al. The kinetics of two-dimensional TCR and pMHC interactions determine T-cell responsiveness. Nature. 2010;464:932–936. doi: 10.1038/nature08944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huppa JB, et al. TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature. 2010;463:963–967. doi: 10.1038/nature08746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anikeeva N, et al. Quantum dot/peptide-MHC biosensors reveal strong CD8-dependent cooperation between self and viral antigens that augment the T cell response. Proc Natl Acad Sci USA. 2006;103:16846–16851. doi: 10.1073/pnas.0607771103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yachi PP, Ampudia J, Gascoigne NR, Zal T. Nonstimulatory peptides contribute to antigen-induced CD8-T cell receptor interaction at the immunological synapse. Nat Immunol. 2005;6:785–792. doi: 10.1038/ni1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.