Abstract
Cell size varies greatly among different types of cells, but the range in size that a specific cell type can reach is limited. A long-standing question in biology is how cells control their size. Escherichia coli adjusts size and growth rate according to the availability of nutrients so that it grows larger and faster in nutrient-rich media than in nutrient-poor media. Here, we describe how, using classical genetics, we have isolated a remarkably small E. coli mutant that has undergone a 70% reduction in cell volume with respect to wild type. This mutant lacks FabH, an enzyme involved in fatty acid biosynthesis that previously was thought to be essential for the viability of E. coli. We demonstrate that although FabH is not essential in wild-type E. coli, it is essential in cells that are defective in the production of the small-molecule and global regulator ppGpp. Furthermore, we have found that the loss of FabH causes a reduction in the rate of envelope growth and renders cells unable to regulate cell size properly in response to nutrient excess. Therefore we propose a model in which fatty acid biosynthesis plays a central role in regulating the size of E. coli cells in response to nutrient availability.
Keywords: AccD, inner membrane, lipopolysaccharide, outer membrane, SpoT
Bacteria regulate their size and growth rate in response to nutrient availability. For example, Escherichia coli, Salmonella typhimurium, and Bacillus subtilis cells grow larger and faster in nutrient-rich medium than in nutrient-poor medium (1–6). Changes in temperature can alter growth rate but not size (2). Therefore, the size a cell attains depends on the nutritional composition of the growth medium, suggesting that nutrients affect a rate-limiting step(s) that controls size and the rate of growth.
Bacteria must coordinate cell size, growth rate, and division in response to nutrient availability. Indeed, when E. coli changes its size, it also changes its generation time inversely; however, it maintains the cell mass-to-DNA ratio constant because it initiates DNA replication whenever it reaches a particular cell mass or a multiple of that mass (6). Interestingly, recent studies have shown that B. subtilis and E. coli use different regulatory mechanisms to couple cell size and DNA replication (4, 7). In E. coli DNA replication is not initiated until the cell reaches an appropriate size, but size does not affect the timing of replication in B. subtilis. Nevertheless, the amount of active DnaA, which unwinds DNA at the origin and thereby triggers replication (8, 9), is relevant in controlling the initiation of replication in both bacteria (7). Furthermore, a metabolic pathway for glucolipid biosynthesis regulates cell size in B. subtilis in response to nutrients under conditions that promote rapid growth (4). In this pathway, the UDP-glucose transferase UgtP inhibits the assembly of the divisome, the division machinery. The levels and localization of UgtP vary with nutrient availability so that assembly of the divisome is delayed under nutrient-rich conditions, resulting in longer cells.
We do not understand how nutrients regulate cell size in E. coli, but this regulation must be connected in some way to envelope biogenesis, because the bacterial cell envelope constitutes the cell boundary. In E. coli, the cell envelope contains several compartments: the cytoplasmic or inner membrane (IM); the outer membrane (OM); the periplasm, an aqueous compartment that separates the IM and OM; and the peptidoglycan layer, a cell-wall polymer sacculus that is located in the periplasm and is covalently attached to the OM (10). The OM contains lipids that are asymmetrically arranged in the bilayer so that the inner leaflet is composed of phospholipids, and the outer leaflet contains LPS (11). LPS is essential for the viability of E. coli and is composed of lipid A, the core oligosaccharide, and the O-antigen (12–14). Biogenesis of LPS has been most studied in E. coli K-12 strains, which produce an LPS molecule that lacks O-antigen. Complete synthesis of this LPS occurs at the inner leaflet of the IM; therefore LPS must be transported across the entire cell envelope to reach its final location, the cell surface. Eight proteins are required for LPS transport: the ATP-binding cassette (ABC) transporter MsbA, which flips LPS across the IM (15–17), and seven Lpt factors, which transport LPS from the IM to the cell surface (Fig. 1A). LptB/F/G form an ABC transporter that is believed to extract LPS from the IM (18–20); LptC associates with LptB/F/G and periplasmic LptA (21, 22); and LptD/E form a complex at the OM (23–25). All Lpt factors are required for LPS transport and viability in E. coli (19, 20, 24, 26), and they form a transenvelope protein bridge that is thought to mediate transport of LPS from the IM to the OM (27, 28).
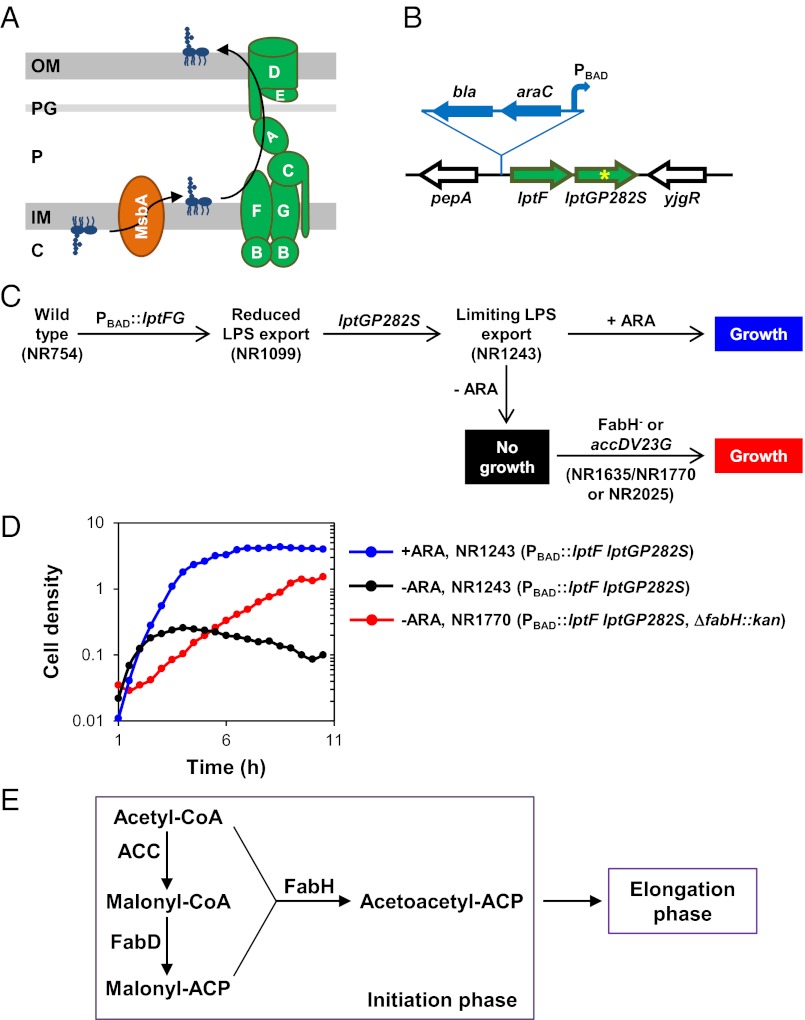
Fig. 1.
Defects in FA biosynthesis suppress lethality caused by insufficient transport of LPS to the cell surface. (A) Pathway for the transport of LPS across the E. coli cell envelope. MsbA flips LPS across the IM, and the LptABCDEFG proteins transport to and assemble LPS at the OM (see the Introduction for details). (B) Chromosomal lptFG locus in NR1243. Insertion in the native chromosomal lptFG locus of a bla-araC-PBAD cassette (blue) 101 bp upstream of lptF resulted in strain NR1099. Localized mutagenesis of NR1099 yielded NR1243, which carries a CCG-to-TCG mutation in codon 282 of lptG (yellow asterisk). (C) Evolution and phenotypes of strains defective in LPS transport used in this study. NR1099 (reduced LPS transport) was derived from wild-type strain NR754. NR1243 (limiting LPS export) containing lptGP282S was derived from NR1099, and its growth is dependent on the presence of arabinose (ARA). Lethality of NR1243 in the absence of arabinose is suppressed by mutations in fabH (strains NR1635, which carries sup4, and NR1770, which carries ∆fabH::kan) or accD (strain NR2025, which carries accDV23G). (D) Growth of strain NR1243 with limiting LPS transport in LB in the presence (blue) or absence (black) of arabinose, and of suppressor strain NR1770 (red) in the absence of arabinose. Growth was monitored by optical density at 600 nm. (E) AccD and FabH are enzymes that participate in the initiation phase of FA biosynthesis. AccD is a subunit of ACC, which carboxylates acetyl-CoA to produce malonyl-CoA. FabH is a condensing enzyme that synthesizes the acetoactyl-ACP precursor necessary for the elongation phase in FA biosynthesis.
In this study, we searched for suppressors of a lethal defect in the Lpt pathway and found mutations in genes encoding enzymes involved in fatty acid (FA) biosynthesis. These mutations greatly reduce cell size and growth rate. Therefore, we propose that suppression results from a significant decrease in the growth rate of the cell envelope. In addition, we show that, contrary to previous reports (29), FabH is not essential for FA biosynthesis and viability in E. coli. Furthermore, we show that FabH is required to adjust cell size properly and propose a model for how E. coli regulates size and growth rate in response to nutrient availability.
Results
Defects in FA Biosynthesis Suppress Lethality Caused by Defects in LPS Biogenesis.
The cell surface of most Gram-negative bacteria is covered with LPS, which is transported from the IM to the OM by the Lpt system (Fig. 1A) (13, 14, 30). Mild defects in LPS transport increase the sensitivity to many antibiotics, because wild-type levels of LPS at the cell surface are required to maintain the barrier-like quality of the OM (31, 32). Severe defects in LPS transport can be lethal, because the presence of LPS at the OM is essential for the viability of E. coli (12–14). Interestingly, other Gram-negative bacteria, such as Neisseria meningitidis and Acinetobacter baumannii, produce LPS but also can survive without it (33–35). How some bacteria are able to compensate for the loss of LPS is unknown.
We wondered if it would be possible to find mutants of E. coli that can survive with limited amounts of LPS in their OM. To investigate this question, we searched for suppressor mutations that would allow the survival of a mutant strain that has a severely compromised Lpt pathway. Depletion of any Lpt protein leads to the same phenotypes (13, 14, 26), so there is no specific reason to choose a defect in a particular Lpt factor. In our studies, we used a strain (NR1243) that has two defects in the Lpt pathway (Fig. 1B). One is that the production of the LptF/G components has been placed under the control of a promoter that is inducible with arabinose (Fig. 1 A and B). This change alone is not lethal, because even without inducer there is still enough expression of lptFG to support viability (Fig. 1C, NR1099). In addition, the NR1243 mutant strain carries a partial loss-of-function mutation in lptG (i.e., lptGP282S). The combination of these two changes renders the growth of this mutant strain dependent on the presence of inducer, which is required to attain high levels of expression of the mutant lptGP282S allele (Fig. 1C, NR1243). Without inducer, LPS transport is limiting for growth (Fig. 1 C and D), resulting in the same phenotypes as reported for depletion of LptF/G (19). Therefore, strain NR1243 is a conditionally lethal lpt mutant that is dependent on the presence of inducer to survive.
We isolated spontaneous suppressor mutations that allow survival of this mutant strain in the absence of inducer (Fig. 1C). Two suppressor strains form small colonies in both the presence and the absence of inducer; using this phenotype, we mapped the suppressor alleles sup4 and sup11 to fabH and accD. The sup4 allele contains a duplication in tandem of the ATGGCG sequence that is located at bp 476–481 of fabH, resulting in the insertion of Asp-Gly in a conserved loop in FabH, an enzyme involved in FA biosynthesis (Fig. 1E) (36–40). sup11 corresponds to accDV23G, which results from a T-to-G transversion at position 68 in accD. AccD is a subunit of acetyl-CoA carboxylase (ACC) (Fig. 1E). Therefore, both suppressor mutations are in genes specifying enzymes of the initiation phase of FA biosynthesis. These suppressors are loss-of-function alleles, because both the small-colony morphology phenotype and suppression are abolished in the presence of their respective wild-type fabH and accD alleles encoded by the pCA24NFabH and pCA24NAccD plasmids (41). Because AccD is essential, accDV23G is a partial loss-of-function allele (42). However, replacement of fabH with a kanamycin-resistance cassette also suppresses the requirement for inducer and confers the small-colony phenotype, suggesting that sup4 is a total loss-of-function allele (Fig. 1D). Because sup4 can revert easily without selection, but deletions never can, we continued our studies with strains that carry ΔfabH::kan (42). The remainder of this study focuses on this ΔfabH suppressor unless indicated otherwise.
Suppression by ΔfabH of the inducer dependency of the lpt mutant strain NR1243 is not the result of either increased LPS synthesis or induction of the σE or Cpx envelope stress responses (Fig. S1), which up-regulate factors involved in OM biogenesis (43, 44). Importantly, we could not introduce a ΔlptB::kan, ΔlptF::kan, ΔlptG::kan, or ΔlptFG::kan allele into a ΔfabH mutant, demonstrating that ΔfabH cannot suppress the lethality caused by the total loss of LPS transport to the OM. Thus, ΔfabH does not suppress in NR1243 by bypassing the need for LPS at the cell surface. Instead, ΔfabH allows the survival of cells that have very limited Lpt function.
FabH Is Not Essential for FA Biosynthesis in E. coli.
FabH is a condensing enzyme involved in the initiation cycle of type II FA biosynthesis, the pathway that bacteria use to synthesize FAs (for reviews, see refs. 40 and 45). FabH synthesizes acetoacetyl-acyl carrier protein (ACP), the precursor needed for the elongation cycle during which, in each round, two carbons are added to the acyl-ACP intermediate until the FA reaches the proper length (Fig. 1E). FabH has been reported to be essential for E. coli viability and is believed to be the only enzyme that can synthesize the elongation precursor acetoacetyl-ACP in vivo (29). Because this result contradicts our report that ΔfabH::kan mutants are viable, as well as previous reports from a systematic gene deletion study (42), we investigated the reason for this discrepancy.
Previous attempts to delete fabH were done using a strain that carries both relA1 and spoT1 alleles (29). SpoT and RelA synthesize ppGpp, a small molecule that regulates a plethora of functions, including transcription, translation, and replication (for review, see ref. 46). SpoT also can function as a ppGpp hydrolase. The relA1 defect results in reduced synthase activity of RelA, whereas spoT1 is a loss-of-function allele that reduces both the synthase and hydrolase activities of SpoT (47, 48). Therefore, a relA1 spoT1 mutant cannot regulate ppGpp levels properly. In contrast, our fabH mutant strains are relA1spoT+ (46, 49). Given that ppGpp can affect many cellular processes, we investigated whether the discrepancy regarding the essentiality of FabH could be explained by the difference in spoT alleles present in the strains used. We hypothesized that ΔfabH and ΔspoT could be a synthetically lethal pair.
Notably, relA+ ΔspoT single mutants are not viable, but relA1ΔspoT and ΔrelA ΔspoT double mutants are (50). Therefore, to test whether ΔfabH and ΔspoT are synthetically lethal, we used relA1 and ΔrelA backgrounds. We could not introduce ΔfabH::kan into strains that are relA1 ΔspoT207::cam, suggesting that ΔfabH and ΔspoT are synthetically lethal. To confirm this synthetic lethality, we conducted a linkage-disruption test as follows. The ΔspoT207::cam pyrE60 zib563::Tn10 alleles are genetically linked. Therefore, if such a triple mutant is used as a donor in a P1 transduction cross where transfer of zib563::Tn10 is selected for in tetracycline-resistant transductants, the ΔspoT207::cam and pyrE60 alleles also are transferred at high frequency. Indeed, these three markers were cotransduced at the expected frequency when the recipient was a relA1 FabH+ strain (Table 1). In contrast, none of the tetracycline-resistant transductants acquired the ΔspoT207::cam allele when the recipient was a relA1 ΔfabH mutant. Thus, in crosses using ΔfabH cells, linkage between zib563::Tn10 and ΔspoT207::cam is disrupted because ΔfabH and ΔspoT are synthetically lethal. Linkage between pyrE and zib563::Tn10 also is skewed in this cross because ΔspoT207::cam and pyrE60 are very close to each other on the chromosome. The pyrE60 allele is not responsible for the aforementioned linkage disruption, because crosses using wild-type pyrE donors yielded similar results. Moreover, we obtained similar results with ΔrelA::kan ΔfabH recipients.
Table 1.
fabH and spoT null alleles are synthetically lethal in relA1 strains
| % transductants |
|||
| Recipient | zib563::Tn10 | ΔspoT207::cam | pyrE60 |
| relA1 | 100 | 68 | 64 |
| relA1 ΔfabH::kan | 100 | 0 | 26 |
Tetracycline-resistant transductants were selected and screened for chloramphenicol resistance (ΔspoT207::cam) and inability to grow on minimal glucose medium (pyrE60).
These results show that ΔspoT and ΔfabH are synthetically lethal. Consequently, FabH is essential in mutants where ppGpp synthesis is compromised, and this essentiality is likely the reason previous attempts to delete fabH failed (29). We must clarify that the presence of relA1 is not required for viability of the fabH mutant, because we also could introduce the ΔfabH::kan allele into the relA+spoT+ E. coli K-12 strains MG1655 and W3110 (51, 52). Thus, FabH is not essential in wild-type E. coli and is not the only enzyme that can synthesize in vivo the precursor required for the elongation cycle in FA biosynthesis.
Loss of FabH Results in Altered FA Composition.
Because fabH mutants are viable even in minimal medium, another condensing enzyme must substitute for FabH in the initiation cycle of FA biosynthesis. The small-colony phenotype displayed by fabH mutants suggests that this complementation is not perfect and that FA biosynthesis is defective in these mutants. To investigate this suggestion, we compared FA composition in wild-type and ΔfabH cells using gas chromatography analysis of FA methyl esters (GC-FAME) (53, 54).
GC-FAME analyses showed that the loss of FabH results in an overall increase in the content of unsaturated FA by ∼10%, mostly because of a similar decrease in C16:0 (Table 2). Levels of C18:1ω7c also increase, probably in part because of an additional round of elongation of C16:1ω7c, whose levels are decreased in the fabH mutant. The increase in C18 species and the concomitant decrease in C16 species is the opposite of the phenotype reported for cells that produce increased levels of FabH, where a decrease in C18:1 and an increase in C14:0 and C16:0 were observed (39). Thus, our data show that strains lacking FabH show altered FA composition.
Table 2.
Profile of major FA in wild-type (NR754) and ΔfabH::kan (NR1769) strains
| FA species | Wild type* | ΔfabH |
| 12:0 | 3.91 ± 0.13 | 2.42 ± 0.11 |
| 14:0 | 7.17 ± 0.17 | 2.76 ± 0.04 |
| 14:0 3OH | 8.70 ± 0.47 | 9.99 ± 0.21 |
| 16:1 ω7c | 25.87 ± 0.81 | 12.59 ± 0.28 |
| 16:0 | 26.70 ± 0.15 | 17.23 ± 0.17 |
| 17:0 cyclo | 1.74 ± 0.31 | 4.36 ± 0.18 |
| 18:1 ω 7c | 19.42 ± 1.70 | 43.03 ± 0.87 |
| 18:0 | 0.91 ± 0.10 | 1.65 ± 0.12 |
| Total | 94.41 | 94.04 |
*Values represent the per cent of the total FA species (those reported here and some minor species). Data are the average ± SD of three independent analyses.
The changes in FA composition observed in the ΔfabH mutant are absent in an accDV23G mutant (Table S1). Because ΔfabH and accDV23G alleles both suppress the lethality caused by low levels of LPS transport to the OM, and they both confer slow-growth and small-size phenotypes (see below), we conclude that none of these phenotypes results from the aforementioned changes in FA composition. However, the altered FA content might explain why the ΔfabH allele increases the permeability of the OM to hydrophobic antibiotics but the accDV23G mutant does not (Table 3).
Table 3.
ΔfabH, but not accDV23G, increases OM permeability
| Relative change in MIC (MICWT/MICmutant)† |
|||||
| Strain | Relevant genotype | ARA* | Bacitracin | Erythromycin | Rifampin |
| NR1099 | PBAD::lptF lptG | − | 2 | 1 | 4 |
| NR1099 | PBAD::lptF lptG | + | 1 | 1 | 2 |
| NR1243 | PBAD::lptF lptGP282S +ARA | + | 4 | 2 | 4 |
| NR1770 | PBAD::lptF lptGP282S ΔfabH::kan | + | 32 | 8 | 4 |
| NR1770 | PBAD::lptF lptGP282S ΔfabH::kan | − | 128 | 8 | 16 |
| NR1769 | ΔfabH::kan | − | 16 | 2 | 2 |
| NR1771 | ΔfabH::FRT | − | 16 | 2 | 2 |
| NR2025 | PBAD::lptF lptGP282S accDV23G | + | 1 | 1 | 4 |
| NR2025 | PBAD::lptF lptGP282S accDV23G | − | 4 | 1 | 16 |
| NR2020 | purF::tet | − | 1 | 1 | 1 |
| NR2025 | purF::tet accDV23G | − | 1 | 1 | 1 |
*ARA refers to the presence (+) or absence (−) of arabinose in the growth medium.
†Minimal inhibitory concentrations (MIC) were determined for wild-type strain NR754 and mutant strains in LB in the presence or absence of arabinose (ARA). Values shown represent the ratio of the MIC of wild-type NR754 grown in LB in the absence of arabinose to the MIC of each mutant.
Defects in FA Biosynthesis Reduce Cell Size.
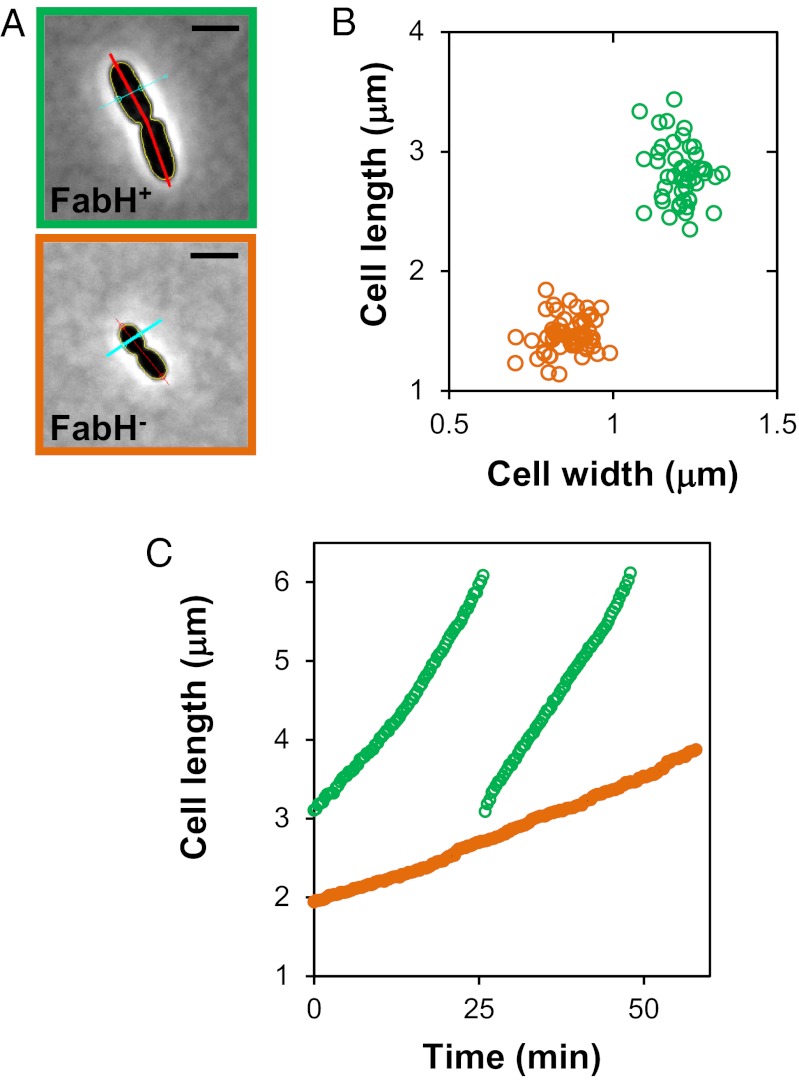
A striking phenotype of cells that lack FabH is that they are much smaller than their wild-type parent (Fig. 2). Measurements of wild-type and ΔfabH::kan cells growing logarithmically in LB revealed that wild-type cells are ∼50% longer and ∼30% wider than ΔfabH cells (Table 4). As a result, the average surface area and volume of ΔfabH cells are 11.55 (± 1.03) μm2 and 1.21 (± 0.22) μm3, respectively. In contrast, the average surface area and volume of wild-type cells are 22.28 (± 1.5) μm2 and 4.16 (± 0.45) μm3, respectively. Strikingly, the loss of FabH reduces cell surface area by ∼50% and cell volume by ∼70%. It is likely that this remarkable reduction in cell size in a ΔfabH mutant is the result of limited biosynthesis of membranes. FAs are required to synthesize the lipid component of membranes. Because cells are delimited by membranes, it is possible that FA biosynthesis is involved in determining cell size by controlling the amount of membranes synthesized.
Fig. 2.
Loss of FabH causes a decrease in cell size and cell envelope growth rate. (A) Microscope images and image analysis of representative wild-type (NR754) and ∆fabH::kan (NR1769) cells. (Scale bars, 2 μm.) Red and cyan lines intersecting with yellow cell contours are used to measure cell length and width, respectively. (B) Scatter plot of cell length and width of wild-type (green) and ∆fabH::kan (orange) cells. (C) Cell length was monitored during growth of wild-type (green, two cell cycles) and ∆fabH::kan (orange, one cell cycle) cells on LB-agarose pads at 37 °C.
Table 4.
Regulation of cell size in response to nutrient availability is compromised in fabH mutants
| Strain | Relevant genotype | Growth medium* | Length† (μm) | Width† (μm) | Generation time (min)‡ | Surface§ (μm2) | Volume§ (μm3) | Surface area/ volume |
| NR754 | Wild type | LB | 2.81 ± 0.24 | 1.21 ± 0.05 | 26 | 22.26 | 4.16 | 5.3 |
| Minimal glucose medium | 1.84 ± 0.15 | 0.89 ± 0.07 | 61 | 14.04 | 1.51 | 9.3 | ||
| NR1769 | ΔfabH::kan | LB | 1.46 ± 0.15 | 0.87 ± 0.06 | 52 | 11.55 | 1.21 | 9.6 |
| Minimal glucose medium | 1.36 ± 0.09 | 0.81 ± 0.04 | 86 | 10.61 | 0.97 | 10.6 |
*Cells were grown at 37 °C in either LB or glucose M63 minimal medium.
†Average dimensions (length and width) and SD of cells were obtained from at least 48 cells per strain and condition.
‡Average generation time of the culture.
§Surface area and volume were calculated from the average length and width.
To test this hypothesis, we determined whether cell size would be reduced in cells in which FA biosynthesis is decreased by means other than ΔfabH, namely by treatment with a small-molecule inhibitor and the accDV23G allele. Indeed, treatment of wild-type cells with a subinhibitory concentration of cerulenin, which inhibits FA elongation (55), reduces cell size (2.07 ± 0.32 μm in length × 1.01 ± 0.06 μm in width). In addition, we found that the size of ΔfabH and accDV23G mutant cells is similar. These results confirm our hypothesis that alterations in FA biosynthesis affect cell size. In addition, they show that the effect on cell size is not specific to the loss of FabH, because compromising FA biosynthesis at different steps with either a small molecule or a mutation causes a reduction in cell size.
Loss of FabH Function Compromises Regulation of Cell Size in Response to Nutrient Availability.
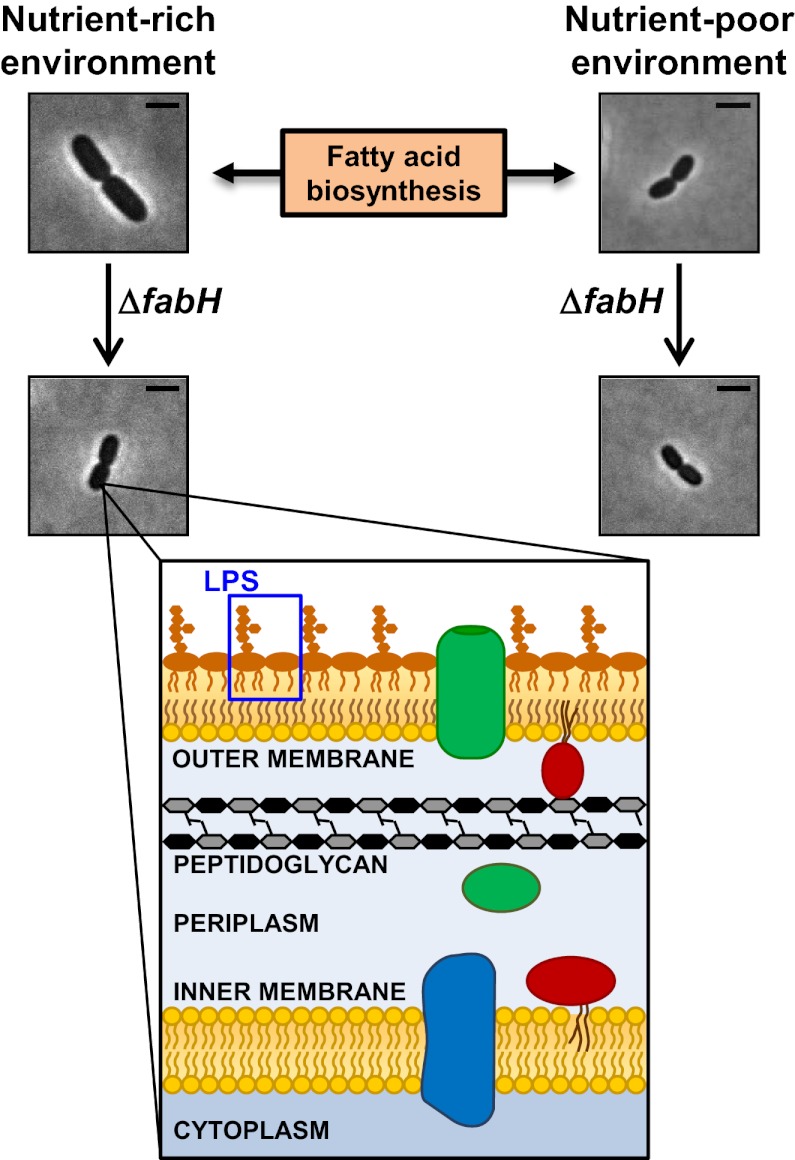
E. coli regulates cell size in response to the availability of nutrients so that it grows larger in rich medium than in minimal medium (1, 2, 5). Because FA biosynthesis is altered in response to nutrient levels (56–58), we wondered whether the loss of FabH would interfere with the ability of E. coli to regulate cell size in response to nutrient availability. To test this question, we compared cell size of cells grown in rich or minimal medium. As expected, wild-type cells grown in glucose minimal medium are shorter and thinner than those grown in rich medium; in contrast, the size of ΔfabH cells does not change significantly (Table 4). Thus, cell size regulation in response to nutrient availability is impaired in cells that lack FabH.
Loss of FabH Function Suppresses Lethality Caused by Defects in LPS Biogenesis by Reducing the Rate of Cell Envelope Growth.
The lpt mutant NR1243 dies without inducer because transport of LPS to the cell surface is compromised. ΔfabH and accDV23G suppress this lethality. Both these mutations affect FA biosynthesis, decrease cell size, and increase doubling time. One hypothesis to explain these observations is that LPS transport in NR1243 might be too slow to cope with the rate at which the rest of the cell envelope grows and that suppressors act by slowing down envelope growth rate. To probe this notion, we first compared the rate of elongation of the cell envelope in wild-type cells and in the fabH mutant. ΔfabH cells are shorter, and they divide at a slower rate than wild-type cells (Fig. 2C). Together, these phenotypes result in an ∼75% reduction in the rate of elongation of the cell envelope in the fabH mutant (∼0.12 μm/min for the wild type and ∼0.03 μm/min for the fabH mutant). This finding is consistent with the hypothesis that a reduction in the rate of envelope growth suppresses the lpt defect in NR1243.
If this model is correct, slowing down envelope growth by other means also should suppress this lethality. Because cells growing in minimal medium are smaller and have a longer doubling time than when they grow in rich medium (Table 4), their cell envelope also must grow at a lower rate. We therefore tested whether the lpt mutant NR1243 could grow in minimal medium in the absence of inducer and found that it can. Simply increasing doubling time to a level similar to that observed in minimal medium without altering cell size does not suppress lethality. For example, the lpt mutant cannot grow in LB at room temperature (∼20 °C); under this condition, cells divide at the same rate as in minimal medium but without reducing cell size (2). Likewise, the ΔrrmJ::kan allele, which affects ribosomal maturation and increases doubling time to ∼50 min without affecting cell size (59), is not a suppressor in the lpt mutant strain NR1243. Together, these results support a model in which ΔfabH suppresses the lethality that results from severely defective LPS transport by decreasing the rate of envelope growth and thereby restoring a balance between the rates of LPS assembly at the cell surface and envelope biogenesis.
Discussion
In E. coli, severe defects in the biogenesis of LPS can be lethal. Here, we have shown that lethality caused by a severe defect in LPS transport to the cell surface can be suppressed by defects in FA biosynthesis. Specifically, this lethality is suppressed by mutations in fabH and accD, two genes that encode enzymes involved in FA biosynthesis (40). We propose that this suppression is a consequence of the slower rate of growth of the cell envelope that results from a reduction of cell size and slower growth rate. Importantly, we also have demonstrated that, contrary to previous reports, FabH is not required for FA biosynthesis and, consequently, is not essential for the viability of E. coli (29). However, we show that FabH is required to adjust cell size properly in response to nutrient availability, and we propose that FA biosynthesis is involved in the regulation of cell size by nutrients in the environment (Fig. 3).
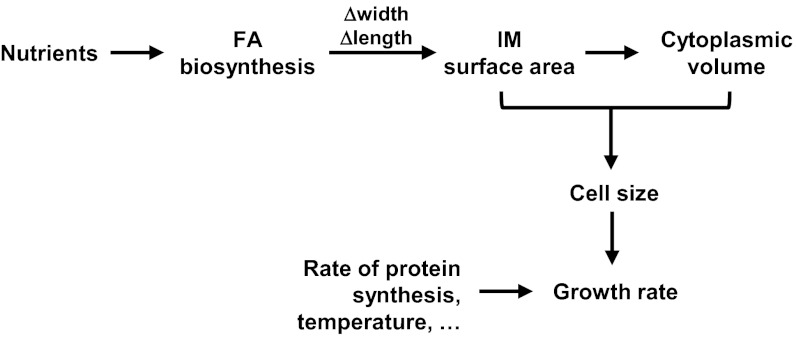
Fig. 3.
Model for the regulation of cell size and growth rate in response to nutrient availability. The flux of nutrients in the cell controls FA biosynthesis, and in some way FA biosynthesis controls both cell width and length. Cell width and length define the IM surface area of the cell, which in turn limits the cytoplasmic volume (∼cell size). The biosynthetic capacity of the cell, which controls the rate of mass increase (i.e., growth rate), depends on both the IM surface area and the cytoplasmic volume (cell size), because all cellular building blocks are synthesized at the IM and in the cytoplasm. Other factors can alter growth rate without affecting size. See Discussion for details.
FA Biosynthesis Regulates Cell Size in Response to Nutrient Availability.
In E. coli, chromosomal DNA replication is not initiated until the cell reaches an appropriate mass (cell size) (6, 7). Because E. coli is rod-shaped, how fast this cell size is reached in a particular environment depends on how fast the cell elongates. In turn, the rate of elongation depends on the biosynthetic capacity of the cell to convert nutrients into building blocks, namely lipids, proteins, nucleic acids, and carbohydrates. In E. coli, the synthesis of these building blocks occurs in the cytoplasm and the IM. Therefore, the size of these cellular compartments limits the biosynthetic capacity of the cell. According to this analysis, ΔfabH cells grow slowly in rich medium because they have significantly less cytoplasm and IM than wild type, being 70% smaller in volume and 50% smaller in surface area.
The small size of ΔfabH cells results from a decrease in the rate of FA biosynthesis. Cell size also can be reduced by decreasing the rate of FA biosynthesis at different steps in the pathway, as illustrated by treatment with subinhibitory concentrations of cerulenin or by the presence of the accDV23G allele. The small size of cells deficient in FA biosynthesis likely results from a decrease in the rate of IM lipid biogenesis because the cytoplasm, which contributes to most of the cellular volume (or size), is constrained by the amount of IM surface area (Fig. 3). The decrease in cell size appears to be specific to defects in FA biosynthesis and not a consequence of a reduction in the synthesis of any major membrane component, such as proteins. Indeed, we decreased the rate of protein synthesis by introducing a ΔrrmJ::kan allele; this mutation slows growth rate, and therefore envelope biogenesis, but does not alter cell size (59 and 60).
Why does a fabH mutant cell not grow to the same size as a wild-type cell in rich medium albeit at a slower rate? We propose that FA biosynthesis is a central process in controlling cell size and that wild-type E. coli changes cell size in response to nutrients by altering FA biosynthesis through metabolism (Fig. 3). Supporting this model is the fact that ΔfabH cells are defective in regulating their size in response to nutrient availability. ΔfabH cells are blind to nutrient excess and are constitutively small because the regulatory mechanism that couples nutrient availability and cell size in E. coli is compromised. Thus, the loss of FabH is epistatic to the step at which nutrient availability regulates FA biosynthesis, which in turn is crucial for determining cell size.
The molecular mechanism by which FA biosynthesis regulates cell size is unknown. However, we believe that it involves a change in both cell width and length. When rod-shaped E. coli and Salmonella change cell size in response to nutrients, they change both length and width (Table 4 and refs. 2 and 61), a more efficient way to reduce cell volume than simply changing length. In contrast, at steady state, when E. coli cells grow and divide, they change only their length (62). Changing only cell length does not alter the surface area-to-volume ratio, but changing cell width does (Table 4). It is possible that E. coli uses this surface area-to-volume ratio to distinguish a change in size caused by a shift in nutrients from the change in size that the cell undergoes during division at steady state. This distinction could be important to coordinate size, rate of mass increase, and division. It is worth noting that E. coli and B. subtilis coordinate cell size, growth rate, and initiation of DNA replication differently, although both bacteria use DnaA to control the initiation of replication (7). Interestingly, cell width does not change in the rod-shaped B. subtilis bacterium in response to nutrient availability (4, 7). It remains to be determined whether this is a fundamental difference that exists between rod-shaped Gram-positive (e.g., B. subtilis) and Gram-negative (e.g., E. coli and Salmonella) bacteria and whether changing the surface area-to-volume ratio itself can alter the effective concentration of a factor such as DnaA. It also remains to be determined whether ppGpp is involved in regulating cell size at a step downstream of FA biosynthesis. The synthetic lethality we have observed between fabH and spoT could be the result of this regulatory function.
How nutrient availability is coupled to FA biosynthesis also is unknown, although it is clear that the rate of FA biosynthesis increases with nutrient availability (56–58). The mechanism for this control has been attributed to the effect that nutrients have on growth rate, and it is thought that growth rate itself regulates FA biosynthesis (58, 63). In contrast, we propose that nutrient availability determines the rate of FA biosynthesis; FA biosynthesis then controls cell size, which determines the overall biosynthetic capacity of the cell and therefore growth rate (Fig. 3). Placing growth rate downstream of FA biosynthesis in this pathway reflects how the biosynthetic capacity of the cell depends on both the surface area of the IM and the volume of the cytoplasm. In addition, this placement is in agreement with the fact that growth rate can be slowed by environmental factors such as temperature and by mutations (e.g., ΔrrmJ::kan) without altering cell size (2, 59). Proposing a central role for FA biosynthesis also is appealing because it physically links metabolism and cell size through IM biogenesis. Furthermore, it offers a mechanism to coordinate growth of the entire cell envelope, because all envelope components must be produced, assembled, and/or transported across the IM.
Revisiting the Pathway for Type II FA Biosynthesis.
The current model for type II FA biosynthesis designates FabH as the enzyme required to synthesize in vivo acetoacetyl-ACP, the final product of the initiation phase and precursor of the elongation cycle (Fig. 1E) (40), because FabH was reported to be essential for viability of E. coli and Lactococcus lactis (29). However, we have demonstrated that FabH is not essential in E. coli. This discrepancy is explained by the fact that previous attempts to delete fabH were conducted in a spoT mutant, and we have shown that fabH is synthetically lethal with spoT. Therefore, the loss of SpoT function makes FabH essential. Cells lacking SpoT are defective in the production of ppGpp, a small molecule that regulates many essential functions including division, transcription, and the biosynthesis of proteins, peptidoglycan, phospholipids, and FA (46). This synthetic lethality could stem from a misregulation of FA biosynthesis in a spoT mutant. Interestingly, FA starvation triggers a SpoT-mediated accumulation of ppGpp, which in turn regulates several essential functions (46, 64, 65). This regulatory role of ppGpp might be essential in the absence of FabH. Alternatively, ppGpp might be necessary for proper production of whichever enzyme(s) are substitutes for FabH. In any event, the current model for type II FA biosynthesis in E. coli needs revision. In addition, these findings have important implications for whether FabH is an effective target for antibiotic intervention (55, 66–69). At the moment, whether any of the condensing enzymes involved in FA elongation (FabF/B) can substitute for FabH in E. coli is unknown. Alternatively, it is possible that another type of β-ketoacyl-ACP synthase exists, as has been shown very recently in Pseudomonas (70).
Materials and Methods
Bacterial Strains and Growth Conditions.
All strains are derivatives of NR754, an araD+ revertant of MC4100 (19, 49). Alleles were transferred using P1 transduction (71). LB and M63 minimal broth and agar were prepared as previously described (71). When appropriate, growth medium was supplemented with 0.2% glucose or l-arabinose (wt/vol), kanamycin (25 μg/mL), ampicillin (25 μg/mL), chloramphenicol (20 μg/mL), and cerulenin (25 μg/mL). The ΔfabH::kan and ΔrelA::kan alleles and the pCA24NFabH and pCA24NAccD plasmids were obtained from the Keio and ASKA collections, respectively (41, 42). The ΔfabH::kan allele was introduced into NR754 and NR1243 to construct NR1769 and NR1770, respectively. The kan cassette in NR1769 was excised to construct the fabH::FRT allele in NR1771 (72). Strain CF11311, a gift from M. Cashel (National Institutes of Health, Bethesda, MD), was the source for the ΔspoT207::cam, pyrE60, and zib563::Tn10 alleles. Except for recombineering and when indicated, strains were grown with aeration at 37 °C, and their growth was monitored by optical density at 600 nm (OD600).
Construction of NR1099.
The primers 5yjgPPbad (5′-AACTCATGTCCGCTGTTGCGATGACTTCGTGTTAATCTTAACGTTATTACGGCATTGGCACGTCAGAACAGTATATATGAGTAAACTTGGTCTG) and 3yjgPPbad (5′-GCAGGATTTTAGCTTGTTTCATGGCTTAAACGTCATTTATTCTCTTGAGTCGTCGAAATCGTCGCTAAGATTCCTTAGAGCTCGAATTCCC) were used to amplify a 2,234-bp PCR product containing bla-araC- PBAD from pKD46 (73). 5yjgPPbad anneals 5–28 bp (underlined) downstream of bla, and 3yjgPPbad anneals 20–40 bp (underlined) downstream of the PBAD promoter in pKD46. The PCR product was electroporated into DY378 for recombineering to replace the fragment −139 to −101 bp upstream of lptF with bla-araC- PBAD (74). Transformants were selected in medium with 25 μg/mL ampicillin and 0.02% arabinose. The resulting bla-araC- PBAD::lptFG was introduced into NR754 using P1 transduction by selection on medium containing ampicillin and arabinose to yield NR1099.
Construction of NR1243.
Strain NR1099 was mutagenized with N-methyl-N'-nitro-N-nitrosoguanidine (NTG) (71). Wild-type NR754 was used as a recipient in a cross with a P1 lysate raised in NTG-mutagenized NR1099, and ampicillin-resistant transductants were obtained in the presence of arabinose. These transductants were screened for arabinose-dependent growth. One mutant strain that was dependent on arabinose for growth was used in a P1 cross as a donor with NR754 as recipient. An ampicillin-resistant transductant whose growth is dependent on the presence of arabinose was designated “NR1243.” DNA sequence revealed that NR1243 carries a C-to-T transition at bp 844 of lptG, which changes a conserved Pro at residue 282 of LptG to Ser.
Isolation of Suppressor Strains.
Arabinose-independent suppressors of NR1243 were obtained by plating NR1243 on LB without arabinose. Strain NR1635, which carries sup4, and strain NR2025, which carries sup11, arose at a frequency of ∼10−7. We mapped sup4 and sup11 by P1 transduction using a pool of mini-Tn10 insertional mutants and determined the location of linked mini-Tn10 insertions using arbitrary PCR as previously described (75). Kanamycin-marked alleles from the Keio collection in the region of interest also were used in mapping (42). DNA sequencing revealed the identity of sup4 and sup11. The accDV23G allele was linked to purF::tet to transduce it into wild-type strain NR754 to generate NR2020 (purF::tet) and NR2022 (purF::tet accDV23G).
Characterization of Cell Size and Shape.
Cells were grown into early exponential phase and transferred onto agarose pads for phase-contrast imaging on an inverted Nikon TE2000-E microscope. Details of the imaging acquisition were described previously (76). Computational analysis of single-cell size is implemented in MATLAB. Because cell length varies during the cell cycle, cell length and width were measured only on dividing cells with an “8” morphology. Surface was calculated as 4πr2 + 2πl and volume as 4/3 πr3 + πr2l, where r represents radius or width/2, and l represents the length of the cell.
Sensitivity to Antibiotics.
To measure minimal inhibitory concentration (MIC) values, wild-type and mutant cells were grown to early exponential phase (OD600 ∼0.05), diluted with LB to a cell density of 104 cells/mL, and transferred to a 96-well flat-bottomed plate with polystyrene wells and a lid (3596; Corning Costar). Appropriate concentrations of antibiotics were prepared by 1:2 serial dilutions and added to the plate with cell cultures. Plates were incubated overnight at 37 °C. The first clear well with the lowest concentration of drug indicated the MIC value for each antibiotic.
Determining Generation Time.
OD600 growth curves were used to derive generation time and growth rate (doublings per hour). Overnight cultures of wild-type NR754 and fabH mutant NR1769 were diluted 1:100 in 3 mL fresh medium and allowed to grow to early exponential phase (OD600 ∼0.05) in 14-mL BD Falcon tubes with vigorous shaking at 37 °C. Optical density then was monitored every 45 min for ∼4 h and was plotted against time on a logarithmic scale. Linear regression analysis was carried out on this plot, and its slope was used to calculate generation time and growth rate.
FA Content Analysis.
NR754 and NR1769 were grown in LB broth to OD600 ∼0.8, washed in LB broth, pelleted, and frozen at −80 °C. Lipids were saponified in sodium hydroxide and methanol, methylated in acidified methyl alcohol, extracted in hexane and methyl tertiary-butyl ether, and base washed by Microbial ID, Inc. using their Sherlock Microbial Identification System standard protocol. Samples were subjected to FA methyl ester analysis using gas chromatography equipped with a flame ionization detector. Sherlock Microbial Identification System software was used to identify species by comparing retention times with an external calibration standard manufactured by Microbial ID, Inc. (53, 54). Although usually not reported in E. coli, we also detected C18:1ω9c and C19:cyclo ω8c as minor species in some samples.
Supplementary Material
Acknowledgments
We thank Michael Cashel for providing strain CF1131 and Mark Mandel and Emily Butler for helpful discussions on this work. This work was supported by Grants AI081059 (to D.K.) and GM081617 (to R.K.) and by start-up funds from The Ohio State University (to N.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 15097 (volume 109, number 38).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209742109/-/DCSupplemental.
References
- 1.Woldringh CL, Grover NB, Rosenberger RF, Zaritsky A. Dimensional rearrangement of rod-shaped bacteria following nutritional shift-up. II. Experiments with Escherichia coli B/r. J Theor Biol. 1980;86:441–454. doi: 10.1016/0022-5193(80)90344-6. [DOI] [PubMed] [Google Scholar]
- 2.Schaechter M, Maaloe O, Kjeldgaard NO. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J Gen Microbiol. 1958;19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- 3.Sargent MG. Control of cell length in Bacillus subtilis. J Bacteriol. 1975;123:7–19. doi: 10.1128/jb.123.1.7-19.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weart RB, et al. A metabolic sensor governing cell size in bacteria. Cell. 2007;130:335–347. doi: 10.1016/j.cell.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donachie WD, Begg KJ, Vicente M. Cell length, cell growth and cell division. Nature. 1976;264:328–333. doi: 10.1038/264328a0. [DOI] [PubMed] [Google Scholar]
- 6.Donachie WD. Relationship between cell size and time of initiation of DNA replication. Nature. 1968;219:1077–1079. doi: 10.1038/2191077a0. [DOI] [PubMed] [Google Scholar]
- 7.Hill NS, Kadoya R, Chattoraj DK, Levin PA. Cell size and the initiation of DNA replication in bacteria. PLoS Genet. 2012;8:e1002549. doi: 10.1371/journal.pgen.1002549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bramhill D, Kornberg A. Duplex opening by DnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988;52:743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- 9.Katayama T, Ozaki S, Keyamura K, Fujimitsu K. Regulation of the replication cycle: Conserved and diverse regulatory systems for DnaA and oriC. Nat Rev Microbiol. 2010;8:163–170. doi: 10.1038/nrmicro2314. [DOI] [PubMed] [Google Scholar]
- 10.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamio Y, Nikaido H. Outer membrane of Salmonella typhimurium: Accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium. Biochemistry. 1976;15:2561–2570. doi: 10.1021/bi00657a012. [DOI] [PubMed] [Google Scholar]
- 12.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruiz N, Kahne D, Silhavy TJ. Transport of lipopolysaccharide across the cell envelope: The long road of discovery. Nat Rev Microbiol. 2009;7:677–683. doi: 10.1038/nrmicro2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sperandeo P, Dehò G, Polissi A. The lipopolysaccharide transport system of Gram-negative bacteria. Biochim Biophys Acta. 2009;1791:594–602. doi: 10.1016/j.bbalip.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 15.Doerrler WT, Gibbons HS, Raetz CR. MsbA-dependent translocation of lipids across the inner membrane of Escherichia coli. J Biol Chem. 2004;279:45102–45109. doi: 10.1074/jbc.M408106200. [DOI] [PubMed] [Google Scholar]
- 16.Polissi A, Georgopoulos C. Mutational analysis and properties of the msbA gene of Escherichia coli, coding for an essential ABC family transporter. Mol Microbiol. 1996;20:1221–1233. doi: 10.1111/j.1365-2958.1996.tb02642.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Z, White KA, Polissi A, Georgopoulos C, Raetz CR. Function of Escherichia coli MsbA, an essential ABC family transporter, in lipid A and phospholipid biosynthesis. J Biol Chem. 1998;273:12466–12475. doi: 10.1074/jbc.273.20.12466. [DOI] [PubMed] [Google Scholar]
- 18.Narita S, Tokuda H. Biochemical characterization of an ABC transporter LptBFGC complex required for the outer membrane sorting of lipopolysaccharides. FEBS Lett. 2009;583:2160–2164. doi: 10.1016/j.febslet.2009.05.051. [DOI] [PubMed] [Google Scholar]
- 19.Ruiz N, Gronenberg LS, Kahne D, Silhavy TJ. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2008;105:5537–5542. doi: 10.1073/pnas.0801196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sperandeo P, et al. Characterization of lptA and lptB, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J Bacteriol. 2007;189:244–253. doi: 10.1128/JB.01126-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sperandeo P, et al. New insights into the Lpt machinery for lipopolysaccharide transport to the cell surface: LptA-LptC interaction and LptA stability as sensors of a properly assembled transenvelope complex. J Bacteriol. 2011;193:1042–1053. doi: 10.1128/JB.01037-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tran AX, Dong C, Whitfield C. Structure and functional analysis of LptC, a conserved membrane protein involved in the lipopolysaccharide export pathway in Escherichia coli. J Biol Chem. 2010;285:33529–33539. doi: 10.1074/jbc.M110.144709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chng SS, Ruiz N, Chimalakonda G, Silhavy TJ, Kahne D. Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc Natl Acad Sci USA. 2010;107:5363–5368. doi: 10.1073/pnas.0912872107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu T, et al. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2006;103:11754–11759. doi: 10.1073/pnas.0604744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freinkman E, Chng SS, Kahne D. The complex that inserts lipopolysaccharide into the bacterial outer membrane forms a two-protein plug-and-barrel. Proc Natl Acad Sci USA. 2011;108:2486–2491. doi: 10.1073/pnas.1015617108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sperandeo P, et al. Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J Bacteriol. 2008;190:4460–4469. doi: 10.1128/JB.00270-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tefsen B, Geurtsen J, Beckers F, Tommassen J, de Cock H. Lipopolysaccharide transport to the bacterial outer membrane in spheroplasts. J Biol Chem. 2005;280:4504–4509. doi: 10.1074/jbc.M409259200. [DOI] [PubMed] [Google Scholar]
- 28.Chng SS, Gronenberg LS, Kahne D. Proteins required for lipopolysaccharide assembly in Escherichia coli form a transenvelope complex. Biochemistry. 2010;49:4565–4567. doi: 10.1021/bi100493e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai CY, Cronan JE. Beta-ketoacyl-acyl carrier protein synthase III (FabH) is essential for bacterial fatty acid synthesis. J Biol Chem. 2003;278:51494–51503. doi: 10.1074/jbc.M308638200. [DOI] [PubMed] [Google Scholar]
- 30.Narita S. ABC transporters involved in the biogenesis of the outer membrane in gram-negative bacteria. Biosci Biotechnol Biochem. 2011;75:1044–1054. doi: 10.1271/bbb.110115. [DOI] [PubMed] [Google Scholar]
- 31.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruiz N, Kahne D, Silhavy TJ. Advances in understanding bacterial outer-membrane biogenesis. Nat Rev Microbiol. 2006;4:57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- 33.Bos MP, Tefsen B, Geurtsen J, Tommassen J. Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc Natl Acad Sci USA. 2004;101:9417–9422. doi: 10.1073/pnas.0402340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steeghs L, et al. Meningitis bacterium is viable without endotoxin. Nature. 1998;392:449–450. doi: 10.1038/33046. [DOI] [PubMed] [Google Scholar]
- 35.Moffatt JH, et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother. 2010;54:4971–4977. doi: 10.1128/AAC.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davies C, Heath RJ, White SW, Rock CO. The 1.8 A crystal structure and active-site architecture of beta-ketoacyl-acyl carrier protein synthase III (FabH) from escherichia coli. Structure. 2000;8:185–195. doi: 10.1016/s0969-2126(00)00094-0. [DOI] [PubMed] [Google Scholar]
- 37.Jackowski S, Rock CO. Acetoacetyl-acyl carrier protein synthase, a potential regulator of fatty acid biosynthesis in bacteria. J Biol Chem. 1987;262:7927–7931. [PubMed] [Google Scholar]
- 38.Qiu X, et al. Refined structures of beta-ketoacyl-acyl carrier protein synthase III. J Mol Biol. 2001;307:341–356. doi: 10.1006/jmbi.2000.4457. [DOI] [PubMed] [Google Scholar]
- 39.Tsay JT, Oh W, Larson TJ, Jackowski S, Rock CO. Isolation and characterization of the beta-ketoacyl-acyl carrier protein synthase III gene (fabH) from Escherichia coli K-12. J Biol Chem. 1992;267:6807–6814. [PubMed] [Google Scholar]
- 40.Zhang YM, Rock CO. Membrane lipid homeostasis in bacteria. Nat Rev Microbiol. 2008;6:222–233. doi: 10.1038/nrmicro1839. [DOI] [PubMed] [Google Scholar]
- 41.Kitagawa M, et al. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 42.Baba T, et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol 2:2006.0008.
- 43.Ades SE. Regulation by destruction: Design of the sigmaE envelope stress response. Curr Opin Microbiol. 2008;11:535–540. doi: 10.1016/j.mib.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 44.Vogt SL, Raivio TL. Just scratching the surface: An expanding view of the Cpx envelope stress response. FEMS Microbiol Lett. 2012;326:2–11. doi: 10.1111/j.1574-6968.2011.02406.x. [DOI] [PubMed] [Google Scholar]
- 45.Cronan JE, Thomas J. Bacterial fatty acid synthesis and its relationships with polyketide synthetic pathways. Methods Enzymol. 2009;459:395–433. doi: 10.1016/S0076-6879(09)04617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Potrykus K, Cashel M. (p)ppGpp: Still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 47.Metzger S, Schreiber G, Aizenman E, Cashel M, Glaser G. Characterization of the relA1 mutation and a comparison of relA1 with new relA null alleles in Escherichia coli. J Biol Chem. 1989;264:21146–21152. [PubMed] [Google Scholar]
- 48.Laffler T, Gallant JA. Stringent control of protein synthesis in E. coli. Cell. 1974;3:47–49. doi: 10.1016/0092-8674(74)90036-1. [DOI] [PubMed] [Google Scholar]
- 49.Casadaban MJ. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 50.Xiao H, et al. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- 51.Blattner FR, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 52.Hayashi K, et al. 2006. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol Syst Biol 2:2006.0007.
- 53.Haack SK, Garchow H, Odelson DA, Forney LJ, Klug MJ. Accuracy, reproducibility, and interpretation of Fatty Acid methyl ester profiles of model bacterial communities. Appl Environ Microbiol. 1994;60:2483–2493. doi: 10.1128/aem.60.7.2483-2493.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stager CE, Davis JR. Automated systems for identification of microorganisms. Clin Microbiol Rev. 1992;5:302–327. doi: 10.1128/cmr.5.3.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heath RJ, White SW, Rock CO. Inhibitors of fatty acid synthesis as antimicrobial chemotherapeutics. Appl Microbiol Biotechnol. 2002;58:695–703. doi: 10.1007/s00253-001-0918-z. [DOI] [PubMed] [Google Scholar]
- 56.Tao H, Bausch C, Richmond C, Blattner FR, Conway T. Functional genomics: Expression analysis of Escherichia coli growing on minimal and rich media. J Bacteriol. 1999;181:6425–6440. doi: 10.1128/jb.181.20.6425-6440.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takamura Y, Nomura G. Changes in the intracellular concentration of acetyl-CoA and malonyl-CoA in relation to the carbon and energy metabolism of Escherichia coli K12. J Gen Microbiol. 1988;134:2249–2253. doi: 10.1099/00221287-134-8-2249. [DOI] [PubMed] [Google Scholar]
- 58.Li SJ, Cronan JE., Jr Growth rate regulation of Escherichia coli acetyl coenzyme A carboxylase, which catalyzes the first committed step of lipid biosynthesis. J Bacteriol. 1993;175:332–340. doi: 10.1128/jb.175.2.332-340.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bügl H, et al. RNA methylation under heat shock control. Mol Cell. 2000;6:349–360. doi: 10.1016/s1097-2765(00)00035-6. [DOI] [PubMed] [Google Scholar]
- 60.Caldas T, Binet E, Bouloc P, Richarme G. Translational defects of Escherichia coli mutants deficient in the Um(2552) 23S ribosomal RNA methyltransferase RrmJ/FTSJ. Biochem Biophys Res Commun. 2000;271:714–718. doi: 10.1006/bbrc.2000.2702. [DOI] [PubMed] [Google Scholar]
- 61.Rosenberger RF, Grover NB, Zaritsky A, Woldringh CL. Control of microbial surface-growth by density. Nature. 1978;271:244–245. doi: 10.1038/271244a0. [DOI] [PubMed] [Google Scholar]
- 62.Marr AG, Harvey RJ, Trentini WC. Growth and division of Escherichia coli. J Bacteriol. 1966;91:2388–2389. doi: 10.1128/jb.91.6.2388-2389.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marini PE, Perez CA, de Mendoza D. Growth-rate regulation of the Bacillus subtilis accBC operon encoding subunits of acetyl-CoA carboxylase, the first enzyme of fatty acid synthesis. Arch Microbiol. 2001;175:234–237. doi: 10.1007/s002030100256. [DOI] [PubMed] [Google Scholar]
- 64.Heath RJ, Jackowski S, Rock CO. Guanosine tetraphosphate inhibition of fatty acid and phospholipid synthesis in Escherichia coli is relieved by overexpression of glycerol-3-phosphate acyltransferase (plsB) J Biol Chem. 1994;269:26584–26590. [PubMed] [Google Scholar]
- 65.Battesti A, Bouveret E. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol Microbiol. 2006;62:1048–1063. doi: 10.1111/j.1365-2958.2006.05442.x. [DOI] [PubMed] [Google Scholar]
- 66.Castillo YP, Pérez MA. Bacterial beta-ketoacyl-acyl carrier protein synthase III (FabH): An attractive target for the design of new broad-spectrum antimicrobial agents. Mini Rev Med Chem. 2008;8:36–45. doi: 10.2174/138955708783331559. [DOI] [PubMed] [Google Scholar]
- 67.Li HQ, et al. Synthesis of C(7) modified chrysin derivatives designing to inhibit beta-ketoacyl-acyl carrier protein synthase III (FabH) as antibiotics. Bioorg Med Chem. 2009;17:6264–6269. doi: 10.1016/j.bmc.2009.07.046. [DOI] [PubMed] [Google Scholar]
- 68.Lv PC, Sun J, Luo Y, Yang Y, Zhu HL. Design, synthesis, and structure-activity relationships of pyrazole derivatives as potential FabH inhibitors. Bioorg Med Chem Lett. 2010;20:4657–4660. doi: 10.1016/j.bmcl.2010.05.105. [DOI] [PubMed] [Google Scholar]
- 69.Pérez-Castillo Y, Froeyen M, Cabrera-Pérez MA, Nowé A. Molecular dynamics and docking simulations as a proof of high flexibility in E. coli FabH and its relevance for accurate inhibitor modeling. J Comput Aided Mol Des. 2011;25:371–393. doi: 10.1007/s10822-011-9427-z. [DOI] [PubMed] [Google Scholar]
- 70.Yuan Y, Sachdeva M, Leeds JA, Meredith TC. Fatty acid biosynthesis in Pseudomonas aeruginosa is initiated by FabY: A new class of β-ketoacyl acyl carrier protein synthases. J Bacteriol. 2012 doi: 10.1128/JB.00792-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Silhavy TJ, Berman ML, Enquist LW. Experiments with Gene Fusions. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 72.Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 73.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu D, et al. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Malinverni JC, Silhavy TJ. An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc Natl Acad Sci USA. 2009;106:8009–8014. doi: 10.1073/pnas.0903229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chung HS, et al. Rapid beta-lactam-induced lysis requires successful assembly of the cell division machinery. Proc Natl Acad Sci USA. 2009;106:21872–21877. doi: 10.1073/pnas.0911674106. [DOI] [PMC free article] [PubMed] [Google Scholar]