Abstract
The RNA ligase RtcB splices broken RNAs with 5′-OH and either 2′,3′-cyclic phosphate or 3′-phosphate ends. The 3′-phosphate ligase activity requires GTP and entails the formation of covalent RtcB-(histidinyl)-GMP and polynucleotide-(3′)pp(5′)G intermediates. There are currently two models for how RtcB executes the strand sealing step. Scheme 1 holds that the RNA 5′-OH end attacks the 3′-phosphorus of the N(3′)pp(5′)G end to form a 3′,5′-phosphodiester and release GMP. Scheme 2 posits that the N(3′)pp(5′)G end is converted to a 2′,3′-cyclic phosphodiester, which is then attacked directly by the 5′-OH RNA end to form a 3′,5′-phosphodiester. Here we show that the sealing of a 2′,3′-cyclic phosphate end by RtcB requires GTP, is contingent on formation of the RtcB–GMP adduct, and involves a kinetically valid RNA(3′)pp(5′)G intermediate. Moreover, we find that RtcB catalyzes the hydrolysis of a 2′,3′-cyclic phosphate to a 3′-phosphate at a rate that is at least as fast as the rate of ligation. These results weigh in favor of scheme 1. The cyclic phosphodiesterase activity of RtcB depends on GTP and the formation of the RtcB–GMP adduct, signifying that RtcB guanylylation precedes the cyclic phosphodiesterase and 3′-phosphate ligase steps of the RNA splicing pathway.
INTRODUCTION
Escherichia coli RtcB exemplifies a recently discovered family of RNA ligases implicated in tRNA splicing and RNA repair (1–4). Unlike classic RNA and DNA ligases, which join 3′-OH and 5′-phosphate ends, RtcB seals broken RNAs with 5′-OH and either 2′,3′-cyclic phosphate (>p) or 3′-phosphate ends. Previously, we proposed that RtcB executes a multi-step ligation pathway (5,6) entailing the following: (i) hydrolysis of the 2′,3′-cyclic phosphate to a 3′-phosphate; (ii) reaction of the enzyme with GTP to form a covalent RtcB-(histidinyl-N)-GMP intermediate; (iii) transfer of guanylate to the polynucleotide 3′-phosphate to form a polynucleotide-(3′)pp(5′)G intermediate; and (iv) attack of a 5′-OH on the N(3′)pp(5′)G end to form the splice junction and liberate GMP (Figure 1, scheme 1). In support of this pathway, we showed that GTP was required for 3′-phosphate/5′-OH sealing (5), identified His337 as the site of RtcB guanylylation (6), and demonstrated the formation of polynucleotide-(3′)pp(5′)G and its kinetic intermediacy during a 3′-phosphate/5′-OH ligation reaction (6).
Figure 1.
Alternative models for RtcB-catalyzed RNA ligation. See text for details.
Desai and Raines (7) have proposed an alternative pathway for RtcB sealing of 3′-phosphate and 5′-OH ends, whereby (i) GTP is used as an energy source to recyclize the 3′-phosphate end; and (ii) strand sealing occurs via a direct one-step attack of the 5′-OH nucleophile on the 2′,3′-cyclic phosphate with expulsion of the ribose O2′ (Figure 1, scheme 2). However, the predicted intermediates in the putative GTP-dependent cyclization reaction were not demonstrated, nor was it shown that recyclization kinetically precedes sealing.
Given the wide phylogenetic distribution of RtcB enzymes in bacteria, archaea and metazoa and their imputed role in archaeal and metazoan tRNA splicing (1–3)—supported by the fact that E. coli RtcB can perform the essential tRNA splicing functions in budding yeast (4)—it is a matter of importance to clarify the RtcB reaction mechanism. Fortunately, the two models engender different predictions about the role of GTP. In scheme 1, GTP and covalent guanylylation of the enzyme are necessary for ligation of both 2′,3′-cyclic phosphate and 3′-phosphate ends because formation of the activated RNA(3′)pp(5′)G intermediate is an obligate step (Figure 1). By contrast, scheme 2 posits a role for GTP in recyclization and predicts that GTP and covalent guanylylation of the enzyme ought to be dispensable for “direct” ligation of a substrate with a 2′,3′-cyclic phosphate end (Figure 1). Here we put these and other predictions to the test and find that the results militate against the scheme entailing recyclization and direct ligation, in favor of a pathway in which the conversion of an RNA 2′,3′-cyclic end to 3′-phosphate is a necessary (and kinetically valid) step in the ligation of 2′,3′-cyclic phosphate and 5′-OH ends. By installing a ligation-blocking 5′-phosphate on the RNA substrate, we show that GTP and RtcB guanylylation are critical for the RNA 2′,3′-cyclic phosphodiesterase (CPDase) activity.
MATERIALS AND METHODS
RtcB purification
Wild-type E. coli RtcB and mutant H337A were purified to homogeneity as described previously (6). Protein concentrations were determined using the BioRad dye reagent with BSA as the standard.
RNA substrates
The 20-mer HORNAp oligonucleotide labeled with 32P at the penultimate phosphate was prepared by T4 Rnl1-mediated addition of [5′-32P]pCp to a 19-mer synthetic oligoribonucleotide as described (5). The HORNAp was treated with E. coli RNA 3′-terminal phosphate cyclase (RtcA) and ATP to generate a 2′,3′-cyclic phosphate derivative, HORNA > p (5). The HORNAp and HORNA > p substrates were gel-purified prior to use in RtcB assays. These RNAs were also 5′-phosphorylated by reaction with T4 Pnkp-D167N and ATP to form pRNAp and pRNA > p, respectively, and then gel-purified (5).
RtcB activity assays
Reaction mixtures containing 50 mM Tris-HCl (pH 8.0), 2 mM MnCl2, 0.1 mM GTP, 0.1 µM radiolabeled RNA substrate and 1 µM RtcB were incubated at 37°C. The reactions were quenched at the times specified by adjusting the samples to 50 mM EDTA. The samples were either (i) mixed with an equal volume of 90% formamide, 50 mM EDTA and then analyzed by electrophoresis through a 40-cm 20% polyacrylamide gel containing 7.5 M urea in 45 mM Tris-borate, 1 mM EDTA or (ii) digested for 30 min at 37°C with 1000 U RNase T1 (Fermentas) prior to PAGE. The 32P-labeled RNAs were visualized by autoradiography of the gel and, where specified, quantified by scanning the gel with a Fuji Film BAS-2500 imager.
RESULTS
RtcB requires GTP to seal a 2′,3′-cyclic phosphate end
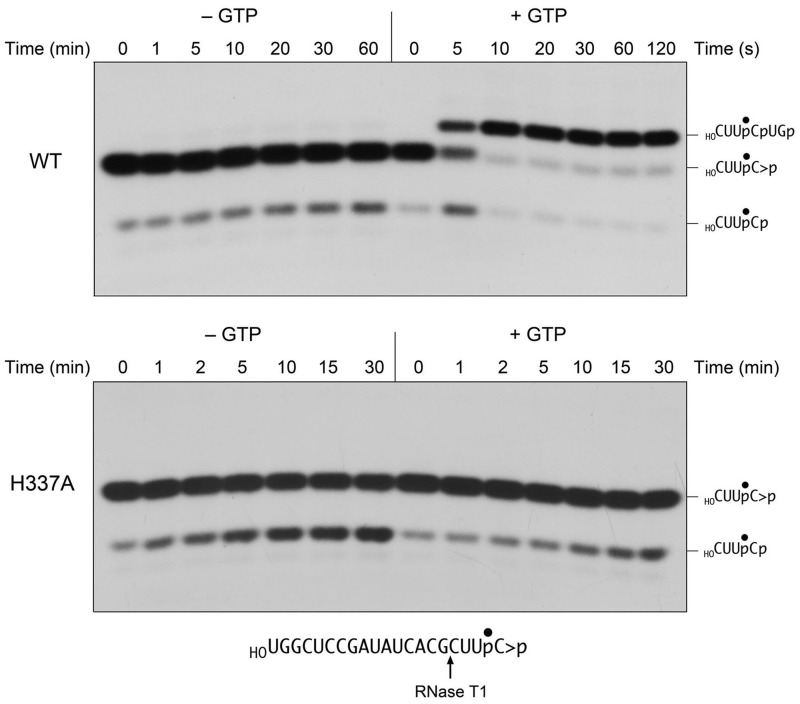
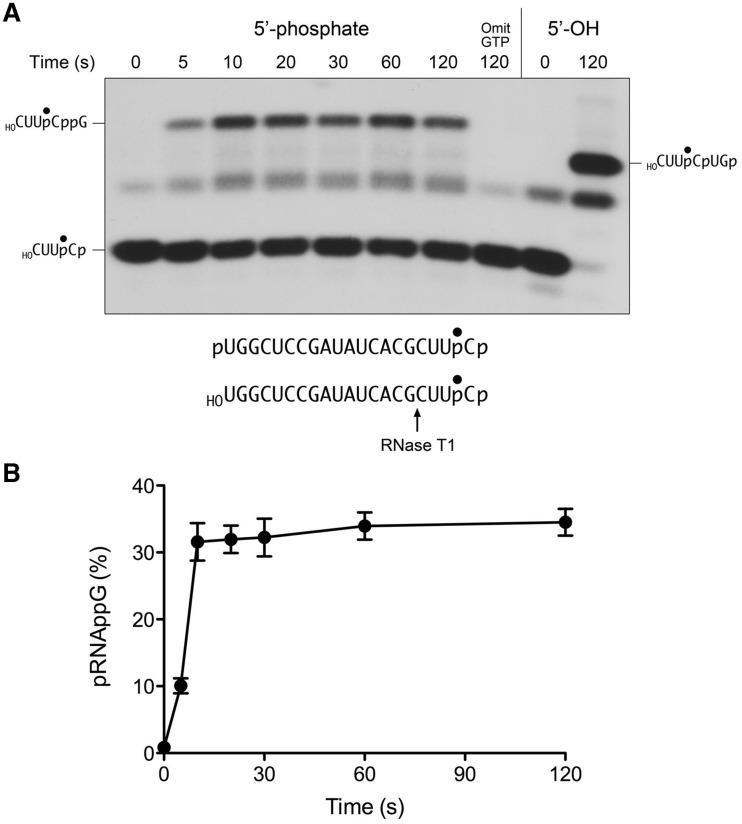
The requirement for GTP is interrogated as shown in Figure 2, where the substrate for ligation is a 20-mer RNA strand with 5′-OH and 2′,3′-cyclic phosphate ends and a single radiolabel between the 3′-terminal and penultimate nucleosides. To prepare this substrate, we used T4 RNA ligase 1 to transfer [5′-32P]pCp to the 3′ end of a 19-mer RNA oligonucleotide to generate a 20-mer strand with 5′-OH and 3′-phosphate ends. We then used E. coli RNA 3′-phosphate cyclase RtcA (8,9) to convert the 3′-phosphate end of the labeled 20-mer RNA into a 2′,3′-cyclic phosphodiester. The HORNA > p substrate was reacted with a 10-fold molar excess of wild-type RtcB and 2 mM manganese (the obligate metal cofactor) in the presence of 0.1 mM GTP or in the absence of exogenous nucleotide. The reactions were quenched with EDTA at the times specified. The mixtures were digested with RNase T1 and then analyzed by denaturing PAGE (Figure 2). RNase T1 incised the substrate 3′ of the most distal guanosine to yield the 32P-labeled tetranucleotide HOCUUpC > p (Figure 2, lanes 0). This analysis revealed that the efficiency of cyclization of the 3′-phosphate end of the 20-mer during the preparative reaction with RtcA was 92-93%, i.e. 7-8% of the RNase T1 product consisted of the more rapidly migrating tetranucleotide 3′-monophosphate, HOCUUpCp (Figure 2, lanes 0). In the presence of GTP, RtcB caused a depletion of the HOCUUpC > p T1 fragment and the appearance of a more slowly migrating T1 fragment that corresponded to a 6-mer oligonucleotide, HOCUUpCpUGp, released by T1 incision at the guanosines flanking the ligation junction (Figure 2, top right panel). The reaction endpoint was attained in 20-30 s with 87% of the input substrate being ligated. At the 5-s time point, we noted a transient increase in the abundance of the 3′-phosphate T1 fragment HOCUUpCp, which comprised 21% of the labeled material, while the 6-mer ligation product comprised 35% of the labeled material. These results are consistent with scheme 1 in which hydrolysis of the 2′,3′ > p end to a 3′-phosphate precedes the ligation of the 3′-phosphate and 5′-OH ends.
Figure 2.
GTP requirement for 2′,3′-cyclic phosphate ligation. Reaction mixtures containing 50 mM Tris-HCl (pH 8.0), 2 mM MnCl2, 0.1 µM HORNA > p substrate (depicted at bottom), 1 µM wild-type RtcB (top panel) or H337A mutant (bottom panel), and either 0.1 mM GTP (+GTP) or no added nucleotide (−GTP) as specified were incubated at 37°C. The reactions were quenched with EDTA at the times specified. The RNAs were digested with RNase T1 and the products were analyzed by Urea–PAGE and visualized by autoradiography. The 32P-labeled T1 fragments derived from the input 2′,3-cyclic phosphate substrate (HOCUUpC > p), a transient 3′-phosphate derivative (HOCUUpCp) and the ligated product (HOCUUpCpUGp) are indicated on the right.
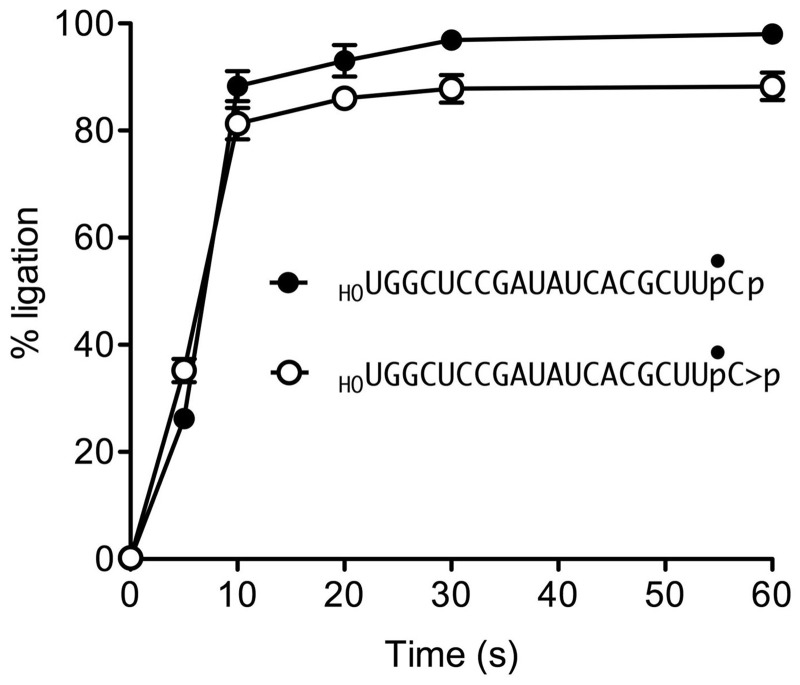
The kinetic profile of the HORNA > p ligation reaction is plotted in Figure 3, with each datum being the average of three separate experiments ± SEM. Also shown in Figure 3 is the time course of ligation of an otherwise identical 20-mer HORNAp substrate with a 3′-phosphate end, which attained an endpoint in 20-30 s with 98% of the input substrate being sealed. The kinetics of ligation of the two substrates were virtually superimposable, suggesting that there is no unique rate-limiting step that distinguishes the pathway for joining a 2′,3′-cyclic phosphate versus a 3′-phosphate. Accordingly, if scheme 1 is correct, then the rate of the CPDase reaction must be as fast or faster than the rate of the slowest of the subsequent steps of 3′-phosphate ligation. If scheme 2 is correct, then the rates of each of the proposed steps of the GTP-dependent cyclization reaction must be as fast or faster than the rate of the proposed final “direct ligation” step.
Figure 3.
Kinetic profiles of 2′,3′-cyclic phosphate and 3′-phosphate ligation. Reaction mixtures containing 50 mM Tris-HCl (pH 8.0), 2 mM MnCl2, 0.1 mM GTP, 1 µM RtcB and 0.1 µM HORNA > p (open circle) or HORNAp (closed circle) substrates were incubated at 37°C. The reactions were quenched with EDTA at the times specified. The RNAs were digested with RNase T1 and analyzed by Urea–PAGE. The extent of ligation is plotted as a function of time. Each datum is the average of three independent experiments ± SEM.
A parsimonious reading of the direct ligation mechanism posited in scheme 2 predicts that sealing of a 2′,3′ > p substrate ought not to require GTP. Yet, we found that omission of GTP prevented HORNA > p ligation, i.e. there was virtually no accumulation of the T1 fragment diagnostic of end joining even after a 60-min reaction (Figure 2, top left panel). By contrast, there was a slow but appreciable conversion of the input 2′,3′ > p end to a 3′-phosphate; i.e. the 3′-phosphate species increased from 7% of the total labeled material at time 0 to 19% after 60 min. These results weigh against the GTP-independent direct ligation mechanism proposed by Desai and Raines (7), while also adding an unanticipated feature to scheme 1, whereby the initial CPDase reaction appears to depend on GTP (see later in the text).
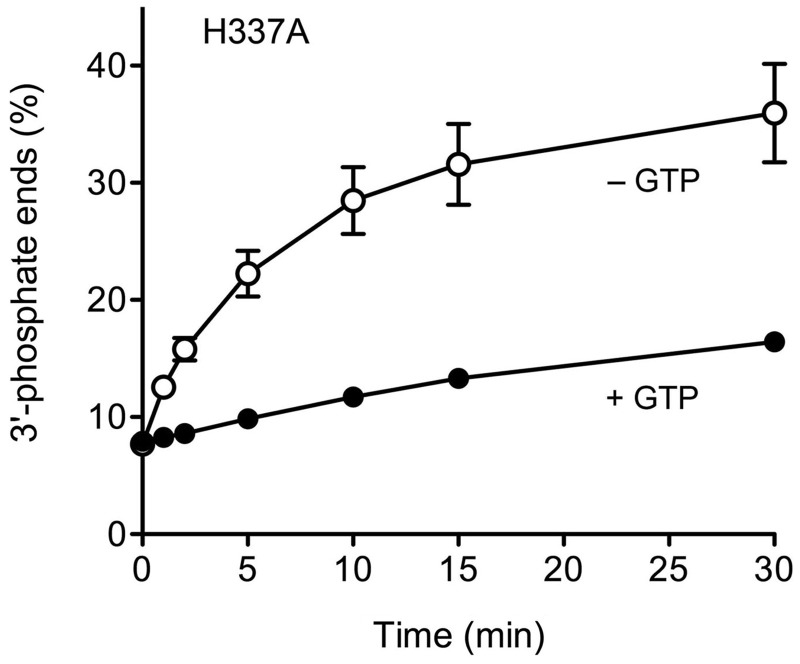
To consolidate our conclusions about the GTP requirement for the 2′,3′ > p ligation and CPDase steps, we tested the effects of the RtcB H337A mutation on both activities. His337 is the site covalent guanylylation of RtcB, and its mutation to alanine abolishes both formation of the RtcB-GMP adduct and the sealing of 5′-OH and 3′-phosphate ends (6). Here we found that the H337A protein was inert in ligating the HORNA > p substrate in the presence of GTP, even when the reaction was extended to 30 min (Figure 2, lower right panel), implying that formation of the RtcB-GMP adduct is a necessary step in the pathway of 2′,3′ > p ligation. This outcome is mandated by scheme 1 but not by scheme 2. Although H337A failed to ligate the 2′,3′ > p end in the presence of GTP, it nonetheless did slowly convert the 2′,3′ > p to a 3′-phosphate (Figure 2, lower panel). The kinetic profile of the H337A CPDase reaction in the presence of GTP (quantified in Figure 4) mirrored that of wild-type RtcB in the absence of GTP (Figure 2 and data not shown). A notable finding was that the rate and extent of the H337A CPDase reaction was greater in the absence of GTP than in its presence (Figure 4). A likely basis for this effect will be discussed.
Figure 4.
2′,3′-CPDase activity of RtcB H337A. The H337A mutant was reacted with the HORNA > p substrate in the presence of 0.1 mM GTP (+GTP) or in the absence of added nucleotide (−GTP) as described in Figure 2. The abundance of the 3′-phosphate-terminated T1 fragment HOCUUpCp, expressed as the percent of the total radiolabeled material, is plotted as a function of reaction time. Each datum is the average of three independent experiments ± SEM.
RtcB transfers GMP to the RNA 3′-phosphate end
We previously demonstrated the transfer of [32P]GMP from the covalent RtcB–GMP complex to the 3′-phosphate terminus of a DNA polynucleotide to form a polynucleotide-(3′)pp(5′)G species (6). We also showed that when RtcB was reacted with a 32P-labeled 5′-OH/3′-phosphate-terminated 20-mer substrate composed of 19 ribonucleotides and a single 3′-terminal deoxynucleoside, a predominant radiolabeled species—HORNA19dC(3′)pp(5′)G—accumulated at early times (comprising 35% of the labeled material) and then declined thereafter, concomitant with the progressive formation of a ligated circular RNA product (6). By contrast, a kinetic analysis of the reaction of RtcB with an all-RNA 32P-labeled 5′-OH/3′-phosphate-terminated 20-mer revealed only trace levels of HORNA(3′)pp(5′)G at early times, comprising ≤1% of the labeled RNA. These results were consistent with the following ideas: (i) that the polynucleotide-3′-guanylate is a covalently activated intermediate in the RtcB strand sealing pathway; (ii) that the rate of phosphodiester synthesis during ligation of an all-RNA 3′-phosphate end is much faster than the preceding rate of guanylylation of the 3′-phosphate and (iii) that the ability to detect the accumulation of the HORNA19dC(3′)pp(5′)G intermediate reflects slowing of the rate of the phosphodiester synthesis step in the absence of the 2′-OH moiety of 3′-terminal nucleoside of the substrate.
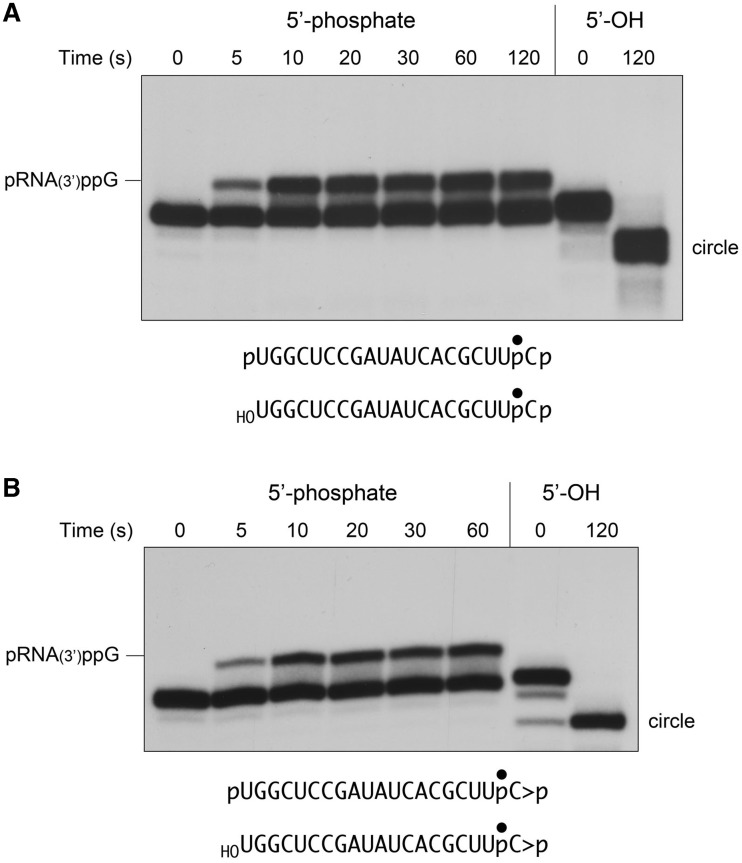
Here we aimed to trap the elusive RNA(3′)pp(5′)G intermediate by placing a ligation-blocking phosphate group at the 5′ terminus of the RNA 3′-phosphate substrate (Figure 5A), by reaction of the labeled HORNAp strand with cold ATP and a mutated (phosphatase-dead) version of T4 polynucleotide 5′-kinase/3′-phosphatase (10). The resulting non-ligatable pRNAp substrate was reacted with a 10-fold molar excess of RtcB in the presence of GTP, and the products were analyzed directly by denaturing PAGE (i.e. without RNase T1 digestion). A control reaction containing the ligatable HORNAp substrate was analyzed in parallel. The control ligation affirmed that RtcB converted the linear 20-mer HORNAp oligonucleotide into a sealed circular product that migrated ∼2 nucleotide steps faster than the input substrate (Figure 5A). As expected, there was little or no circularization of the pRNAp substrate. Rather, pRNAp was converted by RtcB into a novel species that migrated ∼2 nucleotide steps slower than the input substrate (Figure 5A), and which was not detected when RtcB reacted with the ligatable HORNAp substrate. This novel product corresponds to pRNA(3′)pp(5′)G. To further characterize the reaction products, we digested the samples with RNase T1 prior to denaturing PAGE (Figure 6A). Here we saw that the reaction with pRNAp yielded a novel T1 fragment (designated as HOCUUpCppG) that migrated slower than the ligation junction fragment HOCUUpCpUGp (Figure 6A). A key control showed that the formation of the novel T1 fragment was strictly dependent on the inclusion of GTP in the reaction mixture (Figure 6A), consistent with this species being a 3′-guanylylated derivative of the input substrate. (Note that only a trace amount of ligation junction fragment was formed during the reaction with pRNAp, which is attributable to a low level of residual 5′-OH ends that were not phosphorylated by polynucleotide kinase during substrate preparation.) The rate and extent of the appearance of the slower-migrating intact guanylylated RNA product in Figure 5A matched that of the guanylylated T1 fragment in Figure 6A. The kinetic profile of the RNA 3′ guanylylation reaction is shown in Figure 6B. The reaction reached an endpoint in 10-20 s with one-third of the input pRNAp being converted to pRNAppG. The fact that not all of the RNA 3′-ends were guanylylated at the endpoint when ligation was precluded by a 5′-phosphate can be rationalized if the RNAppG end remains engaged by RtcB and there is an equilibrium established between the forward reaction of GMP transfer from RtcB-(His337)–GMP to the RNA 3′-phosphate and the reverse reaction of GMP transfer from RNAppG back to the enzyme. The key point, however, is that the rate profile for the RNA 3′-phosphate guanylylation reaction (Figure 6B) was nearly identical to those of the single-turnover RNA 3′-phosphate and 2′,3′-cyclic phosphate ligation reactions (Figure 3). This signifies that RNAppG is a kinetically valid intermediate in the RtcB RNA ligation pathway.
Figure 5.
Formation of RNA(3′)pp(5′)G. Reaction mixtures containing 50 mM Tris-HCl (pH 8.0), 2 mM MnCl2, 0.1 mM GTP, 1 µM RtcB and 0.1 µM pRNAp or HORNAp (panel A) or pRNA > p or HORNA > p (panel B) substrates as specified were incubated at 37°C. The reactions were quenched with EDTA/formamide at the times specified and then analyzed by Urea–PAGE. The radiolabeled RNAs were visualized by autoradiography.
Figure 6.
Kinetic profile of guanylate transfer to an RNA 3′-phosphate. (A) RtcB was reacted with pRNAp or HORNAp in the presence of 0.1 mM GTP as described in Figure 5. The reactions were quenched with EDTA at the times specified. (A control mixture containing RtcB and pRNAp but no added GTP was incubated for 120 s). The RNAs were digested with RNase T1, and the products were analyzed by Urea–PAGE and visualized by autoradiography. (B) The extent of conversion of the pRNAp substrate to pRNAppG, expressed as the percent of the total radiolabeled material, is plotted as a function of reaction time. Each datum is the average of four independent experiments ± SEM. (Three of the experiments were performed without RNase T1 treatment as in Figure 5 and one experiment was performed with RNase T1 treatment as in panel A).
Kinetics and GTP-dependence of the 2′,3′-CPDase reaction
We prepared a 5′-phosphorylated version of the 2′,3′ > p RNA substrate in order to study the fate of the cyclic phosphodiester in the absence of ligation. According to scheme 1, the cyclic ends must be hydrolyzed for ligation to ensue, in which case the rate of the CPDase reaction must be at least as fast as the overall rate of 2′,3′ > p RNA sealing. By contrast, scheme 2 posits that the RNA 5′-OH is the relevant nucleophile for attack on the 2′,3′-cyclic phosphate during the final step of 3′-5′ phosphodiester formation, in which case the 2′,3′ > p end ought to be stable if the RNA 5′ end is a monophosphate. Here we found that the reaction of RtcB with pRNA > p in the presence of GTP resulted in the rapid appearance of the same slower-migrating guanylylated species, designated pRNA(3′)ppG in Figure 5B, that was observed in the pRNAp reaction (Figure 5A). The route to this outcome is shown in scheme 1, in which RtcB hydrolyzes the 2′,3′ > p end to a 3′-phosphate and then performs the RNA 3′ guanylylation step. Scheme 2 does not account for such an outcome. Note that the occurrence of the CPDase reaction is not observed in the experiment in Figure 5B because the 20-mer pRNA > p and pRNAp strands are not resolved by the PAGE procedure.
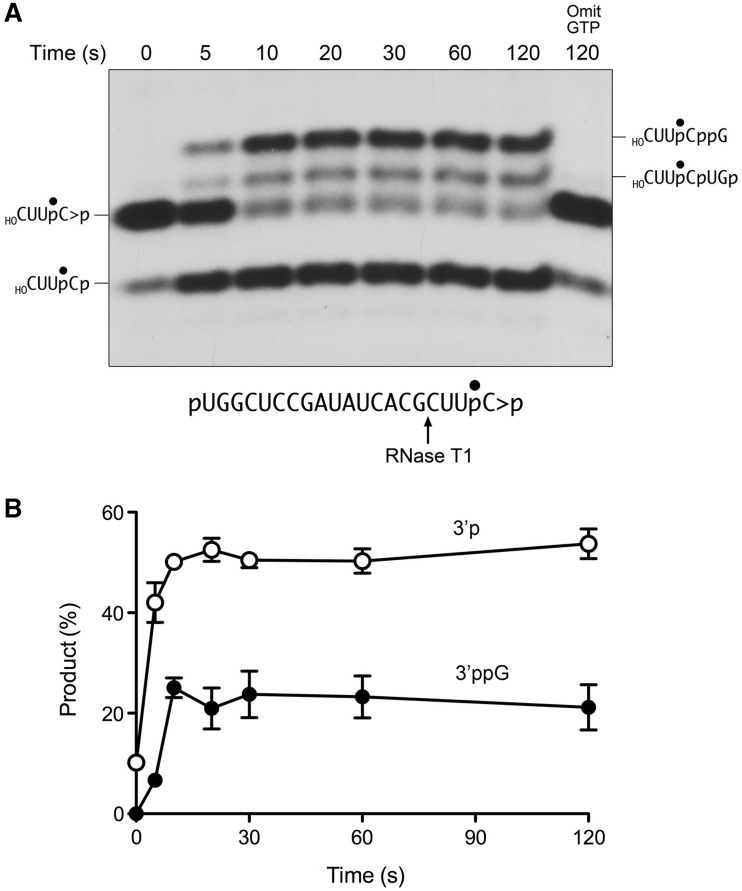
The reaction outcomes were clarified by treating the samples with RNase T1 prior to PAGE (Figure 7A). In this experiment, we saw that the initial 2′,3′ > p end was consumed within 10 s, being converted predominantly to the more rapidly migrating 3′-phosphate derivative (Figure 7A). The 3′-phosphate comprised 42% of the total radiolabeled material after 5 s (compared with 10% at time 0) and 53% at the reaction endpoint (Figure 7B). The appearance of the 3′ guanylylated T1 fragment lagged behind the generation of the 3′-phosphate end, as expected according to scheme 1. At the reaction endpoint/equilibrium, the 3′ guanylylated species comprised 23% of the radiolabeled material (Figure 7B). These results show that the CPDase reaction is at least as fast as the overall 2′,3′ > p ligation reaction (compare Figures 3 and 7B). We detected a low level of formation of the ligation junction fragment (Figure 7A), comprising ∼10% of the radiolabeled material, which we attribute to the presence of residual 5′-OH ends in the substrate preparation. Neither the 3′ guanylylated RNA nor the ligated species was generated when GTP was omitted from the reaction mixture (Figure 7A). The salient finding was that GTP omission also effaced the hydrolysis of the 2′,3′ > p to a 3′-phosphate during a 2-min reaction (Figure 7A), thereby fortifying the inferences from the experiments in Figure 2 that the RtcB CPDase activity is GTP-dependent. Tracking the kinetics of processing of the 2′,3′-cyclic phosphate end during a longer reaction of RtcB with pRNA > p in the absence of GTP revealed a steady accumulation of 3′-phosphate ends over 30 min, to an extent of 22% of the total labeled material (not shown). This increase was dependent on the inclusion of RtcB in the reaction mixtures. The rate of the CPDase reaction was 1000-fold greater in the presence of GTP than in its absence.
Figure 7.
Kinetics of the hydrolysis of RNA 2′,3′-cyclic phosphate and subsequent 3′-phosphate guanylylation. (A) RtcB was reacted with pRNA > p in the presence of 0.1 mM GTP as described in Figure 5. The reactions were quenched with EDTA at the times specified. (A control mixture containing RtcB and pRNAp but no added GTP was incubated for 120 s). The RNAs were digested with RNase T1 and the products were analyzed by Urea–PAGE and visualized by autoradiography. (B) The levels of the 3′-phosphate T1 fragment (open circle) and the 3′-ppG T1 fragment (closed circle), expressed as the percentage of the total radiolabeled material, are plotted as a function of reaction time. Each datum is the average of three independent experiments ± SEM.
Purine NTP analogs illuminate nucleobase recognition by RtcB
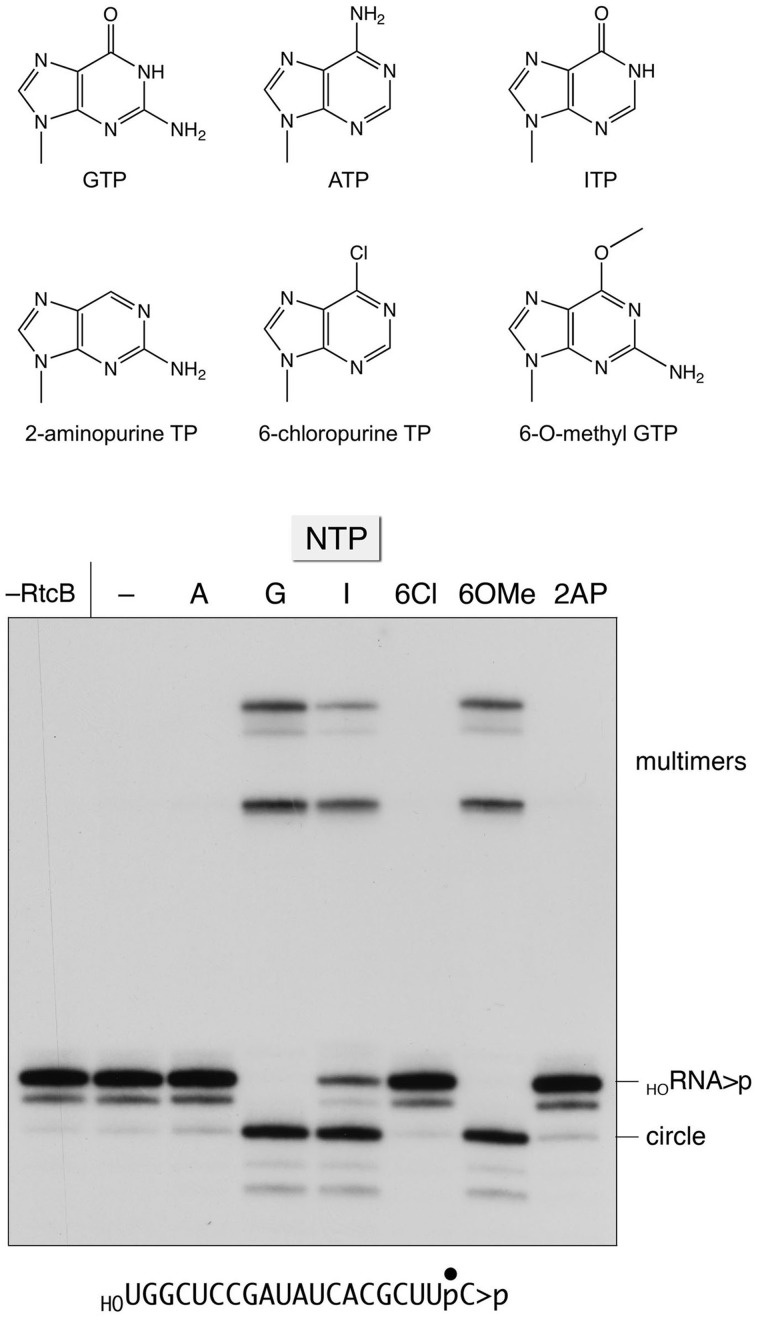
The requirement for GTP as a substrate for covalent activation of the ligase enzyme and the polynucleotide monophosphate substrate distinguishes RtcB from the classic ATP-dependent RNA and DNA ligases, which comprise a superfamily of structurally homologous covalent nucleotidyltransferases that act via an enzyme-(lysyl-Nζ)-AMP intermediate (11). Indeed, RtcB is unable to utilize ATP in lieu of GTP to form the RtcB-NMP adduct or to perform RNA 3′-phosphate ligation (5). Here we sought to gain insights to the nucleobase specificity of RtcB by testing various purine ribonucleoside triphosphate analogs (Figure 8) as substrates for HORNA > p ligation. The reaction products were analyzed by PAGE without prior RNase T1 digestion (Figure 8B). RtcB converted the linear substrate RNA into a more rapidly migrating circular RNA species as a consequence of intramolecular end-joining. More slowly migrating multimers (products of intermolecular ligation) were also generated. We found that the NTP requirement for ligation was satisfied by GTP and 6-O-methyl guanosine triphosphate and inosine triphosphate. However, 6-chloropurine ribonucleoside triphosphate, and 2-aminopurine ribonucleoside triphosphate were inactive, as was ATP (Figure 8). We conclude that (i) the 6-oxo atom is essential because it acts as a hydrogen-bond acceptor, presumably from an amino acid side chain atom on the enzyme (denoted as −XH in Figure 9) and (ii) the exocyclic 2-amino group is not strictly essential.
Figure 8.
Purine nucleobase specificity for RNA ligation. Reaction mixtures containing 50 mM Tris-HCl (pH 8.0), 2 mM MnCl2, 0.1 µM HORNA > p substrate, 1 µM RtcB and either 0.1 mM of the indicated purine NTP (ATP, GTP, ITP, 6-chloropurine TP, 6-O-methyl GTP, or 2-aminopurine TP) or no added NTP (−) were incubated for 10 min at 37°C. The products were analyzed by Urea–PAGE and visualized by autoradiography. RtcB was omitted from a control reaction in the leftmost lane. The labeled HORNA > p substrate and the ligated circle and multimer products are indicated on the right.
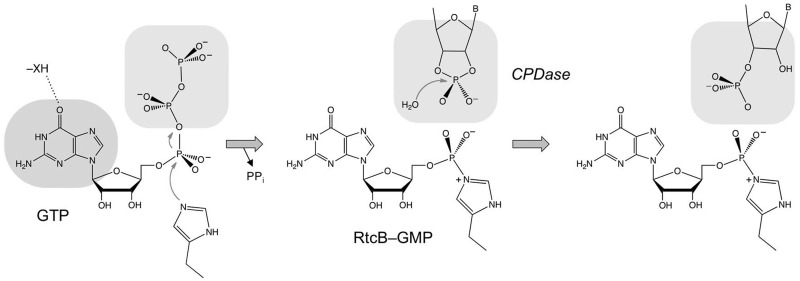
Figure 9.
Scheme for the GTP-dependent 2′,3′-CPDase activity of RtcB. The model posits that guanylylation of RtcB on His337 precedes the CPDase reaction. The histidine-GMP adduct is depicted with a P–Nε covalent bond, although it is not yet established whether the nucleophilic atom is histidine Nε or Nδ. The nucleobase specificity for a 6-oxopurine is likely dictated by hydrogen-bond donation from an enzymic moiety—XH. The model assumes that the binding pocket for the GTP β and γ phosphates (the PPi leaving group in the RtcB guanylylation reaction) overlaps that for the RNA 2,′3′-cyclic phosphate terminus in the CPDase reaction.
DISCUSSION
By studying the reaction of RtcB with a 2′,3′-cyclic phosphate substrate, we provide evidence that supports the ligation pathway in scheme 1 and weighs against the alternative model in scheme 2. Scheme 2 does not predict or explain the stringent requirement for GTP in the ligation of 2′,3′-cyclic phosphate ends, and it does not include or consider the intrinsic CPDase activity of RtcB. Whereas the hydrolysis of the 2′,3′-cyclic phosphate is an obligate first step en route from a 2′,3′-cyclic phosphodiester (which is the product of the incision step of tRNA splicing) to a 3′-phosphate end (the immediate substrate for 3′ guanylylation and subsequent end joining according to scheme 1); such hydrolysis would compete with and thwart the direct ligation mechanism that defines scheme 2. The observed rates of the CPDase and RNA 3′-guanylylation steps under single-turnover conditions studied here are consistent with these reactions being “on-pathway” with respect to 2′,3′-cyclic phosphate and 3′-phosphate ligation. Our results have implications concerning the spectrum of RNA repair reactions that RtcB might perform, insofar as sealing need not be limited to RNAs with 2′,3′-cyclic phosphate ends. RNA-damaging ribotoxins and tRNA splicing endonucleases generate 2′,3′-cyclic phosphate termini as their end-products, and these broken RNAs have been considered the most plausible substrates for splicing or repair by RtcB ligases (1–4). However, many other ribonucleases generate 3′-phosphates, which can now be considered potential RtcB substrates.
The unexpected finding here was that the rate of the CPDase reaction of wild-type RtcB was enhanced by three orders of magnitude by GTP, even though GTP is not chemically transformed during the hydrolysis reaction. Because the H337A mutation reduces 2′,3′ > p hydrolysis to a basal rate, we surmise that RtcB–GMP formation precedes cyclic phosphodiester hydrolysis, as illustrated in the scheme in Figure 9. A potential explanation for the dependence of the CPDase activity on RtcB guanylylation is that occupancy of the guanylate site on the enzyme facilitates engagement of the 3′ end of the RNA substrate in the RtcB active site. There is precedent for such a requirement the case of exemplary ATP-dependent DNA and RNA ligases, where recognition of a 5′-phosphate nicked nucleic acid substrate is contingent on prior formation of the covalent ligase-AMP intermediate, and nick recognition is effaced by mutation of the lysine residue to which AMP is attached (12–15). The rationale for this mechanism, which is suggested by the structures of ATP-dependent polynucleotide ligases bound to their nicked nucleic acid substrates (16–20), is that the reactive ends of the broken nucleic acid are bound on the surface of the ligase overlying the covalent AMP adduct, so that prior engagement of the damaged nucleic acid would preclude the binding of ATP and formation of ligase-AMP. Also, the assignment of damage recognition to pre-adenylylated ligase avoids the problem of non-productive occlusion of sites of nucleic acid damage by inactive ligase apoenzyme. Whereas an analogous mechanism in RtcB makes sense, we do not exclude alternative scenarios involving substrate-assisted catalysis, whereby the covalent guanylate somehow assists CPDase reaction chemistry.
The crystal structure of an archaeal RtcB apoenzyme revealed a distinctive fold with no similarity to any known ligases or phosphotransferases (21). RtcB has a deep and wide active site pocket lined by several conserved and essential histidines and a cysteine, in addition to the nearby histidine nucleophile, that are presumed to comprise the manganese-binding site (4–6,21). The RtcB active site also includes important basic amino acids (5), some of which might interact with either the GTP β and γ phosphates or the RNA 3′-terminal phosphate. Our observation here that GTP inhibits the basal H337A CPDase activity provides a clue to the issue of phosphate interactions. The GTP effect can be explained if the binding sites for the GTP β and γ phosphates (i.e. the PPi leaving group during the RtcB guanylylation step) and the RNA terminal phosphate overlap (Figure 9). In the wild-type RtcB reaction pathway, the dissociation of the PPi leaving group after formation of the histidinyl-GMP adduct would allow the binding of the RNA 2′,3′-cyclic phosphate end, which is hydrolyzed in situ to a 3′-phosphate and then (we presume) instantly guanylylated via nucleophilic attack of an RNA 3′-phosphate oxygen on the histidinyl-GMP phosphorus. However, in the case of H337A, which cannot execute GMP transfer from GTP to RtcB, the β and γ phosphates impede engagement of RNA > p. Note that in the classic ATP-dependent DNA and RNA ligases, the positions and atomic contacts of the PPi leaving group extensively overlap that of the 5′-phosphate end of the broken polynucleotide substrate (14,16–20,22–25).
FUNDING
Funding for open access charge: National Institutes of Health [GM46330].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
S.S. is an American Cancer Society Research Professor.
REFERENCES
- 1.Tanaka N, Shuman S. RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J. Biol. Chem. 2011;286:7727–7731. doi: 10.1074/jbc.C111.219022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Popow J, Englert M, Weitzer S, Schleiffer A, Mierzwa B, Mechtler K, Trowitzsch S, Will CL, Lürhmann R, Söll D. HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science. 2011;33:760–764. doi: 10.1126/science.1197847. [DOI] [PubMed] [Google Scholar]
- 3.Englert M, Sheppard K, Aslanian A, Yates JR, Söll D. Archaeal 3′-phosphate RNA splicing ligase characterization identified the missing component in tRNA maturation. Proc. Natl. Acad. Sci. USA. 2011;108:1290–1295. doi: 10.1073/pnas.1018307108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka N, Meineke B, Shuman S. RtcB, a novel RNA ligase, can catalyze tRNA splicing and HAC1 mRNA splicing in vivo. J. Biol. Chem. 2011;286:30253–30257. doi: 10.1074/jbc.C111.274597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka N, Chakravarty AK, Maughan B, Shuman S. A novel mechanism of RNA repair by RtcB via sequential 2′,3′-cyclic phosphodiesterase and 3′-phosphate/5′-hydroxyl ligation reactions. J. Biol. Chem. 2011;286:43134–43143. doi: 10.1074/jbc.M111.302133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakravarty AK, Subbotin R, Chait BT, Shuman S. RNA ligase RtcB splices 3′-phosphate and 5′-OH ends via covalent RtcB-(histidinyl)-GMP and polynucleotide-(3′)pp(5′)G intermediates. Proc. Natl. Acad. Sci. USA. 2012;109:6072–6077. doi: 10.1073/pnas.1201207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desai KK, Raines RT. tRNA ligase catalyzes the GTP-dependent ligation of RNA with 3′-phosphate and 5′-hydroxyl termini. Biochemistry. 2012;51:1333–1335. doi: 10.1021/bi201921a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Genschik P, Drabikowski K, Filipowicz W. Characterization of the Escherichia coli RNA 3′-terminal phosphate cyclase and its σ54-regulated operon. J. Biol. Chem. 1998;273:25516–25526. doi: 10.1074/jbc.273.39.25516. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka N, Smith P, Shuman S. Structure of the RNA 3′-phosphate cyclase–adenylate intermediate illuminates nucleotide specificity and covalent nucleotidyl transfer. Structure. 2010;18:449–457. doi: 10.1016/j.str.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang LK, Shuman S. Mutational analysis defines the 5′ kinase and 3′ phosphatase active sites of T4 polynucleotide kinase. Nucleic Acids Res. 2002;30:1073–1080. doi: 10.1093/nar/30.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shuman S, Lima CD. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr. Opin. Struct. Biol. 2004;14:757–764. doi: 10.1016/j.sbi.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 12.Sekiguchi J, Shuman S. Nick sensing by DNA ligase requires a 5′ phosphate at the nick and occupancy of the adenylate binding site on the enzyme. J. Virol. 1997;71:9679–9684. doi: 10.1128/jvi.71.12.9679-9684.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sriskanda V, Shuman S. Chlorella virus DNA ligase: nick recognition and mutational analysis. Nucleic Acids Res. 1998;26:525–531. doi: 10.1093/nar/26.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Odell M, Sriskanda V, Shuman S, Nikolov D. Crystal structure of eukaryotic DNA ligase–adenylate illuminates the mechanism of nick sensing and strand joining. Mol. Cell. 2000;6:1183–1193. doi: 10.1016/s1097-2765(00)00115-5. [DOI] [PubMed] [Google Scholar]
- 15.Nandakumar J, Shuman S. How an RNA ligase discriminates RNA damage versus DNA damage. Mol. Cell. 2004;16:211–221. doi: 10.1016/j.molcel.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 16.Pascal JM, O’Brien PJ, Tomkinson AE, Ellenberger T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004;432:473–478. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- 17.Nandakumar J, Shuman S, Lima CD. RNA ligase structures reveal the basis for RNA specificity and conformational changes that drive ligation forward. Cell. 2006;127:71–84. doi: 10.1016/j.cell.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 18.Nandakumar J, Nair PA, Shuman S. Last stop on the road to repair: structure of E. coli DNA ligase bound to nicked DNA-adenylate. Mol. Cell. 2007;26:257–271. doi: 10.1016/j.molcel.2007.02.026. [DOI] [PubMed] [Google Scholar]
- 19.Nair PA, Nandakumar J, Smith P, Odell M, Lima CD, Shuman S. Structural basis for nick recognition by a minimal pluripotent DNA ligase. Nature Struct. Mol. Biol. 2007;14:770–778. doi: 10.1038/nsmb1266. [DOI] [PubMed] [Google Scholar]
- 20.Cotner-Gohara E, Kim IK, Hammel M, Tainer JA, Tomkinson AE, Ellenberger T. Human DNA ligase III recognizes DNA ends by dynamic switching between two DNA-bound states. Biochemistry. 2010;49:6165–6176. doi: 10.1021/bi100503w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okada C, Maegawa Y, Yao M, Tanaka I. Crystal structure of an RtcB homolog protein (PH1602-extein protein) from Pyrococcus horikoshii reveals a novel fold. Proteins. 2006;63:1119–1112. doi: 10.1002/prot.20912. [DOI] [PubMed] [Google Scholar]
- 22.Subramanya HS, Doherty AJ, Ashford SR, Wigley DB. Crystal structure of an ATP-dependent DNA ligase from bacteriophage T7. Cell. 1996;85:607–615. doi: 10.1016/s0092-8674(00)81260-x. [DOI] [PubMed] [Google Scholar]
- 23.Deng J, Schnaufer A, Salavati R, Stuart KD, Hol WG. High resolution crystal structure of a key editosome enzyme from Trypanosoma brucei: RNA editing ligase 1. J. Mol. Biol. 2004;343:601–613. doi: 10.1016/j.jmb.2004.08.041. [DOI] [PubMed] [Google Scholar]
- 24.El Omari K, Ren J, Bird LE, Bona MK, Klarmann G, LeGrice SF, Stammers DK. Molecular architecture and ligand recognition determinants for T4 RNA ligase. J. Biol. Chem. 2006;281:1573–1579. doi: 10.1074/jbc.M509658200. [DOI] [PubMed] [Google Scholar]
- 25.Pascal JM, Tsodikov OV, Hura GL, Song W, Cotner EA, Classen S, Tomkinson AE, Tainer JE, Ellenberger T. A flexible interface between DNA ligase and PCNA supports conformational switching and efficient ligation of DNA. Mol. Cell. 2006;24:279–291. doi: 10.1016/j.molcel.2006.08.015. [DOI] [PubMed] [Google Scholar]