Abstract
DNA methylation plays a key role in epigenetic regulation of eukaryotic genomes. Hence the genome-wide distribution of 5-methylcytosine, or the methylome, has been attracting intense attention. In recent years, whole-genome bisulfite sequencing (WGBS) has enabled methylome analysis at single-base resolution. However, WGBS typically requires microgram quantities of DNA as well as global PCR amplification, thereby precluding its application to samples of limited amounts. This is presumably because bisulfite treatment of adaptor-tagged templates, which is inherent to current WGBS methods, leads to substantial DNA fragmentation. To circumvent the bisulfite-induced loss of intact sequencing templates, we conceived an alternative method termed Post-Bisulfite Adaptor Tagging (PBAT) wherein bisulfite treatment precedes adaptor tagging by two rounds of random primer extension. The PBAT method can generate a substantial number of unamplified reads from as little as subnanogram quantities of DNA. It requires only 100 ng of DNA for amplification-free WGBS of mammalian genomes. Thus, the PBAT method will enable various novel applications that would not otherwise be possible, thereby contributing to the rapidly growing field of epigenomics.
INTRODUCTION
DNA methylation at the C5 position of cytosine plays a pivotal role in epigenetic regulation of eukaryotic genomes. Accordingly, the genome-wide distribution of 5-methylcytosine (5mC), or the methylome, has been attracting intense attention, leading to development of various methods for its interrogation. While these methods have been shown to produce largely consistent results (1,2), whole-genome bisulfite sequencing (WGBS) is currently the sole method that can attain both single-base resolution and genome-wide coverage. It has been successfully applied to elucidation of the methylomes of Arabidopsis thaliana (3,4), silkworm (5), honeybee (6) and 20 other species in various branches of eukaryotic phylogenetic tree (7,8), as well as that of human embryonic stem cells (9,10), induced pluripotent stem cells (11), peripheral blood mononuclear cells (12), colon cancer cells (13) and so on. These WGBS data have led to novel discoveries that could not have been attained by other methods. As the cost of sequencing decreases, WGBS is increasingly becoming the method of choice for methylome analysis.
While WGBS has been proven to be powerful, the current method has some practical limitations: it typically requires 5 μg of DNA as starting material, as well as global PCR amplification. It is often difficult, or sometimes prohibitive, to prepare this amount of DNA from many biologically interesting samples, such as early embryos, embryonic tissues and eggs of mammals. Furthermore, global PCR amplification inevitably invites ‘clonal’ reads and skewed representation. While the former can be removed in silico, their presence reduces the net sequencing throughput. The latter may cause inaccurate estimation of the methylation level. While WGBS libraries have been made even from submicrogram quantities of DNA (14,15), the reliance on global PCR amplification is even more absolute when starting with a limited amount of DNA, thereby further increasing the risk of artifacts. It would therefore be ideal to have a PCR-free method that is applicable to a minute amount of DNA.
By circumventing bisulfite-induced degradation of sequencing templates inherent to the current WGBS protocols, we developed a novel method that requires only submicrogram quantities of DNA for amplification-free WGBS of mammalian genomes. The method will provide an efficient alternative to conventional ones, enabling various novel applications that would not otherwise be possible.
MATERIALS AND METHODS
DNA
Genomic DNA was prepared from Neurospora crassa using a standard protocol (16). Arabidopsis thaliana seedling DNA isolated using DNeasy PLANT Mini Kit (Qiagen) and mouse astrocyte DNA isolated using a standard SDS-Proteinase K method were generous gifts from Hiroshi Shiba and Kinichi Nakashima, respectively. We treated genomic DNA with RNase ONE (Promega) followed by Proteinase K (Qiagen). The enzyme-treated DNA was purified using AMPure XP SPRI beads (Beckman). The amount of DNA was quantified using Quant-iT dsDNA kit (Invitrogen) and Qubit fluorometer (Invitrogen).
Bisulfite treatment
We used reagents supplied in Imprint DNA modification kit (Sigma) for bisulfite treatment, according to the one-step modification procedure recommended by the manufacturer. Briefly, 10 μl of DNA solution (125 pg–100 ng) was combined with 110 μl of Imprint Bisulfite Modification Reagent, denatured at 99°C for 6 min and incubated at 65°C for 90 min. For purification of bisulfite-treated DNA, we used PureLink PCR micro kit (Invitrogen) but not the Spin column in Imprint DNA modification kit (Sigma). To the 120 μl solution of bisulfite-treated DNA, we added 1 μg of DNA-free yeast RNA and 480 μl of PureLink binding buffer. We captured the DNA on PureLink PCR micro column using QIAvac 24 (Qiagen) and washed the column with 750 μl of PureLink wash buffer. We performed desulfonation of the bisulfite-treated DNA on the column by loading 100 μl of buffer BD in EpiTect Bisulfite Kit (Qiagen). Following incubation at room temperature for 8 min, we washed the column twice with 300 μl of PureLink wash buffer. Finally, we loaded 20 μl of 10 mM Tris–HCl (pH 8.5) to the column, incubated it at room temperature for 2 min and centrifuged it at 10 000 rpm for 1 min to elute the DNA. This procedure achieved 99.3% conversion rate, judging from the data on the chloroplast genome in Arabidopsis. Consistently, Kobayashi et al. (15) reported ∼99% conversion rates for λ DNA spiked in mouse samples. This is presumably because the bisulfite treatment with heat denaturation induces much more prominent DNA fragmentation than that with alkaline denaturation, thereby facilitating denaturation and efficient conversion (Supplementary Figure S1).
First-strand DNA synthesis
To the bisulfite-treated DNA solution prepared as above, we added 5 μl of 10 × NEBuffer2 (NEB), 5 μl of 2.5 mM dNTPs (Takara) and 4 μl of 100 μM BioPEA2N4 (5′-biotin-ACA CTC TTT CCC TAC ACG ACG CTC TTC CGA TCT NNN N-3′) and adjusted the total volume to 50 μl with water. Following incubation at 94°C for 5 min and 4°C for 5 min, we started DNA synthesis by adding 1.5 μl of 50 U/μl Klenow fragment (3′→5′ exo−) (NEB) to the solution. We kept the reaction temperature at 4°C for 15 min, raised it to 37°C at a rate of 1°C/min and finally kept it at 37°C for 90 min. Then, we killed the enzyme activity by heating the tube at 70°C for 10 min.
Purification of first-strand DNA
To the first-strand DNA synthesis reaction described above, we added 50 μl of AMPure XP SPRI beads (Beckman) and incubated the solution at room temperature for 10 min. We collected the SPRI beads and rinsed them with 75% (v/v) ethanol. We then suspended the beads in 45 μl of 10 mM Tris–acetate (pH 8.0) and transferred the supernatant to a new tube containing 5 μl of 10× ExTaq buffer (Takara). We added 50 μl of AMPure XP to the solution and repeated the DNA purification step described above, except for increasing the elution volume to 50 μl. We took 20 μl of Dynabeads M280 Streptavidin (Invitrogen), washed them well with 2× B&W buffer [10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 3 M LiCl] and finally resuspended them in 50 μl of 2× B&W buffer. Then, we combined the DNA solution eluted from AMPure XP (50 μl, see above) and the washed Dynabeads M280 Streptavidin (50 μl). Following incubation at room temperature for 30 min with constant rotation, we collected the beads and washed them with 180 μl of 2× B&W buffer. We suspended the washed beads in 180 μl of 0.1 N NaOH and stood the suspension at room temperature for 2 min. We repeated the alkaline wash step again and washed the beads with 180 μl of 2 x B&W buffer followed by 180 μl of 10 mM Tris–HCl (pH 7.5).
Second-strand DNA synthesis
We suspended the washed beads in 50 μl of 1× NEBuffer2 (NEB) containing 0.25 mM dNTPs and 8 μM PE-reverse-N4 (5′-CAA GCA GAA GAC GGC ATA CGA GAT NNN N-3′). Following incubation at 94°C for 5 min and 4°C for 5 min, we started DNA synthesis by adding 1.5 μl of 50 U/μl Klenow fragment (3′→5′ exo−) (NEB) to the solution. We kept the reaction temperature at 4°C for 15 min, raised it to 37°C at a rate of 1°C/min and finally kept it at 37°C for 30 min. Following heat inactivation of the enzyme at 70°C for 10 min, we collected and suspended the beads in 50 μl of 1× ThermoPol Reaction Buffer (NEB) containing 0.25 mM dNTPs and 8 U of Bst DNA polymerase large fragment (NEB). We incubated the tube at 65°C for 30 min to complete the second-strand DNA synthesis. Then, we collected the beads and immediately used them for the following elution step.
Elution and size fractionation
We suspended the collected beads in 50 μl of 1× Phusion HF buffer containing 0.25 mM dNTPs, 2 U Phusion Hot Start High-Fidelity DNA Polymerase (Finnzyme) and 0.8 μM Primer-3 (5′-AAT GAT ACG GCG ACC ACC GAG ATC TAC ACT CTT TCC CTA CAC GAC GCT CTT CCG ATC T-3′). We incubated the suspension at 94°C for 5 min, 55°C for 10 min and 72°C for 30 min. This step not only makes the eluted DNA double stranded to be precisely size-selected by SPRI beads, but also synthesizes the sequence required for bridge PCR. We transferred the supernatant to a new tube, added 1 μl of 20 U/μl Exonuclease I (NEB) and incubated the tube at 37°C for 15 min. Following incubation at 70°C for 10 min to inactivate Exonuclease I, we purified the double-stranded template DNAs with 50 μl of AMPure XP twice as described above, except for reducing the volume of final elution to 20 μl. This size selection step effectively removed DNA fragments smaller than 200 bp; the average length and size range of the templates were typically 300–400 bp and 200–700 bp, respectively (Supplementary Figure S2).
Real-time PCR quantification of sequencing template
We performed real-time PCR on ABI7000 SDS system (Applied Biosystems) using SYBR Premix ExTaq (Takara) according to manufacturer’s instruction. Primers used were PE-forward (5′-AAT GAT ACG GCG ACC ACC GAG ATC TAC AC-3′) and PE-reverse (5′-CAA GCA GAA GAC GGC ATA CGA GAT-3′). We used PhiX v2 Control Kit (Illumina) as the standard for quantification.
Illumina sequencing
Based on the qPCR quantification, we diluted appropriate amount of template DNA to 19 μl with Buffer EB (QIAGEN) and added 1 μl of 2 N NaOH (Illumina) for denaturation. To this solution (20 μl), we added 100 μl of Hybridization Buffer A [900 mM NaCl, 180 mM Tris–HCl (pH 7.4)] and used the solution for cluster generation. Single-end reads were generated by GAIIx and HiSeq2000 at RIKEN Omics Science Center and by GAIIx at Kyushu University Genome Analysis Consortium, according to the manufacturer's instruction.
Mapping and data analysis
In principle, current PBAT method sequences the strand complementary to the bisulfite-converted, C-poor DNA. Accordingly, the obtained reads are generally G-poor. We converted every residual G in the reads to A in silico (G-to-A conversion) to make the sequences fully composed of A, C and T. On the other hand, we prepared two reference genome sequences, one by converting every G to A and the other by converting every C to T, the latter of which represents the G-to-A-converted version of the complementary strand of the reference genome sequence. We then mapped the G-to-A converted reads to G-to-A-converted both strands of the target genome by SOAP2 aligner (17). Following this three-letter alignment, we reversed all the conversions applied to the reads and reference genome sequences and then refined each alignment using the Smith–Waterman algorithm. We used a score matrix that does not penalize a mismatch between A in the read and G in the top strand of reference genome sequence.
Note that we occasionally encountered C-poor reads corresponding to the bisulfite-treated DNA strands (see Supplementary Figure S3 for a plausible mechanism for their generation). If such C-poor reads are processed as above, they lead to alignments rich in apparent methylation and mismatches between T in the reads and C in the top strand of reference genome sequence. These reads should be subjected to C-to-T conversion prior to mapping and evaluated using a score matrix that does not penalize a mismatch between T in the read and C in the top strand of reference genome sequence. To take the presence of such reads into account, we subjected every read to both G-to-A and C-to-T conversions followed by mapping using respective scoring matrices and finally selected the alignment with the highest score.
While we used the above-mentioned strategy with fixed-length seeds and Smith–Waterman algorithm for the Neurospora and Arabidopsis genomes (Supplementary Tables S1 and S2), we took a different one with adaptive seeds (18) and pair-wise alignment for the mouse and human genomes (Supplementary Table S3).
We used only uniquely mapped reads to calculate methylation rates. We developed a genome browser in house to visualize genome scale data.
RESULTS
Post-bisulfite adaptor tagging to circumvent bisulfite-induced degradation of WGBS templates
To develop a highly efficient WGBS method, we first questioned why the yield of sequencing templates by current protocols should be so low as to necessitate global PCR amplification. It has been reported that bisulfite treatment leads to a dramatic loss of DNA, but the recovery rate, when using recently developed reagents, has improved markedly: we could recover 30–80% of the input DNA using several commercially available kits (Supplementary Figures S1A and S4A). It thus seems that the quantity per se does not matter.
We then investigated available WGBS protocols and noticed that, without exception, they include bisulfite treatment of template DNAs that are ligated to the sequencing adaptors at both their ends. Bisulfite treatment induces DNA breakage; this would inevitably eliminate a certain fraction of the template DNAs (Figure 1A). This adverse effect is well exemplified by bisulfite treatment of a DNA size standard, which resulted in good recovery in amounts (40–70%) but remarkable degradation observed as a smear on gel electrophoresis (Supplementary Figure S4C). While PCR amplification of bisulfite-treated DNA templates can increase the total mass of DNA, it can only multiply undamaged templates, not damaged ones (Figure 1A).
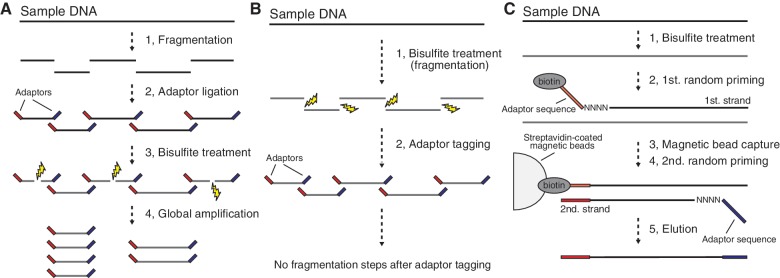
Figure 1.
WGBS and PBAT. (A) Schematic of the conventional WGBS protocols. Bisulfite treatment follows adaptor tagging, thereby leading to bisulfite-induced fragmentation of adaptor-tagged template DNAs. (B) Schematic of PBAT strategy. Bisulfite treatment precedes adaptor tagging, thereby circumventing bisulfite-induced fragmentation of adaptor-tagged template DNAs. (C) Random priming-mediated PBAT method. Two rounds of random priming on bisulfite-treated DNA generate directionally adaptor-tagged template DNAs.
To circumvent the bisulfite-induced fragmentation of sequencing templates, we conceived a novel strategy by simply reversing the order of adaptor tagging and bisulfite treatment, reasoning that if adaptor tagging follows bisulfite treatment, adaptor-tagged templates would escape destructive conditions and could then be fully used for sequencing (Figure 1B). Therefore, this ‘post-bisulfite adaptor tagging (PBAT)’ strategy should, in principle, achieve a wider coverage than current protocols that include bisulfite treatment of adaptor-tagged templates (Figure 1B).
Efficient preparation of WGBS templates by random priming-mediated PBAT
To implement the PBAT strategy, we developed a simple method based on random primer extension (Figure 1C). In this method, we used bisulfite-treated DNA as a template for first-strand DNA synthesis that is primed from a 5′-biotinylated adaptor primer bearing a random tetramer sequence (N4) at its 3′-end. Following the removal of residual primers and primer-dimers, we purified the first-strand DNA using streptavidin-coated magnetic beads, followed by alkaline denaturation. We next synthesized the second-strand DNA using the immobilized first-strand DNA as the template and another adaptor primer that also bears N4 at its 3′-end. Finally, we eluted the second-strand DNA from the beads and used them for sequencing after appropriate size selection.
Following extensive optimization of each step in the method outlined above, we prepared WGBS templates for Illumina GAIIx using from 100 ng to 125 pg of N. crassa genomic DNA, without using any global amplification (Table 1, rows 1–6). We successfully obtained 47 million reads using 6.5% of the templates generated from 100 ng of DNA (i.e. equivalent to 6.5 ng of input DNA) in a single lane of the Illumina GAIIx (Table 1, row 1). Notably, we also obtained 2.2 and 4.0 million uniquely mapped reads from 125 and 250 pg of DNA, respectively (Table 1, rows 5 and 6). These reads led to 6.2- and 11.0-fold coverage of 95.3 and 98.2% of the Neurospora genome (i.e. 5.9- and 10.8-fold coverage of the entire genome), respectively (Supplementary Figure S5 and Supplementary Table S1). Similarly, we obtained 7.3 million reads per 1 ng of mouse DNA by GAIIx (Table 1, row 7). This high efficiency enabled amplification-free WGBS of mouse astrocyte from 100 ng of DNA (see below).
Table 1.
Performance of PBAT method
| Sample (Illumina platform) | Template preparationa |
Illumina sequencingb |
Total yieldc |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Amount of starting DNA | Yield in template preparation |
dsDNA copy number injected per lane (M) (% prepared template) | Number of obtained reads per lane (M) |
Yield in Illumina sequencing (%) | Reads per ng of starting DNA (M) | Gb per ng of starting DNA | |||||
| Percent amount | dsDNA copy number (M) | Total (%) | Uniquely mapped (%) | Multiply mapped (%) | Unmapped (%) | ||||||
| Neurospora crassa (GAIIx) | 100 ng | 4.6 | 13 950 | 907 (6.5) | 47.1 (100) | 27.9 (59.2) | 1.2 (2.5) | 18.1 (38.3) | 5 | 7.3 | 0.84 |
| Neurospora crassa (GAIIx) | 10 ng | 9.4 | 2857 | – | – | – | – | – | – | – | – |
| Neurospora crassa (GAIIx) | 1 ng | 19.6 | 596 | – | – | – | – | – | – | – | – |
| Neurospora crassa (GAIIx) | 500 pg | 23.9 | 363 | – | – | – | – | – | – | – | – |
| Neurospora crassa (GAIIx) | 250 pg | 13.2 | 101 | 101 (100) | 7.2 (100) | 4.0 (56.0) | 0.2 (2.6) | 3.0 (41.4) | 7 | 28.9 | 3.35 |
| Neurospora crassa (GAIIx) | 125 pg | 12.3 | 47 | 47 (100) | 5.4 (100) | 2.2 (41.1) | 0.1 (2.4) | 3.0 (56.5) | 11 | 42.7 | 4.95 |
| M. musculus, astrocyte (GAIIx) | 100 ng | 7.2 | 22 000 | 928 (4.2) | 31.0 (100) | 20.3 (64.3) | 1.6 (5.2) | 9.7 (30.5) | 3 | 7.3 | 0.89 |
| M. musculus, neuron (HiSeq2000) | 100 ng | 8.8 | 26,800 | 930 (3.5) | 223.4 (100) | 148.0 (66.3) | 12.3 (5.5) | 63.0 (28.2) | 24 | 64.3 | 6.50 |
aAmount of DNA was quantified using Quant-iT dsDNA kit (Invitrogen) and Qubit fluorometer (Invitrogen). Copy number of template DNA was determined by qPCR using the PhiX v2 Control Kit (Illumina) as standard, where the length of templates was assumed to be 300 bp on average. M; million.
bIllumina sequencing generated 116-nt single-end reads by GAIIx (rows 1, 5, 6), 121-nt single-end reads by GAIIx (row 7) and 101-nt single-end reads by HiSeq2000 (row 8). Yield of Illumina sequencing was calculated by dividing total read number (output) by injected DNA copy number (input).
cTotal yield per ng of starting DNA was estimated through the multiplication of read length by total number of reads per ng of starting DNA. Gb; gigabase.
Evaluation of WGBS data obtained by PBAT
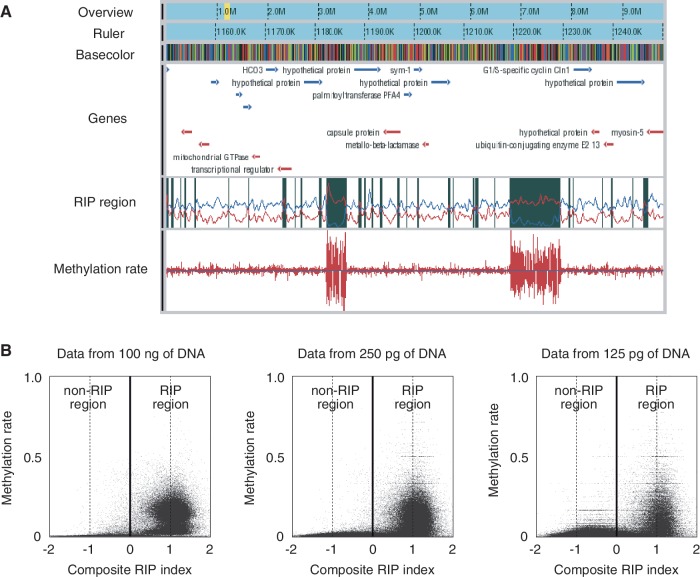
We next examined the data obtained. DNA methylation in N. crassa is concentrated in the relics of a genome defense system termed repeat-induced point mutation (RIP) (19). The regions subject to RIP (RIP region) can be predicted by the RIP index calculated from the characteristic nucleotide composition induced by RIP (20). MeDIP-chip data for linkage group VII confirmed the cooccurrence of DNA methylation and RIP on a chromosome-wide scale (20). Our WGBS data, including those obtained from 250 and 125 pg of DNA, were consistent with their finding, thereby extending it to a genome-wide scale (Figure 2A and B).
Figure 2.
WGBS of N. crassa by PBAT. (A) A snapshot of WGBS data obtained from 100 ng of DNA. The browser contains six tracks for overview, ruler, basecolor (nucleotide sequence), gene, RIP region and methylation rates from the top to the bottom. The yellow band in the overview track indicates the position of the region displayed in the other five tracks. Genes on the top and bottom strands are colored in blue and red, respectively. A RIP region colored in black in the RIP region track was defined as a region with a positive value of the composite RIP index obtained by subtracting the RIP substrate index (CpA + TpG/ApC + GpT, blue line plot in the track) from the RIP product index (TpA/ApT, red line plot in the track) (19). Methylation rates are indicated for both top and bottom DNA strands. Note that two large RIP regions are heavily methylated. (B) Cooccurrence of methylation and RIP. The moving averages (window size, 500 bp; step size, 100 bp) of methylation rate were plotted against those of the composite RIP index. WGBS data obtained from 100 ng and 250 and 125 pg of DNA were used for the analysis (Supplementary Table S1).
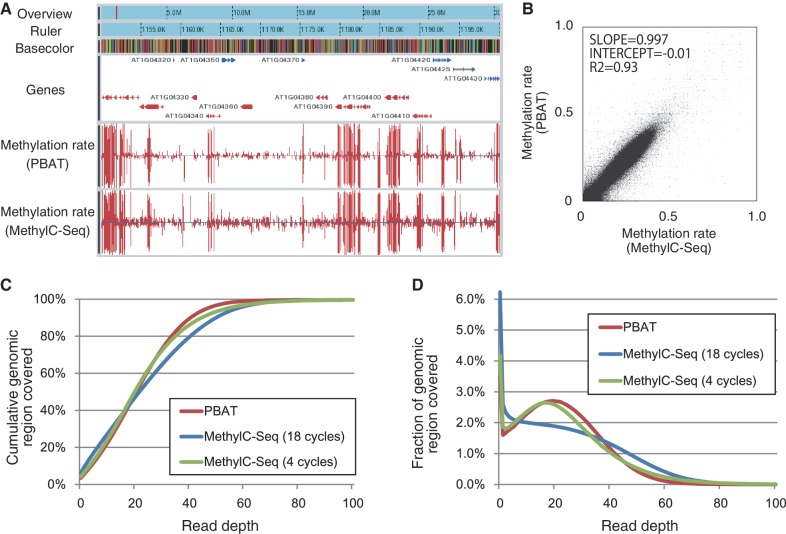
We also applied the PBAT method to A. thaliana seedlings and compared the data with those obtained by the conventional method MethylC-Seq (Figure 3A; Supplementary Figure S6 and Supplementary Table S2) (4). Both methods gave largely consistent results in terms of the distribution of 5mC (R2 = 0.93, Figure 3B and Supplementary Figure S6). However, the PBAT method covered the genome in a less biased manner than did MethylC-Seq (Figure 3C and D). The bias is presumably due to 18 cycles of global PCR amplification used to generate the MethylC-Seq data (4). Consistently, recent MethylC-Seq data from the same group using four cycles of PCR (21) covered the genome in a less biased manner comparable to the PBAT data (Figure 3C and D).
Figure 3.
Comparison of A. thaliana WGBS data obtained by PBAT and MethylC-Seq. (A) A snapshot of WGBS data for seedlings of A. thaliana ecotype Col-0. The browser contains six tracks for overview, ruler, basecolor (nucleotide sequence), gene and methylation rates for the PBAT and MethylC-Seq data from the top to the bottom. The red band in the overview track indicates the position of the region displayed in the other five tracks. Genes on the top and bottom strands are colored in blue and red, respectively. The two bottom tracks display methylation rates for both strands calculated from the PBAT data obtained from 100 ng of DNA and the MethylC-Seq data obtained from 5 µg of DNA using 18 cycles of PCR enrichment (4). (B) Correlation between the PBAT and MethylC-Seq data. The moving averages (window size, 1000 bp; step size, 200 bp) of methylation rate were calculated from the two data sets and plotted for comparison. (C) Cumulative coverage of the A. thaliana genome by the PBAT and MethylC-Seq data (Supplementary Table S2). The percentage of the genome covered by differing maximum depth of reads is shown for the two data sets. The MethylC-Seq data on MA line 19 obtained from 2 µg of DNA using 4 cycles of PCR enrichment (20) is also included for comparison. (D) Coverage of the A. thaliana genome by the PBAT and MethylC-Seq data. The percentage of the genome covered at the indicated read depth is shown for the three data sets (Supplementary Table S2).
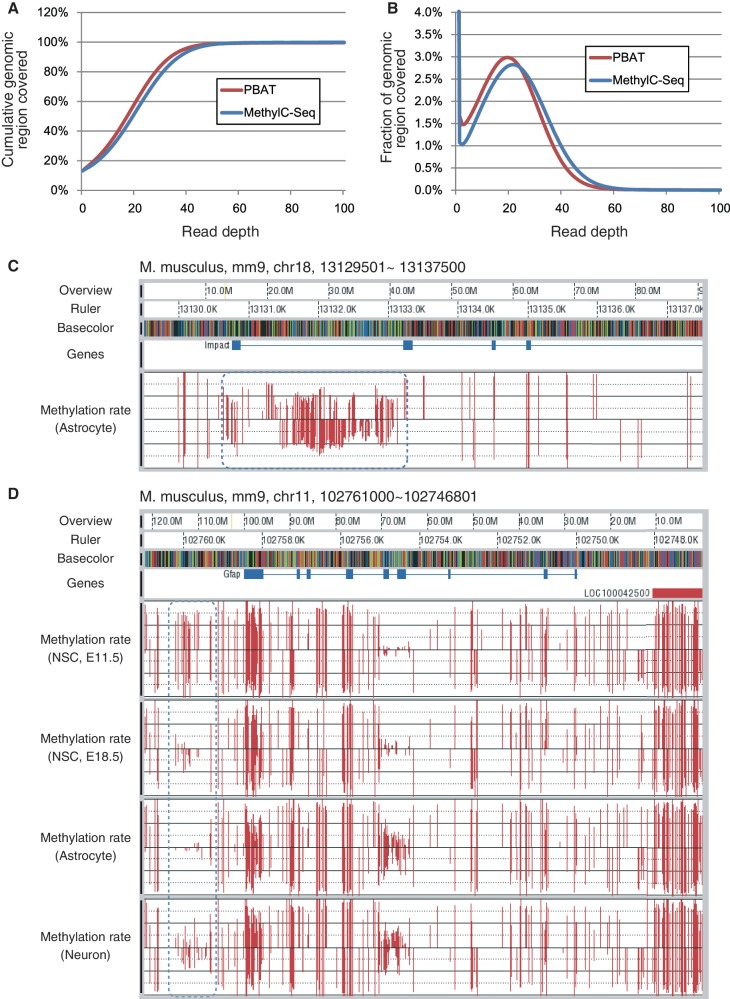
We next applied the PBAT method to 100 ng of mouse astrocyte DNA. Using the obtained templates, we ran 23 lanes of Illunima GAIIx in total to obtain 949 million, 121-nt single-end reads and succeeded in amplification-free, 24.4-fold coverage of 86.8% of the mouse genome (i.e. 21.1-fold coverage of the entire genome) (Supplementary Table S3). While we intended to compare the status of genome coverage between PBAT and MethyC-Seq, we found no publicly available mouse data of comparable size. Accordingly, we instead used the MethylC-Seq data of the human IMR90 cell line, which was generated using four cycles of global PCR (9) to achieve 23.9-fold coverage of 86.8% of the human genome (i.e. 20.8-fold coverage of the entire genome) (Supplementary Table S3). The two data showed a comparable performance in terms of genome coverage (Figure 4A and B; Supplementary Figure S7).
Figure 4.
WGBS of mouse astrocyte by PBAT. (A) Cumulative coverage of the mouse genome by the PBAT data. The percentage of the genome covered by differing maximum depth of reads is shown. The PBAT data were obtained from 100 ng of astrocyte DNA without using any global PCR amplification (Supplementary Table S3). For comparison, cumulative coverage of the human genome by the MethylC-Seq data obtained from 5 µg of the IMR90 cell DNA with four cycles of PCR amplification (9) is also included (Supplementary Table S3). (B) Coverage of the mouse genome by the PBAT data. The percentage of the genome covered at the indicated read depth by the PBAT data is shown (Supplementary Table S3). For comparison, coverage of the human genome by the MethylC-Seq data (9) is also included (Supplementary Table S3). (C) Imprinted DMR of a paternally expressed gene Impact. The blue dotted round rectangle indicates methylation status of the DMR. The black horizontal lines in the methylation rate track indicate 50% methylation level for top and bottom strands of DNA. Note that apparently hemi-methylated stretches are derived from the regions with less than five reads, which were omitted from the calculation of methylation rates. (D) DMR in the upstream of Gfap gene. The blue dotted round rectangle indicates differential methylation among neural stem cells at embryonic day 11.5 and 18.5, astrocyte and neuron. Methylation rates were calculated for the cytosine residues covered by at least five reads.
To evaluate the relevance of PBAT data, we examined the methylation status of imprinted differentially methylated regions (DMRs) (Figure 4C and Supplementary Figure S8). For instance, the DMR of a paternally expressed gene Impact, which spans its promoter to the first intron (22,23), showed ∼50% methylation level, in good contrast to the other regions with a high level of methylation (Figure 4C). Other imprinted DMRs also showed ∼50% methylation levels (Supplementary Figure S8). These results suggest that the PBAT method covered methylated and unmethylated alleles in an unbiased manner. In addition to astrocytes, we applied the PBAT method to neuron and neural stem cells prepared from mouse telencephalon at embryonic day 11.5 and 18.5, and successfully determined their methylomes (to be published elsewhere). We confirmed that differential methylation of Gfap promoter among these cells (24,25) was correctly recapitulated in the PBAT data set (Figure 4D).
Taken together, these results indicate that the PBAT method can generate biologically relevant methylome data comparable to those obtained by MethylC-Seq, but, notably, starting from a much smaller amount of DNA and not requiring any global amplification.
DISCUSSION
We have developed a novel WGBS method with unprecedented efficiency using the PBAT strategy that circumvents bisulfite-induced fragmentation of sequencing templates (Figure 1 and Table 1). We assume that total elimination of steps for DNA ligation and gel purification also contributes to the high efficiency. The method enables mammalian WGBS from submicrogram quantities of DNA, notably, without using any global amplification: we indeed succeeded in amplification free, 21.1-fold coverage of the mouse genome starting from 100 ng of astrocyte DNA (Figure 4 and Supplementary Table S3). This is in good contrast to previous mammalian WGBS studies (9–13), which require microgram quantities of DNA and global PCR amplification. Since extensive amplification inevitably tends to bias the coverage (Figure 3C and D), PBAT would serve as an effective alternative to conventional methods, especially when the amount of DNA is so limited as to necessitate a higher number of PCR cycles. Notably, a previous study reported reduced-representation bisulfite sequencing (RRBS) from 30 ng of DNA with 12 cycles of PCR to achieve 25-fold coverage of selected genomic regions (26). Whereas RRBS is a cost-effective method for genome-scale analysis from limited amounts of DNA, PBAT would be the choice for those who prefer genome-wide analysis; the use of HiSeq2000, which can generate a much larger number of reads than GAIIx from the same amount of injected template DNA (Table 1, row 8), should achieve amplification-free mammalian WGBS even from 30 ng of DNA.
The PBAT method would also be particularly suitable for genome-scale methylome scanning of samples consisting of less than 1000 cells. For instance, a pioneering work from 150 ng of DNA with 15 cycles of PCR generated 5.4 million reads to scan the methylome of mouse primordial germ cells (14). The PBAT method can generate a similar number of reads even from subnanogram quantities of DNA without using any global amplification (Table 1, rows 5 and 6). Indeed, it has been successfully applied to 400 germinal vesicle-stage mouse oocytes (i.e. ∼4.8 ng of DNA) to achieve 19.3 million amplification-free uniquely mapped reads (15). Notably, GAIIx was able to capture ∼5% of denatured DNA molecules injected into the flowcell (Table 1, column 10); ≥95% were lost in the cluster generation step. Therefore, to fully exploit the sequencing templates prepared from very precious samples, one may prefer to use, at the risk of biased representation, minimal cycles of PCR or linear amplification to generate ‘clonal’ copies, which can serve as ‘back-ups’ against the drop-off at this sequencing step.
The PBAT method would have two potential drawbacks. One is site preferences in the random priming steps, leading to ‘pile-ups’ of reads. The PBAT data contained such piled-ups (Supplementary Figure S9) but covered the Arabidopsis and mouse genomes as well as, or even better than, those obtained by conventional methods. This is presumably because the length of Illumina reads exceeds the distance between adjacent preferential priming sites. Nevertheless, efforts to increase the randomness of priming would further enhance this method. The other concern is differential priming between methylated and unmethylated alleles, leading to inaccurate estimation of methylation level. With this concern in mind, we assumed that it is not so prominent in practice, judging from the data on mouse imprinted DMRs (Figure 4 and Supplementary Figure S8), the accordance of methylation levels between the PBAT and MethylC-Seq data on Arabidopsis (Figure 3) and qPCR confirmation of methylation levels estimated from the Neurospora PBAT data (Supplementary Figure S10).
In conclusion, we developed the PBAT method as a highly efficient alternative to conventional WGBS methods. We expect that it can enable various novel applications that would not otherwise be possible. Furthermore, the random priming procedure described here can be applied to the sequencing of a minute amount of genomic/metagenomic DNAs as well as RNAs.
ACCESSION NUMBERS
DRX000759–000763, DRX001198.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3 and Supplementary Figures 1–10.
FUNDING
Research Program of Innovative Cell Biology by Innovative Technology (Cell Innovation) and Genome Network Project of the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan and CREST, JST (to T.I.). Funding for open access charge: MEXT.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Yoshiyuki Sakaki for continuous encouragement, Kinichi Nakashima for mouse DNA, Hiroshi Shiba for Arabidopsis DNA and RIKEN Omics Science Center and Kyushu University Genome Analysis Consortium for Illumina sequencing. We are also grateful to Tomohiro Kono and Hisato Kobayashi for useful comments and sharing unpublished data.
REFERENCES
- 1.Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat. Biotechnol. 2010;28:1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bock C, Tomazou EM, Brinkman AB, Müller F, Simmer F, Gu H, Jäger N, Gnirke A, Stunnenberg HG, Meissner A. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat. Biotechnol. 2010;28:1106–1114. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiang H, Zhu J, Chen Q, Dai F, Li X, Li M, Zhang H, Zhang G, Li D, Dong Y, et al. Single base-resolution methylome of the silkworm reveals a sparse epigenomic map. Nat. Biotechnol. 2010;28:516–520. doi: 10.1038/nbt.1626. [DOI] [PubMed] [Google Scholar]
- 6.Lyko F, Foret S, Kucharski R, Wolf S, Falckenhayn C, Maleszka R. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol. 2010;8:e1000506. doi: 10.1371/journal.pbio.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. Conservation and divergence of methylation patterning in plants and animals. Proc. Natl Acad. Sci. USA. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 9.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O’Malley R, Castanon R, Klugman S, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Zhu J, Tian G, Li N, Li Q, Ye M, Zheng H, Yu J, Wu H, Sun J, et al. The DNA methylome of human peripheral blood mononuclear cells. PLoS Biol. 2010;8:e1000533. doi: 10.1371/journal.pbio.1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, et al. Increased methylation variation in epigenetic domains across cancer types. Nat. Genet. 2011;43:768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi H, Sakurai T, Imai M, Takahashi N, Fukuda A, Obata Y, Sato S, Nakabayashi K, Hata K, Sotomaru Y, et al. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet. 2012;8:e1002440. doi: 10.1371/journal.pgen.1002440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis RH. Neurospora: Contributions of a Model Organism. Oxford, UK: Oxford University Press; 2000. [Google Scholar]
- 17.Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 18.Kiełbasa SM, Wan R, Sato K, Horton P, Frith MC. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011;21:487–493. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rountree MR, Selker EU. DNA methylation and the formation of heterochromatin in Neurospora crassa. Heredity. 2010;105:38–44. doi: 10.1038/hdy.2010.44. [DOI] [PubMed] [Google Scholar]
- 20.Lewis ZA, Honda S, Khlafallah TK, Jeffress JK, Freitag M, Mohn F, Schübeler D, Selker EU. Relics of repeat-induced point mutation direct heterochromatin formation in Neurospora crassa. Genome Res. 2009;19:427–437. doi: 10.1101/gr.086231.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitz RJ, Schultz MD, Lewsey MG, O’Malley RC, Urich MA, Libiger O, Schork NJ, Ecker JR. Transgenerational epigenetic instability is a source of novel methylation variants. Science. 2011;334:369–373. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okamura K, Hagiwara-Takeuchi Y, Li T, Vu TH, Hirai M, Hattori M, Sakaki Y, Hoffman AR, Ito T. Comparative genome analysis of the mouse imprinted gene Impact and its nonimprinted human homolog IMPACT: toward the structural basis for species-specific imprinting. Genome Res. 2000;10:1878–1889. doi: 10.1101/gr.139200. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi H, Suda C, Abe T, Kohara Y, Ikemura T, Sasaki H. Bisulfite sequencing and dinucleotide content analysis of 15 imprinted mouse differentially methylated regions (DMRs): paternally methylated DMRs contain less CpGs than maternally methylated DMRs. Cytogenet. Genome Res. 2006;113:130–137. doi: 10.1159/000090824. [DOI] [PubMed] [Google Scholar]
- 24.Takizawa T, Nakashima K, Namihira M, Ochiai W, Uemura A, Yanagisawa M, Fujita N, Nakao M, Taga T. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev. Cell. 2002;1:749–758. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- 25.Juliandi B, Abematsu M, Nakashima K. Epigenetic regulation in neural stem cell differentiation. Dev. Growth Diff. 2010;52:493–504. doi: 10.1111/j.1440-169X.2010.01175.x. [DOI] [PubMed] [Google Scholar]
- 26.Gu H, Bock C, Mikkelsen TS, Jäger N, Smith ZD, Tomazou E, Gnirke A, Lander ES, Meissner A. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat. Methods. 2010;7:133–136. doi: 10.1038/nmeth.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.