Abstract
Due to advances in high-throughput biotechnologies biological information is being collected in databases at an amazing rate, requiring novel computational approaches that process collected data into new knowledge in a timely manner. In this study, we propose a computational framework for discovering modular structure, relationships and regularities in complex data. The framework utilizes a semantic-preserving vocabulary to convert records of biological annotations of an object, such as an organism, gene, chemical or sequence, into networks (Anets) of the associated annotations. An association between a pair of annotations in an Anet is determined by the similarity of their co-occurrence pattern with all other annotations in the data. This feature captures associations between annotations that do not necessarily co-occur with each other and facilitates discovery of the most significant relationships in the collected data through clustering and visualization of the Anet. To demonstrate this approach, we applied the framework to the analysis of metadata from the Genomes OnLine Database and produced a biological map of sequenced prokaryotic organisms with three major clusters of metadata that represent pathogens, environmental isolates and plant symbionts.
INTRODUCTION
In many branches of scientific information is collected in tables, forms or questionnaires. Most biological databases, for example, accumulate knowledge by annotating or curating different biological objects or their relationships (1). This information includes, but is not limited to, characteristics of sequenced genomes (2), genes (3–5), chemicals (6,7) and enzymes/metabolic pathways (8–10). With advances in high-throughput sequencing and omics technologies, the number of such resources is growing at an unprecedented rate (11–13). To facilitate their usage, a dedicated academic journal that introduces their description (14) and even a new resource, BioDBCore, to collect attributes of the databases, has emerged (15). While databases help scientists to gather and integrate massive amounts of information by downloading various types of data, the task of identifying hidden regularities in the data is left open (16). For this reason, computational approaches that sift non-spurious associations hidden in large and complex data and discover clusters of these annotations are needed.
One known approach to mining associations in large data sets is association rule (Arule) learning (17). This algorithm was initially designed to find frequently associated products in supermarket-sale data to understand consumer purchasing behaviors. Recently, the technique was applied to mine biological associations: to identify a predictive combinations of genes in the genotype–phenotype relationships (18), to discover adjacent amino acids on a binding site of a protein complex (19), to analyze disordered proteins in prokaryotes (20) and to extract combinations of gene annotations from a list of over-expressed genes (20,21). Association rule learning, however, has serious drawbacks for extracting hidden regularities among biological annotations. First, it generates a large number of spurious rules that are largely redundant. These rules are not easy to use for further analysis, and they are difficult to filter, cluster and visualize. Secondly, association rule learning captures associations between annotations only when they directly co-occur in the data. Consequently, all indirect associations that may underlie important regularities are lost. Thirdly, since the algorithm is blind to the semantic structure of data, the produced rules do not reflect the initial hierarchy or type of each annotation, it makes the results difficult to interpret and cluster.
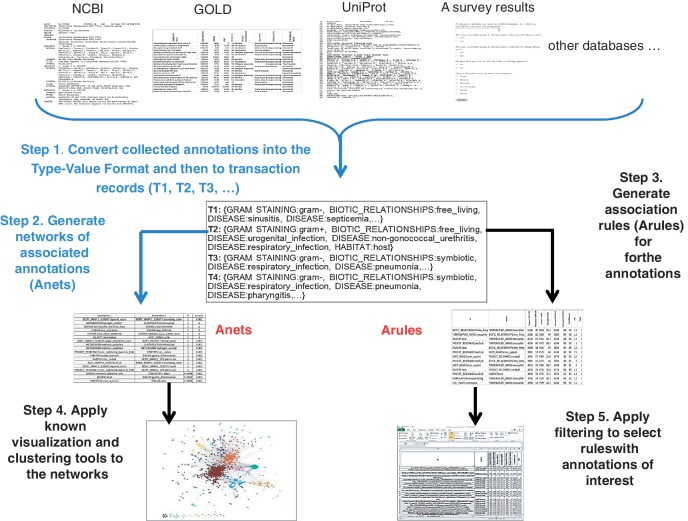
This paper introduces a computational framework (Figure 1) that embraces both classical association rule learning and a novel approach to identify indirect associations and hidden biological regularities within a large data set. We address drawbacks discussed above by introducing two new concepts, the type-value format of biological annotations and the association network (Anet). The type-value format is a flattened representation of a controlled vocabulary. It helps to restore important semantic relationships of the annotations after their processing by computational algorithms. This format simplifies filtering and grouping of annotations in Anets and in Arules. An association in an Anet is computed by considering both direct and indirect associations in the data. As in other biological network representations, an Anet allows researchers to engage network analysis tools including various types of clustering (22–25) and visualization (26,27) techniques. In addition, since in each result from classical Arule learning and Anets, an association retains annotation hierarchies, the analysis of subsequently inferred knowledge (e.g. biological groups or clusters) is greatly enhanced. In this paper, we apply this framework to the data collected in the Genomes OnLine Database (GOLD) (2) and present the analysis procedures and biological regularities inferred from the data.
Figure 1.
Computational framework for analysis of annotations collected in biological databases. Steps 1 and 2 (blue) are described in the text in more detail. Step 3 uses a classic ‘Apriori’ algorithm for learning Arules from the type-value formatted transactions. Step 4 employs known visualization and clustering tools to analyze the generated Anets. Step 5 uses filtering tools available in spreadsheet applications.
MATERIALS AND METHODS
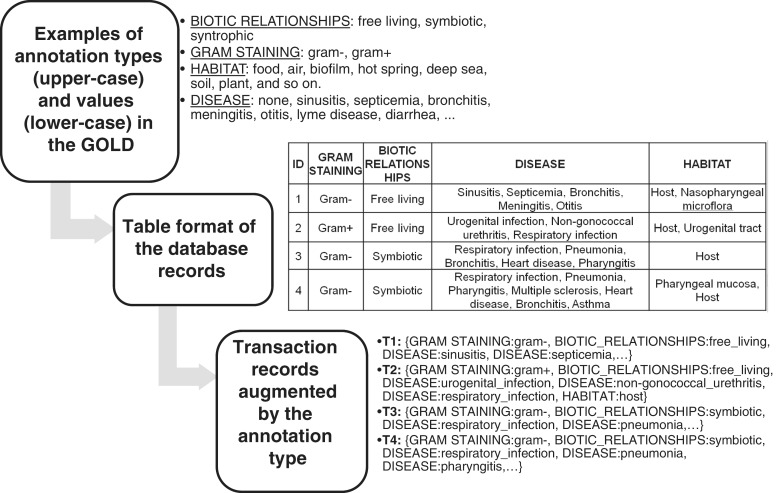
The proposed algorithm to convert a data table with annotations into type-value transaction records (Step 1 in Figure 1) was implemented as a Perl program called ‘t2t.pl’. The novel algorithm for generation of Anets (Step 2 in Figure 1) was implemented in C++ as a program called ‘anet’. Both programs and their documentation are available for download at http://sourceforge.net/projects/anets. The programs were applied to process a set of annotations provided as metadata by the GOLD research team in a table format (Figure 2). GOLD is a comprehensive resource of biological annotations for sequenced bacterial and archaeal organisms (2). On the date of this analysis (March 17, 2011), it included 7331 prokaryotic genomes (rows) with each genome annotated by 105 features or types (columns). The table included numerous annotations represented phylogenetic information, sequencing project information, phenotypic features of the organisms and their general environmental characteristics. Although the metadata are not meant to represent well-developed ontologies, we found that most annotation types (columns) in the metadata are based on a controlled vocabulary so that an annotation of a certain type can be easily converted into the type-value format. For this study, we selected 26 features or annotation types reflecting (i) phenotypic, phylogenetic and genomic features of the organisms, such as gram-staining, phenotypes, oxygen requirement, salinity tolerance, sporulation, metabolic features, motility, cell shape, arrangement, temperature range, genome size and GC content, and classifications of the organism at the level of phylum; (ii) general environmental characteristics, such as biotic and symbiotic relationships, habitat, associated hosts and diseases; and (iii) classification in terms of practical relevance of the project (human pathogen, plant pathogen, bioremediation, agricultural and others). Sets of quantitative values representing a range, for example the GC contents or genome size, were mapped into three discrete levels: ‘low’ (‘small’), ‘medium’ and ‘high’ (‘large’), respectively (Supplementary Figure S1). The resulting data set of annotations was then converted into type-value formatted transactions and used to produce Anets and Arules.
Figure 2.
An example of the conversion of annotation records given as a table into type-value formatted transactions using 4 truncated (4 columns only) database records in the GOLD. Each row in the table provides metadata for a sequenced organism, and each column groups the metadata by the type.
The type-value formatted transactions produced by ‘t2t.pl’ for the GOLD data set were then used as an input for the program ‘anet’ to generate the Anets. The data set was used to evaluate three measures of similarity when generating Anets: Pearson correlation, Spearman's rank correlation coefficient and Jaccard coefficient (or cosine). We also tested a normalization of the support profile by dividing each support value in the profile of an annotation by the total number of database records with the annotation. We found no significant difference in the resulting biological inferences in the case study. The generated Anet (Supplementary Data S1) was further analyzed using Markov clustering algorithm (22) (Supplementary Table S1) and visualized using Cytoscape (26).
Arules (Step 3 in Figure 1) were produced by applying ‘Apriori’ (28) to type-value formatted transactions generated by ‘t2t.pl’ for the GOLD data set. Each Arule is interpreted as an ‘if-then’ statement with the confidence, support and a set of auxiliary statistics provided in Supplementary Data S2 and Supplementary Table S2. An example of the statement from the GOLD is: if ‘GRAM_STAINING:gram+|SIZE(KB):large|MOTILITY:nonmotile’, then ‘GC_CONTENT:high’. This Arule means that if three annotations of a bacterium in the ‘if’ part of the Arule co-occur then it is frequently annotated by the annotation given in the ‘then’ part of the Arule. The support for an Arule is a probability that a randomly selected record in the database will contain annotation values from both parts of the Arule. The confidence is a conditional probability that a randomly selected record of a bacterial organism in the GOLD will have the annotation from ‘then’ part of the Arule given that the record has all annotations from ‘if’ part of the Arule.
RESULTS
Type-value format for biological annotations
To simplify computational processing, filtering and grouping of biological annotations in a data set, we convert them into a list of transaction-like records (Figure 2). A transaction is a list of items selected together. A typical example is a list of items bought by a customer on a single purchase. A traditional transaction record, however, does not associate items with their types, e.g. dairy products or bakery products. In our study, a record has the same format as a transaction used in conventional association rule learning (28), but each item is also supplemented with a prefix describing a more general level of the conceptual hierarchy inferred from the database. In other words, a transaction record is not a list of appearances of annotations only, but a list of composite information; an annotation with its associated class or type. A need of the more structure of the transaction stems from two-level organization of information in many biological databases where each annotation is usually based on a controlled vocabulary and includes not only annotations but also their types. Each type of annotation has its own set of allowed annotation items, terms or values. Information in survey forms, questionnaires, tables and many biological databases, including GenBank (5), UniProt (4), MetaCyc (10), KEGG (8), also has a controlled vocabulary and a similar two-level structure (type-value). We supplement each annotation in the transaction records by its type and in this way preserve the two-level structure of biological information in the generated networks and the Arules. Figure 2 demonstrates how database records with annotation values given in a table are converted to transactions augmented by the annotation type using an example from the GOLD.
Association network
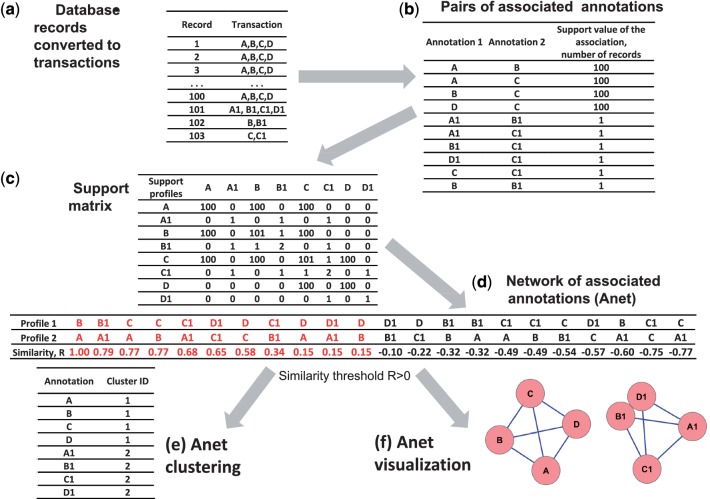
To represent annotations collected in the database as a network of their association, or Anet, we analyze each pair-wise association among all unique annotations in the database. Both direct and indirect associations are found to be important in this research. In the genome-wide association studies (GWAS) databases (29), for example, two phenotypes do not always coincide, or associate directly, in any single transaction, but they may be linked indirectly by a set of single nucleotide polymorphisms (SNPs) reported in the same set of genes. To capture not only direct associations but also indirect associations between annotations, we compute an association of two annotations by calculating a correlation between their co-occurrence profiles, that is, their co-occurrences with all other annotations in the data. In this fashion, both direct and indirect co-occurrences are considered in the computation (Figure 3). More specifically, suppose that we have found n annotations {A1,…,An}, where each annotation Ai co-occurs with one or more other annotations. We characterize such a direct association between two annotations Ai and Aj by a support value Aij. If Ai and Aj co-occur then Aij is equal to the number of records in the database where Ai and Aj co-occur; otherwise the support value Aij is zero. The support value of the annotation with itself, Aii, is equal to the number of records in the database that include annotation Ai. A matrix comprised of all support values {Aij}, where I = 1,…,n, j = 1,…,n, is referred as a support matrix; and Aii denotes the entry at the i-th row and the j-th column in the matrix. We define a ‘support profile’ of an annotation Ai as a vector of support values for pairwise associations that include Ai (the association with itself is also included). A support profile, therefore, is just the row i of the support matrix Aij. Similarity between two annotations Ai and Aj is estimated by similarity of their profiles, or a pair of corresponding rows from the support matrix Aij, using a similarity measure, such as Pearson correlation coefficient, Spearman's rank correlation coefficient or Jaccard coefficient. The resulting pairs of annotations, along with the value of similarity of their support profiles, represent a weighted network, Anet.
Figure 3.
A workflow for revealing associated annotations in the database using an example of 103 database records converted to transactions (a). The algorithm includes (b) calculation of support values for each pairs of unique annotations in the database (associations with 0 support values are not shown); (c) transformation of all support values into a support matrix with each row/column representing a support profile of an annotation; (d) generation of the Anet using Pearson correlation coefficient (R) as the similarity measure for each pair of profiles, (e) clustering of the Anet using a threshold for the correlation coefficient and (f) the Anet visualization.
Figure 3 gives a simple example of an Anet built from 103 database records with 8 unique annotations (A, B, C, D, A1, B1, C1 and D1). The records are constructed to present two communities, ABCD (supported by 100 records) and A1B1C1D1 (supported by 1 record), intersected in two records: one record with annotations B and B1 and one record with annotations C and C1. The example demonstrates how the Anet helps to identify the communities by considering the similarity of the support profiles instead of the direct co-occurrences of the annotations. The threshold value for the profiles similarity measured by the Pearson correlation was set to 0 so only pairs of annotations with a positive similarity value are included in the Anet. In the example, although two pairs of annotations, B and B1 and A1 and B1, each are supported by one database record, the significance of their associations computed in terms of the support profiles are very different because of indirect associations. As a result, while annotations A1 and B1 associate significantly with the similarity value R = 0.79, annotations B and B1 do not associate (R = −0.32), and this is not included in the Anet. The same is true for annotations C and C1.
Setting Anet resolution using a Monte Carlo simulation
We set the level of resolution for an Anet from the statistical significance of similarity between support profiles of the annotations. To assess this significance, we calculated the P-value of a similarity score using a Monte Carlo simulation approach (Supplementary Figure S2) (30). The P-value was calculated by randomly selecting two annotations Ai and Aj from a set of co-occurring annotations {A1,…,An}, extracting the support profile for each of them from the support matrix, and then calculating a similarity measure of the profiles. These calculations were repeated for 10 000 random pairs of annotations. The P value for a given value of the similarity measure was then calculated as the fraction of the random pairs with the value of similarity greater than the given value. By setting a threshold for the P-value, we limit the number of pairs of associated annotations and generate a network of desired granularity and resolution.
Applying the framework
We applied the framework to analyze metadata (1176 unique annotation values classified into 26 types) from 7017 prokaryotic genomes. We used the annotations in the GOLD that were available in a table format as described in Figure 2. Since many different annotation types and their values often co-occur in metadata from an organism, or in one row of the table, the structure of the collected information is complex, with pervasive connections among the annotation values. This complicates the discovery of regularities in the data. On the other hand, the complex structure of the data provides a good case study for the proposed framework. Here, our goal was to uncover modular structures and general regularities underlying inter-relationships among phenotypic, genomic and environmental characteristics of sequenced prokaryotes and then to explore individual relationships among specific annotations using Arules.
The Anets were produced at two different levels of granularity, P-values of 0.05 and 0.01 (Figure 4, Supplementary Dataset S1). The numbers of vertices (annotations) and edges (associations) are 1136 and 34 545, and 949 and 6944, respectively. About half of the identified associations in the Anets are indirect, i.e. between annotations that do not co-occur directly in any record of the database. We find that 55% (19 015 out of 34 545) and 50% (3467 out of 6744) of associations are indirect in the Anets constructed with P-values 0.05 and 0.01, respectively. Most indirect associations were found to connect annotations of a small number of closely related organisms of the same genus but annotated with different diseases or isolated from different hosts. In the latter case, the related organisms have similar or even identical annotations except the host name. Therefore, it is logical to consider the different host names annotated for the related genomes, even if they never belong to one record, as associated annotations. Another example includes the annotations ‘HABITAT:oral_microflora’ (99 organisms) and ‘DISEASE:opportunistic_infection’ (97 organisms). They never co-occur, but have similar co-occurrence profiles, thus were found associated with high significance (P = 0.0098).
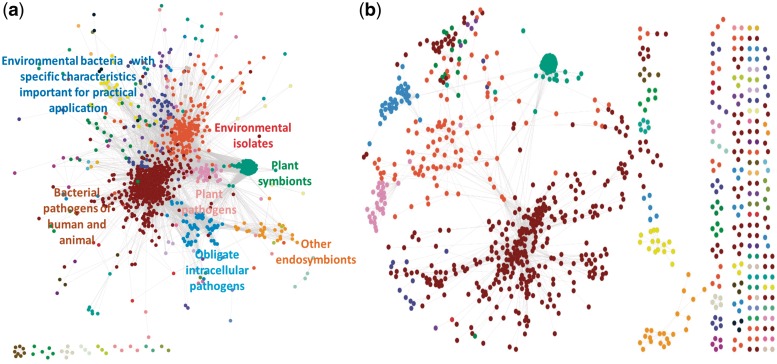
Figure 4.
Biological maps of sequenced prokaryotic organisms based on their metadata collected in the GOLD. The maps are based on the Anet (Supplementary Dataset S1) generated from the metadata using Pearson correlation as the similarity measure, and two P-value thresholds: 0.05 (a) and 0.01 (b). The maps link environmental, physiological, genomic and phenotypic characteristics based on similarity of profiles of their co-occurrences in the sequenced prokaryotic organisms and reveal similar communities/clusters of the annotations (Supplementary Table S1) indicated by color. Names of seven most populated clusters were assigned by manual curation of ‘PROJECT_RELEVANCE’ annotations within each cluster.
Since the Anets included not only direct but also indirect associations, they provide clusters that cannot be identified if only direct associations are considered. To empirically validate this, we constructed the equivalent network using only direct associations. The network was generated using Arules with two annotations (Type to be equal 1 in Supplementary Dataset S2). The network is found to be very dense, from which we could not find any distinct cluster (Supplementary Figure S3). On the other hand, we were able to find distinct biologically meaningful clusters of annotations (Figure 4) from the Anets using both direct and indirect associations. In both cases, we applied the same clustering algorithm and visualized the clustering results using the same Cytoscape layout. We conclude that the Anet not only incorporated indirect associations but also removed insignificant direct associations by the new similarity measure and the statistical significance test.
Post-analysis of the Anets shows that similar clusters of annotations are identified even when Anets are generated at different levels of resolution (different P-value cutoffs). The full list of identified clusters is available in Supplementary Table S1. Annotations in large clusters suggest a strong connection between phenotypic, genomic and environmental characteristics of the organism on one hand and the relevance of the organism to human needs on the other. The type-value format of the annotations enabled the discovery of these connections. Annotations in an individual cluster or across multiple clusters are easily aligned by their types making relationships among annotations are readily interpreted. The relevance of organisms to human needs, for example, was identified by values of the annotation type ‘PROJECT_RELEVANCE’ and phenotypic characteristics of organisms by values of types ‘METABOLISM’, ‘OXYGEN_REQUIREMENT’, ‘SYMBIOTIC_RELATIONSHIP’ and ‘TEMPERATURE_RANGE’. Annotation types also play an important role in defining signature characteristics of the bacterial pathogens. The type ‘PROJECT_RELEVANCE’ found in this cluster includes pathogen related values such as ‘animal pathogen’, ‘human pathogen’, ‘medical’ and ‘dental pathogen’. Likewise three annotation types ‘DISEASE’, ‘HOST_NAME’ and ‘HABITAT’ are also found to extract characteristics of pathogens. A close investigation of the cluster also reveals that a pathogen may have a few limited cell shapes and arrangements and may be characterized in general as a non-sporulating free living mesophile with facultative, aerobic or anaerobic respiration and of low or medium GC content in the genome.
The second largest cluster represents characteristics of environmental isolates that are reflected in ‘PROJECT_RELEVANCE’ with 36 annotation values of ‘environmental’, ‘evolutionary’, ‘bioremediation’, ‘ecological’ and ‘carbon cycle’. The other annotations in the cluster include a diverse set of different environmental habitats (36 annotations), metabolic activities (56 annotations) and phylogenetic groups (17 annotations), along with such characteristics as obligate aerobic respiration and high genomic GC content. The third largest cluster of annotations represents characteristics of plant symbionts isolated from diverse plant hosts (88 annotations), roots and root nodules, and characterized by nitrogen fixing metabolic activity. Four other large clusters denote obligate intracellular pathogens, mainly from the phylum Chlamydiae; plant pathogens; environmental bacteria with specific characteristics important for practical applications; and other endosymbionts, such as symbionts of insects and nematodes.
Using Arules to find frequently co-occurred annotations and to examine regularities inferred from Anets
Discovering regularities from Arules is rather challenging. A key characteristic of Arules is redundancy. Lower order rules (rules with smaller number of items) are largely subsumed by higher order rules (rules with larger number of items). As a result, the number of generated Arules is usually huge requiring methods to select the most important or interesting Arules. Clusters of annotations produced from an Anet can provide the necessary guidance for such selection. These clusters contain annotations with significantly correlated support profiles and, therefore, more likely represent important regularities hidden in the data. Selection of Arules for clustered annotations can also provide comprehensive statistics on how frequently the annotations associate directly and, thus, supplement the information revealed by Anet with additional evidence of a direct association.
We decided to use Arules to further investigate two interesting regularities discovered in two major clusters: an association of high genomic GC content with annotations of environmental isolates and medium and low GC content with annotations of pathogens. For example, in each of the clusters, the type PROJECT_RELAVENCE is found but with different values. While ‘GC_CONTENT:low’ and ‘PROJECT_RELEVANCE:human_pathogen’ belong to cluster pathogen, ‘GC_CONTENT:high’ and non-human pathogen related values such as ‘PROJECT_RELEVANCE:biotechnological’ belong to cluster environmental isolates (Figure 4 and Supplementary Table S1). For the analysis, we gathered the statistics of 102 381 Arules, where each rule is of at least 80% confidence and the support value is of 0.05%, which amounts to at least four database records (Supplementary Dataset S2). We then selected Arules that contain ‘GC_CONTENT:low’ or ‘GC_CONTENT:high’ with the minimum support of 50 records (Supplementary Table S2). The resulting sets were 51 and 22 Arules for low and high GC content, respectively. Nine rules out of 51 for low GC content included ‘PROJECT_RELEVANCE:human_pathogen’, and 5 out of 22 rules for high GC content included ‘PROJECT_RELEVANCE: biotechnological’, or ‘agricultural’. None of the high GC content rules included ‘PROJECT_RELEVANCE:human_pathogen’, and none of the low GC content rules included ‘PROJECT_RELEVANCE:biotechnological’ or agricultural. Two other important associations found by Anet are between the type of cellular respiration and the GC content and between the genome size and the GC content. The associations are also confirmed in the Arules. The latter relationship between the genome size and GC content was also confirmed by computing the correlation between genome sizes in terms of kilo base pairs and GC content for complete prokaryotic genomes. We found a medium level of correlation R = 0.53 between these characteristics (Supplementary Figure S4).
We further analyzed relationships identified by Anets and Arules in the context of published observations on the genomic GC content in different organisms. Lower GC content in obligatory pathogens/symbionts, as well as in phages, plasmids and insertions elements, was described before and linked to the higher energy cost and limited availability of G and C over A and T/U (31). Associations of GC content with the type of cellular respiration and with genome size are also reported previously from an analysis of smaller sets of organisms (32–34). Our data generated, from a significantly greater number of organisms, show a similar trend. Importantly, our analysis associates high GC content, larger genome and obligate aerobic respiration with complex environmental habitats and with a diversity in metabolic activities and physiological characteristics of prokaryotic organisms.
DISCUSSION
Considering the amazing rate at which data are accumulated in natural and social sciences, new methods that process and interpret large and complex data are increasingly important. The proposed approach makes a step in this direction providing a way to transform a combination of numerical and nominal data collected in tables, survey forms, questionnaires or type-value annotation records into networks of associations. After the transformation, different statistical and algorithmic tools can be applied for further analysis and visualization of the data. The case study shows how the approach discovers hidden regularities in annotation data from bacterial genomes through the data transformation, computation of associations, clustering, statistical evaluation and visualization. The application domain of the proposed framework is not limited to biological data. It can, for example, be applied to approximate the meaning of texts documents, to analyze social communities, to visualize results of surveys and even to facilitate clustering of densely nested weighted networks. In the latter case, the nested network could be converted into a support matrix and then into Anets for further clustering and visualization (steps c, d, e and f in Figure 3).
Like with any statistical analysis, the proposed approach has some limitations. First, it cannot automatically generate a comprehensive output by processing a collection of type-value formatted annotation records with incorrect syntax or semantics. Syntactically, each record in the dataset must conform to the required format. Semantically, each record must include characterizations of the same object such as a protein, genome, gene or person. Furthermore, a proper selection of annotation types with controlled vocabularies that are independent and relevant to the goal of the analysis is required to produce meaningful results. In the GOLD study, for example, we had to exclude 78 annotation types that fail to meet the criteria. Also, we had to introduce two nominal ranges for two types, genome size and the GC content, which were relevant to our analysis. Another caveat is that the approach is blind to bias potentially inherent in the collected data. Such bias can affect regularities discovered by Anet. For example, due to the difficulty in sequencing and phenotypic characterization of non-cultured organisms, the analyzed GOLD data set is obviously dominated by cultured prokaryotes. Threshold parameters used to produce and to cluster Anets must also be carefully adjusted not only for a given data set but also for a chosen similarity measure. Recently developed novel clustering algorithms, like linkcomm (link communities) (23), and measures of similarity, like maximal information-based non-parametric exploration (MINE) statistics (35), may help to uncover a modular structures in the collected data and hidden regularities. Finally, it is important to note that the time required to process a data set is rather dependent on the number of unique annotations in the data, not simply the data volume.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–4 and Supplementary Datasets 1 and 2.
FUNDING
Genomic Science Program, U.S. Department of Energy, Office of Science, Biological and Environmental Research as part of the Plant Microbe Interfaces Scientific Focus Area and the BioEnergy Science Center. The BioEnergy Science Center is a U.S. Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science. Funding for open access charge: Office of Biological and Environmental Research in the DOE Office of Science; Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the U.S. Department of Energy under [contract DE-AC05-00OR22725].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Genomes OnLine Database research team for providing data for the study, J. S. Foster from the UT Medical Center for assistance in preparation the manuscript, and anonymous reviewers for helpful suggestions that allow us to improve the manuscript and software.
REFERENCES
- 1.Howe D, Costanzo M, Fey P, Gojobori T, Hannick L, Hide W, Hill DP, Kania R, Schaeffer M, St Pierre S, et al. Big data: the future of biocuration. Nature. 2008;455:47–50. doi: 10.1038/455047a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernal A, Ear U, Kyrpides N. Genomes OnLine Database (GOLD): a monitor of genome projects world-wide. Nucleic Acids Res. 2001;29:126–127. doi: 10.1093/nar/29.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Magrane M, Consortium U. UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011;2011:bar009. doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2011;39:D52–D57. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiegers TC, Davis AP, Cohen KB, Hirschman L, Mattingly CJ. Text mining and manual curation of chemical-gene-disease networks for the comparative toxicogenomics database (CTD) BMC Bioinformatics. 2009;10:326. doi: 10.1186/1471-2105-10-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis AP, King BL, Mockus S, Murphy CG, Saraceni-Richards C, Rosenstein M, Wiegers T, Mattingly CJ. The Comparative Toxicogenomics Database: update 2011. Nucleic Acids Res. 2011;39:D1067–D1072. doi: 10.1093/nar/gkq813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanehisa M. The KEGG database. Novartis Found Symp. 2002;247:91–101. discussion 101–103, 119–128, 244–152. [PubMed] [Google Scholar]
- 9.Schomburg I, Chang A, Ebeling C, Gremse M, Heldt C, Huhn G, Schomburg D. BRENDA, the enzyme database: updates and major new developments. Nucleic Acids Res. 2004;32:D431–D433. doi: 10.1093/nar/gkh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2010;38:D473–D479. doi: 10.1093/nar/gkp875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson AD, O'Donnell CJ. An open access database of genome-wide association results. BMC Med. Genet. 2009;10:6. doi: 10.1186/1471-2350-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forbes SA, Tang G, Bindal N, Bamford S, Dawson E, Cole C, Kok CY, Jia M, Ewing R, Menzies A, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38:D652–D657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, et al. NCBI GEO: archive for functional genomics data sets–10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landsman D, Gentleman R, Kelso J, Francis Ouellette BF. DATABASE: a new forum for biological databases and curation. Database (Oxford) 2009;2009:bap002. doi: 10.1093/database/bap002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaudet P, Bairoch A, Field D, Sansone SA, Taylor C, Attwood TK, Bateman A, Blake JA, Bult CJ, Cherry JM, et al. Towards BioDBcore: a community-defined information specification for biological databases. Database (Oxford) 2011;2011:baq027. doi: 10.1093/database/baq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Attwood TK, Kell DB, McDermott P, Marsh J, Pettifer SR, Thorne D. Calling International Rescue: knowledge lost in literature and data landslide! Biochem. J. 2009;424:317–333. doi: 10.1042/BJ20091474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agrawal R, Imieliński T, Swami A. ACM SIGMOD Proceedings of the 1993 ACM SIGMOD international conference on Management of data. Vol. 22. New York, NY, USA: ACM Press; 1993. Mining association rules between sets of items in large databases; pp. 207–216. [Google Scholar]
- 18.Tamura M, D'Haeseleer P. Microbial genotype-phenotype mapping by class association rule mining. Bioinformatics. 2008;24:1523–1529. doi: 10.1093/bioinformatics/btn210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo HC, Ong PL, Lin JC, Huang JP. Discovering amino acid patterns on binding sites in protein complexes. Bioinformation. 2011;6:10–14. doi: 10.6026/97320630006010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pavlovic-Lazetic GM, Mitic NS, Kovacevic JJ, Obradovic Z, Malkov SN, Beljanski MV. Bioinformatics analysis of disordered proteins in prokaryotes. BMC Bioinformatics. 2011;12:66. doi: 10.1186/1471-2105-12-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hackenberg M, Matthiesen R. Annotation-Modules: a tool for finding significant combinations of multisource annotations for gene lists. Bioinformatics. 2008;24:1386–1393. doi: 10.1093/bioinformatics/btn178. [DOI] [PubMed] [Google Scholar]
- 22.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalinka AT, Tomancak P. linkcomm: an R package for the generation, visualization, and analysis of link communities in networks of arbitrary size and type. Bioinformatics. 2011;27:2011–2012. doi: 10.1093/bioinformatics/btr311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahn YY, Bagrow JP, Lehmann S. Link communities reveal multiscale complexity in networks. Nature. 2010;466:761–764. doi: 10.1038/nature09182. [DOI] [PubMed] [Google Scholar]
- 25.Jurisica I, King AD, Przulj N. Protein complex prediction via cost-based clustering. Bioinformatics. 2004;20:3013–3020. doi: 10.1093/bioinformatics/bth351. [DOI] [PubMed] [Google Scholar]
- 26.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu ZJ, Hung JH, Wang Y, Chang YC, Huang CL, Huyck M, DeLisi C. VisANT 3.5: multi-scale network visualization, analysis and inference based on the gene ontology. Nucleic Acids Res. 2009;37:W115–W121. doi: 10.1093/nar/gkp406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sriphaew K, Theeramunkong T. Fast algorithms for mining generalized frequent patterns of generalized association rules. Ieice T. Inf. Syst. 2004;E87d:761–770. [Google Scholar]
- 29.O'Donnell CJ, Johnson AD. An open access database of genome-wide association results. BMC Med. Genet. 2009;10:6. doi: 10.1186/1471-2350-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westfall PH, Young SS. New York, NY: John Wiley and Sons, Inc.; 1993. Resampling-based multiple testing: Examples and methods for p-value adjustment. [Google Scholar]
- 31.Rocha EPC, Danchin A. Base composition bias might result from competition for metabolic resources. Trends Genet. 2002;18:291–294. doi: 10.1016/S0168-9525(02)02690-2. [DOI] [PubMed] [Google Scholar]
- 32.Naya H, Romero H, Zavala A, Alvarez B, Musto H. Aerobiosis increases the genomic guanine plus cytosine content (GC%) in prokaryotes. J. Mol. Evol. 2002;55:260–264. doi: 10.1007/s00239-002-2323-3. [DOI] [PubMed] [Google Scholar]
- 33.Bohlin J, Snipen L, Hardy SP, Kristoffersen AB, Lagesen K, Donsvik T, Skjerve E, Ussery DW. Analysis of intra-genomic GC content homogeneity within prokaryotes. BMC Genomics. 2010;11:464. doi: 10.1186/1471-2164-11-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bentley SD, Parkhill J. Comparative genomic structure of prokaryotes. Annu. Rev. Genet. 2004;38:771–792. doi: 10.1146/annurev.genet.38.072902.094318. [DOI] [PubMed] [Google Scholar]
- 35.Reshef DN, Reshef YA, Finucane HK, Grossman SR, McVean G, Turnbaugh PJ, Lander ES, Mitzenmacher M, Sabeti PC. Detecting novel associations in large data sets. Science. 2011;334:1518–1524. doi: 10.1126/science.1205438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.