Abstract
Cockayne syndrome (CS) is a rare human disorder characterized by pathologies of premature aging, neurological abnormalities, sensorineural hearing loss and cachectic dwarfism. With recent data identifying CS proteins as physical components of mitochondria, we sought to identify protein partners and roles for Cockayne syndrome group B (CSB) protein in this organelle. CSB was found to physically interact with and modulate the DNA-binding activity of the major mitochondrial nucleoid, DNA replication and transcription protein TFAM. Components of the mitochondrial transcription apparatus (mitochondrial RNA polymerase, transcription factor 2B and TFAM) all functionally interacted with CSB and stimulated its double-stranded DNA-dependent adenosine triphosphatase activity. Moreover, we found that patient-derived CSB-deficient cells exhibited a defect in efficient mitochondrial transcript production and that CSB specifically promoted elongation by the mitochondrial RNA polymerase in vitro. These observations provide strong evidence for the importance of CSB in maintaining mitochondrial function and argue that the pathologies associated with CS are in part, a direct result of the roles that CSB plays in mitochondria.
INTRODUCTION
Cockayne syndrome (CS) is a rare autosomal recessive segmental progeria characterized by cachectic dwarfism, cutaneous photosensitivity, sensorineural hearing loss, cataracts, loss of subcutaneous fat and neuropathologies (white matter hypomyelination, central nervous system calcification and microcephaly (1,2)). Mutations in the Cockayne syndrome group B (CSB) gene (ERCC6) account for ∼80% of CS cases (3) and have also been found in de Sanctis–Cacchione syndrome (4), Cerebro-oculo-facio-skeletal syndrome (5) and UV sensitive syndrome (6); the remaining causative CS mutations reside in the CSA gene (ERCC8, CKN1) (7). CSA is a WD40 repeat-containing protein that is part of an ubiquitin ligase complex with Cul4A and Roc1 (8), though no direct biochemical function has yet been ascribed to CSA. The CSB protein contains seven sequence motifs characteristic of superfamily 2 (SF2) adenosine triphosphatases (ATPases)/helicases/nucleic acid translocases (9).
CSB stably interacts with a range of double-stranded nucleic acids (e.g. DNA/DNA, DNA/RNA and RNA/RNA) and possesses an adenosine triphosphate (ATP)-independent complementary strand annealing activity to generate double-stranded molecules (10,11). CSB has been shown to wrap DNA around itself (12), change plasmid topology, remodel mononucleosomes (13) and remove or rearrange a pre-bound protein from DNA (11), although the in vivo roles of these biochemical activities are as of yet unclear. The most prominent biochemical activity of CSB is its ability to hydrolyze ATP in the presence of duplex DNA and Mg2+ (14–16) or uniquely Ca2+ (11), yet no associated motor activity (helicase/translocase) coupled to ATP hydrolysis has been identified (11,14,15) and no protein partners of CSB have been identified that stimulate this activity in the presence of double-stranded DNA (dsDNA).
CSB is essential for nuclear transcription-coupled nucleotide excision repair (TC-NER)—a sub-pathway of NER that operates to remove RNA polymerase blocking lesions (17)—and is considered the human transcription-coupled repair factor (14). CSB also plays roles in general nuclear transcription, interacting with and stimulating elongation by RNA polymerases I (∼10-fold) and II (∼3-fold) (18–23). In addition, CSB acts as an auxiliary factor in nuclear base excision repair (BER) (24–27) and participates in mitochondrial BER, facilitating efficient repair of oxidative mitochondrial DNA (mtDNA) lesions (28) and promoting incision activities for additional types of base lesions (uracil and 5-hydroxyuracil) in mitochondrial extracts (29).
Consistent with functions in mitochondria, both CSA and CSB have been shown to localize within this organelle (29,30). mtDNA repair abnormalities associated with CSB deficiency have been revealed as accumulation of 8-oxoguanine (31) and formamidopyrimidine (27) base lesions in mtDNA from csbm/m mice, accelerated accumulation of mtDNA mutations in CSB-deficient human cells (30) and increased mitochondrial mutagenesis in CSB knockdown human cells (29). csbm/m mouse embryonic fibroblasts were also shown to be hypersensitive to agents that inhibit mitochondrial bioenergetics (e.g. rotenone, 3-nitroproprionic acid, KCN and 2-deoxyglucose) through a mechanism that did not involve mtDNA damage, and altered mitochondrial respiratory complex organization was found in fibroblasts and brain of csbm/m mice (31), indicating roles for CSB outside of mtDNA repair (31). In addition, CSB specifically has been shown to co-localize with the essential mitochondrial protein factor TFAM (mitochondrial transcription factor A (mtTFA), TCF6) (29), which functions in multiple aspects of mitochondrial metabolism.
TFAM is a 29-kDa nuclear-encoded high-mobility group (HMG) family protein that is an abundant and central protein in mtDNA maintenance and was originally purified from HeLa cells as an activator of mitochondrial transcription (32). In addition, TFAM binds mtDNA and organizes it into nucleoid-like structures (33), appears to be important for mitochondrial genome replication (34) and is vital for early development as homozygous disruption leads to embryonic lethality in mice (E10.5), with embryos lacking mtDNA (35). In addition to TFAM, mitochondrial transcription factor B2 (TFB2M) and mitochondrial RNA polymerase (POLRMT) comprise the apparatus required for efficient mitochondrial transcription initiation in vitro (36,37). Previously, it was thought that the core human mitochondrial transcriptional initiation apparatus was a three-component system, but recent work has determined that it is a two-component system comprised of POLRMT and TFB2M, with TFAM acting as a transcriptional activator (38).
Transcription of the ∼16.5 kb mitochondrial genome is vital, with large polycistronic transcripts, that arise from the light and heavy strand promoters (LSP and HSP), encoding components necessary for maintaining proper mitochondrial function and cellular energy generation. The LSP transcript (∼11 kb) codes for eight transfer RNAs, a single subunit of the oxidative phosphorylation (OXPHOS) system and generates the primer required for mtDNA replication. The HSP transcript (∼16 kb) encodes 12 subunits of the OXPHOS system, 2 ribosomal RNAs and 14 transfer RNAs. Interestingly, POLRMT produces only short transcripts (∼20–100 nt) using single-stranded DNA (ssDNA) as a template in vitro (39). Recently, a protein factor (c17orf42/transcription elongation factor of mitochondria (TEFM)) has been identified that promotes elongation by POLRMT (using ssDNA as a template). TEFM was found to enhance formation of transcripts to ∼400 nt in length and to influence the ratio of long to short RNAs produced by POLRMT, with production of long transcripts over short transcripts increased ∼2-fold (40). Mitochondrial ribosomal protein L12 (MRPL12) has also recently been characterized as a factor that influences mitochondrial transcript production, increasing the amount of transcripts produced ∼3-fold, though whether MRPL12 exerts this effect at the initiation, elongation or both phases of transcription is as of yet unclear (41). It is, nevertheless, apparent that additional factors are required for production of the large mtDNA-encoded transcripts.
The ‘free radical theory of aging’ postulates that the cause of aging is the accumulation of reactive oxygen species (ROS)-induced damage (42,43). Since ROS can damage mtDNA, a vicious cycle may be propagated whereby the increased mtDNA damage leads to impaired mitochondrial function (replication, transcription and OXPHOS), thereby promoting increased ROS and further mitochondrial dysfunction. Logically, it follows that it is therefore especially important to maintain proper mitochondrial function. We hypothesize that certain pathologies of the premature aging disease CS may be rooted in mitochondrial dysfunction caused by loss of critical activities performed by the CS proteins. Herein, we sought to examine the role(s) that CSB plays in mitochondria, focusing on potential contributions to transcription.
MATERIALS AND METHODS
CS1AN cell lines
CS1AN is a SV40 transformed fibroblast cell line derived from a patient with CS from complementation group B and contains one CSB allele truncating the protein at amino acid 337 (K337Stop) and the other allele containing a 100-nt deletion in exon 13 (44). The parental cell line was stably transfected with pcDNA3.1 vector (CS1AN-V) or vector that either stably expresses wild-type CSB protein (CS1AN-CSBWT) or an ATPase-deficient CSB protein (CS1AN-CSBE646Q) (25). Cells were cultured in Dulbecco’s Modified Eagle Media (DMEM) supplemented with 10% fetal bovine serum (FBS), 1 × penicillin/streptomycin and 400 μg/ml geneticin at 37°C in a humidified 5% CO2 incubator.
Mitochondrial transcript measurement
Total RNA was isolated from CS1AN-CSBWT, CS1AN-CSBE646Q and CS1AN-V cell lines using TRIzol reagent (Invitrogen, Carlsbad, CA). Purified RNA was quantitated using a Nanodrop ND-1000 spectrophotometer. Complementary DNA (cDNA) was prepared from 2 µg of total RNA by random hexameric priming and MultiScribe reverse transcriptase (Applied Biosystems, Foster City, CA). Dilutions of cDNA (between 2 and 200 pg) were used as input template for quantitative polymerase chain reaction (Q-PCR) with gene-specific probes for mitochondrial-encoded MTCO1, MTA6 or MTND4L using 7900HT fast real-time PCR system (Applied Biosystems). Thermocycling conditions were 2 min at 50°C, 10 min at 95°C, 15 s at 95°C and 1 min at 60°C, cycled 40 times. Gene amplification was measured by an onboard fluorescence detector in real time. MTCO1, MTA6 and MTND4L transcript levels were normalized to transcript levels for the nuclear-encoded gene glyceralde-3-phosphate dehydrogenase (GAPDH) as per (45). Averages and standard deviations of transcript levels were calculated from triplicate experiments from three independent RNA isolations.
Quantitative long-range PCR measurement of mtDNA damage and mtDNA copy number
The long amplification Q-PCR assay was performed as described in (46) with minor changes. In brief, DNA was extracted from CS1AN-CSBWT, CS1AN-CSBE646Q and CS1AN-V cell lines using the QIAamp DNA blood extraction kit (QIAGEN, CA) and quantified using the Pico Green dsDNA Quantitation kit (Molecular Probes, Invitrogen, CA). The relative amount of initial mtDNA or nuclear DNA added to the Q-PCR reaction was verified using a real-time PCR TAQMAN® probe for the mitochondrial gene MTCO1 and GAPDH for nuclear DNA (Applied Biosystems, CA). We used the human mitochondrial primer set 14 841 and 5999 and the human nuclear primer set 48 510 and 62 007 to amplify 8.9 and 13.5 kb products, respectively (46). PCR product formation was verified by agarose gel electrophoresis. Amplification was conducted using an Eppendorf Mastercycler (Eppendorf, NY). The amount of PCR product was quantitated using the PicoGreen procedure and normalized against input mtDNA and genomic DNA. The quantitation of the mitochondrial copy number relative to a nuclear housekeeping gene was conducted in an identical manner to the mtDNA quantitation described above using GAPDH TAQMAN® probe (Applied Biosystems) as the internal genomic control. One sample t-test was performed on each group using the theoretical mean of 100%.
TFAM knockdown and immunofluorescence
ON-TARGETplus SMARTpool siRNAs directed against TFAM messenger RNA (mRNA) (M-019734-00) and siGENOME non-targeting siRNA1 (scrambled control, D-001210-01) were obtained from Dharmacon RNAi Technologies, Lafayette, CO. HeLa cells, in DMEM supplemented with 10% FBS, were seeded at a density of 5 × 104 cells in Lab-Tek chambered slides (Nunc, Rochester, NY) and allowed to adhere for 24 h at 37°C. Cells were then transfected with appropriate siRNAs and incubated at 37°C for 24 h. Subsequently, media were replaced with DMEM supplemented with 10% FBS and 1 × penicillin/streptomycin, and cells were incubated for an additional 48 h. Cells were fixed with 4% (vol/vol) formaldehyde, permeabilized with 0.2% (vol/vol) TritonX-100 and blocked with 3% (wt/vol) bovine serum albumin (BSA). For CSB detection, cells were incubated with rabbit α-CSB antibody (H-300, Santa Cruz Biotechnologies, Santa Cruz, CA) at a dilution of 1/200 for 1 h at 37°C. For recognition of COX4, cells were incubated with goat α-COX4 antibody (Q-17, Santa Cruz Biotechnologies) at a dilution of 1/200 for 1 h at 37°C. Slides were washed with PBS for 30 min at room temperature (RT) three times. For visualization of CSB and COX4, slides were incubated with Alexa Fluor-conjugated donkey α (rabbit and goat, respectively)-secondary antibodies (Sigma-Aldrich, St Louis, MO) at a dilution of 1/1000 for 1 h at 37°C. Slides were washed with PBS for 30 min at RT three times. Indirect immunofluorescence of CSB was performed by exciting slides at a wavelength of 514 nm and for COX4, indirect immunofluorescence was performed by exciting slides at a wavelength of 640 nm. All slides were imaged on a Nikon Eclipse TE2000E Spinning-disk Confocal Microscope. Co-localization analysis of CSB and COX4 was performed using Volocity software (Perkin-Elmer, Waltham, MA).
Proteins
Recombinant CSBWT and CSBE646Q proteins were expressed in Sf9 insect cells through a baculoviral system as described (11,15). Purification was performed by Ni2+ affinity, heparin and anion exchange (source Q) chromatography, which allowed for isolation of protein to near homogeneity. Recombinant TFAMWT and TFAML58A were expressed in Escherichia coli BL21(DE3) cells and purified by Ni2+ affinity chromatography to near homogeneity as described (47). Recombinant POLRMT and TFB2M were purchased from Enzymax, LLC (Lexington, KY). Recombinant XPG was a kind gift from Dr Orlando Scharer (State University of New York, Stony Brook).
CSB:TFAM co-immunoprecipitation
Recombinant HA-tagged CSB and His6-tagged TFAM were incubated in 20 mM HEPES pH 7.9, 4 mM MgCl2, 0.05 mM ATP, 40 µg/ml BSA and 1 mM DTT at 4°C overnight in the presence of 5-fold excess of monoclonal mouse α-HA antibody (12CA5, Novus Biologicals, Littleton, CO) and protein A/G agarose (Thermo Scientific, Pierce Protein Research Products, Rockford, IL). Protein A/G agarose and associated material were pelleted by centrifugation at 1500 × g for 1 min, washed with 25 mM HEPES pH 7.9, 300 mM KCl, 10% (vol/vol) glycerol and 0.01% NP40, and pelleted by centrifugation at 1500 × g for 1 min. Pelleted material was resuspended in 2 × sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) loading dye, incubated at 95°C for 5 min and loaded onto a 4–20% Tris–glycine–SDS polyacrylamide gel. Protein was transferred to a PVDF membrane at 90 V for 90 min. TFAM was detected using mouse monoclonal α-His6 antibody (Calbiochem, Gibbstown, NJ).
Mitochondrial isolation and detection of CSB and TFAM
GM1030 lymphoblastoid cells were obtained from ATCC and cultured in DMEM supplemented with 10% FBS and 1 × penicillin/streptomycin at 37°C in a shaking incubator with 5% CO2. Mitochondria were isolated by harvesting cells by centrifugation and resuspending in MSHE buffer (0.21 M mannitol, 0.07 M sucrose, 10 mM HEPES (pH 7.4), 1 mM EDTA, 1 mM EGTA, 0.15 mM spermine, 0.75 mM spermidine, 5 mM DTT and protease inhibitor cocktail). Cells were homogenized on ice in a Potter–Elvehjem homogenizer. Homogenate was subjected to centrifugation at 500 × g for 10 min then 10 000 × g for 12 min. Pelleted material was layered on a 1:1 mixture of percol and 2 × MSHE buffer gradient and centrifuged at 130 000 × g for 1 h. The fraction containing mitochondria was washed in MSHE buffer and resuspended in a mixture of 100 μl of buffer A (10 mM Tris–HCl, pH 7.8 and 200 mM KCl) and 200 μl of buffer B (10 mM Tris–HCl, 600 mM KCl, 2 mM DTT, 2 mM EDTA, 40% glycerol, 0.2% NP-40 and 0.5 mM PMSF) containing protease inhibitors (Roche, Indianapolis, IN). Resuspended mitochondria were sonicated three times at power level 3 with a 10-s pulse time separated by 1 min intervals on ice using a Microson Ultrasonic Cell Disruptor. The sonicated mixture was incubated at 4°C for 1.5 h on a rocker then centrifuged at 130 000 × g for 1 h at 4°C. Supernatant was dialyzed against buffer C (25 mM HEPES–KOH, pH 8.0, 100 mM KCl, 1 mM DTT, 1 mM EDTA, 17% glycerol and 12 mM MgCl2) followed by another round of centrifugation for 10 min at 16 000 × g (47).
Forty micrograms of GM1030 mitochondrial extract was mixed with 2 × SDS–PAGE loading dye, incubated at 95°C for 5 min and loaded on a 6 or 12% Tris–glycine–SDS polyacrylamide gel along with recombinant CSB or TFAM at concentrations of 10, 30, 100, 300 or 1000 ng, respectively. Protein was transferred to a PVDF membrane at 90 V for 90 min. CSB was detected using rabbit polyclonal α-CSB antibody, raised against the C-terminal portion of the protein, at a dilution of 1/500. TFAM was detected using mouse polyclonal α-TFAM antibody (ab89818; Abcam, Cambridge, MA) at a dilution of 1/500. Donkey α-rabbit (711-035-152) or goat α-mouse (115-035-003) peroxidase conjugated secondary antibodies were used (Jackson ImmunoResearch Laboratories, West Grove, PA) at dilutions of 1/1000.
ATPase assay
Buffers for standard ATPase reactions contained 20 mM HEPES–OH, pH 8.0, 0.05 mM ATP, 40 µg/ml BSA and 1 mM DTT. Reactions were supplemented with 1 mM Ca2+ or 4 mM Mg2+ as indicated. ATPase reactions used 30 nM CSB and 12.5 µCi γ-32P ATP. Reactions were incubated for 1 h at 30°C with 150 ng of pUC19 supercoiled plasmid DNA in a final volume of 10 µl. Reactions were stopped by the addition of 5 µl 0.5 M EDTA. ATP hydrolysis was analyzed by polyethyleneimine-thin layer chromatography (PEI-TLC) using 1 M formic acid/0.8 M LiCl. PEI-TLC plates were analyzed and quantitated on a Typhoon phosphorimager using OptiQuant TL software. For reactions with mitochondrial transcription proteins, 30, 60 or 120 nM of TFB2M, POLRMT or TFAM were included.
Restriction enzyme site accessibility assay
Buffer for restriction enzyme site accessibility assays contained 10 mM Tris–Cl, pH 7.4, 20 mM NaCl, 25 mM KCl, 0.5 mM EDTA, 5 mM DTT and 5% (vol/vol) glycerol. Fifty femtomoles of substrate (42BamHI/42Comp, Supplementary Table S1) was incubated with 1 µg (40 pmol, 1 µM) TFAM at 30°C for 15 min in a final volume of 30 µl. Subsequently, CSBWT (50, 150, 250, 350 or 500 fmol) was added to the reaction, followed immediately by addition of 1 U BamHI enzyme (New England Biolabs, Ipswich, MA) and incubation at 30°C for 30 min. An equal volume of stop buffer (95% formamide, 20 mM EDTA, 0.5% bromophenol blue and 0.5% xylene cyanol) was then added. Samples were heated at 95°C for 3 min, and subsequently loaded onto a 15% polyacrylamide-urea denaturing gel and electrophoresed at 225 V for 90 min. Substrates and products were visualized and quantitated by phosphorimager analysis using OptiQuant TL.
Electrophoretic mobility shift assay
Buffer for CSB protein removal assays was the same as used for restriction enzyme site accessibility assays. Fifty femtomoles of substrate (42BamHI/42Comp) was incubated with 150 ng (5.5 pmol, 275 nM) TFAM at 30°C for 30 min in a final volume of 20 µl. CSBWT (3, 9, 15, 18 or 25 nM) was then added to the reactions and allowed to proceed at 30°C for 30 min. Reactions were loaded on a native 4% polyacrylamide gel at 4°C and run at 15 mA for 90 min. Substrates and products were visualized and quantitated as above.
Mitochondrial transcription initiation assay
Reaction conditions consisted of 10 mM HEPES–OH, pH 7.5, 100 mM KCl, 10 mM MgCl2, 1 mM DTT and 0.1 mg/ml BSA. The rNTP mix contained 400 µM ATP, 150 µM CTP, 150 µM GTP, 10 µM UTP and 10 µCi α-32P UTP. 2.5 µM HSP dsDNA template (hmtHSPTS/hmtHSPNTS, Supplementary Table S1) was included in the reactions with 250 nM TFB2M, 250 nM TFAM, 250 POLRMT and 250, 500 or 1000 nM CSB. TFB2M, TFAM, POLRMT and oligonucleotide substrate were incubated together at 32°C for 5 min, then CSB was added and incubated at 32°C for 5 min before addition of rNTP mix to initiate the reactions. Transcription was allowed to proceed for 30 min at 32°C, and reactions were then quenched by addition of formamide stop buffer. Samples were heated at 95°C for 3 min and then loaded onto a 20% polyacrylamide-urea denaturing gel and electrophoresed at 200 V for 3 h. Substrates and products were visualized and quantitated as above.
POLRMT elongation assay
Reaction buffer contained 10 mM Tris–Cl, pH 8.0, 20 mM MgCl2, 1 mM DTT and 0.1 mg/ml BSA. The rNTP mix was as above, but with 1 µCi α-32P UTP. Two hundred nanograms single-stranded M13mp18 DNA (New England Biolabs) was included in the reactions with 40 nM POLRMT and 40, 80 or 160 nM CSB. Transcription was allowed to proceed for 1 h at 32°C, and then reactions were quenched by addition of formamide stop buffer. Samples were heated at 95°C for 5 min and then loaded onto a 5% polyacrylamide-urea denaturing gel and electrophoresed at 20 W for 2.5 h. Substrates and products were visualized and quantitated as above.
RESULTS
CSB-deficient cells have a defect in mitochondrial transcription
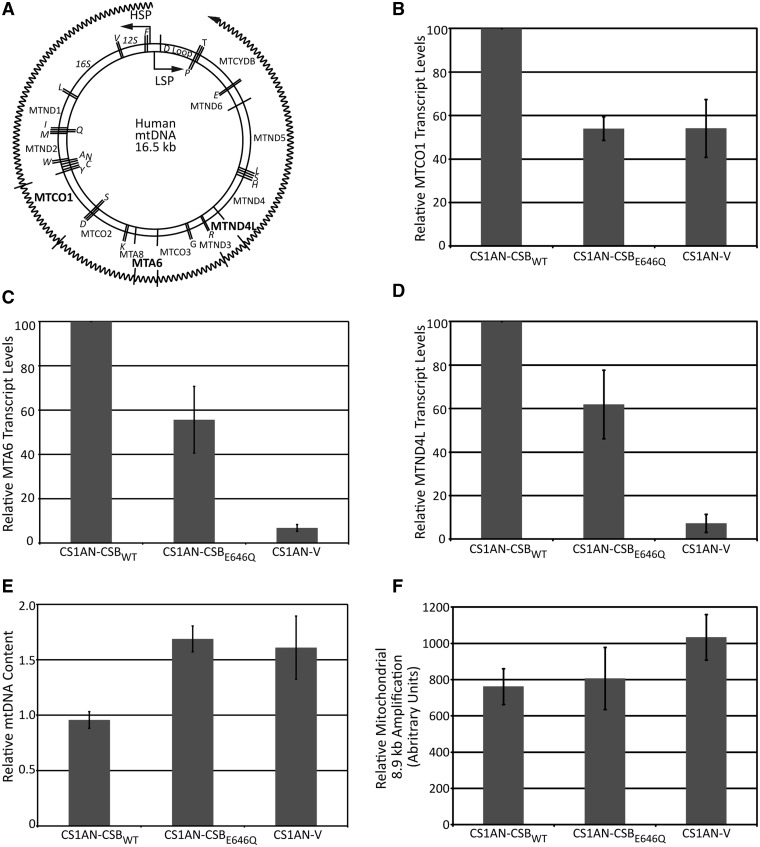
In the nucleus, CSB is involved in both DNA repair (TC-NER and BER) and transcription. A role for CSB in mtDNA repair has been intimated (28,29), yet no role in mitochondrial transcription has been ascribed. Using cells derived from a patient with CS (25), either stably expressing wild-type CSB protein (CS1AN-CSBWT) or an ATPase-deficient CSB protein (CS1AN-CSBE646Q), or stably transfected with vector alone (CS1AN-V), we investigated whether CSB status had an effect on transcription of mitochondrial genes. Using quantitative reverse transcriptase (RT)–PCR, we measured relative transcript levels of three differentially positioned mitochondrially encoded genes transcribed from the HSP: MTCO1 (mtDNA nucleotides 5904–7445, coding for cytochrome c oxidase I, complex IV), MTA6 (mtDNA nucleotides 8527–9207, coding for ATP synthase F0 subunit 6, complex V) and MTND4L (mtDNA nucleotides 10 470–10 766, coding for NADH dehydrogenase 4 L, complex I) (Figure 1A). In cells lacking CSB or expressing an ATPase dead version of the protein (CSBE646Q), MTCO1 transcript levels were ∼50% lower than in isogenic cells expressing CSBWT (Figure 1B). For MTA6 and MTND4L, the transcript reduction in CSB-deficient cells was even more dramatic, with levels reduced to <10% of that seen with the CSBWT complemented cells (Figure 1C and D). CS1AN cells expressing CSBE646Q produced more MTA6 and MTND4L transcript than CSB-deficient cells (∼60% versus ∼10% total), but still ∼40% less than that of CSBWT complemented cells (Figure 1C and D). These results indicate that the ATPase activity of CSB is required to maintain full transcription of MTCO1, MTA6 and MTND4L, yet that there is an ATP-independent contribution of CSB to mitochondrial transcription, as CSBE646Q was able to promote ∼40% more MTA6 and MTND4L transcript formation than measured in CSB-deficient cells.
Figure 1.
CSB-deficient cells display a defect in mitochondrial transcription. (A) Human mitochondrial genome schematic. Genes expressed from the LSP are denoted on the interior, whereas genes expressed from the HSP are denoted on the exterior. Genes for rRNAs and tRNAs (denoted by the single amino acid letter that they code for) are italicized. The exterior wavy line represents the polycistronic transcript generated from the HSP. Transcript segments that were measured are noted with lines through the transcript and gene names listed in bold. (B) Graph displaying relative mitochondrial MTCO1 transcription levels, quantified by Q-RT-PCR, in cells from a patient with CS stably expressing wild-type CSB (CS1AN-CSBWT), stably expressing CSB ATPase dead protein (CS1AN-CSBE646Q) or stably transfected with an empty vector (CS1AN-V). (C) Graph displaying relative mitochondrial MTA6 transcription levels, quantified by Q-RT-PCR, in CS1AN-CSBWT, CS1AN-CSBE646Q or CS1AN-V cells. (D) Graph displaying relative mitochondrial MTND4L transcription levels, quantified by Q-RT-PCR, in CS1AN-CSBWT, CS1AN-CSBE646Q or CS1AN-V cells. (E) Graph displaying relative mtDNA content, quantified by Q-PCR, in CS1AN-CSBWT, CS1AN-CSBE646Q or CS1AN-V cells. (F) Graph displaying relative amplification levels of an 8.9 kb segment of the mitochondrial genome from CS1AN-CSBWT, CS1AN-CSBE646Q or CS1AN-V cells.
We considered that the reduced mitochondrial transcript levels could be due to deceased mitochondrial content. To address this, we measured the relative mtDNA content in CS1AN-CSBWT, CS1AN-CSBE646Q or CS1AN-V cells by real-time Q-PCR. For this, mitochondrial target sequence amplification is normalized to the amplification of a nuclear genome target sequence and copy number calculated from the threshold cycle values. Consistent with recent data (48), we observed that cells expressing an ATPase-deficient CSB or lacking CSB protein displayed increased mtDNA content relative to cells expressing wild-type CSB protein (Figure 1E), indicating that the diminished steady-state levels of MTCO1, MTA6 and MTND4L transcripts were not because of lower mtDNA levels.
As CSB has previously been shown to function in the repair of mtDNA damage, we wondered whether the reduction in transcription was due to increased polymerase blocking lesions within mtDNA. Quantitative long-range PCR was used to compare relative amplification efficiencies of an 8.9 kb portion of the mitochondrial genome in CS1AN-CSBWT, CS1AN-CSBE646Q and CS1AN-V cells. Increased polymerase blocking lesions lead to decreased amplification efficiency, indirectly measuring DNA damage levels (46). We found that cells lacking CSB or expressing CSBE646Q did not show greater levels of polymerase blocking lesions compared with cells expressing CSBWT (Figure 1F). This suggests that the types of oxidative DNA damage that accumulate in the absence of CSB (27,31) do not significantly block polymerase progression and that the transcription defects described above (Figure 1B–D) are not likely due to the roles of CSB in repair of mtDNA damage, but instead a direct function in mitochondrial transcription.
TFAM directly modulates CSB function
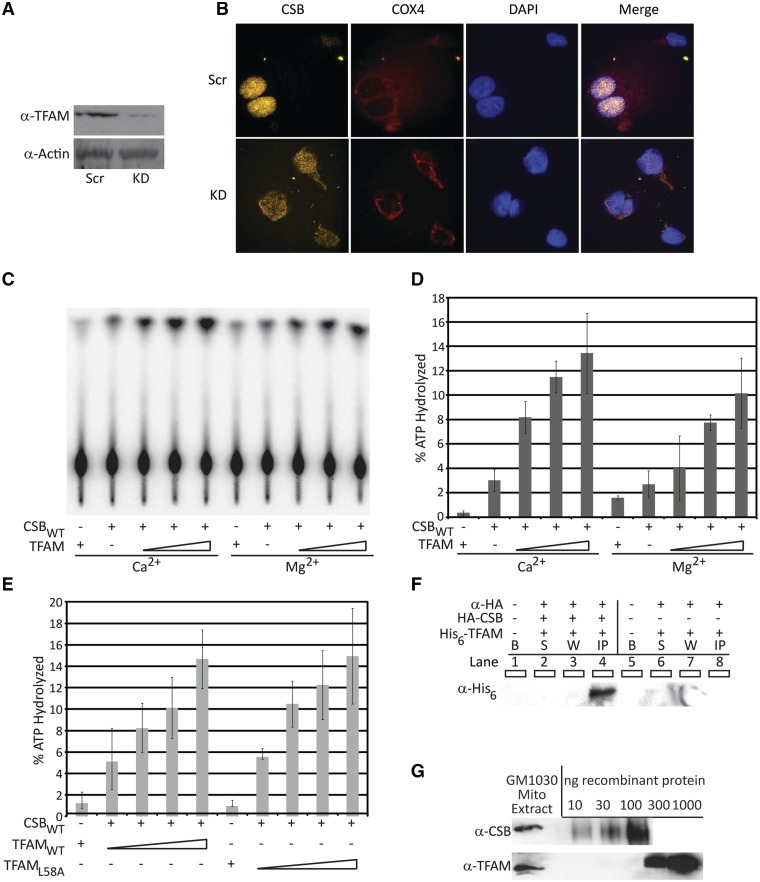
CSB appears to be a dynamic physical component of mitochondria with increased mitochondrial subcellular localization and co-localization with TFAM after oxidative stress (29). We find here that depletion of TFAM in HeLa cells, which decreases cellular TFAM levels to ∼5–10% (Figure 2A), results in increased localization of CSB to mitochondria. Specifically, CSB nuclear staining (yellow) decreases in TFAM knockdown cells (Figure 2B, far left panel) and shows increased co-localization with the mitochondrial protein cytochrome c oxidase subunit IV (COX4, red) (Figure 2B far right panel). This finding is consistent with the idea that CSB intracellular distribution is affected by oxidative stress, which has previously been associated with loss of TFAM (47,49).
Figure 2.
TFAM directly influences CSB localization and ATPase activity. (A) Western blot displaying TFAM knockdown. Scr, scrambled siRNA; KD, knockdown by TFAM-specific siRNA. (B) Immunofluorescence of CSB re-localization to mitochondria in the absence of TFAM. Far left panel, CSB (yellow) localization; middle left panel, mitochondrial marker protein COX4 (red) localization; right middle panel, DAPI staining (blue) of nuclear DNA; far right panel, merged images of CSB (yellow), COX4 (red) and DAPI (blue) in HeLa cells transfected with non-targeted siRNA (Scr) or TFAM siRNA (KD). (C) TLC plate of CSB ATPase activity in the presence of increasing concentrations of TFAM either with Ca2+ or Mg2+. (D) Graph quantifying TFAM concentration-dependent stimulation of CSB ATPase activity in the presence of either Ca2+ or Mg2+. (E) Graph quantifying TFAMWT or TFAML58A concentration-dependent stimulation of CSB ATPase activity in the presence of Mg2+. Average values are plotted with standard deviations of at least 3 independent experiments. (F) Western blot of CSB:TFAM immunoprecipitated material. B represents empty well, S represents supernatant, W represents wash and IP represents immunoprecipitated material, + indicates presence of protein or antibody. (G) Western blots of CSB (top) or TFAM (bottom) protein levels in GM1030 mitochondrial extract. Recombinant CSB (10, 30 or 100 ng) or recombinant TFAM (10, 30, 100, 300 and 1000 ng) were included to estimate efficiency of antibody recognition in order to calculate approximate protein concentration per microgram of mitochondrial extract for each.
As a likely mitochondrial protein partner of CSB and given that many nuclear protein partners of CSB have been tested for stimulation of dsDNA-dependent ATP hydrolysis by CSB and found to be negative (Supplementary Table S2), we assessed whether TFAM had a functional effect on CSB biochemical activity. Stoichiometric ratios of TFAM and CSB (1:1, 2:1 and 4:1) resulted in 2.7-, 3.8- and 4.5-fold increases in CSB ATPase activity in the presence of Ca2+ and 1.5-, 2.9- and 3.8-fold increases in the presence of Mg2+, respectively (Figure 2C and D). Using an ATPase-deficient CSB protein (CSBE646Q) in the dsDNA-dependent ATPase reactions with TFAM, we confirmed that the stimulation was dependent on active ATP hydrolysis by CSB (Supplementary Figure S1). The finding that TFAM robustly stimulated CSB dsDNA-dependent ATP hydrolysis is noteworthy, as the only other protein reported to positively affect CSB ATPase activity in the presence of a 10-nt bubble substrate, XPG (≤2-fold (50)), showed no positive effect when incubated with CSB and dsDNA (Supplementary Figure S2A and B).
To address whether the stimulation of CSB ATPase activity by TFAM was due to a DNA conformation generated by TFAM DNA binding and/or strand destabilization of dsDNA, we used a previously described TFAM DNA binding and strand destabilization mutant (47) containing a substitution in the HMG box A primary DNA intercalation wedge (TFAML58A). Residue L58, located in helix 1 of the HMG1 domain of TFAM, has been found in a co-crystal structure to intercalate between A3 and C4 bases of the LSP, locally distort the DNA and contribute to sequence-specific DNA binding (51). We found that increasing concentrations of wild-type TFAM (TFAMWT) or TFAML58A stimulated CSB dsDNA-dependent ATP hydrolysis activity similarly (Figure 2E), suggesting that the observed activation was due to a direct interaction between TFAM and CSB and not an indirect effect on DNA by TFAM. Consistent with a physical interaction between the two proteins, when HA-tagged CSB was incubated with purified His6-tagged TFAM, we were able to co-precipitate TFAM with an α-HA antibody, whereas no TFAM was present in the precipitated material when CSB was omitted (Figure 2F).
In addition, we performed western blot analysis to determine the in vivo mitochondrial stoichiometries of CSB and TFAM. To do this, we probed for CSB or TFAM in purified mitochondrial extracts from GM1030 human lymphoblastoid cells, alongside titrations of their respective purified recombinant proteins (Figure 2G). Based on densitometry analysis, on average, 0.2 ± 0.08 ng (∼1.2 fmol) of CSB and 4 ± 1.6 ng (∼140 fmol) of TFAM were present per microgram of mitochondrial extract, suggesting a roughly 1:100 CSB to TFAM molecular ratio (Figure 2G), although localized, site-specific concentrations of the two proteins, such as at sites of transcription, cannot be estimated.
CSB removes TFAM bound to DNA
A main function of TFAM is binding mtDNA and organizing it into nucleoid-like structures. Thus, we wondered whether CSB would functionally interact with and modulate the DNA binding activity of TFAM. Using a previously described BamHI restriction endonuclease accessibility assay that monitors CSB protein removal/rearrangement activity of a pre-bound protein (11), we examined whether CSB modified a pre-formed TFAM:DNA complex. An excess of TFAM (∼1 µM) was incubated with 1.5 nM 42mer dsDNA substrate to ensure that nearly all of the centrally positioned BamHI restriction sites were protected (Figure 3A, Lane 1, 53 ± 7.8% versus Lane 3, 24 ± 2.2% incision). Addition of increasing concentrations of CSB (∼2, 6, 12 and 20 nM) in combination with a defined amount of BamHI led to increased substrate cleavage (Figure 3B, Lane 4, 24 ± 1.1%, Lane 5, 24 ± 3.9%, Lane 6, 28 ± 1.1%, Lane 7, 37 ± 5.3% and Lane 8, 39.5 ± 2.2% incision). At the highest CSB concentration, we observed a >1.5-fold increase in BamHI site accessibility (Figure 3B), which is significant given that the concentration of TFAM was between 50 and 500 times that of CSB. This indicated that CSB, in an ATP-independent manner, was able to either remove or rearrange TFAM on DNA to promote restriction site access. We note that when ATP was included in the restriction site accessibility assays, essentially the same result was obtained; increasing concentrations of CSB in combination with a defined amount of BamHI led to increased substrate cleavage (Supplementary Figure S3A and B).
Figure 3.
CSB can remove TFAM from dsDNA. (A) Representative denaturing polyacrylamide gel displaying CSB-mediated TFAM removal/rearrangement. (B) Graph quantifying CSB TFAM removal/rearrangement as measured by BamHI restriction enzyme site accessibility. (C) Representative native polyacrylamide gel illustrating CSB-mediated TFAM removal. (D) Graph quantifying CSB TFAM removal as measured by ratio of bound to unbound substrate. Average values are plotted with standard deviations of at least 3 independent experiments.
To distinguish between TFAM protein removal or rearrangement, we directly assayed for removal of TFAM bound to DNA by CSB using electrophoretic mobility shift assays. In these experiments, incubation of 1.5 nM of the same 42mer substrate used above with 275 nM TFAM resulted in 29 ± 3.9% of the substrate being complexed (Figure 3C, Lane 2). When 3 nM CSB was added, we observed an initial increase in bound substrate to 34 ± 5.9% (1.2-fold, Figure 3C, Lane 3); however, upon further increasing the CSB concentration, decreased dsDNA binding by TFAM was seen, as reflected by an increase in free substrate (Figure 3C, Lanes 4–7). Specifically, 9 nM CSB decreased TFAM dsDNA binding to 16.5 ± 0.7% (a 1.8-fold reduction), 15 nM CSB decreased binding to 7.7 ± 1.3% (a 3.8-fold reduction), 18 nM CSB decreased binding to 2.1 ± 1.6% (a 13.5-fold reduction) and 25 nM CSB decreased TFAM dsDNA binding to 0.5 ± 0.5% (a 59-fold reduction) (Figure 3D). These data collectively suggest that CSB can displace TFAM bound to DNA resulting in increased DNA accessibility.
Components of mitochondrial transcription stimulate CSB biochemical activity
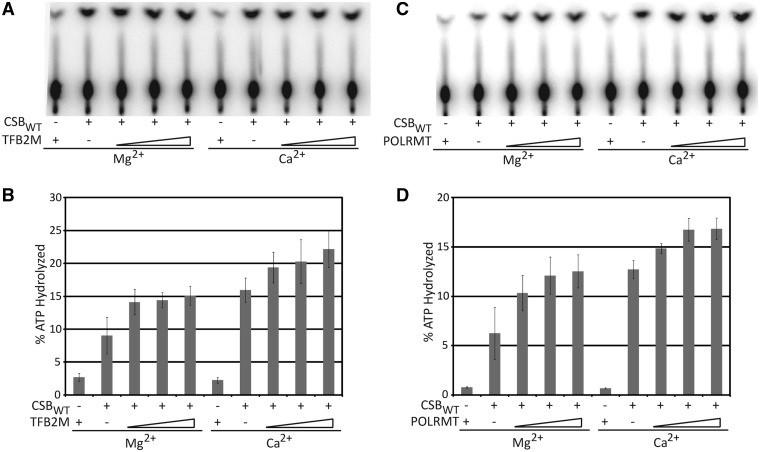
With evidence of a physical CSB:TFAM interaction, reciprocal functional interactions between the two proteins, and the fact that TFAM is a critical component of the mitochondrial transcription apparatus, we examined whether there was a biochemical effect of the core basal mitochondrial transcription components, TFB2M and POLRMT, on CSB. Stoichiometric ratios of TFB2M and CSB (1:1, 2:1 and 4:1) resulted in mild increases in CSB ATPase activity: 1.5-, 1.6- and 1.7-fold in the presence of Mg2+ and 1.2-, 1.3- and 1.4-fold increases in the presence of Ca2+ (Figure 4A and B). When POLRMT was added to the CSB dsDNA-dependent ATPase reactions, we found that increasing concentrations of POLRMT led to a moderate increase in CSB-dependent ATP hydrolysis (Figure 4C). Stoichiometric ratios of POLRMT and CSB (1:1, 2:1 and 4:1) resulted in 1.6-, 1.9- and 2.0-fold increases in CSB ATPase activity in the presence of Mg2+ and 1.2-, 1.3- and 1.4-fold increases in the presence of Ca2+ (Figure 4D). Particularly interesting is the observation that individual proteins (TFAM, POLRMT and TFB2M) comprising a single pathway (mitochondrial transcription) all functionally stimulated the major biochemical activity of CSB.
Figure 4.
TFB2M and POLRMT stimulate the dsDNA-dependent ATPase activity of CSB. (A) TLC plate of CSB ATPase activity in the presence of increasing concentrations of TFB2M either with Mg2+ or Ca2+. (B) Graph quantifying TFB2M concentration-dependent stimulation of CSB ATPase activity in the presence of either Mg2+ or Ca2+. (C) TLC plate of CSB ATPase activity in the presence of increasing concentrations of POLRMT either with Mg2+ or Ca2+. (D) Graph quantifying POLRMT concentration-dependent stimulation of CSB ATPase activity in the presence of either Mg2+ or Ca2+. Average values are plotted with standard deviations of at least 3 independent experiments.
CSB stimulates POLRMT elongation
To elucidate the molecular mechanism behind lower mitochondrial transcription in CSB-deficient cells, we asked what effect CSB had on mitochondrial transcription initiation. Using a reconstituted system comprised of a 90mer dsDNA oligonucleotide substrate encompassing the mitochondrial HSP and 40 bp of template in combination with purified POLRMT, TFB2M and TFAM, as previously described (35), we assayed for native transcript formation by the mitochondrial transcription apparatus in the presence of increasing concentrations of CSB. These studies revealed that addition of CSB to the reconstituted mitochondrial transcription initiation complexes (POLRMT:TFB2M:TFAM-HSP DNA) led to decreased transcript formation (Figure 5A), consistent with CSB possibly removing TFAM from DNA, thereby limiting transcriptional initiation.
Figure 5.
CSB promotes POLRMT elongation. (A) Representative denaturing polyacrylamide gel showing inhibition of mitochondrial transcription initiation by CSB. (B) Representative denaturing polyacrylamide gel displaying CSB enhancement of POLRMT transcriptional elongation. (C) Graph quantifying relative ratios of long/short (>1500 nt/∼20 nt) RNA transcripts produced by POLRMT in the presence of increasing concentrations of CSB. Average values are plotted with standard deviations of at least 3 independent experiments.
Since CSB did not positively affect mitochondrial transcription initiation, we next examined whether CSB could affect elongation by POLRMT. To assay for elongation, 40 nM POLRMT was incubated with M13mp18 ssDNA, rNTPs, α-32P UTP and increasing concentrations of CSB (40, 80 or 160 nM). We found that POLRMT alone produced mainly short RNA fragments (∼20–80 nt, Figure 5B, Lane 3), consistent with previous reports (39,40), whereas stoichiometric ratios of CSB to POLRMT (1:1, 2:1 and 4:1) promoted increased formation of RNAs ≥ 1500 nt in length (Figure 5B, Lanes 4–6). Comparing relative ratios of long (≥1500 nt) versus short (∼20 nt) RNAs revealed a ∼5- to >35-fold increase in long RNA production by POLRMT in the presence of CSB (Figure 5C). This ability of CSB to enhance long transcript formation by POLRMT is consistent with our in vivo data demonstrating a general mitochondrial transcription defect in cells lacking CSB (Figure 1). Lastly, we note that the assays here were performed in the absence of ATP, as we found that inclusion of ATP in the reactions containing POLRMT dramatically reduced polymerization activity, regardless of whether CSB was present or not (unpublished observation).
DISCUSSION
Proper mitochondrial function is critical for cellular energy generation and requires import of nuclear-encoded proteins to perform most mitochondrial activities, including DNA replication, DNA repair and transcription (i.e. the maintenance of mitochondrial genome homeostasis). Mitochondrial dysfunction arising from defects in mtDNA metabolism is causative in many human ailments, including neurological abnormalities, sensorineural hearing loss and myopathy (52,53) and has been hypothesized to be the driving force behind normal human aging (43). CS is a human segmental progeria, characterized by neurological defects, sensorineural hearing loss and muscle atrophy (cachexia) and is caused by mutations in nuclear-encoded genes, whose proteins (CSA and CSB) have recently been found to localize to mitochondria (29,30). We present herein evidence for a direct role of CSB in mitochondrial transcription, supporting the notion that several of the pathologies of CS are related to loss of important mitochondrial functions performed by the CS proteins.
CSB has previously been shown to participate in mitochondrial BER (28,29). In addition to important roles in nuclear and mtDNA repair, CSB has roles in nuclear transcription, interacting with RNAPI and RNAPII (19,21) and stimulating transcriptional elongation 10- and 3-fold, respectively (20,23). We demonstrate herein that, along with its role in mitochondrial BER, CSB functions in mitochondrial transcription. Specifically, our studies found that CSB-deficient cells have a severe reduction (∼50–90%) in mitochondrial transcript levels compared with isogenic cells complemented with CSBWT. Interestingly, for the earlier transcribed gene (MTCO1) of the polycistronic transcript originating from the HSP, CSBE646Q (ATPase-deficient mutant protein) and vector complemented patient cell lines exhibited a similar phenotype, with transcript levels lowered to the same degree (Figure 1B). For the later transcripts, however, the presence of CSBE646Q partially complemented the transcriptional defect (∼60% expression compared with ∼10% expression in CSB-deficient cells; Figure 1C and D), suggesting that CSB may play both an enzymatic and structural role in POLRMT elongation. Measuring mtDNA content by Q-PCR indicated that CSB-deficient and CSB ATPase mutant cells have more mtDNA than cells complemented with wild-type CSB (Figure 1E), consistent with the results of our recent report that found increased mitochondrial content in CSB-deficient cells using an immunofluorescence approach (48). As revealed by long range Q-PCR, CSB status had no significant effect on the level of polymerase blocking lesions in mtDNA (Figure 1F). Together these results indicate that the reduced transcript formation was likely due to a defect in mitochondrial transcription, not reduced mtDNA content nor increased polymerase blocking lesions. Although it has been found that oxidative DNA lesions, such as 8-oxoguanine and formamidopyrimidines, accumulate in mtDNA in the absence of CSB (31) (27), in vitro studies with an RNA polymerase (T7) related to POLRMT revealed efficient transcriptional bypass of these types of lesions (54,55). Upon reconstituting the steps of mitochondrial transcription in vitro, we found that while CSB inhibited transcription initiation (Figure 5A), the protein positively influenced elongation by POLRMT, stimulating formation of long transcripts ∼5- to >35-fold (Figure 5B and C), thus providing evidence of a novel molecular role for CSB in mitochondria.
In addition to delineating the mechanism of the mitochondrial transcription defect in CSB-deficient cells, we identified proteins that stimulated the major biochemical activity of CSB (dsDNA-dependent ATP hydrolysis). A number of mammalian DNA repair and transcription proteins have been tested for their ability to stimulate the dsDNA-dependent ATP hydrolysis activity of CSB, and TFAM, POLRMT and TFB2M are the first proteins reported to positively influence CSB biochemical activity in the presence of fully duplexed DNA (TFAM>>POLRMT > TFB2M; Figures 2C,D and 4). The mitochondrial protein cyclophilin D (CypD, peptidyl-prolyl cis–trans-isomerase D) and nuclear proteins such as XPG (50), APE1 (26), NEIL1, RNA polymerase II and TFIIS (Supplementary Table S2) did not affect CSB dsDNA-dependent ATPase activity. The fact that mitochondrial basal transcription components (POLRMT and TFB2M) and transcription, replication and nucleoid factor TFAM stimulate the ATPase activity of CSB likely underscores the importance of these interactions in maintaining proper mitochondrial function in vivo and implies that the mitochondrial roles of CSB can be more prominent than its roles in the nucleus.
TFAM, a protein involved in most mitochondrial genomic processes, stimulates CSB dsDNA-dependent ATP hydrolysis most robustly (Figure 2C and D) and co-localizes with CSB in mitochondria (29). We show herein that CSB is able to directly remove TFAM from duplex DNA (Figure 3). This protein removal activity of CSB is likely to be important for all aspects of mitochondrial genome metabolism, including the modulation of mitochondrial genome packaging/nucleoid structure to allow for mitochondrial replication, transcription and DNA repair. Unlike the previously reported chromatin remodeling activity of CSB (13), this protein removal activity does not require ATP. This function may directly regulate transcription activation from the mitochondrial LSP and HSP, as both possess canonical TFAM DNA binding sites required for maximal transcription. Our in vitro data reconstituting mitochondrial transcription initiation from the HSP with POLRMT, TFB2M and TFAM in the presence of increasing concentrations of CSB (Figure 5A) indicate that CSB reduces transcript formation, possibly by removing TFAM from the promoter. In addition, given that TFAM exhibits preferential binding to damaged DNA (56,57), CSB may aid in facilitating DNA repair by displacing TFAM from the damage site to allow for efficient recognition and processing. It is tempting to speculate that in addition to functioning as a POLRMT elongation factor and being involved in mitochondrial BER, CSB may be a factor involved in mtDNA nucleoid remodeling.
In closing, our results indicate that, in addition to mtDNA repair, CSB plays an important role in mitochondrial transcription through physical and/or functional interactions with the mitochondrial proteins TFAM, POLRMT and TFB2M. Mitochondrial dysfunction due to a loss of CSB includes roles in mtDNA repair (27–31) as well as roles outside of repair (31), including decreased mitochondrial transcription (Figure 1) caused by a defect in elongation (Figure 5) and modulation of TFAM:DNA complexes (Figure 3), which potentially affects both transcription and mtDNA nucleoid structure. We speculate that loss of these key mitochondrial functions performed by CSB in concert with mitochondrial proteins contributes to the phenotypic characteristics associated with CS and implies an importance of CSB function in mitochondria, in addition to the roles it plays in the nucleus.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–3, Supplementary Methods and Supplementary References [11,26,37,50].
FUNDING
Intramural Research Program of the National Institutes of Health, National Institute on Aging. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Orlando Scharer (State University of New York, Stony Brook) for his kind gift of recombinant XPG protein. We thank Dr Robert Brosh (LMG/NIA/NIH), Dr Morten Schiebye-Knudson (LMG/NIA/NIH) and Dr Melissa Hefferin Berquist (FAZD Center) for critical input on this article.
REFERENCES
- 1.Brooks PJ, Cheng TF, Cooper L. Do all of the neurologic diseases in patients with DNA repair gene mutations result from the accumulation of DNA damage? DNA Repair. 2008;7:834–848. doi: 10.1016/j.dnarep.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Natale V. A comprehensive description of the severity groups in Cockayne syndrome. Am J. Med. Genet. A. 2011;155A:1081–1095. doi: 10.1002/ajmg.a.33933. [DOI] [PubMed] [Google Scholar]
- 3.Mallery DL, Tanganelli B, Colella S, Steingrimsdottir H, van Gool AJ, Troelstra C, Stefanini M, Lehmann AR. Molecular analysis of mutations in the CSB (ERCC6) gene in patients with Cockayne syndrome. Am. J. Hum. Genet. 1998;62:77–85. doi: 10.1086/301686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colella S, Nardo T, Botta E, Lehmann AR, Stefanini M. Identical mutations in the CSB gene associated with either Cockayne syndrome or the DeSanctis–Cacchione variant of xeroderma pigmentosum. Hum. Mol. Genet. 2000;9:1171–1175. doi: 10.1093/hmg/9.8.1171. [DOI] [PubMed] [Google Scholar]
- 5.Meira LB, Graham JM, Jr, Greenberg CR, Busch DB, Doughty AT, Ziffer DW, Coleman DM, Savre-Train I, Friedberg EC. Manitoba aboriginal kindred with original cerebro-oculo-facio-skeletal syndrome has a mutation in the Cockayne syndrome group B (CSB) gene. Am. J. Hum. Genet. 2000;66:1221–1228. doi: 10.1086/302867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horibata K, Iwamoto Y, Kuraoka I, Jaspers NG, Kurimasa A, Oshimura M, Ichihashi M, Tanaka K. Complete absence of Cockayne syndrome group B gene product gives rise to UV-sensitive syndrome but not Cockayne syndrome. Proc. Natl Acad. Sci. USA. 2004;101:15410–15415. doi: 10.1073/pnas.0404587101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henning KA, Li L, Iyer N, McDaniel LD, Reagan MS, Legerski R, Schultz RA, Stefanini M, Lehmann AR, Mayne LV, et al. The Cockayne syndrome group A gene encodes a WD repeat protein that interacts with CSB protein and a subunit of RNA polymerase II TFIIH. Cell. 1995;82:555–564. doi: 10.1016/0092-8674(95)90028-4. [DOI] [PubMed] [Google Scholar]
- 8.Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, Kisselev AF, Tanaka K, Nakatani Y. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 2003;113:357–367. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- 9.Troelstra C, Hesen W, Bootsma D, Hoeijmakers JH. Structure and expression of the excision repair gene ERCC6, involved in the human disorder Cockayne’s syndrome group B. Nucleic Acids Res. 1993;21:419–426. doi: 10.1093/nar/21.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muftuoglu M, Sharma S, Thorslund T, Stevnsner T, Soerensen MM, Brosh RM, Jr, Bohr VA. Cockayne syndrome group B protein has novel strand annealing and exchange activities. Nucleic Acids Res. 2006;34:295–304. doi: 10.1093/nar/gkj410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berquist BR, Wilson DM., 3rd Nucleic acid binding activity of human Cockayne syndrome B protein and identification of Ca(2+) as a novel metal cofactor. J. Mol. Biol. 2009;391:820–832. doi: 10.1016/j.jmb.2009.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beerens N, Hoeijmakers JH, Kanaar R, Vermeulen W, Wyman C. The CSB protein actively wraps DNA. J. Biol. Chem. 2005;280:4722–4729. doi: 10.1074/jbc.M409147200. [DOI] [PubMed] [Google Scholar]
- 13.Citterio E, Van Den Boom V, Schnitzler G, Kanaar R, Bonte E, Kingston RE, Hoeijmakers JH, Vermeulen W. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol. Cell. Biol. 2000;20:7643–7653. doi: 10.1128/mcb.20.20.7643-7653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Selby CP, Sancar A. Human transcription-repair coupling factor CSB/ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J. Biol. Chem. 1997;272:1885–1890. doi: 10.1074/jbc.272.3.1885. [DOI] [PubMed] [Google Scholar]
- 15.Citterio E, Rademakers S, van der Horst GT, van Gool AJ, Hoeijmakers JH, Vermeulen W. Biochemical and biological characterization of wild-type and ATPase-deficient Cockayne syndrome B repair protein. J. Biol. Chem. 1998;273:11844–11851. doi: 10.1074/jbc.273.19.11844. [DOI] [PubMed] [Google Scholar]
- 16.Christiansen M, Stevnsner T, Modin C, Martensen PM, Brosh RM, Jr, Bohr VA. Functional consequences of mutations in the conserved SF2 motifs and post-translational phosphorylation of the CSB protein. Nucleic Acids Res. 2003;31:963–973. doi: 10.1093/nar/gkg164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orren DK, Dianov GL, Bohr VA. The human CSB (ERCC6) gene corrects the transcription-coupled repair defect in the CHO cell mutant UV61. Nucleic Acids Res. 1996;24:3317–3322. doi: 10.1093/nar/24.17.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balajee AS, May A, Dianov GL, Friedberg EC, Bohr VA. Reduced RNA polymerase II transcription in intact and permeabilized Cockayne syndrome group B cells. Proc. Natl Acad. Sci. USA. 1997;94:4306–4311. doi: 10.1073/pnas.94.9.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Gool AJ, Citterio E, Rademakers S, van Os R, Vermeulen W, Constantinou A, Egly JM, Bootsma D, Hoeijmakers JH. The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J. 1997;16:5955–5965. doi: 10.1093/emboj/16.19.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Selby CP, Sancar A. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc. Natl Acad. Sci. USA. 1997;94:11205–11209. doi: 10.1073/pnas.94.21.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradsher J, Auriol J, Proietti de Santis L, Iben S, Vonesch JL, Grummt I, Egly JM. CSB is a component of RNA pol I transcription. Mol. Cell. 2002;10:819–829. doi: 10.1016/s1097-2765(02)00678-0. [DOI] [PubMed] [Google Scholar]
- 22.Yuan X, Feng W, Imhof A, Grummt I, Zhou Y. Activation of RNA polymerase I transcription by cockayne syndrome group B protein and histone methyltransferase G9a. Mol. Cell. 2007;27:585–595. doi: 10.1016/j.molcel.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 23.Lebedev A, Scharffetter-Kochanek K, Iben S. Truncated Cockayne syndrome B protein represses elongation by RNA polymerase I. J. Mol. Biol. 2008;382:266–274. doi: 10.1016/j.jmb.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 24.Tuo J, Muftuoglu M, Chen C, Jaruga P, Selzer RR, Brosh RM, Jr, Rodriguez H, Dizdaroglu M, Bohr VA. The Cockayne syndrome group B gene product is involved in general genome base excision repair of 8-hydroxyguanine in DNA. J. Biol. Chem. 2001;276:45772–45779. doi: 10.1074/jbc.M107888200. [DOI] [PubMed] [Google Scholar]
- 25.Selzer RR, Nyaga S, Tuo J, May A, Muftuoglu M, Christiansen M, Citterio E, Brosh RM, Jr, Bohr VA. Differential requirement for the ATPase domain of the Cockayne syndrome group B gene in the processing of UV-induced DNA damage and 8-oxoguanine lesions in human cells. Nucleic Acids Res. 2002;30:782–793. doi: 10.1093/nar/30.3.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong HK, Muftuoglu M, Beck G, Imam SZ, Bohr VA, Wilson DM., 3rd Cockayne syndrome B protein stimulates apurinic endonuclease 1 activity and protects against agents that introduce base excision repair intermediates. Nucleic Acids Res. 2007;35:4103–4113. doi: 10.1093/nar/gkm404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muftuoglu M, de Souza-Pinto NC, Dogan A, Aamann M, Stevnsner T, Rybanska I, Kirkali G, Dizdaroglu M, Bohr VA. Cockayne syndrome group B protein stimulates repair of formamidopyrimidines by NEIL1 DNA glycosylase. J. Biol. Chem. 2009;284:9270–9279. doi: 10.1074/jbc.M807006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stevnsner T, Nyaga S, de Souza-Pinto NC, van der Horst GT, Gorgels TG, Hogue BA, Thorslund T, Bohr VA. Mitochondrial repair of 8-oxoguanine is deficient in Cockayne syndrome group B. Oncogene. 2002;21:8675–8682. doi: 10.1038/sj.onc.1205994. [DOI] [PubMed] [Google Scholar]
- 29.Aamann MD, Sorensen MM, Hvitby C, Berquist BR, Muftuoglu M, Tian J, de Souza-Pinto NC, Scheibye-Knudsen M, Wilson DM, 3rd, Stevnsner T, et al. Cockayne syndrome group B protein promotes mitochondrial DNA stability by supporting the DNA repair association with the mitochondrial membrane. FASEB J. 2010;24:2334–2346. doi: 10.1096/fj.09-147991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamenisch Y, Fousteri M, Knoch J, von Thaler AK, Fehrenbacher B, Kato H, Becker T, Dolle ME, Kuiper R, Majora M, et al. Proteins of nucleotide and base excision repair pathways interact in mitochondria to protect from loss of subcutaneous fat, a hallmark of aging. J. Exp. Med. 2010;207:379–390. doi: 10.1084/jem.20091834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osenbroch PO, Auk-Emblem P, Halsne R, Strand J, Forstrom RJ, van der Pluijm I, Eide L. Accumulation of mitochondrial DNA damage and bioenergetic dysfunction in CSB defective cells. FEBS J. 2009;276:2811–2821. doi: 10.1111/j.1742-4658.2009.07004.x. [DOI] [PubMed] [Google Scholar]
- 32.Diffley JF, Stillman B. A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc. Natl Acad. Sci. USA. 1991;88:7864–7868. doi: 10.1073/pnas.88.17.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaufman BA, Durisic N, Mativetsky JM, Costantino S, Hancock MA, Grutter P, Shoubridge EA. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell. 2007;18:3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, Rustin P, Gustafsson CM, Larsson NG. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum. Mol. Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 35.Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 36.Litonin D, Sologub M, Shi Y, Savkina M, Anikin M, Falkenberg M, Gustafsson CM, Temiakov D. Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. J. Biol. Chem. 2010;285:18129–18133. doi: 10.1074/jbc.C110.128918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lodeiro MF, Uchida AU, Arnold JJ, Reynolds SL, Moustafa IM, Cameron CE. Identification of multiple rate-limiting steps during the human mitochondrial transcription cycle in vitro. J. Biol. Chem. 2010;285:16387–16402. doi: 10.1074/jbc.M109.092676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shutt TE, Lodeiro MF, Cotney J, Cameron CE, Shadel GS. Core human mitochondrial transcription apparatus is a regulated two-component system in vitro. Proc. Natl Acad. Sci. USA. 2010;107:12133–12138. doi: 10.1073/pnas.0910581107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wanrooij S, Fuste JM, Farge G, Shi Y, Gustafsson CM, Falkenberg M. Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro. Proc. Natl Acad. Sci. USA. 2008;105:11122–11127. doi: 10.1073/pnas.0805399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minczuk M, He J, Duch AM, Ettema TJ, Chlebowski A, Dzionek K, Nijtmans LG, Huynen MA, Holt IJ. TEFM (c17orf42) is necessary for transcription of human mtDNA. Nucleic Acids Res. 2011;39:4284–4299. doi: 10.1093/nar/gkq1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Surovtseva YV, Shutt TE, Cotney J, Cimen H, Chen SY, Koc EC, Shadel GS. Mitochondrial ribosomal protein L12 selectively associates with human mitochondrial RNA polymerase to activate transcription. Proc. Natl Acad. Sci. USA. 2011;108:17921–17926. doi: 10.1073/pnas.1108852108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harman D. Aging: a theory based on free radical and radiation chemistry. J. Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 43.Harman D. The biologic clock: the mitochondria? J. Am. Geriatr. Soc. 1972;20:145–147. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- 44.Troelstra C, van Gool A, de Wit J, Vermeulen W, Bootsma D, Hoeijmakers JH. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne’s syndrome and preferential repair of active genes. Cell. 1992;71:939–953. doi: 10.1016/0092-8674(92)90390-x. [DOI] [PubMed] [Google Scholar]
- 45.Poeggeler B, Knuever J, Gaspar E, Biro T, Klinger M, Bodo E, Wiesner RJ, Wenzel BE, Paus R. Thyrotropin powers human mitochondria. FASEB J. 2010;24:1525–1531. doi: 10.1096/fj.09-147728. [DOI] [PubMed] [Google Scholar]
- 46.Sykora P, Croteau DL, Bohr VA, Wilson DM., 3rd Aprataxin localizes to mitochondria and preserves mitochondrial function. Proc. Natl Acad. Sci. USA. 2011;108:7437–7442. doi: 10.1073/pnas.1100084108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Canugovi C, Maynard S, Bayne AC, Sykora P, Tian J, de Souza-Pinto NC, Croteau DL, Bohr VA. The mitochondrial transcription factor A functions in mitochondrial base excision repair. DNA Repair. 2010;9:1080–1089. doi: 10.1016/j.dnarep.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scheibye-Knudsen M, Ramamoorthy M, Sykora P, Maynard S, Lin PC, Minor RK, Wilson DM, I, Cooper M, II, Spencer R, de Cabo R, et al. Cockayne syndrome group B protein prevents the accumulation of damaged mitochondria by promoting mitochondrial autophagy. J. Exp. Med. 2012;209:855–869. doi: 10.1084/jem.20111721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Balliet RM, Capparelli C, Guido C, Pestell TG, Martinez-Outschoorn UE, Lin Z, Whitaker-Menezes D, Chiavarina B, Pestell RG, Howell A, et al. Mitochondrial oxidative stress in cancer-associated fibroblasts drives lactate production, promoting breast cancer tumor growth: understanding the aging and cancer connection. Cell Cycle. 2011;10:4065–4073. doi: 10.4161/cc.10.23.18254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarker AH, Tsutakawa SE, Kostek S, Ng C, Shin DS, Peris M, Campeau E, Tainer JA, Nogales E, Cooper PK. Recognition of RNA polymerase II and transcription bubbles by XPG, CSB, and TFIIH: insights for transcription-coupled repair and Cockayne Syndrome. Mol. Cell. 2005;20:187–198. doi: 10.1016/j.molcel.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 51.Rubio-Cosials A, Sidow JF, Jimenez-Menendez N, Fernandez-Millan P, Montoya J, Jacobs HT, Coll M, Bernado P, Sola M. Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Nat. Struct. Mol. Biol. 2011;18:1281–1289. doi: 10.1038/nsmb.2160. [DOI] [PubMed] [Google Scholar]
- 52.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 53.Ylikallio E, Suomalainen A. Mechanisms of mitochondrial diseases. Ann. Med. 2012;44:41–59. doi: 10.3109/07853890.2011.598547. [DOI] [PubMed] [Google Scholar]
- 54.Chen YH, Bogenhagen DF. Effects of DNA lesions on transcription elongation by T7 RNA polymerase. J. Biol. Chem. 1993;268:5849–5855. [PubMed] [Google Scholar]
- 55.Tornaletti S, Maeda LS, Kolodner RD, Hanawalt PC. Effect of 8-oxoguanine on transcription elongation by T7 RNA polymerase and mammalian RNA polymerase II. DNA Repair. 2004;3:483–494. doi: 10.1016/j.dnarep.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 56.Yoshida Y, Izumi H, Ise T, Uramoto H, Torigoe T, Ishiguchi H, Murakami T, Tanabe M, Nakayama Y, Itoh H, et al. Human mitochondrial transcription factor A binds preferentially to oxidatively damaged DNA. Biochem. Biophys. Res. Commun. 2002;295:945–951. doi: 10.1016/s0006-291x(02)00757-x. [DOI] [PubMed] [Google Scholar]
- 57.Yoshida Y, Izumi H, Torigoe T, Ishiguchi H, Itoh H, Kang D, Kohno K. P53 physically interacts with mitochondrial transcription factor A and differentially regulates binding to damaged DNA. Cancer Res. 2003;63:3729–3734. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.