Abstract
Neurotrophic factors, such as brain-derived neurotrophic factor (BDNF), are associated with the physiology of the striatum and the loss of its normal functioning under pathological conditions. The role of BDNF and its downstream signaling in regulating the development of the striatum has not been fully investigated, however. Here we report that ablation of Bdnf in both the cortex and substantia nigra depletes BDNF in the striatum, and leads to impaired striatal development, severe motor deficits, and postnatal lethality. Furthermore, striatal-specific ablation of TrkB, the gene encoding the high-affinity receptor for BDNF, is sufficient to elicit an array of striatal developmental abnormalities, including decreased anatomical volume, smaller neuronal nucleus size, loss of dendritic spines, reduced enkephalin expression, diminished nigral dopaminergic projections, and severe deficits in striatal dopamine signaling through DARPP32. In addition, TrkB ablation in striatal neurons elicits a non–cell-autonomous reduction of tyrosine hydroxylase protein level in the axonal projections of substantia nigral dopaminergic neurons. Thus, our results establish an essential function for TrkB in regulating the development of striatal neurons.
Keywords: Huntington disease, neurotrophin
The striatum is a subcortical part of the telencephalon and a major input component of the basal ganglia system. The striatum plays a crucial role in regulating voluntary movements, as well as reward-associated learning and memory. It can be separated into dorsal and ventral parts based on known anatomical and functional distinctions, with the dorsal portion associated with movement regulation and the ventral striatum involved predominantly in mediating neurological functions pertaining to motivation, reward, and emotion. Progressive degeneration of the striatal medium spiny neurons (MSNs) and their main dopaminergic inputs from the substantia nigra are the cardinal symptoms of the neurological disorders Huntington disease (HD) and Parkinson disease, respectively.
Like other telencephalic regions, the striatum contains inhibitory neurons generated from the medial ganglionic eminence. The majority of inhibitory GABAergic neurons are MSNs that project outside of the striatum to neighboring structures, such as the globus pallidus and the substantia nigra. The mechanism regulating development of the striatum involves combinatorial codes of transcription factors and parallels the development of other neural structures; however, the molecular mechanisms underlying the maturation and maintenance of the striatum remain largely unknown. Recent studies have shown that brain-derived neurotrophic factor (BDNF) exerts prosurvival effects on striatal neurons in vitro (1, 2) and delays the onset of degeneration in a transgenic mouse model of HD (3). However, it has been shown that during development and in adulthood, Bdnf mRNA is largely absent from striatal neurons and glia. Instead, BDNF is produced in the cortex and substantia nigra and is transported anterogradely to striatal neurons (4). Indeed, cortical ablation of Bdnf leads to cellular deficits in the striatum (5) and global gene expression impairment (6). These findings suggest an important role for BDNF in the striatum; however, given BDNF’s complex downstream signaling capacities, the nature of its intracellular signaling cascades pertaining to development, maturation, and function of the striatum remains to be delineated.
In this study, we conditionally ablated BDNF or its receptor TrkB in corticostriatal and nigrostriatal neuronal circuits. We found that Bdnf deletion in both cortex and substantia nigra led to complete depletion of BDNF protein in the striatum. Mutant mice displayed dramatic developmental abnormalities and neurological impairments. Furthermore, specific deletion of TrkB from striatal neurons was sufficient to produce this wide range of developmental deficits. Thus, our results demonstrate that BDNF and TrkB play critical paracrine and cell-autonomous roles, respectively, in the development and maintenance of striatal neurons.
Results
Bdnf Ablation in Cortex and Substantia Nigra Depletes Striatal BDNF Protein.
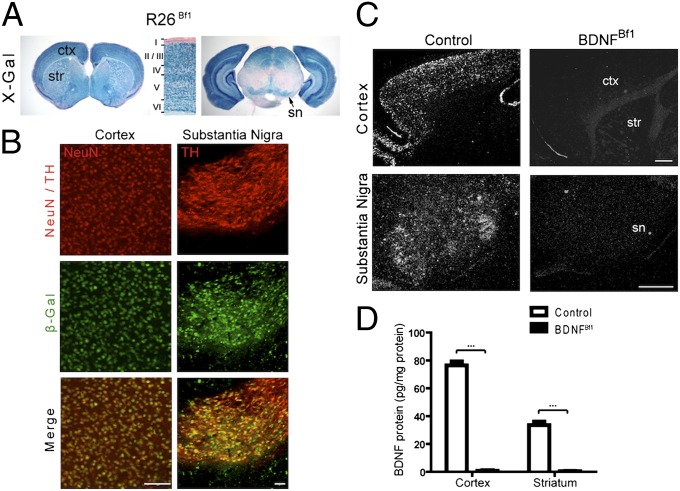
The striatum does not itself produce BDNF, but rather receives BDNF protein through anterograde transport from cortical and nigral neurons (4). To generate conditional knockout (cKO) mice lacking BDNF protein in the striatum, we crossed mice harboring flox alleles of Bdnf with a Cre recombinase line that mediates recombination in both cortex and substantia nigra. It has been shown that mice expressing Cre under the endogenous Bf1 promoter have widespread recombination in the telencephalon (7). By crossing the Bf1-Cre mice with R26 reporter mice (8), we confirmed that Bf1-Cre was active in the cortex and mediated effective recombination in neurons, including those in layer V, where the majority of projections to the striatum originate (R26Bf1 mice; Fig. 1A). X-Gal staining on adult R26Bf1 mice also revealed positive cells in the midbrain region in the proximity of the substantia nigra (Fig. 1A, Right). Double immunostaining for β-gal and tyrosine hydroxylase (TH), a marker for dopaminergic neurons, confirmed that TH-positive neurons also expressed β-gal (Fig. 1B, Right). To definitively demonstrate the effectiveness of Bf1-Cre–mediated deletion of Bdnf, we examined Bdnf mRNA expression by in situ hybridization. We found that although Bdnf was expressed robustly in the cortex and substantia nigra of control mice, it was absent in the Bf1-Cre;Bdnfflox/flox mice (BDNFBf1 mice) at age 1 mo (Fig. 1C). Deletion of Bdnf from these two structures led to complete depletion of BDNF protein from the striatum as measured by ELISA [(Fig. 1D); n = 5–6 for each; P < 0.001 for control–BDNFBf1 comparisons].
Fig. 1.
Bdnf ablation in the cortex and substantia nigra depletes BDNF from the striatum. (A) X-Gal staining on coronal sections of 5-wk-old R26Bf1 mice at the forebrain (Left) and midbrain (Right) levels. Note the robust recombination in the cortex (ctx), striatum (str), and substantia nigra (sn). (Middle) Higher-magnification view of the cortex showing X-Gal staining throughout layers II–VI. (B) Double immunostaining for β-gal (green) and NeuN (red) (Left) or TH (red) (Right) revealed Bf1-Cre-mediated recombination in neurons of the cortex and substantia nigra. (Scale bars: 50 μm.) (C) In situ hybridization for Bdnf mRNA in 4-wk-old control and BDNFBf1 mice. Note the ablation of Bdnf mRNA from the cortex (Upper) and substantia nigra (Lower) of BDNFBf1 mice. (Scale bars: 500 μm.) (D) ELISA showed complete removal of BDNF protein from the cortex and striatum of BDNFBf1 mice. n = 5–6 for each. Results are mean ± SEM. ***P < 0.001.
BDNF Depletion Leads to Motor Dysfunction and Deficits in Striatal Neuron Development.
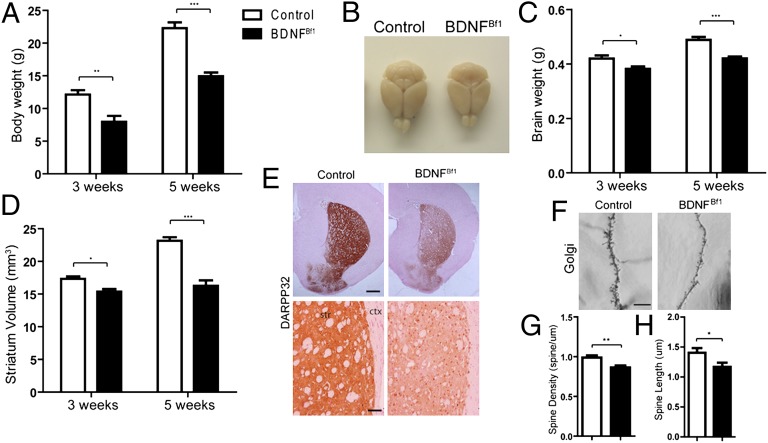
In contrast to Bdnf germline KO mice that die shortly after birth, the BDNFBf1 mice were viable. However, they were runty compared with littermate controls by 3 wk of age (Fig. 2A) and did not survive past 8 wk of age. In addition, the BDNFBf1 mice displayed early-onset behavioral abnormalities, including hindlimb clasping, hyperactivity, and circling (not shown), indicative of neurological dysfunction. Although born with normal-sized brains, the BDNFBf1 mice also showed reduced postnatal brain weight (Fig. 2 B and C; n = 4–6 for each genotype and age; P < 0.05 at 3 wk and P < 0.001 at 5 wk). Analysis of postnatal brain regions revealed significant volume reduction in mutant striatum (Fig. 2D; n = 4–6 for each; P < 0.05 at 3 wk and P < 0.001 at 5 wk), as well as reduction in dentate gyrus (DG) and cortex volume (Fig. S1; n = 5 for each; P < 0.01 and P < 0.05, respectively), consistent with previous reports (5, 9). Notably, the overall architecture of the striatum in BDNFBf1 mice, as evaluated by Nissl and luxol fast blue staining at age 3 wk, was comparable to that in controls (Fig. S1), indicating that BDNF depletion did not affect structural formation of the striatum.
Fig. 2.
BDNF depletion impairs striatal development. (A) BDNFBf1 mice had lower body weight at postnatal 3 wk and 5 wk. n = 4–6 for each. (B and C) At postnatal age 3 wk (C) and 5 wk (B and C), the BDNFBf1 mice had significantly lower brain weight compared with littermate controls. n = 4–6 for each. (D) BDNFBf1 mice had lower striatal volumes at postnatal age 3 wk and 5 wk compared with littermate controls. n = 4–6 for each. (E) (Upper) Immunostaining demonstrated reduced levels of DARPP32 protein in the striatum of BDNFBf1 mice. (Lower) Higher-magnification images showing loss of DARPP32 in both the soma and processes of MSNs. ctx, cortex; str, striatum. (Scale bars: Upper, 500 μm; Lower, 50 μm). (F–H) BDNF depletion led to fewer and shorter dendritic spines on striatal MSNs. (Scale bar: 10 μm.) n = 7 mice for each. Results are mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
To assess whether BDNF depletion resulted in striatum deficits at the cellular level, we evaluated expression levels and distribution of DARPP32 (dopamine- and cAMP-regulated phosphoprotein, 32 kDa), a major target of dopamine-activated adenylyl cyclase and the key mediator of dopamine signaling (10). We saw a dramatic reduction in the intensity of DARPP32 immunoreactivity throughout the striatum of BDNFBf1 mice at age 1 mo (Fig. 2E, Upper). High magnification revealed loss of DARPP32 protein in both the soma and processes of striatal neurons (Fig. 2E, Lower). We further examined striatal MSN morphology using Golgi staining, and found significantly fewer spines on secondary dendritic branches in 3-wk-old BDNFBf1 mice (Fig. 2 F and G; n = 7 mice for each; P < 0.01), as well as shorter spines (Fig. 2H; n = 7 mice for each; P < 0.05). Taken together, these results indicate impaired striatal neuronal maturation in the BDNFBf1 mice.
TrkB Is Required for Striatal Development.
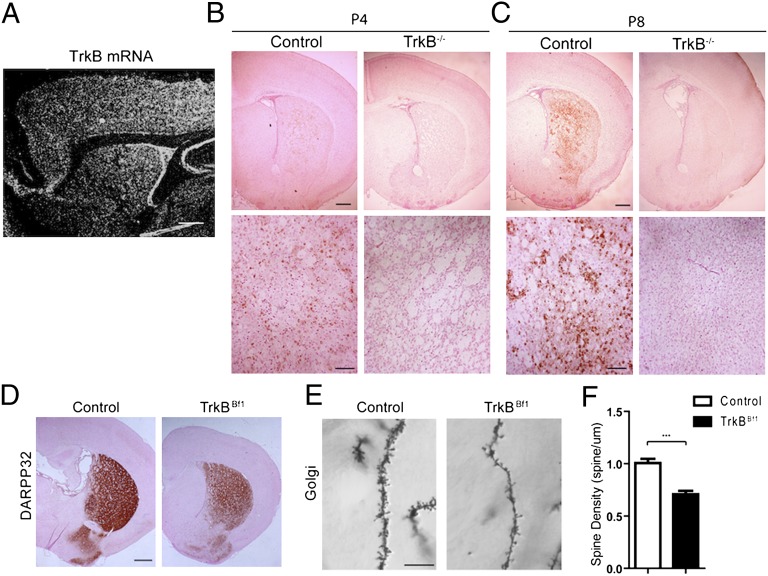
BDNF signals through its high-affinity receptor TrkB, and the pan-neurotrophin, low- affinity receptor p75. Although both receptors are reportedly present in the developing striatum (11), only TrkB is robustly expressed in postnatal and adult striatum (12) (Fig. 3A). To evaluate the requirement for TrkB in striatal development in vivo, we examined TrkB−/− mice at postnatal days 4 and 8. We previously reported that although the majority of TrkB−/− mice die at birth (13), a small percentage are viable in the first postnatal week. Compared with littermate controls, these mice had decreased striatal volume and a dramatic reduction in DARPP32 protein level (Fig. 3 B and C). We also generated a line of TrkB cKO mice using the Bf1-Cre mice, which lack TrkB in the striatum, in addition to other brain regions including cortex, hippocampus, and substantia nigra (TrkBBf1 mice). The TrkBBf1 mice displayed developmental abnormalities similar to those of the BDNFBf1 mice, including reduced brain and body weight, hindlimb clasping, postnatal lethality, circling behavior, and ataxia (not shown). Cellular analysis revealed reduction in DARPP32 immunostaining and decreased spine density on secondary dendrites at 3 wk postnatal age (Fig. 3 D–F; n = 6 mice for each; P < 0.001). Thus, a loss of TrkB impairs normal striatal development. However, whether the requirement for TrkB lies within the striatum or elsewhere could not be assessed in these mutant mice, owing to global deletion of TrkB.
Fig. 3.
Global TrkB ablation impairs striatal development. (A) In situ hybridization for TrkB mRNA detected expression in the cortex and striatum of WT mice. (Scale bar: 500 μm.) (B and C) (Upper) Whereas control mice at postnatal day 4 (B) and day 8 (C) had progressively higher DARPP32 levels in striatum, TrkB−/− mice had dramatically lower DARPP32 levels at both ages. (Lower) Higher-magnification views. P, postnatal. (Scale bars: Upper, 200 μm; Lower, 50 μm.) (D) Bf1-Cre–mediated ablation of TrkB led to reduced DARPP32 protein in striatum. (Scale bar: 500 μm.) (E and F) TrkBBf1 mice had less spine formation on the secondary dendrites of striatal MSNs. (Scale bar: 10 μm.) n = 7 mice for each. Results are mean ± SEM. ***P < 0.001.
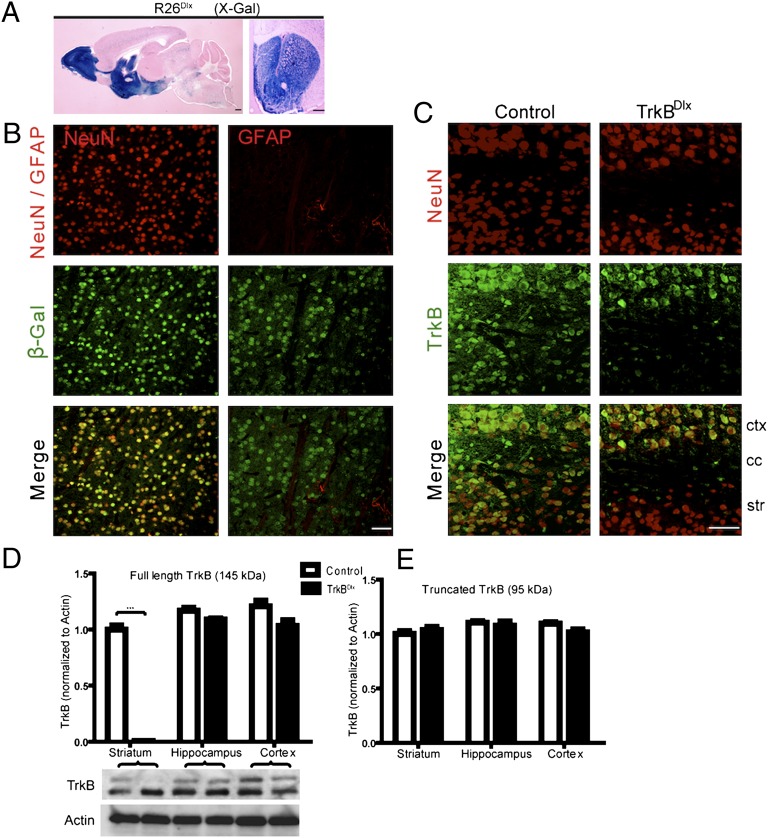
To examine whether striatal-specific deletion of TrkB would recapitulate the BDNFBf1 and TrkBBf1 phenotypes, we used a well-characterized Dlx5/6i-cre transgenic line (14) that mediates robust recombination in the GABAergic cells of the striatum and olfactory bulb (OB) (Fig. 4A). Limited recombination was also observed in the inhibitory neurons of the cortex and hippocampus (Fig. 4A). In striatum, β-gal–expressing cells were positive for the neuronal marker NeuN (Fig. 4B, Left), but did not colocalize with the astrocytic marker GFAP (Fig. 4B, Right). Double immunostaining revealed the presence of TrkB protein in NeuN-positive striatal neurons in control mice (Fig. 4C, Left). Dlx5/6i-cre–mediated ablation of TrkB protein was observed in striatal, but not cortical, neurons (TrkBDlx mice; Fig. 4C). Finally, immunoblotting showed complete absence of the full-length TrkB protein from striatum of 3-wk-old TrkBDlx mice (Fig. 4D; n = 3 for each; P < 0.001). It has been reported that a truncated TrkB splice variant is selectively expressed by astrocytes and mediates neurotrophin-evoked calcium signaling (15). We found that truncated TrkB expression in striatum of TrkBDlx mice was not affected (Fig. 4 D and E; n = 3 for each; P > 0.02 for all control–TrkBDlx comparisons), further confirming the neuron-specific ablation of TrkB by Dlx5/6i-cre. Expression of both forms of TrkB protein was unaffected in cortex and hippocampus (Fig. 4 D and E). These results demonstrate a complete and specific ablation of TrkB from striatal neurons, permitting us to investigate TrkB cell-autonomous function in this cell population.
Fig. 4.
Dlx5/6i-cre–mediated recombination in striatal neurons. (A) R26 reporter revealed Dlx5/6i-cre–mediated recombination in the striatum, OB, and cortex. (Scale bar: 500 μm.) (B) Double immunostaining for β-gal (green) and NeuN (red) (Left) or GFAP (red) (Right) showed Dlx5/6i-cre–mediated recombination in striatal neurons, but not in astrocytes. (Scale bar: 50 μm.) (C) Immunostaining for NeuN (red) (Top) and TrkB (green) (Middle) demonstrated specific ablation of TrkB protein from striatal, but not cortical, neurons of TrkBDlx mice. ctx, cortex; cc, corpus callosum; str, striatum. (Scale bar: 50 μm.) (D and E) Immunoblot analysis for TrkB revealed specific ablation of full-length TrkB protein from striatum of TrkBDlx mice (D). Note that the levels of full-length TrkB in cortex and hippocampus (D), as well as truncated TrkB in all three regions (D and E), were unaffected in TrkBDlx mice. n = 3 for each. Results are mean ± SEM. ***P < 0.001.
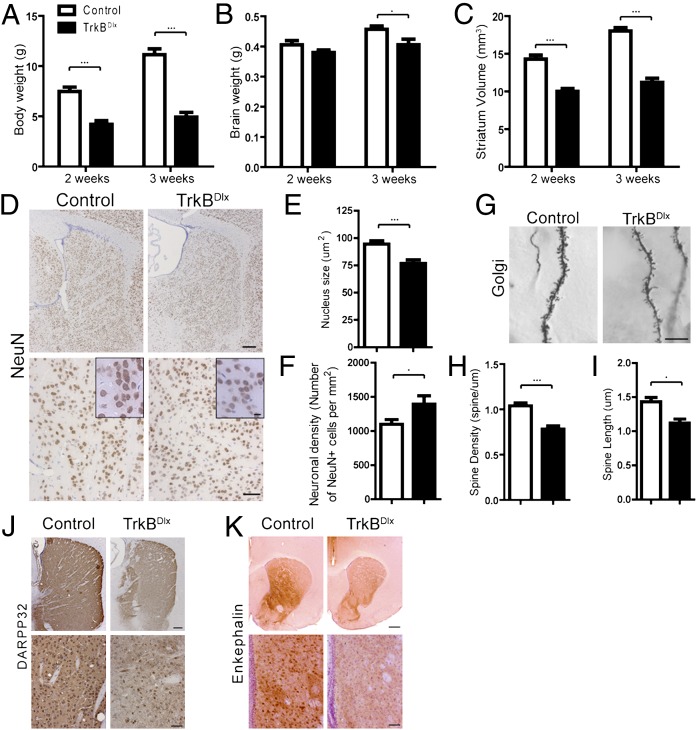
Like BDNFBf1 mice, TrkBDlx mice were runty and did not survive past age 4 wk. Significant decreases in body weight and striatal volume were evident at 2 wk and 3 wk of age (Fig. 5 A and C; n = 5–7 for each; P < 0.001), and a significant reduction in brain weight was evident by 3 wk (Fig. 5B; n = 5–6 for each; P < 0.05). Attesting to the restricted recombination pattern mediated by the Dlx5/6i-cre, we found no overt difference in the size of the DG or cortex (Fig. S2; n = 5–6 for each). Striatal neurons in the TrkBDlx mice had reduced soma size, as evaluated by NeuN immunostaining (Fig. 5 D and E; n = 92–98 neurons from five mice of either genotype; P < 0.001). Neurons in the striatum of TrkBDlx mice were more compact, and quantitative analysis revealed a significant increase in the density of NeuN-positive striatal neurons (Fig. 5 D and F; n = 9 for each; P < 0.5). Along with the reduction in striatal volume (Fig. 5C), TrkBDlx mice had approximately 13% fewer NeuN-positive striatal neurons compared with their littermate controls at age 3 wk (1.58 × 106 vs.1.81 × 106). Golgi staining demonstrated reduced spine density and length in TrkBDlx mice compared with littermate controls (Fig. 5 G–I; n = 8–9 mice for each; P < 0.001 and P < 0.05, respectively). We further assessed the development of striatal MSNs by DARPP32 immunostaining and found reduced protein expression in both the soma and the processes (Fig. 5J). In addition, the density of DARPP32-positive MSNs was reduced in the striatum of TrkBDlx mice (Fig. 5J). Given the increased density of NeuN-positive cells in the TrkBDlx mice (Fig. 5F), along with the fact that DARPP32-positive MSNs represent more than 90% of all neurons in the striatum, the loss of DARPP32 staining likely reflects a decline in gene expression rather than cell loss.
Fig. 5.
Removal of TrkB in striatal neurons impairs striatal development. (A–C) TrkBDlx mice had reduced body weight (A) and brain weight (B), as well as decreased striatal volume (C), at postnatal age 2 wk and 3 wk. n = 5–7 for each. (D) NeuN immunostaining showed a comparable striatal structure in 3-wk-old TrkBDlx mice. (Insets) Higher-magnification views of NeuN-positive striatal cells. (Scale bars: Upper, 200 μm; Lower, 50 μm; Insets, 10 μm). (E) Quantification revealed significantly smaller neuronal nucleus size in striatum of 3-wk-old TrkBDlx mice. n = 92–98 neurons from five mice of each genotype. (F) The density of NeuN-positive striatal neurons was higher in 3-wk-old TrkBDlx mice. n = 9 for each. (G–I) Golgi staining showed that the TrkBDlx mice had fewer and shorter spines on the secondary dendrites of striatal MSNs. (Scale bars: 10 μm.) n = 8–9 mice for each. (J) Immunostaining revealed significant loss of DARPP32 protein in the striatum of TrkBDlx mice. (Scale bars: Upper, 200 μm; Lower, 50 μm.) (K) Enkephalin-positive striatopallidal neurons were reduced in TrkBDlx mice. (Scale bars: Upper, 500 μm; Lower, 50 μm.) Results are mean ± SEM. *P < 0.05; ***P < 0.001.
BDNF has been implicated in regulating the development of a subgroup of striatal MSNs, including the striatopallidal neurons that express the neuropeptide enkephalin (16). To determine whether TrkB loss impairs the development of these neurons, we examined enkephalin expression using an antibody against the mature form of the peptide. By age 3 wk, TrkBDlx mice showed a substantial reduction in the intensity of enkephalin immunostaining in both the soma and the surrounding dendrites (Fig. 5K). Similar to the loss of DARPP32, this reduced enkephalin expression was not accompanied by a comparable loss of NeuN-positive cells (Fig. 5F), and thus most likely reflects impaired terminal maturation and maintenance on the level of individual cells.
Taken together, these findings indicate that a lack of striatal TrkB signaling was responsible for the deficits associated with BDNF depletion in the cortex and substantia nigra.
TrkB Ablation Attenuates Intracellular Signaling.
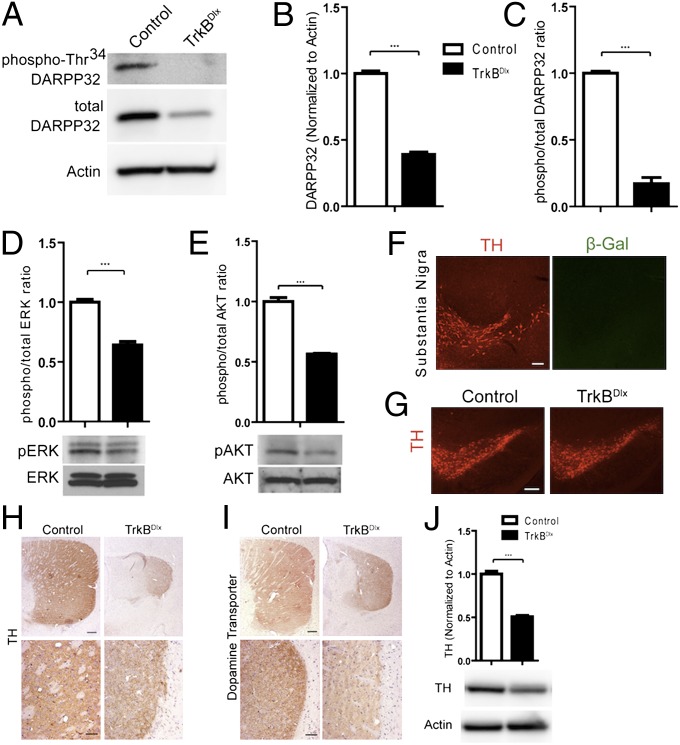
Our findings in the TrkBDlx mice demonstrated impaired cellular striatal development and maturation. The decrease in DARPP32 protein level in the absence of BDNF or TrkB further indicates a disruption in normal dopamine signaling. Consequently, we examined the level of total DARPP32 protein and phospho-Thr34 DARPP32 in striatum of 3-wk-old mice. Recent studies have shown that activation of the D1 class dopamine receptors elicits phosphorylation of DARPP32 on Thr34, which conveys key signal transduction to the protein kinase A cascade (17). Immunoblot analysis revealed a significant loss of total DARPP32 protein in the TrkBDlx mice, confirming the immunohistochemistry results (Fig. 6 A and B; n = 3 for each; P < 0.001), as well as a loss of phospho-Thr34 DARPP32 (Fig. 6A). Quantitative analysis demonstrated a significant reduction in the ratio of phospho to total DARPP32 (Fig. 6C; n = 3 for each; P < 0.001), indicating that the loss of phospho-Thr34 DARPP32 reflects a reduction in total protein, as well as impaired activation of the dopamine signaling pathway in the basal state.
Fig. 6.
TrkBDlx mice have striatal molecular and cellular phenotypes. (A–C) Immunoblot analysis of striatal lysates from control and TrkBDlx mice showed a significant reduction in total DARPP32 levels (A and B), as well as phospho-Thr34 DARPP32 levels (A and C). n = 3 for each. (D and E) TrkB ablation in striatal neurons led to reduced activity of the ERK and AKT pathways, as shown by immunoblot analysis for phospho-Thr202/Thr204 ERK and total ERK proteins, as well as phospho-Ser479 and total AKT proteins. n = 3 for each. (F) Coimmunostaining for TH (red) and β-gal (green) on sections of R26Dlx mice demonstrated lack of recombination in dopaminergic neurons of substantia nigra. (Scale bars: 100 μm.) (G) TH immunostaining in substantia nigra showed comparable numbers of dopaminergic neurons in control and TrkBDlx mice. (Scale bars: 200 μm.) (H and I) (Upper) Three-week-old TrkBDlx mice displayed reduced dopaminergic terminals in striatum, labeled by TH (H) or dopamine transporter (I). (Lower) Higher-magnification views. (Scale bars: Upper, 200 μm; Lower, 50 μm.) (J) Immunoblot analysis showed a 50% reduction in TH protein in the striatum of TrkBDlx mice. n = 3 for each. Results are mean ± SEM. ***P < 0.001.
TrkB activation can be propagated through downstream pathways common to receptor tyrosine kinases, including the ERK and PI3-kinase pathways, which promote neuronal growth, differentiation, and survival. We examined the levels of phospho-ERK1/2 at Thr202/Thr204 and phospho-Ser479 AKT in striatum by immunoblot analysis. At age 3 wk, TrkBDlx mice showed a significant reduction in the levels of both phospho-proteins compared with littermate controls (Fig. 6 D and E; n = 3 for each; P < 0.001 for both). Thus, ablating TrkB in striatal neurons leads to deficits in key intracellular signaling.
TrkB Ablation Leads to Non–Cell-Autonomous Loss of Dopaminergic Projections.
The BDNFBf1 and TrkBDlx mice displayed severe neurological phenotypes, including shortened lifespan and locomotor defects. In particular, the TrkBDlx mice were hypoactive, reminiscent of phenotypes seen in other mutant mice with impaired nigral dopamine signaling, including dopamine-deficient TH mutant mice (18). A previous study suggested that Bdnf ablation in midbrain neurons leads to age-dependent dopaminergic neuron loss (19). We and others have shown that the Dlx5/6i-cre does not mediate recombination in the substantia nigra (Fig. 6F) (14, 20). We examined the substantia nigra dopaminergic neurons in the TrkBDlx mice by TH immunostaining. We found no overt difference in the number of TH-positive dopaminergic neurons at age 3 wk (Fig. 6G); however, in the striatum, we found a substantial reduction in the area and intensity of TH immunostaining in 3-wk-old TrkBDlx mice compared with controls (Fig. 6H). We confirmed this finding by immunostaining for dopamine transporter, a protein residing in dopaminergic terminals (Fig. 6I). Finally, we measured TH levels in striatal protein lysates and found a 50% reduction in TrkBDlx mice (Fig. 6J; n = 3 for each; P < 0.001). These data demonstrate that ablation of TrkB in striatal neurons results in a loss of dopaminergic projections to the striatum through a non–cell-autonomous mechanism.
Discussion
The striatum is the largest CNS structure lacking a detectable endogenous source of BDNF. Instead, striatal neurons receive BDNF from projection neurons of the cortex and substantia nigra via anterograde transport (4). BDNF is important to the survival of striatal neurons in vitro (1, 2), and global ablation of Bdnf in the CNS leads to selective impairment in striatal MSN development (2). Furthermore, modulation of BDNF levels by genetic deletion or overexpression in mice harboring HD-related mutations led to acceleration or amelioration, respectively, of the cellular phenotypes associated with HD (3, 16, 21). Here we demonstrate that Bdnf ablation in cortex and substantia nigra depletes BDNF in the striatum and leads to developmental deficits. Taken together, these findings suggest that striatal neurons are critically dependent on BDNF.
BDNF and its downstream signaling pathways have been implicated in regulating the development and functioning of the rodent brain (22). Removal of BDNF from the cortex and substantia nigra (regions that supply the striatum with BDNF) affects the physiology of these brain regions (19, 23). We used the TrkBDlx mice generated in this study to specifically examine the role of BDNF downstream signaling in striatal neurons, while leaving the cortical and nigra neurons largely unaffected. These mutant mice exhibited a panel of molecular, cellular, and behavioral phenotypes demonstrating severe developmental abnormalities, thus establishing a cell-autonomous role for TrkB in striatal neurons.
Our findings support the notion that neurons in the striatum are highly sensitive to the availability of BDNF. We previously reported that ablating TrkB in neurons of the cerebral cortex, hippocampus, OB, and cerebellum does not lead to drastic cellular degeneration or postnatal lethality (9, 24, 25). Here we demonstrate that TrkBDlx mice, in which TrkB is selectively ablated in striatal neurons, have severe locomotor deficits, reduced body and brain weight, and postnatal lethality, phenotypes almost identical to those seen in mice with broader ablation of TrkB (TrkBBf1 mice). This finding is consistent with a comparatively prominent role for TrkB in the striatum. In addition, a lack of TrkB in striatal MSNs resulted in reduced cell size, fewer dendritic spines, and diminished expression of a key signaling peptide (enkephalin) and a key protein (DARPP32). We further note that DARPP32 reduction coincided with dramatically impaired activation of DARPP32 in the form of Thr34 phosphorylation, indicating reduced basal dopamine signaling. A recent study using a different Dlx5/6-Cre transgenic mouse line reported that deletion of TrkB in the striatum led to profound reduction of striatal volume (by 53%) and NeuN-positive striatal neurons (by 58%) (26). Further investigation revealed normal proliferation but dramatically higher apoptosis. However, we caution that this particular line of Dlx5/6-Cre transgenic mice was reported to have higher cell death levels during embryogenesis compared with control mice lacking the transgene (27). Baydyuk et al. (26) used only mice homozygous for the floxed TrkB alleles as controls for their TrkB mutant mice; thus, whether the observed cell death was caused by the Dlx5/6-Cre transgene alone or was affected by TrkB deletion remains unclear. In the present study, we used a Cre line that did not cause cell death or striatal volume reduction (20) and used littermate TrkBflox/flox, TrkBflox/wt, and TrkBwt/wt;cre/wt mice as controls to eliminate a potential effect of the Dlx5/6-Cre transgene on striatal development. Our results demonstrate that lack of TrkB leads to substantial impairment in striatal cellular functionality.
Our findings are in agreement with previous reports showing that cortical ablation of Bdnf using Emx-Cre (Emx-BDNFKO mice) (5) or global deletion of Bdnf in the postmitotic neurons of the CNS using Tau-Cre (2) leads to a significant reduction in striatal BDNF and striatal impairment. Interestingly, these lines of Bdnf mutant mice were viable and survived into advanced age, whereas the BDNFBf1 mice generated in the current study had an average lifespan of 2 mo. A possible cause of this difference is the complete deletion of Bdnf in the substantia nigra pars compacta mediated by the Bf1-Cre. It has been demonstrated that dopaminergic neurons of the substantia nigra express Bdnf (19) and supply 14% of BDNF protein in the ipsilateral striatum (4). Although this is a smaller contribution compared with the corticostriatal input (66% ipsilateral) (4), the nigrostriatal projection apparently supplies sufficient BDNF to maintain basal striatal function for survival of the Emx-BDNFKO mice. Dopaminergic neurons emit extensive axonal arborizations in the striatum, with each neuron forming axonal terminals that cover up to 5.7% of the total striatal volume (28), thus establishing a strong delivery system for neurotransmitters (dopamine) and trophic factors (e.g., BDNF). Thus, by ablating Bdnf in neurons of both the cortex and substantia nigra, we created a cKO mouse with more severe neurological impairment and demonstrated that striatal BDNF could play a role in postnatal survival.
Taken together, our results demonstrate that ablation of TrkB signaling in striatal neurons impairs the development of these neurons and their associated neural circuits. Future studies on the role of TrkB signaling in the pathophysiology of the striatum and the potential benefits of restoring TrkB signaling may provide insight into preventing, delaying, and reversing the progression of striatal neurodegeneration.
Methods
Mice.
The TrkB+/−, TrkBflox, BDNFflox, Bf1-Cre, and Dlx5/6i-cre mouse lines were generated as described previously (7, 13, 14, 24, 25). R26 mice were obtained from Jackson Laboratory. For generation of the BDNFBf1 mice, BDNFflox/wt;cre/wt mice were interbred with BDNFflox/flox or BDNFflox/wt mice. Littermate BDNFflox/flox, BDNFflox/wt, and BDNFwt/wt;cre/wt mice were indistinguishable and thus were pooled as the control group. TrkB cKO mice (TrkBBf1 and TrkBDlx) were derived by crossing TrkBflox/wt;cre/wt mice with TrkBflox/flox or TrkBflox/wt mice. Littermate TrkBflox/flox, TrkBflox/wt, and TrkBwt/wt;cre/wt mice served as the control group. All animals were maintained on the 129/SvEv and C57BL/6 mixed background. All mouse protocols used in this study were approved by the University of Texas Southwestern Medical Center’s Institutional Animal Care and Research Advisory Committee.
Histology.
Mice were anesthetized and perfused transcardially with PBS followed by 4% (wt/vol) paraformaldehyde (PFA), and the dissected brains postfixed in 4% (wt/vol) PFA at 4 °C. For Golgi staining, mice were perfused with PBS alone, and the dissected brain was incubated in Golgi solution for 12 d, as described previously (24). In situ hybridization for Bdnf and TrkB mRNA was done using 14-μm-thick fresh-frozen sections (24, 29). X-Gal staining was performed on 50-μm-thick floating brain sections prepared on a vibratome (20).
Immunoblot Analysis.
Immunoblot analysis was performed as described previously (30) using the following primary antibodies: actin (Sigma-Aldrich), phospho-Ser479 and total AKT (Cell Signaling), phospho-Thr34 DARPP32 (Cell Signaling), total DARPP-32 (BD Biosciences), phospho-Thr202/Thr204 and total ERK (Cell Signaling), TH (Chemicon), and TrkB (Dr. Louis Reichardt, University of California San Francisco). HRP-conjugated secondary antibodies (Santa Cruz Biotechnology) were used to detect primary antibodies, and visualized using the ChemiGlow kit (Alpha Innotech) in accordance with the manufacturer’s instructions. Membranes blotted for phosphoproteins were stripped and reprobed with antibodies against total proteins. Quantification of bands was performed using Kodak 1D Image Analysis software (30). Values for phosphoproteins were normalized to total proteins, and the levels of actin protein were used as controls for total DARPP32, and TrkB proteins.
BDNF ELISA.
The concentration of BDNF protein in cortical and striatal lysates was measured using a BDNF ELISA Kit (Promega) as described previously (9, 24).
Supplementary Material
Acknowledgments
We thank Shawna Kennedy, Linda McClellan, Steven McKinnon, and Ahmed Mahmoud for technical assistance, and Dr. Louis Reichardt for the TrkB antibody. The Bf1-Cre was a generous gift from Dr. Susan McConnell. We thank Drs. Mark Lush, James Bibb, Serge Nef, and members of the L.F.P. laboratory for sharing reagents and helpful suggestions. This work was supported by National Institute of Neurological Disorders and Stroke Grant R37NS033199, Conte Center Grant 7P50MH066172-07, and the American Cancer Society.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212899109/-/DCSupplemental.
References
- 1.Ivkovic S, Ehrlich ME. Expression of the striatal DARPP-32/ARPP-21 phenotype in GABAergic neurons requires neurotrophins in vivo and in vitro. J Neurosci. 1999;19:5409–5419. doi: 10.1523/JNEUROSCI.19-13-05409.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rauskolb S, et al. Global deprivation of brain-derived neurotrophic factor in the CNS reveals an area-specific requirement for dendritic growth. J Neurosci. 2010;30:1739–1749. doi: 10.1523/JNEUROSCI.5100-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gharami K, Xie Y, An JJ, Tonegawa S, Xu B. Brain-derived neurotrophic factor over-expression in the forebrain ameliorates Huntington’s disease phenotypes in mice. J Neurochem. 2008;105:369–379. doi: 10.1111/j.1471-4159.2007.05137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altar CA, et al. Anterograde transport of brain-derived neurotrophic factor and its role in the brain. Nature. 1997;389:856–860. doi: 10.1038/39885. [DOI] [PubMed] [Google Scholar]
- 5.Baquet ZC, Gorski JA, Jones KR. Early striatal dendrite deficits followed by neuron loss with advanced age in the absence of anterograde cortical brain-derived neurotrophic factor. J Neurosci. 2004;24:4250–4258. doi: 10.1523/JNEUROSCI.3920-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strand AD, et al. Expression profiling of Huntington’s disease models suggests that brain-derived neurotrophic factor depletion plays a major role in striatal degeneration. J Neurosci. 2007;27:11758–11768. doi: 10.1523/JNEUROSCI.2461-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hébert JM, McConnell SK. Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev Biol. 2000;222:296–306. doi: 10.1006/dbio.2000.9732. [DOI] [PubMed] [Google Scholar]
- 8.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, et al. TrkB regulates hippocampal neurogenesis and governs sensitivity to antidepressive treatment. Neuron. 2008;59:399–412. doi: 10.1016/j.neuron.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greengard P, Allen PB, Nairn AC. Beyond the dopamine receptor: The DARPP-32/protein phosphatase-1 cascade. Neuron. 1999;23:435–447. doi: 10.1016/s0896-6273(00)80798-9. [DOI] [PubMed] [Google Scholar]
- 11.Klein R, Martin-Zanca D, Barbacid M, Parada LF. Expression of the tyrosine kinase receptor gene trkB is confined to the murine embryonic and adult nervous system. Development. 1990;109:845–850. doi: 10.1242/dev.109.4.845. [DOI] [PubMed] [Google Scholar]
- 12.Yan Q, Johnson EM., Jr Immunohistochemical localization and biochemical characterization of nerve growth factor receptor in adult rat brain. J Comp Neurol. 1989;290:585–598. doi: 10.1002/cne.902900411. [DOI] [PubMed] [Google Scholar]
- 13.Luikart BW, Nef S, Shipman T, Parada LF. In vivo role of truncated trkb receptors during sensory ganglion neurogenesis. Neuroscience. 2003;117:847–858. doi: 10.1016/s0306-4522(02)00719-4. [DOI] [PubMed] [Google Scholar]
- 14.Zerucha T, et al. A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J Neurosci. 2000;20:709–721. doi: 10.1523/JNEUROSCI.20-02-00709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rose CR, et al. Truncated TrkB-T1 mediates neurotrophin-evoked calcium signalling in glia cells. Nature. 2003;426:74–78. doi: 10.1038/nature01983. [DOI] [PubMed] [Google Scholar]
- 16.Canals JM, et al. Brain-derived neurotrophic factor regulates the onset and severity of motor dysfunction associated with enkephalinergic neuronal degeneration in Huntington’s disease. J Neurosci. 2004;24:7727–7739. doi: 10.1523/JNEUROSCI.1197-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bibb JA, et al. Severe deficiencies in dopamine signaling in presymptomatic Huntington’s disease mice. Proc Natl Acad Sci USA. 2000;97:6809–6814. doi: 10.1073/pnas.120166397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou QY, Palmiter RD. Dopamine-deficient mice are severely hypoactive, adipsic, and aphagic. Cell. 1995;83:1197–1209. doi: 10.1016/0092-8674(95)90145-0. [DOI] [PubMed] [Google Scholar]
- 19.Baquet ZC, Bickford PC, Jones KR. Brain-derived neurotrophic factor is required for the establishment of the proper number of dopaminergic neurons in the substantia nigra pars compacta. J Neurosci. 2005;25:6251–6259. doi: 10.1523/JNEUROSCI.4601-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez-Ortiz E, et al. TrkA gene ablation in basal forebrain results in dysfunction of the cholinergic circuitry. J Neurosci. 2012;32:4065–4079. doi: 10.1523/JNEUROSCI.6314-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pineda JR, et al. Brain-derived neurotrophic factor modulates dopaminergic deficits in a transgenic mouse model of Huntington’s disease. J Neurochem. 2005;93:1057–1068. doi: 10.1111/j.1471-4159.2005.03047.x. [DOI] [PubMed] [Google Scholar]
- 22.Huang EJ, Reichardt LF. Trk receptors: Roles in neuronal signal transduction. Annu Rev Biochem. 2003;72:609–642. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- 23.Gorski JA, Zeiler SR, Tamowski S, Jones KR. Brain-derived neurotrophic factor is required for the maintenance of cortical dendrites. J Neurosci. 2003;23:6856–6865. doi: 10.1523/JNEUROSCI.23-17-06856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luikart BW, et al. TrkB has a cell-autonomous role in the establishment of hippocampal Schaffer collateral synapses. J Neurosci. 2005;25:3774–3786. doi: 10.1523/JNEUROSCI.0041-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He XP, et al. Conditional deletion of TrkB but not BDNF prevents epileptogenesis in the kindling model. Neuron. 2004;43:31–42. doi: 10.1016/j.neuron.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 26.Baydyuk M, et al. TrkB receptor controls striatal formation by regulating the number of newborn striatal neurons. Proc Natl Acad Sci USA. 2011;108:1669–1674. doi: 10.1073/pnas.1004744108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fishell G. Loss of a notch activity in the developing central nervous system leads to increased cell death. Dev Neurosci. 2006;28:517. doi: 10.1159/000096117. [DOI] [PubMed] [Google Scholar]
- 28.Matsuda W, et al. Single nigrostriatal dopaminergic neurons form widely spread and highly dense axonal arborizations in the neostriatum. J Neurosci. 2009;29:444–453. doi: 10.1523/JNEUROSCI.4029-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malkovska I, Kernie SG, Parada LF. Differential expression of the four untranslated BDNF exons in the adult mouse brain. J Neurosci Res. 2006;83:211–221. doi: 10.1002/jnr.20728. [DOI] [PubMed] [Google Scholar]
- 30.Lush ME, Li Y, Kwon CH, Chen J, Parada LF. Neurofibromin is required for barrel formation in the mouse somatosensory cortex. J Neurosci. 2008;28:1580–1587. doi: 10.1523/JNEUROSCI.5236-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.