Abstract
Membrane and secretory proteins fold in the endoplasmic reticulum (ER), and misfolded proteins may be retained and targeted for ER-associated protein degradation (ERAD). To elucidate the mechanism by which an integral membrane protein in the ER is degraded, we studied the fate of the cystic fibrosis transmembrane conductance regulator (CFTR) in the yeast Saccharomyces cerevisiae. Our data indicate that CFTR resides in the ER and is stabilized in strains defective for proteasome activity or deleted for the ubiquitin-conjugating enzymes Ubc6p and Ubc7p, thus demonstrating that CFTR is a bona fide ERAD substrate in yeast. We also found that heat shock protein 70 (Hsp70), although not required for the degradation of soluble lumenal ERAD substrates, is required to facilitate CFTR turnover. Conversely, calnexin and binding protein (BiP), which are required for the proteolysis of ER lumenal proteins in both yeast and mammals, are dispensable for the degradation of CFTR, suggesting unique mechanisms for the disposal of at least some soluble and integral membrane ERAD substrates in yeast.
INTRODUCTION
The endoplasmic reticulum (ER) is the site in which membrane and secretory proteins fold, and only properly folded proteins usually exit the ER. Incompletely folded proteins may be retained in the ER, and if folding cannot be achieved, they may form aggregates or be targeted for ER-associated protein degradation (ERAD) (recent reviews by Bonafacino and Weissman, 1998; Brodsky and McCracken, 1999; Plemper and Wolf, 1999; Römisch, 1999). From studies with both yeast and mammalian cells, the molecular mechanism of ERAD has recently begun to emerge. The selection of ERAD substrates is highly specific because misfolded proteins have to be distinguished from correctly folded and folding-competent proteins. Molecular chaperones may participate in the selection process because they assist in protein folding and a prolonged association between ER lumenal and cytosolic molecular chaperones and misfolded proteins has been observed for several ERAD substrates (Yang et al., 1993; Ping et al., 1994; Knittler et al., 1995; Schmitz et al., 1995; Sawa et al., 1996; de Virgilio et al., 1999). In addition, mutations in some ER lumenal chaperones prevent ERAD in yeast (McCracken and Brodsky, 1996; Plemper et al., 1997; Brodsky et al., 1999; Gillece et al., 1999).
Both in vivo and in vitro data suggest that ERAD substrates are targeted to the proteasome (Biederer et al., 1996; Hampton et al., 1996; Hiller et al., 1996; Qu et al., 1996; Werner et al., 1996; Wiertz et al., 1996), which is a multicatalytic, proteolytic complex in the cytoplasm (reviewed by Voges et al., 1999). For ERAD, ubquitination is necessary for most (Jensen et al., 1995; Ward et al., 1995; Hiller et al., 1996; Biederer et al., 1997; Hampton and Bhakta, 1997; Loayza et al., 1998; Zhou et al., 1998) but not all substrates (McGee et al., 1996; Werner et al., 1996; Yu et al., 1997). Because the proteasome resides in the cytoplasm, soluble ERAD substrates must be retro-translocated from the ER, and Sec61p, the primary component of the translocon, is required for the degradation of soluble proteins (Pilon et al., 1997; Plemper et al., 1997). For the proteolysis of membrane proteins, a role for Sec61p has also been proposed (Wiertz et al., 1996; Bebök et al., 1998; Plemper et al., 1998). Membrane proteins may be extracted from the ER membrane before being degraded by the proteasome, or their cytosolic portions may be “shaved” or “clipped” by the proteasome, and the lumenal loops and transmembrane domains may then be handled either by the proteasome or by an undefined protease. Indeed, there is experimental evidence supporting the involvement of multiple ERAD pathways. Specifically, both signal peptidase (Mullins et al., 1995), and cysteine and serine proteases have been implicated in the degradation of some ER proteins (Wileman et al., 1991; Wikstrom and Lodish, 1992; Gardner et al., 1993; Moriyama et al., 1998; Fayadat et al., 2000, and references therein).
The importance of the ERAD pathway in cellular physiology is underscored by the fact that several disease-associated molecules are ERAD substrates (reviewed by Brodsky and McCracken, 1999). One example is the cystic fibrosis transmembrane conductance regulator (CFTR), the protein in which mutations give rise to cystic fibrosis (Riordan et al., 1989). CFTR is a plasma membrane chloride channel and is composed of two membrane spanning domains (MSD), each of which has six transmembrane segments, two nucleotide-binding domains (NBD1 and NBD2), and a central regulatory (“R”) domain. CFTR folding and maturation in the ER is an inefficient, temperature-sensitive process, as indicated by the fact that ∼80% of wild-type CFTR is degraded via ERAD (Cheng et al., 1990; Denning et al., 1992; Lukacs et al., 1994; Jensen et al., 1995; Ward et al., 1995).
Molecular chaperones have been suggested to facilitate CFTR maturation. Cytosolic chaperones heat shock protein 70 (Hsp70) (Yang et al., 1993), Hdj-2 (Meacham et al., 1999), and Hsp90 (Loo et al., 1998), and the ER lumenal chaperone calnexin (Ping et al., 1994) transiently associate with CFTR in the ER, and their dissociation coincides with CFTR maturation. Purified Hsp70 suppresses the aggregation of the NBD1 in vitro (Strickland et al., 1997) and acts synergistically with Hdj-2 to this end (Meacham et al., 1999). Recently, it was demonstrated that perturbing Hsp90-CFTR association through the use of Hsp90-interacting compounds accelerates CFTR degradation (Loo et al., 1998), suggesting that if Hsp90 cannot fold CFTR, CFTR is instead targeted for degradation.
Because the secretory pathway and ERAD mechanism are conserved between yeast and mammalian cells, and because yeast present powerful genetic tools, we expressed wild-type CFTR in this organism to begin to dissect the pathway by which an integral membrane ERAD substrate is targeted for proteolysis.
MATERIALS AND METHODS
Strains and Transformation
S. cerevisiae strains used were as follows: the ssa1 temperature-sensitive strain JB67 (Matα, his3-11,15, leu2-3112, ura3-52, trp1Δ1, lys2, ssa1-45, ssa2-1, ssa3-1, ssa4-2) and isogenic wild-type JN516 (Matα, his3-11,15, leu2-3112, ura3-52, trp1Δ1, lys2, SSA1, ssa2-1, ssa3-1, ssa4-2) (Becker et al., 1996); Δcne1 (Mata, ade2-1, can1-100, ura3-1, leu2-3112, trp1-1, his3-11,15, cne1::LEU2) and isogenic wild-type W301-1a (Mata, ade2-1, can1-100, ura3-1, leu2-3112, trp1-1, his3-11,15) (Parlati et al., 1995); proteasome mutant WCG4/2 [Mata, leu2-3112, his3-11,15, ura3Δ5, can(s), pre1-1,pre2-2] and isogenic wild-type strain WCG4 (Mata, leu2-3112, his3-11,15, ura3Δ5) (Heinemeyer et al., 1991); ubiquitin-conjugating mutant MHY552 (Matα, his3-Δ200, ura3-52, leu2-3112, lys2-801, trp1-1, ubc6-Δ1::HIS3, ubc7::LEU2) and isogenic wild-type MHY501 (Matα, his3-Δ200, ura3-52, leu2-3112, lys2-801, trp1-1) (Chen et al., 1993); BiP mutant strains MS1111 (Mata, ura3-52, leu2-3112, ade2-101, kar2-1), MS193 (Mata, ura3-52, leu2-3112, ade2-102, kar2-133), and isogenic wild-type RSY801 (Mata, ura3-52, leu2-3112, ade2-101) (Brodsky et al., 1999); vacuolar protease-deficient strain BJ5461 (Mata, ura3-52, trp1, lys2-801, leu2Δ1, his3Δ200, pep4::HIS3, prb1Δ1.6R, can1) and related wild-type BJ5242 (Mata, ura3-52, trp1, leu2-Δ1, his3-Δ200) (Jones, 1991); HRD mutants RHY1952 (Matα, lys2-801, his3Δ200, ura3-52, trp1-1, leu2-3112, hrd1::LEU2) and RHY1903 (Matα, lys2-801, his3Δ200, ura3-52, trp1-1, leu2-3112, hrd3::LEU2) and isogenic wild-type (Matα, lys2-801, his3Δ200, ura3-52, trp1-1, leu2-3112) (Wilhovsky et al., 2000).
Yeast transformation was performed by the lithium acetate procedure as described (Ito et al., 1983).
Plasmids and Antibodies
CFTR expression in yeast was driven by the constitutive phosphoglycerate kinase promoter in a 2 μ plasmid containing URA3 as the selectable marker. Construction of the plasmids used to express untagged and triple-hemagglutinin (HA)-tagged (at the carboxyl terminus) forms of CFTR in yeast will be described elsewhere (Zhang et al., 2001). Yeast containing the 2 μ plasmid pRS426 (Christianson et al., 1992) lacking insert were used as a control where indicated.
Antibodies used in this study were as follows: monoclonal anti-HA mouse (12CA5 clone, 400 μg/ml; Roche Molecular Biochemicals, Indianapolis, IN), monoclonal anti-C mouse (Genzyme, Cambridge, MA), polyclonal anti-Sec61p rabbit (Stirling et al., 1992), and polyclonal anti-binding protein (BiP) rabbit (Brodsky and Schekman, 1993) antibodies. Primary antibodies were detected with sheep anti-mouse IgG horseradish peroxidase-conjugated or donkey anti-rabbit horseradish peroxidase-conjugated antibody (Amersham Pharmacia Biotech, Piscataway, NJ) and the Amersham ECL Western blotting system was used to detect the signal according to the manufacturer's specifications. For quantitative analysis, 125I-protein A was used in place of secondary antibody, and the signal was visualized using a Fuji PhosphorImager and quantified with MacBas software (version 2.4) (Fujiphotofilm; Koshin Graphic Systems, Stanford, CT).
Sodium Carbonate Extraction and Flotation Assay
Yeast microsomes were prepared from a wild-type strain containing the HA-CFTR expression plasmid as described (Brodsky and Schekman, 1993). Then, a 10-μl aliquot of microsomes (∼100 μg of total protein) was mixed with 1 ml of 100 mM Na2CO4 (pH 11.5) and incubated on ice for 30 min (Fujiki et al., 1982). After centrifugation at 230,000 × g at 4°C for 1 h, the pellet was dissolved in 35 μl of 2× SDS-PAGE sample buffer. Proteins in the supernatant were precipitated by incubation on ice for 30 min with trichloroacetic acid (TCA) added to a final concentration of 10%, followed by a 10-min centrifugation at 16,060 × g at 4°C. The pellet was resuspended in 35 μl of 2× SDS-PAGE sample buffer. Proteins from both the pellet and the supernatant were subjected to SDS-PAGE and immunoblot analysis as described above.
To confirm that CFTR was membrane-embedded and could thus float in a sucrose gradient, wild-type and pre1-1pre2-2 yeast transformed with the CFTR-expression vector were grown to midlog phase (OD600 = ∼0.5) in selective medium, and the cells were harvested, washed, and then resuspended in membrane storage buffer/EDTA (MSB: 50 mM HEPES pH 7.6, 150 mM NaCl, 5 mM EDTA, 1 mM dithiothreitol supplemented with phenylmethylsulfonyl fluoride, leupeptin, and pepstatin according to the manufacturer's specifications) to a final concentration of ∼10 OD600/ml. Glass beads were added to the meniscus and the suspension was agitated on a Vortex mixer four times for 30 s with a 1-min incubation on ice between each agitation. The extract was removed, the beads were washed with an equal volume of MSB, and the combined extracts were centrifuged two times at 300 × g for 2 min to remove unbroken cells. A total of 100 μl of the supernatant was mixed with 300 μl of MSB containing 2.3 M sucrose, and this solution was layered onto 300 μl of MSB containing 2.3 M sucrose in a centrifuge tube. MSB supplemented with 1.5 M sucrose (600 μl) and 0.25 M sucrose (500 μl) were then successively layered onto the gradient and the tube was centrifuged in a Beckman SW55 rotor at 100,000 × g for 5 h at 4°C. Aliquots of 150 μl were removed from the top of the gradient and protein profiles were analyzed by SDS-PAGE and immunoblotting.
Indirect Immunofluorescence and Electron Microscopy
Indirect immunofluorescence microscopy was performed essentially as described (Pringle et al., 1989). Log phase cells (OD600 = ∼0.5–0.7) were fixed for 1 h at room temperature by adding formaldehyde to a final concentration of 3.7%. The yeast were washed twice with sorbitol buffer (50 mM potassium phosphate, pH 7.5, 1.2 M sorbitol), and resuspended to a concentration of 106 cells/10 μl in sorbitol buffer containing 0.1% β-mercaptoethanol and 20 μg/ml zymolase 20T (U.S. Biological, Swampscott, MA). After incubation at 37°C for 1 h, cells were washed with sorbitol buffer twice and applied to poly-l-lysine-coated glass slides (20 μl/well) before being fixed for 5 min with 3.7% formaldehyde in phosphate-buffered saline (PBS) (40 mM K2HPO4, 10 mM KH2PO4, pH 7.5, 0.15 M NaCl). A final concentration of 50 mM NH4Cl was used to quench the formaldehyde. The cells were permeabilized with 0.1% NP-40 in PBS supplemented with 0.1% bovine serum albumin (BSA) (PBS-0.1%BSA) and then incubated with anti-HA (1:250 dilution) or anti-C (1:40 dilution), and anti-BiP (1:500) antibodies in PBS-0.1%BSA overnight at 4°C in a humid chamber. After three washes with PBS/BSA and one wash with PBS/BSA/NP-40, the primary antibodies were detected with anti-rabbit IgG TRITC conjugate (1:250 dilution; Sigma, St. Louis, MO) and anti-mouse IgG fluorescein conjugate (1:250 dilution; Roche Molecular Biochemicals, Indianapolis, IN) in PBS-0.1%BSA.
Yeast either containing or lacking the CFTR-expression vector were grown to midlog phase and whole cell electron microscopy was performed as published by Kaiser and Schekman (1990).
CFTR Degradation Assay
Cells expressing CFTR were grown to midlog phase (OD600 = ∼0.5) at 26°C before cycloheximide was added to a final concentration of 50 μg/ml, and were incubated either at 26°C or shifted to 40°C with shaking before they were harvested at the indicated time points to prepare cell extracts. For each time point, a total of 2.5 OD600 of cells was harvested, washed with cold water, and resuspended in 1 ml of cold water. An aliquot of 150 μl of freshly prepared 2 N NaOH/1.12 M β-mercaptoethanol was added, and the yeast were resuspended and left on ice for 15 min. Then, 150 μl of 50% TCA was added, and the extract was incubated on ice for an additional 20 min. After centrifugation at 16,060 × g for 5 min at 4°C, the pelleted proteins were resuspended in TCA sample buffer (80 mM Tris-Cl, pH 8.0, 8 mM EDTA, 120 mM dithiothreitol, 3.5% SDS, 0.29% glycerol, 0.08% Tris base, 0.01% bromphenol blue) and incubated at 37°C for 30 min. Total protein was resolved by SDS-PAGE, followed by immunoblot analysis or quantitative immunoblot analysis (see above).
Pulse-Chase Assay for ERAD
The degradation of the misfolded form of carboxypeptidase Y (CPY*; Hiller et al., 1996) was determined using an HA-epitope-tagged version of CPY* (Ng et al., 2000). In brief, cells transformed with the CPY* expression vector were grown to logarithmic phase in selective medium containing glucose and a pulse-chase analysis by using [35S]methionine was performed as described (Brodsky et al., 1998). CPY* was precipitated from ∼5 × 106 cpm of 35S-labeled protein extract with 10 μg of anti-HA antibody (Roche Molecular Biochemicals, Indianapolis, IN) and protein A-Sepharose (Amersham Pharmacia Biotech). Yeast BiP was precipitated using anti-BiP antibody and protein A-Sepharose.
RESULTS
CFTR Is an ERAD Substrate in Yeast
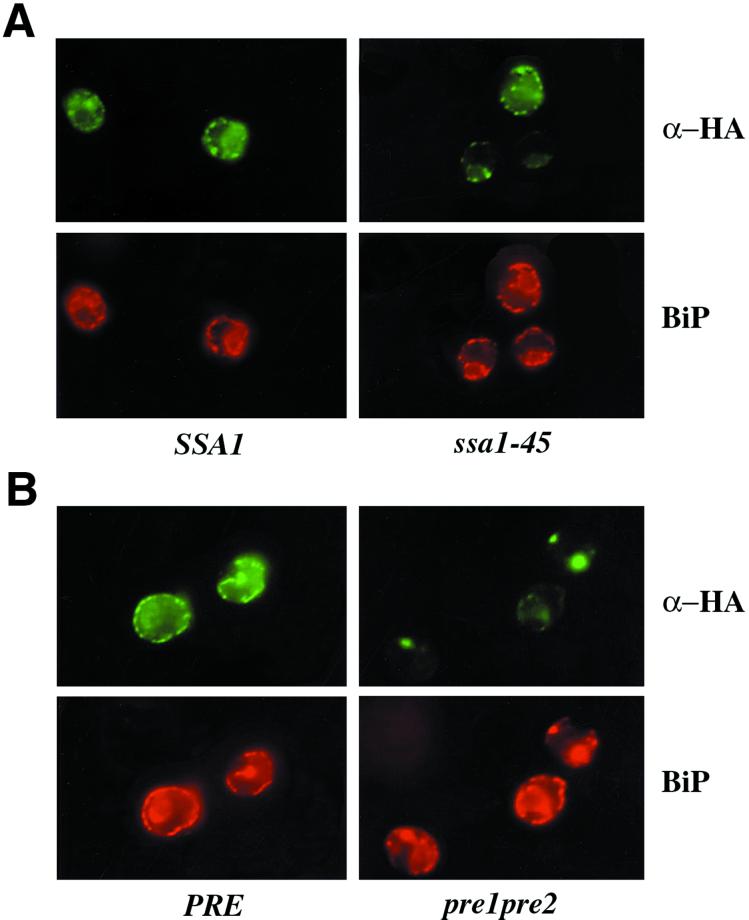
To assess the subcellular localization of CFTR expressed in yeast, indirect immunofluorescence microscopy was performed using cells grown exclusively at 26°C or that had been shifted to 40°C for 1 h. A temperature of 40°C was chosen because CFTR stabilization in temperature-sensitive yeast became maximally pronounced at 40°C (see below). Yeast harboring the CFTR-expressing plasmid (either containing or lacking an HA tag at the C terminus, referred to as HA-CFTR or untagged CFTR below, respectively) were subjected to indirect immunofluorescence microscopy. As shown in Figure 1, A and B, HA-CFTR in wild-type cells (denoted SSA1 and PRE) grown at either 26°C or shifted to 40°C for 1 h exhibit a strong perinuclear punctate pattern and subplasma membrane residency and colocalize with BiP, an ER lumenal Hsp70. For untagged CFTR, an antibody against the C terminus was used instead of α-HA, and identical results were obtained (our unpublished data). These data indicate that CFTR expressed in yeast resides primarily in the ER, as also suggested by others (Huang et al., 1996).
Figure 1.
CFTR colocalizes with BiP in yeast and accumulates at one or two sites in the proteasome mutant strain at 40°C. CFTR-expressing ssa1-45 (A) and pre1-1pre2-2(B) cells and the respective isogenic wild-type cells (SSA1 and PRE) were grown at 26°C, shifted to 40°C for 1 h, and then subjected to indirect immunofluorescence microscopy with anti-HA (to detect HA-CFTR) and anti-BiP antibodies.
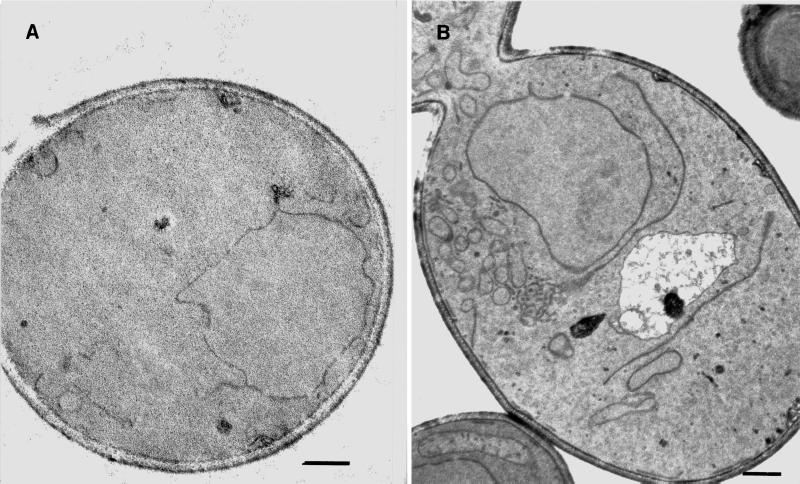
Expression of heterologous proteins in yeast, or overexpression of endogenous proteins may lead to an altered intracellular morphology (Wright et al., 1988; Umebayashi et al., 1997; Becker et al., 1999). To determine whether expression of CFTR similarly affects yeast, logarithmically growing cells containing either a control or the HA-CFTR expression plasmid were prepared for whole cell electron microscopy as described in MATERIALS AND METHODS. As shown in Figure 2, elongated tubular and enlarged vesicular structures were evident only in cells expressing CFTR. These structures are described in greater detail elsewhere (Kuehn, Nijbroek, and Michaelis, unpublished data), and also arise from the expression of mutant forms of the Ste6, the a factor mating pheromone transporter in yeast. They are distinct from those observed when other ER membrane proteins are overexpressed in yeast (Wright et al., 1988; Umebayashi et al., 1997; Becker et al., 1999), or when the secretory pathway is compromised in a sec mutant strain (Nishikawa et al., 1994). In contrast, we failed to observe impaired growth or defects in secretory protein translocation in yeast expressing CFTR (our unpublished results). Because ER membrane proliferation may arise from induction of the unfolded protein response in the ER (Cox et al., 1997), we introduced an unfolded protein response (UPR) reporter plasmid into yeast already containing the CFTR-expression plasmid or the plasmid lacking an insert, but found that the UPR was not induced by CFTR expression. Thus, the molecular basis for the membrane proliferation observed in the CFTR-expressing yeast is unknown.
Figure 2.
Intracellular membranes proliferate in the CFTR-expressing yeast. Wild-type yeast containing either an empty control vector (A) or the CFTR-expression plasmid (B) were examined by thin section electron microscopy according to the method of Kaiser and Schekman (1990). Bar, 0.5 μm.
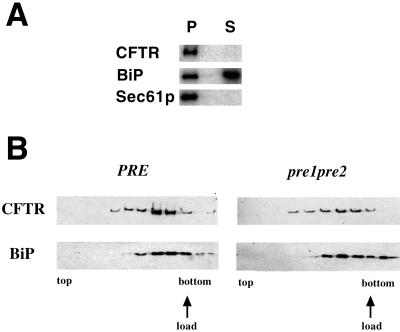
To verify that CFTR expressed in yeast inserts into the ER membrane, ER-derived microsomes (Brodsky and Schekman, 1993) were prepared from HA-CFTR–expressing cells and treated with sodium carbonate (Fujiki et al., 1982). As presented in Figure 3A, HA-CFTR and Sec61p, the ER translocation channel, were found exclusively in the membrane pellet, whereas BiP, an ER lumenal protein, is primarily (∼66%) located in the supernatant. Incomplete extraction of BiP might have been observed from its interaction with components of the translocation machinery (Brodsky and Schekman, 1993).
Figure 3.
CFTR is an integral membrane protein. (A) ER-derived microsomes were prepared from CFTR-expressing cells, extracted with sodium carbonate, and then centrifuged. Total protein from the membrane pellet (P) and supernatant (S) was resolved by SDS-PAGE and subjected to immunoblot analysis. BiP and Sec61p serve as soluble and membrane protein controls, respectively. (B) Cell extracts from CFTR-expressing wild-type and pre1-1pre2-2 cells were prepared, layered at the bottom of a centrifuge tube, and subjected to ultracentrifugation as described in MATERIALS AND METHODS. Immunoblots to detect the indicated proteins in fractions from the top to the bottom of the gradient are shown. The position in the gradient at which the cell extract was loaded is also indicated.
To confirm that CFTR integrates into the membrane, cell extracts from CFTR-expressing cells were prepared and mixed with a dense sucrose solution, overlayed with sucrose-containing buffers of lower densities, and then subjected to ultracentrifugation. As shown in Figure 3B, we observed that CFTR floated in the sucrose gradient coincident with BiP. We did note that some of the BiP failed to float, suggesting that a portion of it became liberated from the vesicles, most likely during the preparation of the cell extracts. These combined immunofluorescence and biochemical data indicate that CFTR is integrated into the ER membrane in yeast.
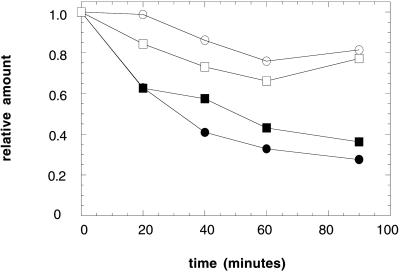
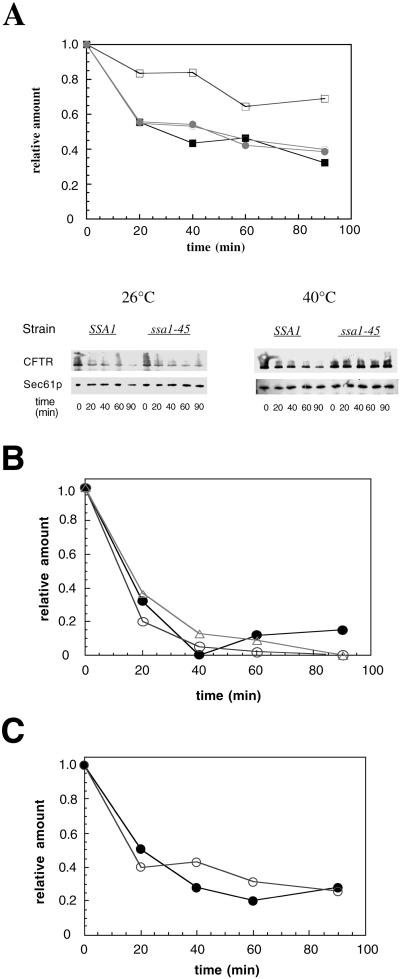
We next addressed whether wild-type CFTR is degraded via the ubiquitin-proteasome pathway in yeast, as in mammalian cells (Jensen et al., 1995; Ward et al., 1995). The degradation of CFTR was assayed in growing cells after the addition of cycloheximide. The amount of CFTR remaining at various time points was quantified by immunoblot analysis by using 125I-protein A. We found that HA-CFTR was stabilized in the pre1-1pre2-2 mutant (Figure 4), a strain with mutations that abrogate ∼95% of the activity of the proteasome (Heinemeyer et al., 1993). HA-CFTR was also stabilized in the ubc6,7 strain, which had been deleted for the ubiquitin conjugation enzymes Ubc6p and Ubc7p (Figure 4). We note in this and other experiments (see below) that the extent of CFTR degradation in unique wild-type yeast strains differs, indicating the necessity of using isogenic strains to measure ERAD in vivo. Regardless, our results indicate that the proteasome and ubiquitin conjugation facilitate the degradation of CFTR in yeast.
Figure 4.
CFTR degradation in yeast is compromised in strains defective for proteasome function and ubiquitin conjugation. CFTR-expressing cells grown at 26°C and then incubated for 1 h at 40°C were harvested at the indicated time points after the addition of cycloheximide, and cell extracts were prepared and subjected to SDS-PAGE and immunoblot analysis as described in MATERIALS AND METHODS. Results from a quantitative immunoblot analysis are shown. The amount of CFTR at time zero was set to 1. ●, PRE1PRE2; ○, pre1-1pre2-2; ▪, UBC6UBC7; □, ubc6ubc7.
The intracellular degradation of many proteins in yeast occurs in the vacuole, and misfolded secretory proteins can be targeted to this organelle (Hong et al., 1996, and references therein). To confirm that the degradation of CFTR was independent of vacuolar proteases, we examined the fate of HA-CFTR in a Δpep4 mutant in which most vacuolar proteases are inactivated (Jones, 1991), but found that CFTR degradation was unaffected (Zhang et al., 2001).
ERAD of Soluble and Membrane Proteins Requires Unique Chaperones
The ER lumenal chaperones calnexin and BiP are required for the efficient degradation of soluble ERAD substrates (Le et al., 1994; Knittler et al., 1995; Schmitz et al., 1995; McCracken and Brodsky, 1996; Qu et al., 1996; Plemper et al., 1997; Brodsky et al., 1999), and both ER lumenal (e.g., calnexin) and cytosolic chaperones (e.g., Hsp70 and Hsp90) associate with CFTR in mammalian cells (Yang et al., 1993; Ping et al., 1994; Loo et al., 1998; Meacham et al., 1999). S. cerevisiae has two classes of cytosolic Hsp70s, encoded by the SSA and SSB genes (Boorstein et al., 1994). Although the Ssb proteins are involved in protein translation (Pfund et al., 1998), the Ssa proteins are more similar to mammalian Hsp70 and are required for a variety of chaperone-dependent activities (Miao et al., 1997). There are four Ssa proteins, Ssa1-4p, of which the expression of at least one is essential for viability (Werner-Washburne et al., 1987). Therefore, to explore whether Hsp70 is required for CFTR degradation, we used a strain containing an SSA1 temperature-sensitive allele, ssa1-45, and in which ssa2-4 had been inactivated (Becker et al., 1996). The isogenic “wild-type” strain contains SSA1 and similarly lacks functional ssa2-4. We found that CFTR degradation is robust at both 26 and 40°C in wild-type cells (Figure 5A); ∼60% of CFTR is degraded after 90 min in wild-type cells, regardless of the temperature at which the experiment was performed. In contrast, in the ssa1-45 mutant strain, CFTR degradation is proficient at 26°C, but the protein is significantly stabilized at 40°C. When the degradation of untagged CFTR was examined using an antibody against the C terminus of the protein, CFTR was also stabilized in the ssa1-45 strain at 40°C (our unpublished results). We conclude that Hsp70 is required to facilitate CFTR degradation in yeast, although it is dispensable for the ERAD of two soluble proteins both in vivo and in vitro (Brodsky et al., 1999).
Figure 5.
CFTR is stabilized only in the ssa1-45 strain at 40°C. CFTR-expressing cells grown exclusively at 26°C or shifted to 40°C for 1 h were harvested at the indicated times after the addition of cycloheximide, and cell extracts were prepared and subjected to immunoblot analysis. (A) Results from a quantitative immunoblot analysis are plotted and the corresponding PhosphorImager analysis is shown. The amount of CFTR at time zero was set to 1. ●, SSA1 cells expressing CFTR at 26°C; ○, ssa1- expressing CFTR at 26°C; ▪, SSA1-expressing CFTR at 40°C; □, ssa1-45 cells expressing CFTR at 40°C. Sec61p serves as a loading control. (B) Degradation of HA-CFTR was assayed in wild-type (●), kar2-1 (○), or kar2-133 (▵) yeast as described in A, and the results were analyzed and quantified. (C) HA-CFTR degradation was measured in wild-type (●) or Δcne1 (○) yeast.
Because the Ydj1p cochaperone stimulates the ATPase activity of Ssa1p (Cyr et al., 1992) and YDJ1 and SSA1 interact genetically (Becker et al., 1996), Ydj1p may also facilitate CFTR degradation. In addition, the ydj1-151 mutant strain is defective for the ubiquitin-dependent degradation of some cytoplasmic substrates (Lee et al., 1996). We found, however, that CFTR was degraded efficiently in ydj1-151 yeast (our unpublished results).
To determine whether BiP and calnexin play a role in the degradation of CFTR, we examined CFTR turnover in kar2-1, kar2-133, and Δcne1 strains in which the degradation of the soluble ERAD substrate protein proalpha factor was debilitated in vitro (McCracken and Brodsky, 1996; Brodsky et al., 1999). Strains containing the kar2-1 or kar2-133 mutations are also defective for the degradation of the soluble substrate A1PiZ in vivo (Brodsky et al., 1999). As shown in Figure 5, B and C, we observed that the rate of CFTR proteolysis was identical in the kar2 and cne1 mutant strains and their corresponding isogenic wild-type strains. Together, these results indicate that different sets of chaperones are required to degrade several membrane and soluble proteins, and suggest that the respective ERAD pathways are distinct.
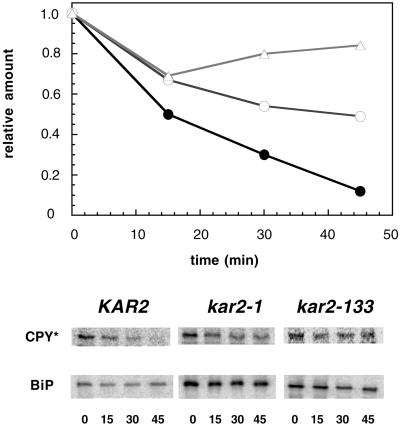
To confirm that the degradation of soluble ERAD substrates is compromised in the kar2-1 and kar2-133 strains, we examined the fate of CPY* in the wild-type and kar2 mutants by chase analysis after the addition of cycloheximide to cells. The ERAD of CPY* was established by Wolf and colleagues (Hiller et al., 1996), and using a distinct kar2 mutant, Plemper et al. (1997) found that the degradation and retro-translocation of CPY* from the ER required BiP. When we measured the proteolysis of CPY* in wild-type and the kar2 mutants used in this study, stabilization of CPY* was observed in the kar2 strains (Figure 6). Even after a 45-min chase at a semipermissive temperature of 30°C, the amounts of CPY* remaining as a percentage of the initial levels were 12, 49, and 84% in the wild-type strain and the kar2-1 and kar2-133 mutants, respectively.
Figure 6.
CPY* degradation is attenuated in kar2-1 and kar2-133 mutant strains. The proteolysis of CPY* in wild-type, kar2-1, and kar2-133 cells harboring an HA-tagged CPY* expression vector was examined by pulse-chase analysis at 30°C after cycloheximide addition as described in MATERIALS AND METHODS. The relative amount of CPY* at time zero was set to 1. Wild-type (KAR2), ●; kar2-1, ○; kar2-133, (▵). The original gel is also shown and the fate of BiP in each strain was examined as a loading control. The time (min) of the chase is indicated below each lane.
Two HRD Gene Products Are Not Required for the ERAD of CFTR
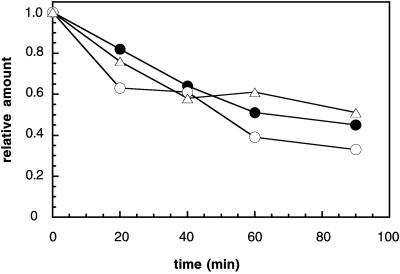
Hampton and colleagues (1996) have isolated a number of mutants that are defective for the regulated degradation of HMG-CoA reductase in yeast. Two of the proteins encoded by the corresponding genes, Hrd1/Der3p and Hrd3p, form a stoichiometic complex and cooperate during ERAD (Gardner et al., 2000). Although the hrd1 and hrd3 mutants display a mild defect in the degradation of two integral membrane ERAD substrates (Wilhovsky et al., 2000), other ERAD substrates are significantly stabilized in the hrd1/der3 mutant (Bordallo et al., 1998; Plemper et al., 1998). To determine whether CFTR degradation is affected by the hrd1 and hrd3 mutations, we introduced the CFTR expression plasmid into these mutants and an isogenic wild-type strain and measured the levels of CFTR over time, as described above. As shown in Figure 7, we observed no significant difference in the overall rate of CFTR degradation in wild-type, hrd1, and hrd3 yeast. Thus, CFTR, like UP* (Wilhovsky et al., 2000) is UBC6/7-dependent but HRD1-independent.
Figure 7.
CFTR degradation is unaffected in cells deleted for the HRD1/DER3 and HRD3 genes. The degradation of CFTR in wild-type (○) and hrd1 (●) and hrd3 (▵) mutant cells was examined as described in MATERIALS AND METHODS. Shown are the means of two independent data sets. SDs are within 20% of the means.
CFTR Concentrates in Cells Mutated for the Proteasome
To gain insight into how CFTR is selected and targeted to the proteasome, we examined CFTR localization under conditions in which CFTR degradation is impeded. Indirect immunofluorescence microscopy was performed with the pre1-1pre2-2 and ssa1-45 mutant strains and isogenic wild-type cells expressing CFTR. As presented above, in wild-type cells, CFTR appears in a strong punctate pattern and colocalizes with BiP (Figure 1A). In ssa1-45 cells, at both 26 and 40°C (Figure 1A), similar results were observed. However, when proteasome function is attenuated and the cells are shifted to 40°C, CFTR can concentrate to one or two “dots” in some cells; a representative section of the cells is shown in Figure 1B. We do not believe that these spots represent “aggresomes, ” extracted CFTR that accumulates in a perinuclear site in mammalian cells when CFTR is overexpressed or cells are grown in the presence of proteasome inhibitors (Johnston et al., 1998), for the following reasons. First, CFTR and the lumenal chaperone BiP colocalize in these images. Second, CFTR floats in sucrose gradients regardless of whether it derived from wild-type or the proteasome-mutant strain (Figure 3B) in which degradation is attenuated (Figure 4A). The fact that not all pre cells expressing CFTR display one or two dots (Figure 1B) may represent heterogenous levels of CFTR expression. In fact, we have noted that this pattern arises in cells expressing the greatest amount of CFTR (our unpublished results), suggesting that lower levels of CFTR may be “handled” by residual ERAD activity or other proteases. Nevertheless, these results indicate that the residency of CFTR is differentially affected when its proteolysis is abrogated by defects in Hsp70 or the proteasome.
DISCUSSION
The work described here establishes CFTR as an ERAD substrate in the yeast S. cerevisiae. We found that CFTR expressed in yeast is an integral membrane protein retained in the ER, and that proteasome activity and ubiquitin conjugation systems are necessary for maximal degradation. Furthermore, we found that the cytoplasmic Hsp70 Ssa1p, but neither the lumenal Hsp70 BiP nor lumenal chaperone calnexin is required for CFTR proteolysis. This is opposite to the chaperone requirements for the degradation of soluble ERAD substrates: the ER lumenal proteins calnexin and BiP are required, but the cytoplasmic chaperone Ssa1p is not (McCracken and Brodsky, 1996; Plemper et al., 1997; Brodsky et al., 1999; Figure 6). Together, these results suggest unique mechanisms for the quality control of at least some integral membrane and soluble ER proteins.
Our results are consistent with work from other laboratories investigating the chaperone requirements for the degradation of integral membrane proteins in the yeast ER. First, Plemper et al. (1998) observed that BiP function is dispensable for the degradation of Pdr5p*, an integral membrane ERAD substrate. Second, while this manuscript was in preparation, Hill and Cooper (2000) reported that Ssa1p was necessary for the degradation of Vph1p, an integral membrane subunit of the vacuolar ATPase that becomes an ERAD substrate when the VMA22 gene product is absent, whereas mutations in KAR2 had no effect on the degradation of Vph1p. Third, we found that Ssa1p is required, and BiP is dispensable for the destruction of an ER-retained, mutated form of Ste6p (“Ste6p*”), the integral membrane a-factor transporter in yeast that is homologous to CFTR (Harper, Brodsky, and Michaelis, unpublished data).
Because cytosol prepared from the ssa1-45 mutant strain shifted to the nonpermissive temperature supports the degradation of soluble ERAD substrates (Brodsky et al., 1999), the defect in CFTR degradation that we observed in the ssa1-45 mutant cannot arise from proteasome inactivation. Instead, we favor a model in which Ssa1p retains cytoplasmic domains of ER membrane proteins in a protease-accessible conformation, an activity that would be essential if the proteasome or other protease initially shaves or clips this domain. We previously found that approximately equal amounts of Ssa1p are associated with ER-derived microsomes and “free” in yeast cytosol (Brodsky et al., 1999), suggesting that much of this chaperone may be positioned to associate with ERAD substrates. Several other observations support this model. First, each of the integral membrane ERAD substrates (i.e., Vph1p, CFTR, Ste6p*) that require Ssa1p activity for degradation contain large, cytoplasmic domains. If this model is correct, one might expect that Ssa1p prevents the formation of protein aggregates and promotes protein folding. In fact, we found previously that cytoplasm prepared from the ssa1-45 mutant is unable to refold heat-denatured firefly luciferase in vitro, whereas cytosol prepared from the isogenic wild-type strain refolds luciferase (Brodsky et al., 1999). Second, others have observed that mammalian Hsp70 suppresses the aggregation of the NBD1 of CFTR in vitro (Strickland et al., 1997; Meacham et al., 1999). Third, if Ssa1p “holds” CFTR in a conformation that is accessible to the proteasome or unidentified protease, then cleavage may initiate within the large, cytosolically disposed NBD and/or R domains. Indeed, upon overexposure of gels in which degradation was assayed, we noted CFTR degradation intermediates of molecular weights ∼80–120,000 (our unpublished results), a size consistent with cleavage within the NBD and R domains. CFTR degradation intermediates in this molecular weight range have also been observed when CFTR biogenesis was examined in mammalian cells (Lukacs et al., 1994; van Oene et al., 2000). And fourth, Ssa1p may not be essential for the degradation of integral membrane ERAD substrates presenting less prominent cytoplasmic domains. Indeed, only a minor effect on the proteolysis of Sec61-2p was observed when its stability was assessed in the ssa1-45 strain (S. Nishikawa, S. Fewell, Y. Kato, J. Brodsky, T. Endo, unpublished data).
To explain the requirement for BiP in the degradation of soluble but not integral membrane ERAD substrates, we suggest that lumenal domains must be preserved in an aggregation-free state, an activity that BiP is known to exhibit (reviewed by Gething, 1997). In contrast, the lumenal and transmembrane domains of integral membrane proteins may be removed independent of BiP, perhaps through direct extraction by the proteasome (Mayer et al., 1998; Xiong et al., 1999). Finally, it is possible that BiP may be required to “unlock” the translocation channel to permit ERAD substrates to retro-translocate after their complete import into the ER (Plemper et al., 1999). This model arises from the demonstration by Johnson and coworkers that BiP might gate the translocation pore in the mammalian ER (Hamman et al., 1998).
Inherent in these models, and because of the different CFTR immunofluorescence staining patterns observed in the ssa1-45 and pre1-1pre2-2 strains (Figure 1), we suggest that CFTR degradation in yeast is a multistep process. Only when proteasome function was attenuated at 40°C did we observed CFTR localization to one or two sites in the ER. Similar structures have been detected in yeast expressing Ste6p* and are termed ER-associated bodies (ERABs). As shown here for CFTR, Ste6p*-induced ERAB formation does not involve induction of the UPR, and a detailed immunofluorescence and electron microscopic analysis of the morphology of ERABs has been undertaken and will be described elsewhere (Kuehn, Nijbroek, and Michaelis, unpublished data). Unique localization patterns in the ssa1 and pre mutants may arise if Ssa1p acts upstream of the proteasome in the CFTR degradation pathway: The spots we observe in the proteasome mutant strain could represent the final staging points before CFTR proteolysis, whereas CFTR delivery to these sites is halted when Hsp70 is inactivated. Because the CFTR that concentrates to these spots is membrane-associated, as determined by the floatation analysis (Figure 3B), our results also suggest that the catalytic activity of the proteasome is required to degrade or extract CFTR at or from the yeast ER membrane. Such a role for the proteasome was also suggested by Mayer et al. (1998) when the degradation of a hybrid ER membrane protein was examined in strains lacking a functional proteasome.
Because CFTR remains membrane-associated when proteolysis is compromised in yeast, we believe that CFTR does not reside in aggresomes. In contrast, aggresomes form in CFTR-expressing mammalian cells when overexpressed or when proteasome activity is blocked (Johnston et al., 1998; Wigley et al., 1999). Thus, there may be unique mechanisms to handle accumulated and undegraded CFTR in yeast and mammals. In fact, it has been suggested that aggresome formation may be cell-type specific, because aggresomes have not been observed in every CFTR-expressing mammalian cell line when proteasome function is attenuated (Chen et al., 2000). Finally, it is possible that aggresome formation requires high levels of CFTR than cannot be produced in yeast.
Although ∼20% of wild-type CFTR in mammalian cells escapes degradation and transits to the plasma membrane, CFTR in yeast appears to reside primarily in the ER. However, we cannot exclude the possibility that a fraction of CFTR in yeast escapes ERAD and migrates through the secretory pathway. Relevant to this hypothesis, we often find that a fraction of CFTR resists degradation (see for example, Figure 5C), and that a minor fraction of CFTR (∼10%) comigrates with Pma1p, a plasma membrane protein, upon velocity sucrose gradient analysis (our unpublished results).
Finally, the data presented in this article may be pertinent to the study of membrane protein degradation in the mammalian ER. In agreement with our results, Fisher et al. (1997) reported that the degradation of ApoB100 in HepG2 cells is enhanced approximately twofold when cytoplasmic Hsp70 is overexpressed, suggesting that the chaperone facilitates ERAD. A role for both the cytoplasmic Hsp70 and Hsc70 molecular chaperones during CFTR biogenesis in mammalian cells has also been uncovered by the use of modulators of chaperone activity. First, Rubenstein and Zeitlin (2000) showed that sodium phenylbutyrate decreases the amount of Hsc70-ΔF508 CFTR complexes in mammalian cells and that there is a concomitant rescue of the ΔF508 mutant phenotype; however, Hsp70 levels increase under these conditions (P. Zeitlin, personal communication). Second, Jiang et al. (1998) discovered that ΔF508 CFTR-expressing cells treated with the Hsp70/Hsc70-interacting drug deoxyspergualin exhibited a partial restoration in cAMP-stimulated chloride channel activity. These investigators hypothesized that altering the interaction of the chaperone with CFTR may promote maturation, although it is possible that DSG stimulates Hsp70, thus enhancing CFTR folding and increasing the yield of “active” ΔF508-CFTR. Interpreting the conclusions from these diverse studies is further complicated by the fact that mammalian Hsp70 has been shown to facilitate ubiquitin conjugation onto several proteins substrates in vitro (Bercovich et al., 1997). Clearly, further work will be directed to better understand the roles of Hsp70/Hsc70 in eucaryotic protein turnover.
ACKNOWLEDGMENTS
We thank Drs. Raymond Frizzell and Robert Bridges for reagents, their invaluable support, and for many helpful discussions, and Dr. Gergely Lukacs for critical reading of the manuscript. We also thank Drs. Elizabeth Craig, Dieter Wolf, Mark Hochstrasser, Caroline Slayman, Elizabeth Jones, Peter Walter, Randy Hampton, and Davis Ng for generously supplying reagents, and Tom Harper for help with imaging technology. This work was supported by a Pilot-Feasibility grant from the Cystic Fibrosis Foundation's Research Development Program at the University of Pittsburgh (to J.L.B.), by Grant MCB-9722889 from the National Science Foundation (to J.L.B. and A.A.M.), and by Grants GM-51508 and DK-58029 from the National Institutes of Health (to S.M.).
REFERENCES
- Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in Yeast Genetics. A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1997. [Google Scholar]
- Bebök Z, Mazzochi C, King SA, Hong JS, Sorscher EJ. The mechanism underlying cystic fibrosis transmembrane conductance regulator transport from the endoplasmic reticulum to the proteasome includes Sec61β and a cytosolic, deglycosylated intermediary. J Biol Chem. 1998;273:29873–29878. doi: 10.1074/jbc.273.45.29873. [DOI] [PubMed] [Google Scholar]
- Becker F, Block-Alper L, Nakamura G, Harada J, Wittrup KD, Meyer DI. Expression of the 180-kD ribosome receptor induces membrane proliferation and increased secretory activity in yeast. J Cell Biol. 1999;146:273–284. doi: 10.1083/jcb.146.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J, Walter W, Yan W, Craig EA. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol Cell Biol. 1996;16:4378–4386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercovich B, Stancovski I, Mayer A, Blumenfeld N, Laszlo A, Schwartz AL, Ciechanover A. Ubiquitin-dependent degradation of certain protein substrates in vitro requires the molecular chaperones Hsc70. J Biol Chem. 1997;272:9002–9010. doi: 10.1074/jbc.272.14.9002. [DOI] [PubMed] [Google Scholar]
- Biederer T, Volkwein C, Sommer T. Degradation of subunits of the Sec61p complex, an integral component of the ER membrane, by the ubiquitin-proteasome pathway. EMBO J. 1996;15:2069–2076. [PMC free article] [PubMed] [Google Scholar]
- Biederer T, Volkwein C, Sommer T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science. 1997;278:1806–1809. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- Bonafacino JS, Weissman AM. Ubiquitin and the control of protein fate in the secretory and endocytic compartments. Annu Rev Cell Dev Biol. 1998;14:19–57. doi: 10.1146/annurev.cellbio.14.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorstein WR, Ziegelhoffer T, Craig CA. Molecular evolution of the HSP70 multigene family. J Mol Evol. 1994;38:1–17. doi: 10.1007/BF00175490. [DOI] [PubMed] [Google Scholar]
- Bordallo J, Plemper RK, Finger A, Wolf DH. Der3p-Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol Biol Cell. 1998;9:209–222. doi: 10.1091/mbc.9.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Lawrence JG, Caplan AJ. Mutations in the cytosolic DnaJ homologue YDJ1 delay and compromise the efficient translation of heterologous proteins in yeast. Biochemistry. 1998;37:18045–18055. doi: 10.1021/bi980900g. [DOI] [PubMed] [Google Scholar]
- Brodsky JL, McCracken AA. ER protein quality control and proteasome-mediated protein degradation. Semin Cell Dev Biol. 1999;10:507–513. doi: 10.1006/scdb.1999.0321. [DOI] [PubMed] [Google Scholar]
- Brodsky JL, Schekman R. A Sec63p-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J Cell Biol. 1993;123:1355–1363. doi: 10.1083/jcb.123.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Werner ED, Dubas ED, Goeckeler JL, Kruse KB, McCracken AA. The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation. demonstrates that protein export and import are mechanistically distinct. J Biol Chem. 1999;274:3453–3460. doi: 10.1074/jbc.274.6.3453. [DOI] [PubMed] [Google Scholar]
- Chen EYJ, Bartlett MC, Clarke DM. Cystic fibrosis transmembrane conductance regulator has an altered structure when its maturation is inhibited. Biochemistry. 2000;39:3797–3803. doi: 10.1021/bi992620m. [DOI] [PubMed] [Google Scholar]
- Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT alpha 2 repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- Cheng SH, Gregory RJ, Marshall J, Paul S, Souza DW, White GA, O'Riordan CR, Smith AE. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell. 1990;63:827–834. doi: 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Cox JS, Chapman RE, Walter P. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol Biol Cell. 1997;8:1805–1814. doi: 10.1091/mbc.8.9.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr DM, Lu X, Douglas MG. Regulation of Hsp70 function by a eukaryotic DnaJ homolog. J Biol Chem. 1992;267:20927–20931. [PubMed] [Google Scholar]
- Denning GM, Anderson MP, Amara JF, Marshall J, Smith AE, Welsh MJ. Processing of mutant cystic fibrosis trnasmembrane conductance regulator is temperature-sensitive. Nature. 1992;358:761–764. doi: 10.1038/358761a0. [DOI] [PubMed] [Google Scholar]
- de Virgilio M, Kitzmuller C, Schwaiger E, Klein M, Kreibich G, Ivessa NE. Degradation of a short-lived glycoprotein from the lumen of the endoplasmic reticulum: the role of N-linked glycans and the unfolded protein response. Mol Biol Cell. 1999;10:4059–4073. doi: 10.1091/mbc.10.12.4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayadat L, Siffroi-Fernandez S, Lanet J, Franc JL. Degradation of human thyroperoxidase in the endoplasmic reticulum involves two different pathways depending on the folding state of the protein. J Biol Chem. 2000;275:15948–15954. doi: 10.1074/jbc.M905763199. [DOI] [PubMed] [Google Scholar]
- Fisher EA, Zhou M, Mitchell DM, Wu X, Omura S, Wang H, Goldberg AL, Ginsberg HN. The degradation of apolipoprotein B100 is mediated by the ubiquitin-proteasome pathway and involves heat shock protein 70. J Biol Chem. 1997;272:20427–20434. doi: 10.1074/jbc.272.33.20427. [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner AM, Aviel S, Argon Y. Rapid degradation of an unassembled immunoglobulin light chain is mediated by a serine protease and occurs in a pre-Golgi compartment. J Biol Chem. 1993;268:25940–25947. [PubMed] [Google Scholar]
- Gardner RG, Swarbrick GM, Bays NW, Cronin SR, Wilhovsky S, Seelig L, Kim C, Hampton RY. Endoplasmic reticulum degradation requires lumen to cytosol signaling: transmembrane control of Hrd1p by Hrd3p. J Cell Biol. 2000;151:69–82. doi: 10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething MJ. Mammalian BiP. In: Gething MJ, editor. Guidebook to Molecular Chaperones and Protein-folding Catalysts. Oxford, UK: Oxford University Press; 1997. [Google Scholar]
- Gillece P, Luz JM, Lennarz W J, de la Cruz F J, Römisch K. Export of a cysteine-free misfolded secretory protein from the endoplasmic reticulum for degradation requires interaction with protein disulfide isomerase. J Cell Biol. 1999;147:1443–1456. doi: 10.1083/jcb.147.7.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamman BD, Hendershot LM, Johnson AE. BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocation pore before and early in translocation. Cell. 1998;92:747–758. doi: 10.1016/s0092-8674(00)81403-8. [DOI] [PubMed] [Google Scholar]
- Hampton RY, Bhakta H. Ubiquitin-mediated regulation of 3-hydroxy-3-methylglutaryl-CoA reductase. Proc Natl Acad Sci USA. 1997;94:12944–12948. doi: 10.1073/pnas.94.24.12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton RY, Gardner RG, Rine J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell. 1996;7:2029–2044. doi: 10.1091/mbc.7.12.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer W, Gruhler A, Mohrle V, Mahe Y, Wolf DH. PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chrymotryptic activity and degradation of ubiquitinated proteins. J Biol Chem. 1993;268:5115–5120. [PubMed] [Google Scholar]
- Heinemeyer W, Kleinschmidt JA, Saidowsky J, Escher C, Wolf DH. Proteinase yscE., the yeast proteasome/multicatalytic-multifunctional proteinase: mutants unravel its function in stress induced proteolysis and uncover its necessity for cell survival. EMBO J. 1991;10:555–562. doi: 10.1002/j.1460-2075.1991.tb07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K, Cooper AA. Degradation of unassembled Vph1p reveals novel aspects of the yeast ER quality control system. EMBO J. 2000;19:550–561. doi: 10.1093/emboj/19.4.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller MM, Finger A, Schweiger M, Wolf DH. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- Hong E, Davidson AR, Kaiser CA. A pathway for targeting soluble misfolded proteins to the yeast vacuole. J Cell Biol. 1996;135:623–633. doi: 10.1083/jcb.135.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Stroffekova K, Cuppoletti J, Mahanty SK, Scarborough GA. Functional expression of the cystic fibrosis transmembrane conductance regulator in yeast. Biochem Biophys Acta. 1996;1281:80–90. doi: 10.1016/0005-2736(96)00032-6. [DOI] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen TJ, Loo MA, Pind S, Williams DB, Goldberg AL, Riordan JR. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- Jiang C, Fang SL, Xiao YF, O'Connor SP, Nadler SG, Lee DW, Jefferson DM, Kaplan JM, Smith AE, Cheng SE. Partial restoration of cAMP-stimulated CFTR chloride channel activity in DeltaF508 cells by deoxyspergualin. Am J Physiol. 1998;275:C171–C178. doi: 10.1152/ajpcell.1998.275.1.C171. [DOI] [PubMed] [Google Scholar]
- Johnston AJ, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EW. Tackling the protease problem in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:428–454. doi: 10.1016/0076-6879(91)94034-a. [DOI] [PubMed] [Google Scholar]
- Kaiser CA, Schekman R. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 1990;18:723–733. doi: 10.1016/0092-8674(90)90483-u. [DOI] [PubMed] [Google Scholar]
- Knittler MR, Dirks S, Haas IG. Molecular chaperones involved in protein degradation in the endoplasmic reticulum: quantitative interaction of the heat shock cognate protein BiP with partially folded immunoglobulin light chains that are degraded in the endoplasmic reticulum. Proc Natl Acad Sci USA. 1995;92:1764–1768. doi: 10.1073/pnas.92.5.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le A, Steiner JL, Ferrell GA, Shaker JC, Sifers RN. Association between calnexin and a secretion-incompetent variant of human alpha 1-antitrypsin. J Biol Chem. 1994;268:7514–7519. [PubMed] [Google Scholar]
- Lee DH, Sherman MY, Goldberg AL. Involvement of the molecular chaperone Ydj1 in the ubiquitin-dependent degradation of short-lived and abnormal proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4773–4781. doi: 10.1128/mcb.16.9.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loayza D, Tam A, Schmidt WK, Michaelis S. Ste6p mutants defective in exit from the endoplasmic reticulum (ER) reveal aspects of an ER quality control pathway in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:2767–2784. doi: 10.1091/mbc.9.10.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo MA, Jensen TJ, Cui L, Hou Y, Chang XB, Riordan JR. Perturbation of Hsp90 interaction with nascent CFTR prevents its maturation and accelerates its degradation by the proteasome. EMBO J. 1998;17:6879–6887. doi: 10.1093/emboj/17.23.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs GL, Mohamed A, Kartner N, Chang XB, Riordan JR, Grinstein S. Conformational maturation of CFTR but not its mutant counterpart (ΔF508) occurs in the endoplasmic reticulum and requires ATP. EMBO J. 1994;13:6076–6086. doi: 10.1002/j.1460-2075.1994.tb06954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer TU, Braun T, Jentsch S. Role of the proteasome in membrane extraction of a short-lived ER transmembrane protein. EMBO J. 1998;17:3251–3257. doi: 10.1093/emboj/17.12.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken AA, Brodsky JL. Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. J Cell Biol. 1996;132:291–298. doi: 10.1083/jcb.132.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee TP, Cheng HH, Kumagai H, Omura S, Simoni RD. Degradation of 3-hydroxy-3-methylglutaryl-CoA reductase in endoplasmic reticulum membranes is accelerated as a result of increased susceptibility to proteolysis. J Biol Chem. 1996;271:25630–25638. doi: 10.1074/jbc.271.41.25630. [DOI] [PubMed] [Google Scholar]
- Meacham GC, Lu Z, King S, Sorscher E, Tousson A, Cyr DM. The Hdj-2/Hsc70 chaperone pair facilitates early steps in CFTR biogenesis. EMBO J. 1999;18:1492–1505. doi: 10.1093/emboj/18.6.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao B, Davis J, Craig EA. The Hsp70 family - an overview. In: Gething MJ, editor. Guidebook to Molecular Chaperones and Protein-folding Catalysts. Oxford, UK: Oxford University Press; 1997. pp. 3–13. [Google Scholar]
- Moriyama T, Sather SK, McGee TP, Simoni RD. Degradation of HMG-CoA reductase in vitro. Cleavage in the membrane domain by a membrane-bound cysteine protease. J Biol Chem. 1998;273:22037–22043. doi: 10.1074/jbc.273.34.22037. [DOI] [PubMed] [Google Scholar]
- Mullins C, Lu Y, Campbell A, Fang H, Green N. A mutation affecting signal peptidase inhibits degradation of an abnormal membrane protein in Saccharomyces cerevisiae. J Biol Chem. 1995;270:17139–17147. doi: 10.1074/jbc.270.29.17139. [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Hirata A, Nakano A. Inhibition of endoplasmic reticulum (ER)-to-Golgi transport induces relocalization of binding protein (BiP) within the ER to form the BiP bodies. Mol Biol Cell. 1994;5:1129–1143. doi: 10.1091/mbc.5.10.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DTW, Spear ED, Walter P. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J Cell Biol. 2000;150:77–88. doi: 10.1083/jcb.150.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlati F, Dominguez M, Bergeron JJ, Thomas DY. Saccharomyces cerevisiae CNE1 encodes an endoplasmic reticulum (ER) membrane protein with sequence similarity to calnexin and calreticulin and functions as a constituent of the ER quality control apparatus. J Biol Chem. 1995;270:244–253. doi: 10.1074/jbc.270.1.244. [DOI] [PubMed] [Google Scholar]
- Pfund C, Lopez-Hoyo N, Ziegelhoffer T, Schilke BA, Lopez-Buesa P, Walter WA, Wiedmann M, Craig EA. The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome-nascent chain complex. EMBO J. 1998;17:3981–3989. doi: 10.1093/emboj/17.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon M, Schekman R, Römisch K. Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J. 1997;16:4540–4548. doi: 10.1093/emboj/16.15.4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping S, Riordan JR, Williams DB. Participation of the endoplasmic reticulum chaperones calnexin (p88, IP90) in the biogenesis of the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 1994;269:12784–12788. [PubMed] [Google Scholar]
- Plemper RK, Bohmler S, Bordallo J, Sommer T, Wolf DH. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature. 1997;388:891–895. doi: 10.1038/42276. [DOI] [PubMed] [Google Scholar]
- Plemper RK, Daek PM, Otto RT, Wolf DH. Re-entering the translocon from the lumenal side of the endoplasmic reticulum. Studies on mutated carboxypeptidase yscY species. FEBS Lett. 1999;443:241–245. doi: 10.1016/s0014-5793(98)01724-4. [DOI] [PubMed] [Google Scholar]
- Plemper RK, Egner R, Kuchler K, Wolf DH. Endoplasmic reticulum degradation of a mutated ATP-binding cassette transporter Pdr5 proceeds in a concerted action of Sec61 and the proteasome. J Biol Chem. 1998;273:32848–32856. doi: 10.1074/jbc.273.49.32848. [DOI] [PubMed] [Google Scholar]
- Plemper RK, Wolf DH. Retrograde protein translocation: ERADication of secretory proteins in health and disease. Trends Biochem Sci. 1999;24:266–270. doi: 10.1016/s0968-0004(99)01420-6. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Preston RA, Adams AE, Stearns T, Drubin DG, Haarer BK, Jones EW. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Qu D, Teckman JH, Omura S, Perlmutter DH. Degradation of a mutant secretory protein, alpha1-antitrypsin Z, in the endoplasmic reticulum requires proteasome activity. J Biol Chem. 1996;271:22791–22795. doi: 10.1074/jbc.271.37.22791. [DOI] [PubMed] [Google Scholar]
- Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielinski J, Lok S, Plavsic N, Chou JL, Drumm ML, Iamuzzi MC, Collin FS, Tsui LC. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- Römisch K. Surfing the Sec61 channel: bidirectional protein translocation across the ER membrane. J Cell Sci. 1999;112:4185–4191. doi: 10.1242/jcs.112.23.4185. [DOI] [PubMed] [Google Scholar]
- Rubenstein RC, Zeitlin PL. Sodium 4-phenylbutyrate downregulates Hsc70: implications for intracellular trafficking of ΔF508-CFTR. Am J Physiol Cell Physiol. 2000;278:C259–C267. doi: 10.1152/ajpcell.2000.278.2.C259. [DOI] [PubMed] [Google Scholar]
- Sawa T, Imamura T, Haruta T, Sasaoka T, Ishiki M, Takata Y, Morioka H, Ishihara H, Usui I, Kobayashi M. Hsp70 family molecular chaperones and mutant insulin receptor: differential binding specificities of BiP. and Hsp70/Hsc70 determines accumulation or degradation of insulin receptor. Biochem Biophys Res Commun. 1996;218:449–453. doi: 10.1006/bbrc.1996.0080. [DOI] [PubMed] [Google Scholar]
- Schmitz A, Maintz M, Kehle T, Herzog V. In vivo iodination of a misfolded proinsulin reveals co-localized signals for BiP binding and for degradation in the ER. EMBO J. 1995;14:1091–1098. doi: 10.1002/j.1460-2075.1995.tb07092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling CJ, Rothblatt J, Hosobuchi M, Deshaies R, Schekman R. Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol Biol Cell. 1992;3:129–142. doi: 10.1091/mbc.3.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland E, Qu BH, Millen L, Thomas PJ. The molecular chaperone Hsc70 assists the in vitro folding of the N-terminal nucleotide-binding domain of the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 1997;272:25421–25424. doi: 10.1074/jbc.272.41.25421. [DOI] [PubMed] [Google Scholar]
- Umebayashi K, Hirata A, Fukuda R, Horiuchi H, Ohta A, Takagi M. Accumulation of misfolded protein aggregates leads to the formation of russell body-like dilated endoplasmic reticulum in yeast. Yeast. 1997;13:1009–1020. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1009::AID-YEA157>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- van Oene M, Lukacs GL, Rommens JM. Cystic fibrosis mutations lead to carboxyl-terminal fragments that highlight an early biogenesis step of the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 2000;275:19577–19584. doi: 10.1074/jbc.M002186200. [DOI] [PubMed] [Google Scholar]
- Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- Werner ED, Brodsky JL, McCracken AA. Proteasome-dependent endoplasmic reticulum-associated protein degradation: an unconventional route to a familiar fate. Proc Natl Acad Sci USA. 1996;93:13797–13801. doi: 10.1073/pnas.93.24.13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Washburne M, Stone DE, Craig EA. Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2568–2577. doi: 10.1128/mcb.7.7.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiertz EJ, Tortorella D, Bogyo M, Yu J, Mothes W, Jones TR, Rapoport TA, Ploegh HL. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature. 1996;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- Wigley WC, Fabunmi RP, Lee MG, Marino CR, Muallem S, DeMartino GN, Thomas PJ. Dynamic association of proteasomal machinery with the centrosome. J Cell Biol. 1999;145:481–490. doi: 10.1083/jcb.145.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikstrom L, Lodish HF. Endoplasmic reticulum degradation of a subunit of the asialoglycoprotein receptor in vitro. Vesicular transport from endoplasmic reticulum is unnecessary. J Biol Chem. 1992;267:5–8. [PubMed] [Google Scholar]
- Wileman T, Kane LP, Terhorst C. Degradation of T-cell receptor chains in the endoplasmic reticulum is inhibited by inhibitors of cysteine proteases. Cell Regul. 1991;2:753–765. doi: 10.1091/mbc.2.9.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhovsky S, Gardner R, Hampton R. HRD gene dependence of endoplasmic reticulum associated degradation. Mol Biol Cell. 2000;11:1697–1708. doi: 10.1091/mbc.11.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R, Basson M, D'Ari L, Rine J. Increased amounts of HMG-CoA reductase induce “karmellae”, a proliferation of stacked membrane pairs surrounding the yeast nucleus. J Cell Biol. 1988;107:101–114. doi: 10.1083/jcb.107.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Chong E, Skach WR. Evidence that endoplasmic reticulum (ER)-associated degradation of cystic fibrosis transmembrane conductance regulator is linked to retrograde translocation from the ER membrane. J Biol Chem. 1999;274:2616–2624. doi: 10.1074/jbc.274.5.2616. [DOI] [PubMed] [Google Scholar]
- Yang Y, Janich S, Cohn JA, Wilson JM. The common variant of cystic fibrosis transmembrane conductance regulator is recognized by hsp70 and degraded in a pre-Golgi nonlysosomal compartment. Proc Natl Acad Sci USA. 1993;90:9480–9484. doi: 10.1073/pnas.90.20.9480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Kaung G, Kobayashi S, Kopito RR. Cytosolic degradation of T-cell receptor alpha chains by the proteasome. J Biol Chem. 1997;272:20800–20804. doi: 10.1074/jbc.272.33.20800. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Michaelis S, Brodsky JL. CFTR expression and ER associated degradation in yeast. In: Skach WR, editor. Cystic Fibrosis Methods and Protocols, Methods in Molecular Medicine. Totowa, NJ: Humana Press; 2001. (in press). [DOI] [PubMed] [Google Scholar]
- Zhou M, Fisher EA, Ginsberg HN. Regulated co-translational ubiquitination of Apolipoprotein B100. J Biol Chem. 1998;273:24649–24653. doi: 10.1074/jbc.273.38.24649. [DOI] [PubMed] [Google Scholar]