Abstract
In the living cell, proteins are able to organize space much larger than their dimensions. In return, changes of intracellular space can influence biochemical reactions, allowing cells to sense their size and shape. Despite the possibility to reconstitute protein self-organization with only a few purified components, we still lack knowledge of how geometrical boundaries affect spatiotemporal protein patterns. Following a minimal systems approach, we used purified proteins and photolithographically patterned membranes to study the influence of spatial confinement on the self-organization of the Min system, a spatial regulator of bacterial cytokinesis, in vitro. We found that the emerging protein pattern responds even to the lateral, two-dimensional geometry of the membrane such that, as in the three-dimensional cell, Min protein waves travel along the longest axis of the membrane patch. This shows that for spatial sensing the Min system does not need to be enclosed in a three-dimensional compartment. Using a computational model we quantitatively analyzed our experimental findings and identified persistent binding of MinE to the membrane as requirement for the Min system to sense geometry. Our results give insight into the interplay between geometrical confinement and biochemical patterns emerging from a nonlinear reaction–diffusion system.
Keywords: in vitro reconstitution, microstructures, supported lipid bilayers, spontaneous protein waves, Min oscillations
Pattern formation is one of the most intriguing features of biological systems. It takes place on many different spatial and temporal scales and several levels of complexity (1). Inside of the living cell, nonlinear reaction–diffusion dynamics allow proteins to encode for positional information (2–5), required to coordinate complex processes like cell division (6, 7), cell motility (8, 9), or give rise to information flow within the cell (5, 10). In reverse, cell geometry presumably affects such protein patterns, allowing processes within the cell to respond to changes in cell size and shape (11). However, how these reaction–diffusion systems sense the shape and the dimensions of the cell remains an outstanding question and requires decisive experimental testing.
A well-studied example for a biochemical reaction–diffusion process is the spatiotemporal oscillations of the Min proteins in Escherichia coli (12, 13). The Min system positions the division septum in the cell to its center, such that it divides into two equally sized daughter cells. Oscillations emerge from the interactions of two proteins and the cytoplasmic membrane (14–16): MinD, a membrane-binding ATPase, and MinE, the activator of ATP hydrolysis and thus, antagonist of MinD in its membrane-bound state. In vivo, the intrinsic wavelength of Min protein oscillations is similar to the size of the E. coli cell, about 5 μm versus 3 μm cell length and 1 μm cell diameter (17). The ability of the Min system to sense the enclosed volume of the bacterial cell has been demonstrated in E. coli cells, which lost their normal rod-like shape, either because certain genes had been deleted (18, 19) or after treatment with A22, an inhibitor of MreB activity (20). Even in these cells, the Min proteins were still found to robustly oscillate along the longest cell axis. Only in almost perfectly spherical cells with an aspect ratio larger than 0.96 (width/length) (18), or when the cell diameter exceeded a critical value (20, 21), the oscillation lost its preference for a certain axis. However, the molecular basis for the ability of the Min system to find the longest cell axis is not known.
The ability of the Min proteins to self-organize into mesoscale patterns in vitro has been demonstrated by the reconstitution of traveling protein waves on a supported lipid bilayer, where protein surface waves emerge from repetitive cycles of proteins binding and detaching to and from the membrane (22, 23). In these previous studies, the supported lipid bilayers used to mimic the cell membrane were about 200× larger than the typical periodicity of these waves, which was usually between 50 μm and 100 μm (12, 22, 23). Therefore, in these in vitro experiments, the influence of the geometric boundaries on the overall protein pattern was negligible.
Using a well-controlled in vitro system consisting of purified proteins and micropatterned supported lipid bilayers as reaction compartments, we found that the Min system is able to respond to two-dimensional geometrical boundaries. With the help of a computational model, we analyzed our experimental results and found that the Min system can sense the confining geometry due to persistent binding of MinE to the membrane. We expect that similar mechanisms also apply for other reaction–diffusion systems involved in intracellular geometry sensing.
Results
Min Protein Waves Respond to Their Lateral Confinement.
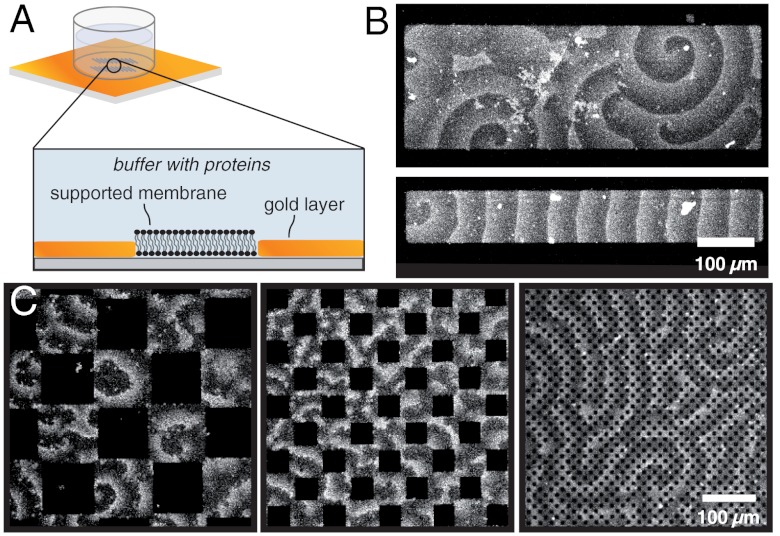
To investigate if and how Min-protein waves are influenced by geometrical confinement in vitro, we fabricated two-dimensional membranes with similar dimensions as the typical length scale of the protein pattern. Motivated by previous studies (24), we used a microstructured layer of gold on glass to create supported membranes of defined dimensions and geometries (Fig. 1A). We then allowed MinD and MinE to self-organize into protein surface waves on these membranes and studied the influence of confinement on the formation and propagation of these waves. Although the space above the membrane was not limited in our experiments, the proteins were located only in a small layer above the membrane during pattern formation (SI Appendix, Fig. S1). Thus, our first aim was to test if scaling down the membrane dimensions by introducing lateral confinement has an effect on the self-organization of Min-protein waves.
Fig. 1.
Influence of confinement on Min protein waves. (A) Schematic view of microstructured membranes. A bilayer is only formed on the areas not covered with gold. (B) Confocal micrograph of Min protein waves formed on microstructured membranes (with 0.8 μm MinD, 0.5 μm MinE with 10% Cy5-labelled MinE). On large membrane patches, the confinement has only a small influence on the protein waves. On smaller membrane patches with widths similar to the wavelength, the waves orient along the long axis. (C) On substrates with large obstacles (100 μm × 100 μm), individual spirals emerge on the membrane. If the obstacles have similar dimensions as the protein waves (50 μm × 50 μm), the wave pattern is irregular. Even smaller obstacles barely influence the protein waves.
On membrane patches much larger than the characteristic length scale of Min-protein waves (larger than 300 μm), geometric confinement did not appear to influence the orientation of Min protein waves (Fig. 1B, Upper). However, when the dimensions of the membrane were similar to the wave’s periodicity (between 50 and 100 μm), waves propagated along the longest axis of the membrane patch and never perpendicular to it (Fig. 1B, Lower). To gain more insight into the role of obstacle size, we performed experiments using checkerboard patterns of quadratic gold and membrane islands: On quadratic membranes with side lengths larger than the intrinsic wavelength of the pattern, we observed individual spirals, which hardly influenced each other (Fig. 1C, Left and Movie S1). However, when the gold and membrane patches had roughly the same dimensions as the characteristic wavelength, only irregular patterns formed, with protein bands moving in many different directions on the membrane patches (Fig. 1C, Center). In the presence of even smaller gold obstacles, which were about one-third of the wavelength, the Min proteins formed regular patterns and propagated almost unimpaired (Fig. 1C, Right).
Thus, lateral geometrical confinement can influence the self-organization of Min protein waves with the waves reacting most strongly when membrane patches or obstacles have roughly the same size as the waves.
The Membrane Geometry Defines the Path of the Traveling Wave.
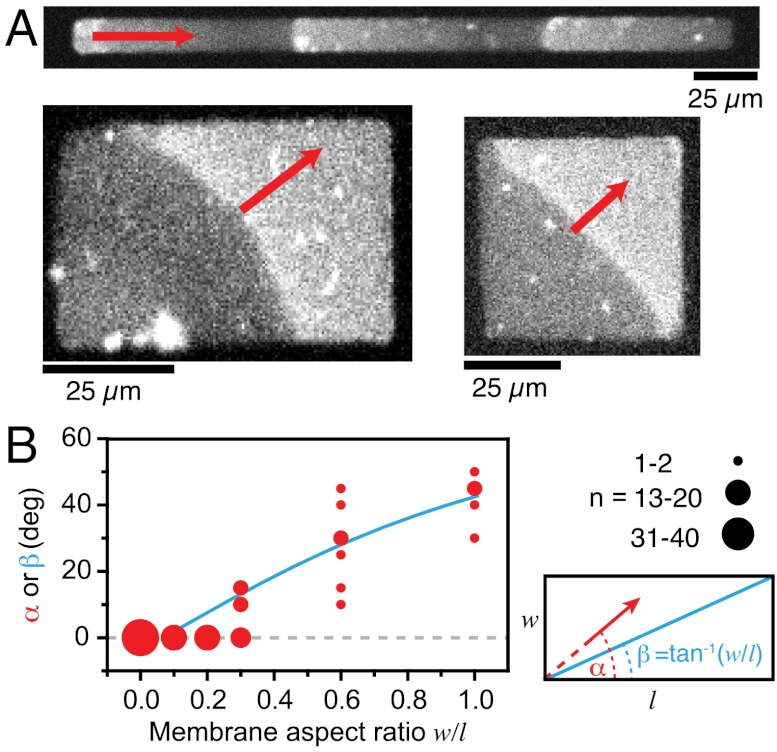
To test the sensitivity of the Min system for the aspect ratio of its reaction compartment in our reconstitution assay, we let protein waves emerge on rectangular membrane patches with different aspect ratios and analyzed the orientation of their traveling direction with respect to this ratio (Fig. 2A and Movie S2). We investigated two sets of samples: small rectangles with an area of 2,500 μm2 and dimensions of 50 μm × 50 μm to 6.25 μm × 400 μm (Fig. 2A), large rectangles with an area of 10,000 μm2 and dimensions of 100 μm × 100 μm to 12.5 μm × 800 μm (SI Appendix, Fig. S2).
Fig. 2.
Influence of membrane patch geometry on Min protein wave orientation. (A) Confocal micrographs of Min protein waves on membrane rectangles of different aspect ratio. (B) With increasing aspect ratio (width/length), the angle of wave propagation (α, red dots) increases corresponding to the angle of the diagonal (β, blue line), which is given by the arctan of the aspect ratio (see lower right graph of 2B).
On long, narrow membranes with an aspect ratio of smaller than 0.3, Min protein waves always travelled parallel to the long axis of the membrane patch, i.e., with a propagation angle of about 0° (Fig. 2 A and B). When incubated on membranes with an aspect ratio larger than 0.3, we found the protein waves to align along the diagonal of the rectangle and, therefore, tend to travel the longest possible path. Accordingly, with increasing aspect ratio the propagation angle increased, until it reached 45° on square-shaped membrane patches. This alignment of protein waves is more precise on small membrane patches than on larger ones: On four times larger rectangles, we found stronger deviations from the diagonal travel path (SI Appendix, Fig. S2).
To closer mimic the rod-like geometry of the E. coli cell, we also used membranes with round ends, denominated as 2D-rods (SI Appendix, Fig. S3). Again, we found that Min protein waves preferentially travel the longest possible path. In 2D-rods the long axis between the two ends of the membrane patch also represents the longest axis. Consistently, the waves also aligned preferentially at an angle of 0° at aspect ratios higher than 0.3. Only for completely round structures, where all possible axes have equal length, the wave propagation axis was random (SI Appendix, Fig. S3).
In summary, we found that Min protein waves respond to differences in membrane geometry and apparently travel along the longest possible path. This finding is consistent with the in vivo finding that the oscillation axis is preserved even in small cells with an aspect ratio close to one, e.g., shortly after cell division (25, 26). Importantly, our observation shows that geometry sensing by the Min system does not require three-dimensional confinement or membrane curvature as in the rod-shaped E. coli cell. Instead, Min protein waves can already sense the geometry of a flat, two-dimensional membrane.
Next, we were interested in how the protein waves behave on a membrane patch where the longest path is not straight but a curved line. Specifically, we were wondering if on a curved membrane path, the traveling protein bands would continue to propagate in the initial direction and run into the boundaries of the membrane patch or change direction to follow the curved path of the membrane.
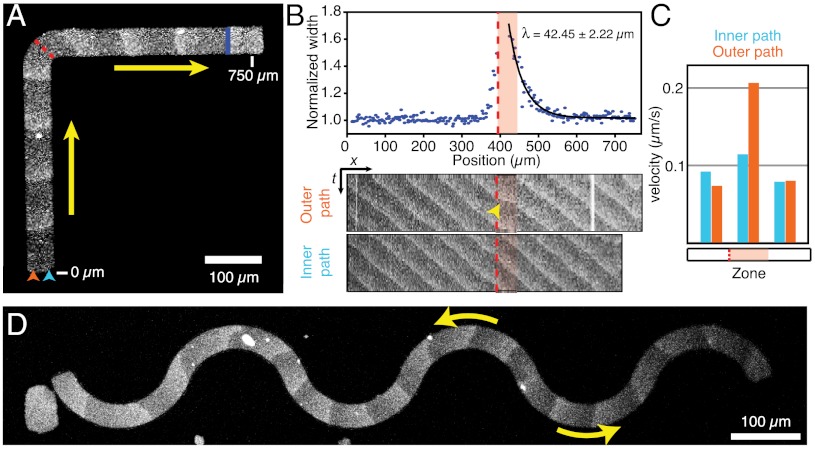
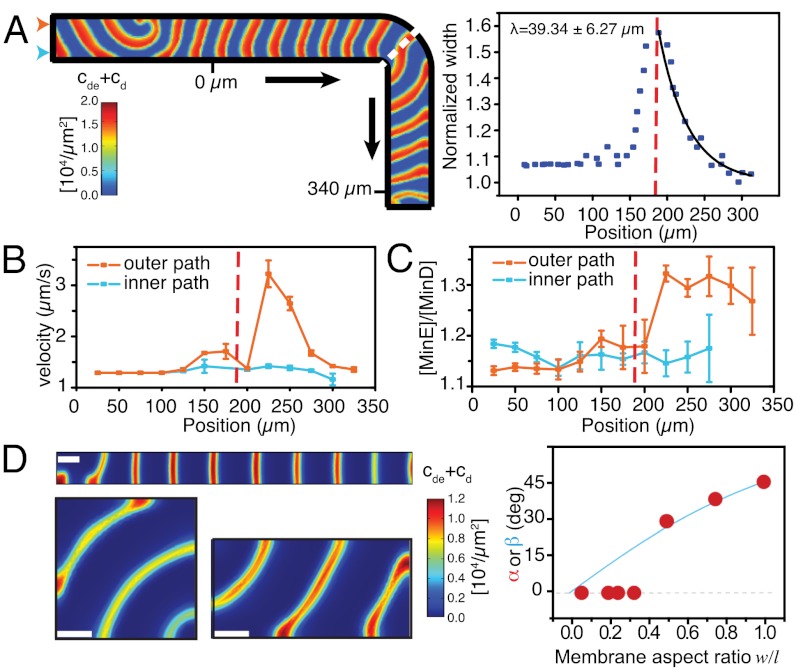
We allowed MinD and MinE to self-organize on a membrane patch shaped like the letter L. In this case, the protein waves would have to turn by 90° to follow the longest possible path. Indeed, we found that at the knee of the membrane patch (Fig. 3A, red dashed line and Movie S3) the protein waves did not persist on their original path but changed direction to travel along the L-shaped path defined by the membrane. Whereas the protein band was aligned perpendicular to the long axis before it reached the knee, it became slightly skewed at the knee because the outer portion of the protein band was lagging behind the inner side. As a result of this distortion, the length of the protein band was increased right after the knee (Fig. 3B, Upper). We quantified this distortion by measuring the relative increase in length depending on the waves position. We found that the protein band extended very abruptly at the location of the knee. Right after the knee, the protein band shortened again, whereas it realigned to travel perpendicular to the new axis (Fig. 3A, blue line). In kymographs obtained from a line either along the outer path or inner path on the membrane (Fig. 3B, Lower), we found that the outer portion of the protein band traveled at the same speed as the inner part before and after the knee but about twice as fast during realignment, thereby quickly catching up with the inner portion. The characteristic length of realignment (λ, from a exponential fit to the decay in wave width) was usually equal or smaller than the typical periodicity of the wave (Fig. 3B and SI Appendix, Fig. S4). This illustrates that a protein band needs less than one complete cycle of collective protein binding and detachment from the membrane to align perpendicular to the leg of the L-shaped membrane.
Fig. 3.
The membrane guides traveling waves. (A) Confocal micrographs of Min protein waves on a L-shaped membrane. Min protein waves turn by 90° following the path of the membrane. (B) Upper: Relative change of protein band width. Exponential fit gives the characteristic travel path length required for realignment of the wave. Lower: Kymographs along the outer (orange arrowhead) or inner (blue arrowhead) path of the membrane shown in A. (C) Velocities within the traveling protein band. During realignment (B, red shaded area), the outer portion of the protein band travels faster than the inner portion, which gives rise to a more horizontal line in the kymograph (B, yellow arrowhead). (D) On a serpentine-shaped membrane protein waves repetitively change direction.
We also allowed MinD and MinE to form waves on membranes of various other shapes. If the membrane was shaped like a serpentine (Fig. 3D), the traveling protein bands quickly and repetitively adjusted their direction to travel along the longest possible path. On a ring-shaped membrane, two sets of wave trains formed, which propagated in opposing directions on the curved membrane path until they met and annihilated each other (SI Appendix, Fig. S5).
These experiments illustrate the ability of the Min system to quickly adapt to changes in the geometry of membrane. Similar steering effects were found for other systems showing chemical waves, e.g., during the catalytic oxidation of carbon monoxide on platinum substrates (27) and the Belousov–Zhabotinsky reaction confined in microstructures (28).
Min Protein Waves Can Couple Across Membrane Gaps.
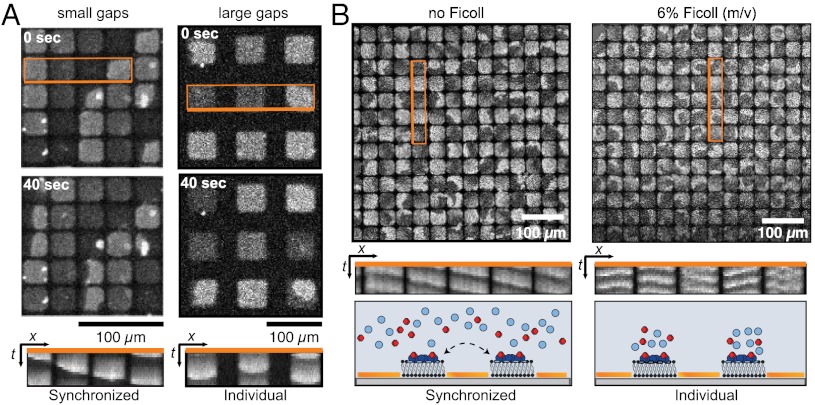
For bacteria, the most dramatic change of its intracellular space happens when the cell divides. Accordingly, during cell division, but before complete constriction into two separate cells, the Min protein pattern changes from normal pole-to-pole oscillations to a transient pattern, where the proteins oscillate between each of the two poles and the closing septum (26, 29). This change of the oscillation pattern does not involve a change in the wavelength. Instead, the constricting septum is thought to limit, but not completely abolish, the diffusive exchange of proteins between the nascent daughter cells, such that two independent, i.e., “split” oscillation patterns appear. To mimic this situation and to study the role of altered diffusive exchange of proteins in our assay, we created grids of quadratic membrane islands, which were separated by gaps of different widths (Fig. 4A).
Fig. 4.
Coupling of waves across membrane gaps. (A) Wave patterns on adjacent membrane patches show synchronized behavior when the distance between the patches is small, but individual behavior on patches separated by large membrane gaps. (B) Whereas waves couple between membrane patches without Ficoll (Left), no coupling is observed after adding Ficoll (Right).
First, we investigated the effect of increasing distances in a grid of membrane patches. We allowed MinD and MinE to self-organize into protein waves on membrane patches whose dimensions were smaller than the periodicity of the waves. At small distances between these patches, we observed a synchronized behavior of the proteins on neighboring membrane patches, giving rise to a traveling wave that swept across individual patches towards a common direction (Fig. 4A, Left and Movie S4). However, when the distance between the patches was larger than 10 μm, protein binding to the membrane patches occurred independently, i.e., with no obvious coupling between to neighboring patches (Fig. 4A, Right and Movie S4).
Next, we limited the diffusive exchange by slowing down the diffusion of proteins in solution. By steadily increasing the concentration of the crowding agent Ficoll, we could continuously reduce the diffusion constant of the proteins in solution, down to a value of about one order of magnitude lower than in the absence of Ficoll (SI Appendix, Fig. S6A). Interestingly, whereas the mobility of reactants usually defines the length-scale of reaction–diffusion patterns, slowing down the diffusion of proteins in solution had only a small effect on the wavelength of the Min protein pattern (SI Appendix, Fig. S6B). To study the effect of reduced protein diffusion on diffusive coupling, we first allowed MinD and MinE to self-organize into waves on membrane patches separated by gaps small enough to still allow for coupling (Fig. 4B, Left). Then, we added Ficoll to the solution (to 6%) and let the system reassemble into a steady state. Now, we found that on the membrane patches, individual protein waves and spirals formed, which did not appear to be coupled to each other (Fig. 4B, Right).
These results show that reducing the diffusion constant of the proteins in solution by one order of magnitude does not significantly influence the length scale of the membrane-bound protein pattern. At the same time it does strongly affect the lateral coupling between membrane patches and thus transmission of spatiotemporal information. The absence of diffusive coupling between patches too far apart or for sufficiently low protein diffusion corresponds to the observation of split protein oscillations in the dividing E. coli cell just before complete fission (26).
Computational Model of Min Protein Surface Waves Under Geometrical Confinement.
We have shown that Min protein waves are able to find the longest path on a membrane patch, to change traveling direction, and to couple across gaps between two membrane patches. To better understand how Min protein waves are influenced by their lateral confinement, we reproduced their dynamic patterns with a computational model (SI Appendix, Dynamic equations).
Recently, two reports have shown that MinE persists at the MinD-membrane surface after activation of the MinD ATPase (23, 30). Although persistent binding of MinE appears to be important for its ability to completely remove MinD from the membrane (23), its possible role for the ability of the Min system to organize the interior of the cell has so far not been addressed. Our model extends previous models by incorporating that MinE transiently interacts with the membrane during the activation of MinD. This description gives a unified account of all currently known stable Min-protein patterns in vivo and in vitro as will be discussed in detail elsewhere.
First, we used our model to understand how the protein wave is able to change its traveling direction and let Min protein waves travel along an L-shaped membrane. Like in our experiments, protein waves are able to change direction to follow the path of the membrane giving rise to an increase in wave width (Fig. 5A; SI Appendix, Fig. S7 and Movies S5 and S6). In agreement with the experimental observation, we found a velocity gradient within the protein band, i.e., the outer part of the protein band travels about twice as fast as the inner part (Fig. 5B). How are the protein waves able to play catchup after they change direction? Our theoretical description is able to give an explanation for this behavior: When we plotted the ratio between the activator MinE and the membrane-bound ATPase MinD along the travel path of the membrane, we found that this ratio is significantly higher at the outer part compared to the inner part of the wave, thereby accelerating the detachment of the proteins from the membrane (23) (Fig. 5C). After realignment, the [MinE/MinD] ratio as well as the velocity gradient reached again the original value. Importantly, we could only reproduce this behavior when we considered transient binding of MinE to the membrane in our model.
Fig. 5.
Computational model reproducing the experimental findings. (A) Left: Min protein waves on a L-shaped membrane. Right: Relative change of protein wave width and exponential fit giving the characteristic travel length required for realignment. (B) Min protein wave velocity and (C) [MinE/MinD] ratio as a function of the distance marked in A. (D) Min protein waves on membrane rectangles with different aspect ratios. With increasing aspect ratio, the angle of wave propagation (α, red dots) increases corresponding to the angle of the diagonal (β, blue curve).
How can the [MinE/MinD] ratio increase during wave realignment? At a convex membrane boundary, the space accessible for bound Min proteins is restricted, implying an increase of the density of membrane-bound proteins. Because MinE is able to transiently interact with the membrane after activation and detachment of MinD (23, 30), the density of membrane-bound MinE increases more than that of MinD. This gives rise to a higher [MinE/MinD] ratio, which locally accelerates the wave (SI Appendix, Fig. S8).
Next, we used our computational model to test protein wave alignment along the diagonal of the rectangular membrane patches with different aspect ratio (Fig. 5D; SI Appendix, Fig. S9 and Movie S7). Our simulations were able to reproduce a distribution of propagation angles at different aspect ratios closely matching our experimental values. What determines the preferred direction of wave propagation? Two effects might play a role: First, due to the given periodicity of the waves, waves traveling along the longest path always outnumber the waves traveling perpendicular to this axis. During synchronization of the pattern, parallel orientation, therefore, always wins over perpendicular orientation, favoring a diagonal alignment of the protein wave. Second, when protein waves run into the border of a membrane patch with an angle larger than 0°, they locally accelerate due to a local increase of the [MinE/MinD] ratio as explained above and align perpendicularly to the membrane border. Whereas the first effect will favor a diagonal alignment of the propagation direction, the second favors a alignment parallel to the long axis of the membrane, especially on membrane patches with dimensions similar as the wave length.
Finally, we used our model to study the coupling between neighboring membrane patches. In our theoretical description, we let protein waves emerge on two neighboring membrane patches. We then altered the diffusive exchange of proteins by either increasing the distance between the two patches or by decreasing mobility of the proteins in solution. Consistent with the experimental findings, waves traveled across the boundary when the diffusive exchange was high enough. When we decreased this exchange, Min protein patterns remained isolated on an individual membrane patch (SI Appendix, Figs. S10 and S11 and Movie S8). This became especially obvious when we visualized the density of the proteins in solution (SI Appendix, Movie S9).
To conclude, using this computational model we were able to get detailed insight into the mechanism of geometry sensing and diffusive coupling of Min protein waves. These data demonstrate that Min protein waves sense the geometry of the flat, two-dimensional membrane, rather than the three-dimensional space of the cell or the curvature of the membrane.
Discussion
In the light of the enormous complexity of living cells, the bottom-up approach of synthetic biology allows us to challenge and significantly aid our understanding of fundamental biological processes. In this study, we combined microsystems technology to structure biomimetic compartments, in vitro reconstitution experiments, and computational modeling to study the mechanisms of how protein patterns are able to organize intracellular space. Despite some fundamental differences compared to the situation in the closed E. coli cell, such as membrane curvature, molecular crowding, and an accordingly smaller length scale of the pattern, we found that the Min system is still capable of detecting the longest axis of the open, two-dimensional membranes used in our reconstituted system.
Protein waves are also thought to organize the interior of eukaryotic cells: For example, spontaneous waves of actin polymerization have been observed in neutrophils and were suggested to arise independently of directed transport by molecular motors (31, 32). As the Min system, these waves are thought to help the cell to sense its geometry, also allowing it to respond to perturbations in cell shape. It will be interesting to see if these actin waves can show a similar ability to sense geometry in a reconstituted in vitro experiment.
Using a computational model we revealed the important role of transient binding of MinE to the membrane and consecutive activation of the membrane-bound ATPase MinD for the ability of the Min system to respond to geometrical confinement. Bacterial DNA segregation machineries encoded on F and P1 plasmids show strong similarities with the Min system (33): a deviant Walker ATPase (ParA), which dimerizes and binds to DNA in the presence of ATP; an activator of the ATPase (ParB), which itself can interact with DNA; and one or more cis-acting DNA regions where ParB binds specifically (parC). These protein systems are thought to move plasmid DNA across the nucleoid surface by employing either a filament-pulling or a diffusion-ratchet mechanism (34–37). In both cases, plasmid DNA segregation would be achieved by the persistent activation of multiple chromosome-bound ParAs by one ParB/parC complex. However, so far it has not been demonstrated whether transient interactions of the ATPase activator ParB with DNA, corresponding to the MinE-membrane interaction, are required to allow ParB to follow retracting regions of high ParA concentration.
In our opinion, in vitro reconstitution experiments together with computational modeling are the most powerful approach to understand how protein systems can organize space much larger than their own dimensions, by either moving large molecules like DNA or, as in the case of the Min system, by generating spatial protein patterns.
Materials and Methods
Microlithography.
Microstructured lipid bilayers were prepared according to Groves et al. (24). Different geometries were produced as two-dimensional gold microstructures using standard photolithography in a class 100 clean room. Fused silica glass wafers (3″’, two sides polished; by Semiconductor Wafer, Inc.) were cleaned in Piranha solution (H2SO4/H2O2) for 30 min at 90 °C, rinsed in water, and dried. Subsequently, the glass was covered by a Cr(5 nm)/Au(50 nm)-layer using electron beam evaporation with condensation rates of 0.1 nm/ sec (Cr) and 0.3 nm/ sec (Au), respectively. This Cr/Au layer was spin-coated with photoresist (ARP 3510, Allresist GmbH, 3,500/ min, 35 s), which was exposed to UV light through a Cr mask (mask aligner MA 56, Suss microtec; 365 nm, 275 W, 8 s) and developed (AR 300-35, Allresist GmbH; diluted with water 1∶1). Exposed reagions of the Cr/Au layer were removed by ion-beam etching. The substrate was decollated using a dicing saw (Disco Corporation) into single square chips of 10–15 mm edge length.
Self-Organization Assay.
Self-organization of Min proteins was achieved as described (22, 23). For all experiments the following concentrations were used: 0.8 μm MinD, 0.5 μm MinE with 10% Cy5-labelled MinE and 2.5 mM ATP. For molecular crowding experiments Ficoll 400 (Sigma Aldrich) to different final concentrations was added to the buffer.
Fluorescence Microscopy.
Samples were imaged using a laser scanning microscope (LSM 510, Zeiss) equipped with a 633 nm laser line and with 10x or 20x objectives. Movies were obtained with a time lapse with 20 s gaps over a period of 5 min at room temperature.
Fluorescence Correlation Spectroscopy.
Diffusion coefficients of Alexa488-labeled MinE proteins in different concentrations of Ficoll were obtained by fluorescence correlation spectroscopy using a Zeiss Confocor 3 system equipped with a 40× objective. The autocorrelation curve of the photon count was fitted using a one-component 3D-diffusion model (38).
Image Analysis.
The propagation angle of Min protein waves was determined using Zeiss LSM software, recorded in bins of 5°, and plotted against the aspect ratio. Aspect ratios were rounded to one-tenth. An angle of zero corresponds to propagation along the longitudinal axis and the aspect ratio was calculated as the ratio of the longitudinal versus the transversal axis. In total, 279 movies were recorded out of which 100 displayed parallel waves, which were included in our analysis. 179 cases showed spiral protein patterns and were not further analyzed.
Computational Model.
The distributions of MinD and MinE in the buffer are given by the densities cD and cE. The distributions of MinD, MinDE-complexes, and MinE on the membrane are, respectively, denoted by cd, cde, and ce. The dynamic equations read
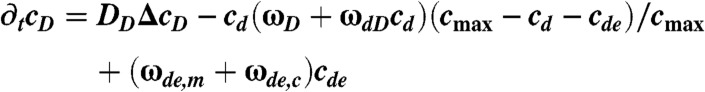 |
[1] |
| [2] |
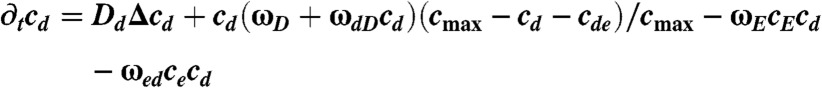 |
[3] |
| [4] |
| [5] |
Here, Δ denotes the Laplace operator in two dimensions. For the meaning of the parameters and their values refer to Table S1; details of the numerical integration of Eqs. 1–5 are given in the SI Appendix (Dynamic equations).
Supplementary Material
ACKNOWLEDGMENTS.
We would like to thank E. Fischer-Friedrich for discussions on the model and Ariadna Martos for protein purification. This work was supported by the Max-Planck-Society (P.S. and M.L.) and by the German Research Foundation as part of the Research Training Group “Nano- and Biotechnologies for Electronic Device Packaging” (GRK 1401) (J.S.) and by the Leibniz-Award (P.S.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206953109/-/DCSupplemental.
References
- 1.Camazine S, et al. Self-Organization in Biological Systems. Princeton, NJ, USA: Princeton University Press; 2003. [Google Scholar]
- 2.Turing A. The chemical basis for morphogenesis. Philos Trans R Soc London Ser B. 1952;237:37–72. [Google Scholar]
- 3.Gierer A, Meinhardt H. A theory of biological pattern formation. Kybernetik. 1972;12:30–39. doi: 10.1007/BF00289234. [DOI] [PubMed] [Google Scholar]
- 4.Muñoz-García J, Neufeld Z, Kholodenko BN. Positional information generated by spatially distributed signaling cascades. PLoS Comput Biol. 2009;5:e1000330. doi: 10.1371/journal.pcbi.1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kholodenko BN. Cell-signalling dynamics in time and space. Nat Rev Mol Cell Biol. 2006;7:165–176. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caudron M, Bunt G, Bastiaens P, Karsenti E. Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science. 2005;309:1373–1376. doi: 10.1126/science.1115964. [DOI] [PubMed] [Google Scholar]
- 7.Athale C, et al. Regulation of microtubule dynamics by reaction cascades around chromosomes. Science. 2008;322:1243–1247. doi: 10.1126/science.1161820. [DOI] [PubMed] [Google Scholar]
- 8.Weiner OD, Marganski WA, Wu LF, Altschuler SJ, Kirschner MW. An actin-based wave generator organizes cell motility. PLoS Biol. 2007;5:e221. doi: 10.1371/journal.pbio.0050221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doubrovinski K, Kruse K. Cell motility resulting from spontaneous polymerization waves. Phys Rev Lett. 2011;107:258103. doi: 10.1103/PhysRevLett.107.258103. [DOI] [PubMed] [Google Scholar]
- 10.Neves SR, et al. Cell shape and negative links in regulatory motifs together control spatial information flow in signaling networks. Cell. 2008;133:666–680. doi: 10.1016/j.cell.2008.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyers J, Craig J, Odde DJ. Potential for control of signaling pathways via cell size and shape. Curr Biol. 2006;16:1685–1693. doi: 10.1016/j.cub.2006.07.056. [DOI] [PubMed] [Google Scholar]
- 12.Loose M, Kruse K, Schwille P. Protein self-organization: lessons from the min system. Ann Rev Biophys. 2011;40:315–336. doi: 10.1146/annurev-biophys-042910-155332. [DOI] [PubMed] [Google Scholar]
- 13.Lutkenhaus L. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev Biochem. 2007;76:539–562. doi: 10.1146/annurev.biochem.75.103004.142652. [DOI] [PubMed] [Google Scholar]
- 14.Meinhardt H, de Boer PAJ. Pattern formation in Escherichia coli: A model for the pole-to-pole oscillations of Min proteins and the localization of the division site. Proc Natl Acad Sci USA. 2001;98:14202–14207. doi: 10.1073/pnas.251216598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howard M, Rutenberg A, de Vet S. Dynamic compartmentalization of bacteria: Accurate division in E. Coli. Phys Rev Lett. 2001;87:278102. doi: 10.1103/PhysRevLett.87.278102. [DOI] [PubMed] [Google Scholar]
- 16.Kruse K. A dynamic model for determining the middle of Escherichia coli. Biophys J. 2002;82:618–627. doi: 10.1016/S0006-3495(02)75426-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raskin DM, de Boer PA. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corbin BD, Yu X-C, Margolin W. Exploring intracellular space: Function of the Min system in round-shaped Escherichia coli. EMBO J. 2002;21:1998–2008. doi: 10.1093/emboj/21.8.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varma A, Huang KC, Young KD. The Min system as a general cell geometry detection mechanism: Branch lengths in Y-shaped Escherichia coli cells affect Min oscillation patterns and division dynamics. J Bacteriol. 2008;190:2106–2117. doi: 10.1128/JB.00720-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischer-Friedrich E. Saarbruecken: Universitaet des Saarlandes; 2009. Pattern formation by the Min system of Escherichia coli. PhD thesis. [Google Scholar]
- 21.Fischer-Friedrich E, Nguyen van yen R, Kruse K. Surface waves of Min-proteins. Phys Biol. 2007;4:38–47. doi: 10.1088/1478-3975/4/1/005. [DOI] [PubMed] [Google Scholar]
- 22.Loose M, Fischer-Friedrich E, Ries J, Kruse K, Schwille P. Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science. 2008;320:789–792. doi: 10.1126/science.1154413. [DOI] [PubMed] [Google Scholar]
- 23.Loose M, Fischer-Friedrich E, Herold C, Kruse K, Schwille P. Min protein patterns emerge from rapid rebinding and membrane interaction of MinE. Nat Struct Mol Biol. 2011;18:577–583. doi: 10.1038/nsmb.2037. [DOI] [PubMed] [Google Scholar]
- 24.Groves JT, Ulman N, Boxer SG. Micropatterning fluid lipid bilayers on solid supports. Science. 1997;275:651–3. doi: 10.1126/science.275.5300.651. [DOI] [PubMed] [Google Scholar]
- 25.Fischer-Friedrich E, Meacci G, Lutkenhaus J, Chate H, Kruse K. Intra- and intercellular fluctuations in Min-protein dynamics decrease with cell length. Proc Natl Acad Sci USA. 2010;107:6134–6139. doi: 10.1073/pnas.0911708107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ventura BD, Sourjik V. Self-organized partitioning of dynamically localized proteins in bacterial cell division. Mol Syst Biol. 2011;7:457. doi: 10.1038/msb.2010.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graham MD, et al. Effects of boundaries on pattern formation: Catalytic oxidation of co on platinum. Science. 1994;264:80–82. doi: 10.1126/science.264.5155.80. [DOI] [PubMed] [Google Scholar]
- 28.Steinbock O, Tóth A, Showalter K. Navigating complex labyrinths: Optimal paths from chemical waves. Science. 1995;267:868–871. doi: 10.1126/science.267.5199.868. [DOI] [PubMed] [Google Scholar]
- 29.Tostevin F, Howard M. A stochastic model of min oscillations in Escherichia coli and Min protein segregation during cell division. Phys Biol. 2006;3:1–12. doi: 10.1088/1478-3975/3/1/001. [DOI] [PubMed] [Google Scholar]
- 30.Park K-T, et al. The Min oscillator uses MinD-dependent conformational changes in MinE to spatially regulate cytokinesis. Cell. 2011;146:396–407. doi: 10.1016/j.cell.2011.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiner OD, et al. An actin-based wave generator organizes cell motility. PLoS Biol. 2007;5:e221. doi: 10.1371/journal.pbio.0050221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doubrovinski K, Kruse K. Cytoskeletal waves in the absence of molecular motors. Europhys Lett. 2008;83:18003. [Google Scholar]
- 33.Gerdes K, Howard M, Szardenings F. Pushing and pulling in prokaryotic DNA segregation. Cell. 2010;141:927–942. doi: 10.1016/j.cell.2010.05.033. [DOI] [PubMed] [Google Scholar]
- 34.Ringgaard S, van Zon J, Howard M, Gerdes K. Movement and equipositioning of plasmids by ParA filament disassembly. Proc Natl Acad Sci USA. 2009;106:19369–19374. doi: 10.1073/pnas.0908347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vecchiarelli AG, et al. ATP control of dynamic P1 ParA–DNA interactions: A key role for the nucleoid in plasmid partition. Mol Microbiol. 2010;78:78–91. doi: 10.1111/j.1365-2958.2010.07314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ptacin JL, et al. A spindle-like apparatus guides bacterial chromosome segregation. Nat Cell Biol. 2010;12:791–798. doi: 10.1038/ncb2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howard M, Gerdes K. What is the mechanism of ParA-mediated DNA movement? Mol Microbiol. 2010;78:9–12. doi: 10.1111/j.1365-2958.2010.07316.x. [DOI] [PubMed] [Google Scholar]
- 38.Schwille P, Oehlenschläger F, Walter NG. Quantitative hybridization kinetics of DNA probes to RNA in solution followed by diffusional fluorescence correlation analysis. Biochemistry. 1996;35:10182–10193. doi: 10.1021/bi960517g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.