Abstract
The human Y box-binding protein-1 (YB-1) is a deoxyribonucleic acid (DNA)/ribonucleic acid (RNA)-binding protein with pleiotropic functions. Besides its roles in the regulation of transcription and translation, several recent studies indicate that YB-1 is a spliceosome-associated protein and is involved in alternative splicing, but the underlying mechanism has remained elusive. Here, we define both CAUC and CACC as high-affinity binding motifs for YB-1 by systematic evolution of ligands by exponential enrichment (SELEX) and demonstrate that these newly defined motifs function as splicing enhancers. Interestingly, on the endogenous CD44 gene, YB-1 appears to mediate a network interaction to activate exon v5 inclusion via multiple CAUC motifs in both the alternative exon and its upstream polypyrimidine tract. We provide evidence that YB-1 activates splicing by facilitating the recruitment of U2AF65 to weak polypyrimidine tracts through direct protein–protein interactions. Together, these findings suggest a vital role of YB-1 in activating a subset of weak 3′ splice sites in mammalian cells.
INTRODUCTION
Recent genome-wide analyses estimate that more than 90% of human multi-exon genes undergo tissue-specific alternative splicing (1,2). Through alternative splicing, a single gene often generates multiple splice variants encoding protein isoforms of different, sometimes even antagonistic functions. Therefore, alternative splicing represents a major source of proteomic diversity and a central mechanism of genetic control in higher eukaryotes (3,4). Misregulation of splicing underlies many human diseases, including cancers (5). Current models propose that both cis-elements and trans-acting factors are important for the regulation of alternative splicing. Trans-acting factors bind to cis-elements identified in either exons or introns, and control the selection of splice sites through modulating the assembly of the splicing machinery (6,7). Among the trans-acting protein factors, members of SR and hnRNP protein families have been extensively studied. Most of them contain the RNA recognition motif (RRM)-type RNA-binding domains. However, we know relatively little about the splicing regulation by trans-acting factors carrying other types of RNA-binding domains.
The Y box-binding protein-1 (YB-1) is a member of the evolutionarily conserved family of cold shock domain (CSD) proteins. It exhibits multiple functions in the regulation of transcription and translation (8). YB-1 is mainly localized to the cytoplasm in normal cells, but is highly expressed in the nucleus of tumor cells, particularly in breast cancer cells (9). A number of studies indicated its role in malignant transformation (10,11). YB-1 protein is composed of an N-terminal alanine- and proline-rich domain, a cold shock domain, and a C-terminal domain composed of alternating basic and acidic clusters. The CSD domain adopts a β-barrel structure with five antiparallel β-strands. It contains the RNA-binding motifs ribonucleoprotein particle domain-1 (RNP-1) and RNP-2, which are characteristic of many RNA-binding proteins. However, so far the RNA-binding specificity of YB-1 has not been well defined. Ray et al. analysed the relative RNA-binding preference of YB-1 using a single binding reaction combined with microarray detection (12). They found that YB-1 prefers binding to short RNAs containing CUGC sequences. Dong et al. reported that selective YB-1 binding targets are CG-rich based on RNA immunoprecipitation results (13). However, these two reports are contrary to two previous studies indicating that YB-1 preferentially recognizes C/A-rich sequences (14,15).
YB-1 has been identified as a spliceosome-associated protein. It could be co-purified with spliceosomal A and B complexes (16,17). Recently, several lines of evidence indicate that YB-1 is also involved in alternative splicing regulation. YB-1 was shown to bind C/A-rich elements in CD44 variable exon v4, and overexpression of YB-1 increases the inclusion of exon v4 (15). YB-1 could also be ultraviolet (UV) crosslinked to an ACAAC enhancer element identified in NF1 exon 37 (14). Phosphatase PP2Cγ, which is capable of promoting spliceosome assembly, interacts with YB-1 and regulates splicing of YB-1 targets (18). Recent work from the Auboeuf's laboratory suggests that YB-1 mediates co-transcriptional exon skipping in response to genotoxic stress (19). However, we know little about the molecular mechanism of YB-1-mediated splicing regulation.
In this study, we report that YB-1 specifically binds to a CAUC consensus. Mutational analyses indicate that the second adenosine (A2) and the fourth cytidine (C4) in the CAUC motif are crucial for YB-1 recognition. We demonstrate that the core CAUC motifs specifically stimulate dsx splicing in vitro and show that CD44 variable exon v5 is a natural target of YB-1. Interestingly, YB-1 is required for CD44 variable exon v5 inclusion through binding to CAUC motifs both in the v5 exon and within its upstream polypyrimidine tract. We provide evidence that YB-1 activates splicing by facilitating the recruitment of U2AF65 to weak polypyrimidine tract through direct protein–protein interactions. Together, these results reveal that YB-1 is able to positively modulate splice site selection via the core CAUC motifs located in both exon and intron.
MATERIALS AND METHODS
Oligonucleotides
The sequences of all oligonucleotides used in this study are listed in the Supplementary Material.
Plasmid construction
After successful cloning, all constructs were sequence verified.
YB-1 expression constructs
The eukaryotic expression construct p3xFLAG-YB-1 (20) was a gift from Dr S. Dunn (University of British Columbia, Canada). To clone the YB-1 shRNA-resistant construct, nucleotides GTC ATC GCA ACG AAG GTT encoding amino acids 54–59 were mutated to GTT ATT GCC ACC AAA GTC by polymerase chain reaction (PCR) using p3xFLAG-YB-1 as the template.
dsx minigenes
pdsx-control was cloned by insertion of 64 nt random sequence into pdsx-XH (21) at the XbaI and HindIII sites. The 64 nt were amplified from pGEM5Zf(+) using primers DSX-ctrl-for and DSX-ctrl-rev. pdsx-#27, pdsx-CATCTG, pdsx-CATCGC, pdsx-GATCTG, pdsx-CATGTG, pdsx-CTTCTG, pdsx-CGTCTG and pdsx-CACCTT were constructed by insertion of annealed oligonucleotides DSX-27-for/-rev, DSX-CATCTG-for/-rev, DSX-CATCGC-for/-rev, DSX-GATCTG-for/rev, DSX-CATGTG-for/rev, DSX-CTTCTG-for/rev, DSX-CGTCTG-for/rev and DSX-CACCTT-for/-rev into XbaI and HindIII sites.
Expression constructs for competitor RNAs ACE sel and ACE sel mut
DNA fragments containing ACE sel and ACE sel mut sequences (15) were generated by annealing oligonucleotides ACE sel-for/-rev and ACE sel mut-for/-rev, respectively. These two fragments were cloned into pComp-control (22) between the XbaI and HindIII sites. The expression of ACE sel or ACE sel mut RNA was driven by T7 promoter.
HA- and GST-tagged U2AF65 and U2AF35 expression constructs
cDNAs containing the full-length open reading frame of U2AF65 and U2AF35 were amplified using primer pairs U2AF65-1/-2, U2AF35-1/-2 and U2AF35-3/-2 and cloned between the BamHI and XbaI or HindIII and XhoI sites of pcDNA3, or BamHI and XhoI sites of pGEX-5X-2. Sequences encoding the human influenza hemagglutinin (HA) tag were included in the antisense primers U2AF65-2 and U2AF35-2. To clone U2AF65 deletion mutants ΔRS, ΔRRM and RRM, PCR fragments generated using primer pairs U2AF65-ΔRS-for/-2, U2AF65-1/-ΔRRM-rev and U2AF65-RRM for/-2 were inserted into the BamHI and XbaI sites of pcDNA3. For U2AF35 deletion mutant ΔRS, PCR fragment amplified using primer pair U2AF35-1/ΔRS-rev was cloned between the HindIII and XhoI sites of pcDNA3.
CD44 v5 minigene pET-v5 and its mutants
The genomic sequence of CD44 gene was amplified using genomic DNA isolated from primary human umbilical vein endothelial cell (HUVEC) cells as template. The genomic fragment from 794 nt upstream of exon v4 to 479 nt downstream of exon v5 was generated using primers v4-794 and v5 + 479 by PCR and cloned into the XhoI and BamHI sites of the Exontrap vector pET01 (MoBiTec, Germany). To simplify our study, exon v4 sequence was removed by two-step PCR method using primer pairs v4-794/Δv4-rev and Δv4-for/v5 + 479. For the v5 mutant constructs, the wild-type construct pET-v5 was used as template for two-step PCRs. Primer pairs v4-794/v5-mut1-rev and v5-mut1-for/v5 + 479 were used for cloning the mut1 construct, v4-794/v5-mut2-rev and v5-mut2-for/v5 + 479 for the mut2 construct, v4-794/v5-mut3-rev and v5-mut3-for/v5 + 479 for the mut3 construct and v4-794/v5-mut4-rev and v5-mut4-for/v5 + 479 for the mut4 construct. To make the double mutant mut2 + 3, primer pairs v4-794/v5-mut3-rev and v5-mut3-for/v5 + 479mut2 were used, and mut2 served as the template for PCR. Then PCR fragments containing mutated sequences were digested with XhoI and BamHI and cloned into pET01.
shRNA expression constructs
Retroviral plasmids containing human YB-1 shRNA target and control sequences were made by annealing oligonucleotides Y-shRNA-for/Y-shRNA-rev and Luc-shRNA-for/Luc-shRNA-rev followed by ligation into the EcoRI and BamHI sites of pSIREN-RetroQ (Clontech).
Protein purification
GST-YB-1, GST-U2AF65, GST-U2AF35, His-YB-1, His-CSD and His-C proteins were purified from Escherichia coli strain BL21 RIL. E. coli cells transformed with expression plasmids were induced with 0.3 mM isopropyl beta-D-1-thiogalactopyranoside (IPTG) for 3 h at 37°C. Purification of GST- and His-tagged proteins was performed using glutathione-Sepharose 4B or Ni-NTA according to manufacturer’s instructions (GE healthcare, USA; QIAGEN, Germany).
SELEX and filter-binding assay
The SELEX and the filter-binding assays were performed as previously described (23).
Gel shift assay
32P-labeled RNAs (130 fmol) were incubated at 30°C under standard splicing conditions with different molar excess of recombinant GST-YB-1 fusion proteins. An aliquot of 5 µl was removed and transferred to a new tube containing 1 µl of heparin (5 mg/ml). RNA-protein complexes were fractionated on 5% native polyacrylamide gels.
In vitro splicing
In vitro splicing reactions were set up in a total volume of 25 µl with 0.5 mM adenosine triphosphate, 3.2 mM MgCl2, 20 mM creatine phosphate, 2.66% polyvinyl alcohol, 60% HeLa cell nuclear extract (Cil Biotech, Belgium) and 10 ng of 32P-labeled RNA substrate. The reactions were incubated at 30°C. The RNAs were extracted and analysed on 8% polyacrylamide gels containing 8 M urea. For splicing competition assays, in vitro splicing reactions were carried out in the presence of unlabeled ACE sel or ACE sel mut RNAs.
Real-time quantitation reverse-transcription PCR (qRT-PCR)
First-strand cDNA was synthesized from 5 µg of total RNAs using SuperScript III reverse transcriptase (Invitrogen, USA) and random primers according to manufacture’s instruction. CD44 constitutive and variable exon-specific primers (listed in the Supplementary Material) were used for qPCR. The real-time qRT-PCR data were analysed using the 2−ΔΔCt method. β-Actin served as a reference gene for qRT-PCR. The mRNA expression levels of YB-1 and CD44 exons in YB-1 knock-down (shYB-1) cells were normalized to those in control knock-down (shLuc) cells.
UV crosslinking and immunoprecipitation
To prepare RNA probes for UV crosslinking, DNA templates for in vitro transcription were obtained by PCR using primer sets T7-DSX-in-for/DSX-ex-rev (from 60 nt upstream of dsx exon 4 to 46 nt downstream of different inserts in exon 4) and T7-v5-70/v5 + 16 (from 70 nt upstream of exon v5 to nt 16 in exon v5). 32P-labeled RNAs were incubated in HeLa cell nuclear extract under standard splicing conditions at 30°C for 15 min. One microliter of tRNA (5 mg/ml) was added into each reaction to remove non-specific binding of proteins. UV crosslinking was done on ice for 20 min with 254-nm UV light. Unprotected RNAs were digested with RNase A (at a final concentration of 1 mg/ml) at 37°C for 20 min. For immunoprecipitation experiment, 10 µl packed volume of protein G-agarose (Roche, Germany) were first incubated with 4 µl YB-1 antibody (1 mg/ml, Sigma, Y0396), 5 µl U2AF65 antibody (2 mg/ml, Abcam, ab5086) or 6.7 µl mouse IgG (1.5 mg/ml, ImB, IL005) in N100 buffer (100 mM NaCl, 50 mM Tris–Cl, pH 8.0, 0.05% NP-40). After rotating at 4°C for 3 h, beads were washed five times with 1 ml of N100 buffer. Fifty microliter of UV-crosslinking reaction (2 standard in vitro splicing reactions) was incubated with protein G beads at 4°C for 3 h with rotation. After washing three times with 1 ml of N300 buffer (300 mM NaCl, 50 mM Tris–Cl, pH 8.0, 0.05% NP-40), bound proteins were fractionated on 12% Sodium dodecyl sulphate (SDS) polyacrylamide gels. For detection of the association between U2AF and YB-1 in HeLa nuclear extract, 25 µl of HeLa cell nuclear extract (pre-treated with RNase A at a final concentration of 2 µg/ml) was used for immunoprecipitation assay. N600 buffer (600 mM NaCl, 50 mM Tris–Cl, pH 8.0, 0.05% NP-40) was used as washing buffer.
Co-immunoprecipitation (Co-IP)
HEK 293 cells that were co-transfected with FLAG-tagged YB-1 and HA-tagged U2AF constructs were lysed in CoIP lysis buffer (50 mM Tris–Cl, pH 7.5, 150 mM KCl, 0.5% NP40, 1 mM PMSF, 2 µg/ml RNase A) at room temperature for 15 min with rotating. The lysate was centrifuged at 13 000 rpm for 10 min at 4°C. The supernatant was collected and incubated with 10 µl anti-Flag M2 beads (Sigma) at 4°C for 3 h with rotating. The beads were subsequently washed three times with 1 ml CoIP washing buffer (50 mM Tris–Cl, pH 7.5, 150 mM KCl, 0.1% NP40, 1 mM PMSF). The bound material was fractionated by SDS- polyacrylamide gel electrophoresis (PAGE) followed by blotting with anti-HA antibody (Roche).
GST pulldown assay
GST, GST-U2AF65 and GST-U2AF35 proteins immobilized on 10 µl of glutathione-Sepharose 4B were incubated with His-YB-1, His-CSD, His-C and a non-related protein for 3 h at 4°C with rotation in the presence of 2 µg/ml RNase A. After incubation, beads were washed four times with 1 ml washing buffer (50 mM Tris–Cl, pH 7.5, 300 mM NaCl, 0.1% NP-40, 1 mM dithiothreitol (DTT) and protease inhibitor cocktail). The bound proteins were fractionated by 12% SDS-PAGE and visualized by Coomassie blue staining.
RNA affinity selection assay
Streptavidin agarose beads were pre-blocked (4 mM HEPES, pH 8.0, 0.2 mM DTT, 2 mM MgCl2, 20 mM KCl, 0.002% NP-40, 0.2 mg/ml tRNA, 1 mg/ml bovine serum albumin [BSA], 0.2 mg/ml glycogen) at 4°C for 1 h and washed with buffer (20 mM HEPES, pH 8.0, 1 mM DTT, 10 mM MgCl2, 400 mM KCl, 0.01% NP-40). Two microgram of biotinylated wild-type- and mut3-derived RNA oligonucleotides (from 70 nt upstream of CD44 exon v5 to nt 16 in exon v5) were bound to 10 μl of streptavidin agarose beads at 4°C for 5 h followed by incubating with either 30 pmol of GST-YB-1 or 30 pmol of GST-U2AF65 or both proteins at 4°C for 3 h. After washing with previously mentioned buffer containing 400 mM KCl, the bound proteins were detected by western blotting using anti-YB-1 or anti-U2AF65 antibody.
Detection of biotinylated RNAs pulldowned by GST fusion proteins
GST and GST-U2AF65 proteins (30 pmol) immobilized on glutathione-Sepharose 4B were incubated with 2 μg of biotinylated wild-type- and mut3-derived RNA oligonucleotides (from 70 nt upstream of CD44 exon v5 to nt 16 in exon v5) in the absence or in the presence of 30 pmol of His-YB-1 or His-CSD. The bound RNAs were extracted with phenol/chloroform, ethanol precipitated and separated on a 10% denaturing polyacrylamide gel followed by detection using North2South® chemiluminescent hybridization and detection kit (Thermo Scientific, USA).
In vivo splicing
The day before transfection, 5 × 105 HEK 293 and MDA-MB-231 cells were seeded into a 3.5-cm culture dish. Wild-type pET-v5 and its mutant constructs were transfected using calcium phosphate method (24) or Lipofectamine 2000 (Invitrogen, USA). Two days after cell transfection, total RNAs were isolated using guanidinium thiocyanate (25). Total RNA (2.5 µg) was annealed to oligo d(T)18 and reverse-transcribed by moloney murine leukemia virus (MMLV) (Promega) according to the manufacturer’s instruction. The resulting first-strand cDNA was further amplified by PCR, using primers pET for and pET rev. The PCR conditions were optimized for each minigene.
Knock-down of YB-1 in MDA-MB-231 cells
MDA-MB-231 cells were grown in Dulbecco’s Modified Eagle’s medium supplemented with 10% fetal bovine serum. Stable knock-down of YB-1 using retrovirus expressed shRNA was established in MDA-MB-231 cells according to manufacturer’s instruction (Clontech). The target sequences of shRNAs are: luciferase control (GL2) 5’-CGUACGCGGAAUACUUCGA-3′ and YB-1 5′-GGUCAUCGCAACGAAGGUU-3′.
RESULTS
Defining the RNA-binding specificity of YB-1 by SELEX
To determine the RNA-binding motif for YB-1, we performed an in vitro SELEX (systematic evolution of ligands by exponential enrichment) experiment. Recombinant GST-YB-1 protein was immobilized on glutathione-Sepharose and incubated with an RNA pool containing a randomized 20-nucleotide (nt) region. After extensive washing followed by proteinase K treatment, the bound RNA was isolated and amplified by RT-PCR. The cDNA was subjected to T7 transcription for the next round of selection. After eight rounds, YB-1-specific RNA ligands were significantly enriched as indicated by the increased RNA binding efficiency from 1.3% (the initial pool) to 12.7% (round 8). We identified C A U/C C as a core consensus using a motif search program MEME (multiple EM for motif elicitation, http://meme.nbcr.net/meme/). The alignment of all 41 RNA sequences obtained is shown in Figure 1. Among the 41 sequences, 28 contain CAUC, and 11 have CACC. This newly identified motif is clearly distinct from CG-rich sequences, but closely resembles the C/A-rich sequences deduced from previous studies.
Figure 1.
Defining the RNA-binding specificity of YB-1 by SELEX. The sequences of 41 SELEX-derived RNAs are listed. The consensus motif C A U/C C is shown in red.
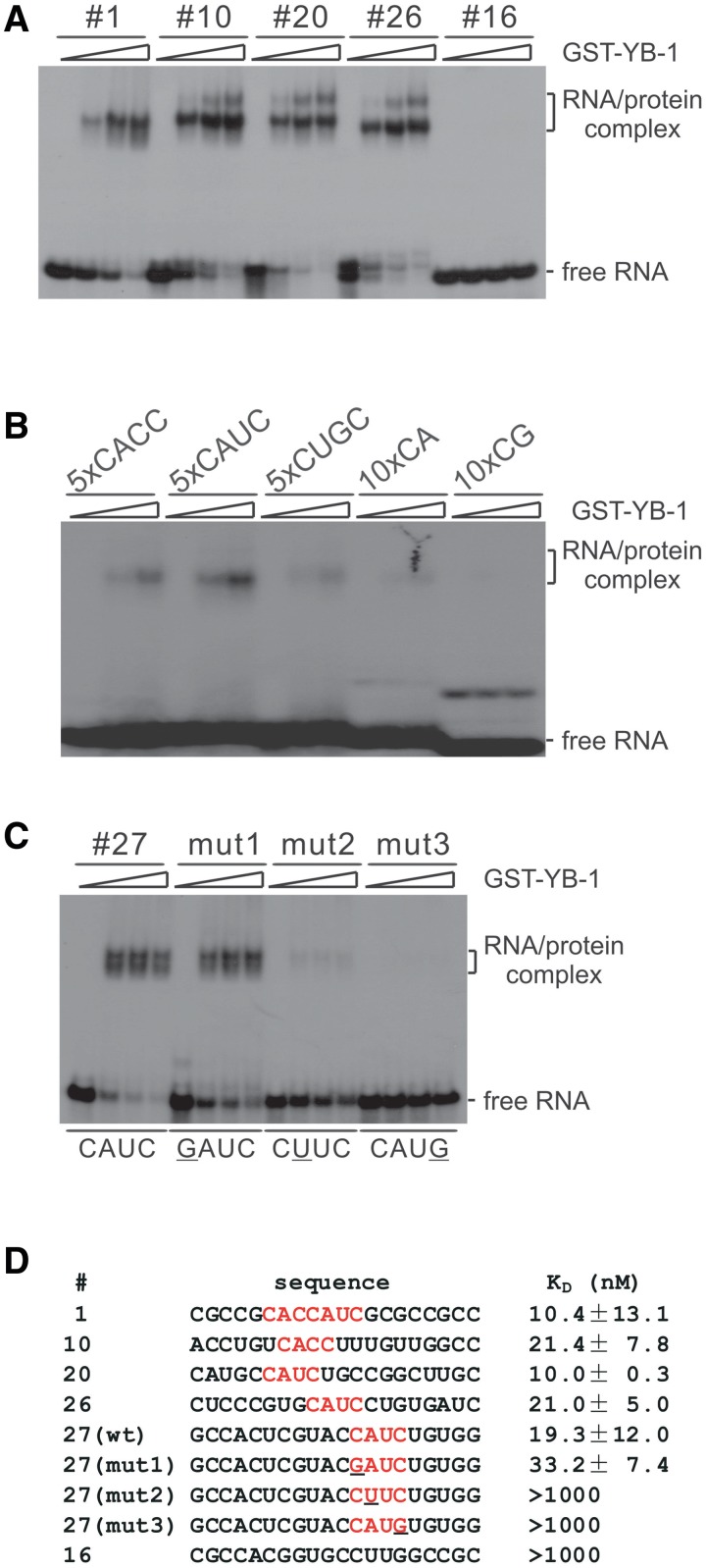
We performed gel mobility shift experiments, confirming that YB-1 indeed binds to SELEX-enriched RNA ligands carrying C A U/C C sequence. As shown in Figure 2A, GST-YB-1 formed two slowly migrating species with SELEX-derived RNAs #1, 10, 20 and 26, which carry either CAUC or CACC sequence. No shift was observed when incubating these RNAs with GST alone (data not shown). Binding of GST-YB-1 was sequence-dependent because GST-YB-1 did not interact with RNA #16, which carries no C A U/C C motif. We also tested the other four sequences (#24, 36, 11 and 17; see Figure 1) lacking C A U/C C motif. No RNA–protein complexes were formed with these transcripts (data not shown).
Figure 2.
YB-1 specifically binds to RNA ligands containing a C A U/C C motif. (A, B, C) Gel shift analyses of the binding activities of selected RNAs with YB-1. 32P-labeled RNAs were incubated with 0, 2.5, 5 and 12.5-fold molar excess of GST-YB-1. RNA-protein complexes were fractionated on a 5% native polyacrylamide gel and visualized by autoradiography. Migration patterns of free RNAs and protein-bound RNAs are indicated on the right. (D) Determination of the KD values of SELEX-derived RNAs tested in panel A and C by filter-binding assays.
It was shown previously that YB-1 binds RNAs containing C/A-rich, CG-rich and CUGC sequences (12–15). To directly compare the binding affinities of YB-1 to these different RNA motifs, we generated short RNAs with five copies of CACC, CAUC, CUGC and 10 copies of CA and CG. We found that GST-YB-1 binds five copies of CACC and CAUC, but not significantly to other RNAs (Figure 2B). Notably, compared with SELEX-derived RNA ligands, YB-1 showed lower binding activity to multimers of RNAs, suggesting that efficient binding of YB-1 to RNA may require properly spaced core motifs or an appropriate context.
To further prove the specific recognition of the core CAUC motif by YB-1, we tested three mutant RNAs derived from SELEX clone #27 (see #27 wild-type and mutant sequences in Figure 2D) by gel shift analysis. We found that YB-1 binds to the mut1 RNA, which carries a mutation at the first cytidine (C1), as efficiently as wild-type RNA. However, when we mutated A2 or C4 in the CAUC motif, both mutant RNAs (mut2 and mut3) could not be recognized by YB-1 (Figure 2C). These results suggest that G is tolerated at certain positions, but does not constitute the central recognition site, explaining why YB-1 binds to RNAs with a high GC content in previous studies (12,13).
Finally, to provide quantitative information on YB-1 binding specificity, we performed filter binding assay to determine the apparent KD values of the RNAs tested previously. Each RNA that carries a CAUC or CACC motif showed apparent KD values within the range of 10–20 nM, indicating high-affinity binding to YB-1 (Figure 2D). Compared with that of #27 wild-type, the KD value of #27 mut1 slightly decreased. Consistent with the gel shift data, the #16, #27 mut2 and #27 mut3 gave rise to very low KD values. In conclusion, YB-1 binds with high affinity to CACC and CAUC motifs in which A2 and C4 are central for YB-1 recognition. These biochemical results provide key criteria for identifying and characterizing YB-1 targets.
YB-1 binding motif CAUC stimulates in vitro splicing of doublesex (dsx) pre-mRNA
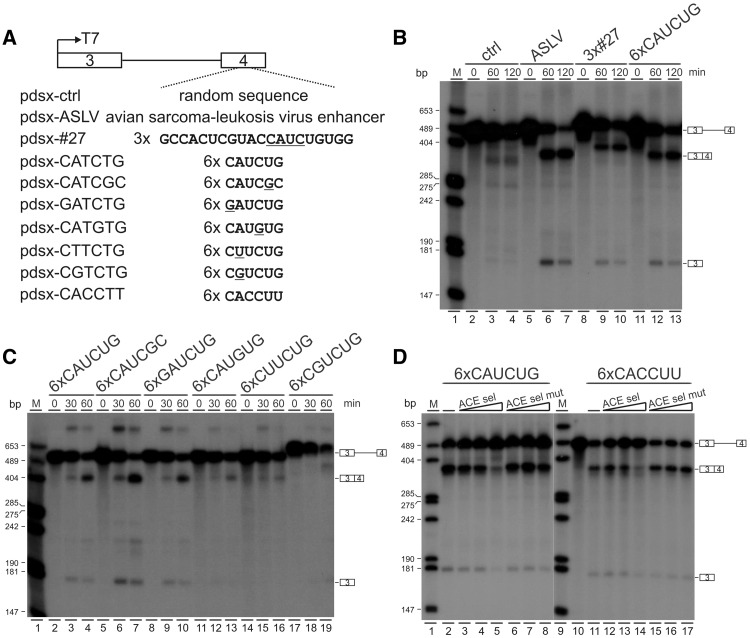
To understand the mechanism by which YB-1 regulates splicing, we first investigated whether the newly defined binding motif CAUC functions as a regulatory cis-element in the splicing of a reporter minigene. It was previously characterized that the 3′ splice site of Drosophila dsx female-specific exon 4 contains a poor polypyrimidine tract interrupted by several purine residues (26). The usage of this 3′ splice site in the dsx pre-mRNA requires a splicing enhancer in exon 4 (27). We made several constructs with different inserts in dsx exon 4 (Figure 3A). 32P-labeled dsx pre-mRNAs were in vitro transcribed and spliced in HeLa nuclear extract. A control construct carrying a random sequence of 64 nt in exon 4 served as a negative control (Figure 3B, lanes 2–4) and the ASLV construct containing a previously characterized enhancer from avian sarcoma-leukosis virus as a positive control (Figure 3B, lanes 5–7). Compared with control pre-mRNAs, insertion of three copies of SELEX winner sequence #27 or six copies of CAUCUG strongly stimulated in vitro splicing of dsx pre-mRNAs (Figure 3B, lanes 8–13). Insertion of six copies of CAUCGC or GAUCUG sequence into dsx exon 4 was also able to efficiently activate dsx splicing (Figure 3C, lanes 5–10). However, when CAUCUG sequence was mutated to CAUGUG, CUUCUG, and CGUCUG, only very low splicing activity was observed (Figure 3C, lanes 11-19). This is consistent with aforementioned gel shift results that A2, but not C1, is important for YB-1 binding. Taken together, these data indicate that the core CAUC motif can specifically stimulate dsx splicing, and the stimulation effect is dependent on YB-1 binding.
Figure 3.
The core binding motif CAUC stimulates in vitro splicing of dsx pre-mRNA. (A) Schematic representation of Drosophila dsx constructs. The dsx exons 3 and 4 are represented by boxes, intron 3 by a solid line. The original enhancer sequences in the dsx female-specific exon 4 were substituted by different sequences indicated below the exon-intron diagram. (B, C) In vitro splicing assay of dsx pre-mRNAs. 32P-labeled pre-mRNAs were spliced in vitro in HeLa cell nuclear extract for the time indicated. The positions of pre-mRNA, first exon and spliced products are indicated on the right. M, 32P-labeled pcDNA3 HpaII restriction fragments. (D) In vitro splicing assay of dsx pre-mRNAs in the presence of specific unlabeled RNAs as competitors. 32P-labeled pre-mRNAs carrying six copies of CAUCUG or CACCUU were spliced in vitro in the absence of any competitors (lanes 2 and 11) or in the presence of ACE sel or ACE sel mut competitors (lanes 3–8 and 12–17). For CAUCUG pre-mRNAs, 600, 1200 and 2400 ng competitor RNAs were used (lanes 3–8). For CACCUU pre-mRNAs, 150, 300 and 600 ng competitor RNAs were added (lanes 12–17).
To provide further evidence that YB-1 mediates CAUC or CACC-dependent splicing activation, we performed in vitro splicing assays supplemented with competitor RNAs. We used the previously characterized short RNAs ACE sel and ACE sel mut as competitors, because ACE sel RNA could be efficiently UV crosslinked to YB-1 and several mutations in the ACE sel mut RNA destroyed YB-1 binding (15). When ACE sel RNA was used as competitor, spliced products generated from dsx pre-mRNAs containing CAUC or CACC motifs decreased significantly (Figure 3D, lanes 2–5, 11–14). In contrast, adding ACE sel mut RNA competitor into the reaction did not affect the in vitro splicing activity (Figure 3D, lanes 6–8, 15–17). In summary, these results demonstrate that the core motif of CAUC functions as a novel exonic splicing enhancer (ESE), and YB-1 acts on this core binding motif to stimulate dsx pre-mRNA splicing in vitro.
YB-1 activates splicing of CD44 exon v5 through binding to a CAUC motif in the v5 exon region
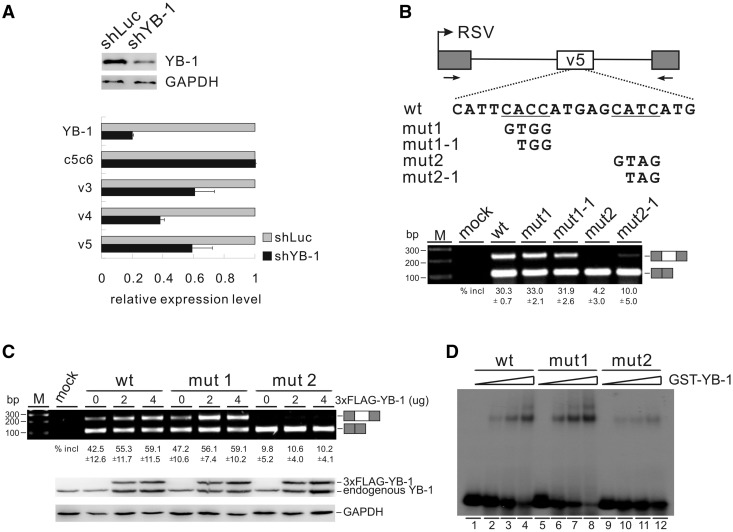
Next, we used CD44 as a model to study the role of the YB-1–CAUC interaction in splicing regulation. CD44 is a group of transmembrane glycoproteins involved in cell–cell and cell–matrix interactions (28). In the human CD44 gene, numerous isoforms are produced by alternative splicing of 9 variable exons, which are located in the middle of 10 constitutive exons. Expression of certain CD44 variable exons correlates with tumorigenesis in a variety of human cancers including breast cancer (29). Because YB-1 is highly expressed in breast cancer cells, we asked whether YB-1 activates splicing of CD44 variable exons. Using shRNA technology, we stably knocked-down YB-1 in breast cancer MDA-MB-231 cells. As determined by western blotting, the knock-down effect was around 80% (Figure 4A). The mRNA expression levels of the constitutively spliced isoform and variable isoforms of CD44 were analysed by RT-PCR using exon-specific primers. As shown in Figure 4A, knock-down of YB-1 did not change the expression level of the constitutive CD44 isoform (c5c6), but strongly reduced the inclusion of variable exons v3, v4 and v5. This result is consistent with the previous report that overexpression of YB-1 increases the inclusion of both v4 and v5 exons in minigene transfection assays (15). To further understand the mechanism of YB-1-regulated CD44 splicing, we used exon v5 as a model to dissect key elements involved in its regulation.
Figure 4.
YB-1 binds to a CAUC motif in CD44 variable exon v5 and stimulates exon v5 splicing. (A) Real-time qRT-PCR analysis of the expression of CD44 variable exons v3, v4 and v5, as well as of constitutive exons c5 and c6 (c5c6) in MDA-MB-231 cells stably transfected with control (luciferase) or YB-1 shRNA. β-Actin served as a reference gene for qRT-PCR. The mRNA expression levels of YB-1 and CD44 exons in YB-1 knock-down cells were normalized to those in control knock-down cells. The YB-1 knock-down efficiency was determined by western blotting with GAPDH as a control. (B) Upper panel: schematic representation of CD44 exon v5 minigene constructs. The CD44 genomic sequence from 794 nt upstream of exon v4 to 479 nt downstream of exon v5 (exon v4 sequence was deleted in this minigene, see Materials and Methods) was inserted into pET01. Exon v5 is represented by a white box, and flanking insulin exons by gray boxes. The potential YB-1-binding sites are underlined. Lower panel: RT-PCR analysis of in vivo splicing pattern of wild-type, mut1, mut1-1, mut2 and mut2-1 constructs transfected into HEK 293 cells. As a control, no DNA was transfected (mock lane). The averages of exon inclusion percentage with standard deviations are shown below (n = 3). (C) RT-PCR analysis of splicing activity of the wild-type, mut1, and mut2 minigene constructs, which were co-transfected with increasing amounts of FLAG-tagged YB-1 expression constructs (0, 2 and 4 µg) in HEK 293 cells. The averages of exon inclusion percentage with standard deviations are shown below (n = 3). The expression level of YB-1 was detected by western blotting, and GAPDH served as a loading control. (D) Gel shift analysis of YB-1 binding affinities to RNAs derived from wt, mut1 and mut2 constructs. Complexes were formed by incubation of 32P-labeled short RNAs with 0-, 25-, 50- and 100-fold molar excess of GST-YB-1 fusion proteins.
We looked carefully in the sequence of exon v5 and found two putative YB-1 binding sites. We cloned exon v5 together with its flanking intron sequence into an exon trap vector, pET01. The two putative RNA-binding sites for YB-1 were mutated separately, resulting in mut1, mut1-1, mut2 and mut2-1 constructs (Figure 4B, upper panel). As shown in Figure 4B (lower panel), mutation of the second putative binding site (a CAUC motif) dramatically inhibited v5 inclusion (mut2 and mut2-1), whereas mutation of the first one (a CACC motif) had no effect (mut1 and mut1-1). To determine whether YB-1 activates v5 splicing through recognizing its binding sites, we co-transfected these minigene constructs with a YB-1 expression vector into HEK 293 cells and determined in vivo splicing patterns by RT-PCR, using primers against the flanking exons. HEK 293 cells were used instead of MDA-MB-231 cells because the v5 inclusion level for the wild-type minigene is lower in HEK 293 than in MDA-MB-231 cells, which is suitable for looking at the stimulation effect by YB-1. Notably, both the wild-type and mut1 showed enhanced v5 inclusion in response to increasing amounts of YB-1. In contrast, co-transfection of YB-1 did not stimulate v5 inclusion of mut2 (Figure 4C). Next, we asked whether the decrease in mutant v5 splicing was caused by a loss of YB-1 binding. Therefore, we assayed the YB-1 binding activity of the wild-type and mutant RNAs containing part of exon v5 sequence by gel shift assay. The RNA probe with the second motif mutated showed a significant reduction of YB-1 binding activity compared with the wild-type and the first mutant probes (Figure 4D, compare lanes 9–12 to lanes 1–8). These data indicate that mutating the YB-1 binding motif CAUC in CD44 v5 resulted in loss of YB-1 binding activity and thereby decreased v5 inclusion. Taken together, CD44 variable exon v5 represents a natural target of YB-1, which activates v5 inclusion through a CAUC motif in exon v5.
The CAUC motifs within the upstream polypyrimidine tract are also required for YB-1-dependent exon v5 inclusion
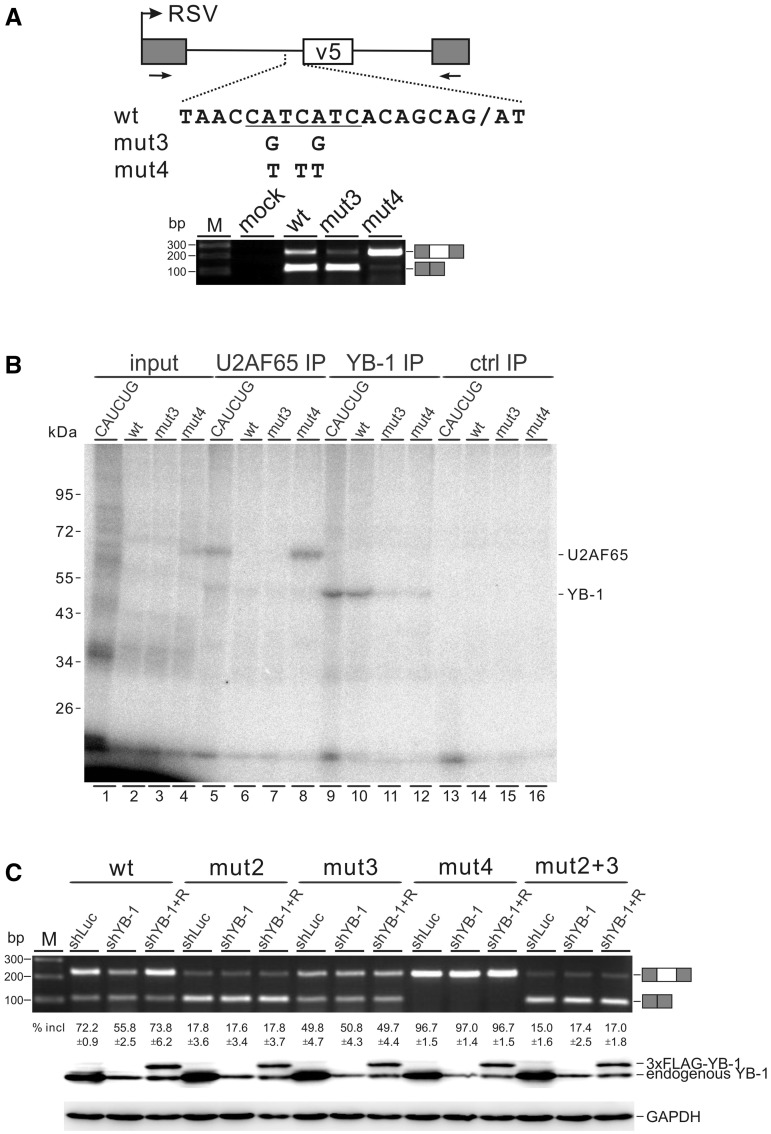
We noted two additional YB-1 binding motifs within the polypyrimidine tract upstream of CD44 exon v5 (Figure 5A). To investigate whether these motifs are required for v5 splicing, we mutated them in construct mut3 by two A-to-G substitutions without changing the purine/pyrimidine composition in this region. The polypyrimidine tract directly upstream of exon v5 is very poor because it is interrupted by purines and does not contain stretches of more than three consecutive pyrimidines. We made another construct, mut4, by introducing three T substitutions to improve the strength of this polypyrimidine tract. After transfecting these constructs into HEK 293 cells, their splicing patterns were assayed by RT-PCR. Compared with the wild-type construct, mut3 led to a strong reduction of v5 inclusion. As expected, mut4 resulted in a significant increase in v5 inclusion (Figure 5A). These data indicate that the CAUC motifs within the poor polypyrimidine tract can also function as an intronic splicing enhancer for efficient splicing of v5.
Figure 5.
The CAUC motifs within the polypyrimidine tract upstream of CD44 v5 are also required for efficient splicing of exon v5. (A) Upper panel: schematic representation of CD44 exon v5 mut3 and mut4 constructs. The labels are the same as in Figure 4B. Lower panel: RT-PCR analysis of in vivo splicing pattern of wild-type, mut3 and mut4 constructs transfected into HEK 293 cells. (B) 32P-labeled RNA transcripts containing 70 nt intron sequence upstream of exon v5 and the first 16 nt of exon v5 were incubated with HeLa nuclear extract and subjected to UV irradiation and RNase digestion. The crosslinked proteins (lanes 1–4) were immunoprecipitated with anti-U2AF65 (lanes 5–8), anti-YB-1 (lanes 9–12) or a control antibody (lanes 13–16). The immunoprecipitates were analysed by SDS-PAGE and visualized by autoradiography. 10% equivalents of the inputs were loaded directly (lanes 1–4). (C) RT-PCR analysis of splicing activity of the wild-type, mut2, mut3, mut4 and mut2 + 3 minigene constructs in MDA-MB-231 cells stably transfected with control (luciferase) shRNA (lanes shLuc), YB-1 shRNA (lanes shYB-1) or co-transfected YB-1 shRNA together with a YB-1-resistant construct (4 μg, lanes shYB-1 + R). The averages of exon inclusion percentage with standard deviations are shown below (n = 3). The expression level of YB-1 was detected by western blotting, and GAPDH served as a loading control.
To address whether the predominant exon-skipping phenotype of mut3 was caused by a loss of YB-1 binding to the mutant RNA, we performed UV crosslinking and immunoprecipitation assays, finding that YB-1 recognized the wild-type RNA probe, but neither the mut3- nor the mut4-derived RNA probe (Figure 5B, lanes 10–12). These data indicate that YB-1 is involved in the recognition of the polypyrimidine tract upstream of v5. This result, coupled with the functional requirement for YB-1 binding in exon v5, suggests that YB-1 may be engaged in a network interaction with its cis-acting elements located in both exonic and intronic regions to enhance splicing, a new regulatory paradigm that is opposite to the well-characterized PTB-mediated interaction network in suppressing 3′ splice site recognition (30,31).
To further test this new regulatory paradigm, we asked whether YB-1 is necessary for the activation of exon v5 splicing through simultaneous recognition of the CAUC elements in both exon v5 and upstream intron. For this purpose, we examined a mut2+3 minigene carrying both mut2 and mut3 mutations in transfected MDA-MB-231 cells after stable knock-down of YB-1. Consistent with the result of in vivo splicing experiment performed in HEK293 cells, mutation of YB-1 binding motifs either in exon v5 (mut2) or within its upstream pyrimidine tract (mut3) decreased the inclusion of exon v5 in MDA-MB-231 control knock-down cells. Mutation of both exonic and intronic CAUC motifs (mut2 + 3) resulted a decreased level of v5 splicing similar to mut2, but did not further reduce v5 inclusion (Figure 5C, shLuc lanes). Knock-down of YB-1 inhibited exon v5 splicing for the wild-type minigene, and expression of a shRNA-resistant YB-1 construct completely restored v5 inclusion. However, knock-down and adding back of YB-1 had no effect on the splicing of v5 for the mut2, mut3, mut2 + 3 and mut4 minigenes (Figure 5C, shYB-1 and shYB-1 + R lanes). Together, these results provide strong evidence that the CAUC binding motifs both in v5 exon and within the upstream polypyrimidine tract are required for activation of v5 splicing by YB-1.
YB-1 enhances the recruitment of U2AF to poor polypyrimidine tracts
Early studies have reported that many natural 3′ splice sites are associated with poor polypyrimidine tracts (32), raising a general question on how these poor 3′ splice sites in the genome are recognized. U2AF65 binding to the polypyrimidine tract is believed to be essential for the early step of spliceosome assembly. Interestingly, we found that U2AF65 bound to the improved polypyrimidine tract of mut4, but not to the wild-type- or mut3-derived RNA probe (Figure 5B, lanes 5–8). The RNA probe derived from pdsx-CATCTG served as a positive control for immunoprecipitation of crosslinked U2AF65 or YB-1 (Figure 5B, lanes 5 and 9). An unrelated antibody was used as a negative control for immunoprecipitation (Figure 5B, lanes 13–16). These results strongly suggest that YB-1 may enhance the recognition of the poor polypyrimidine tract upstream of exon v5 by U2AF65.
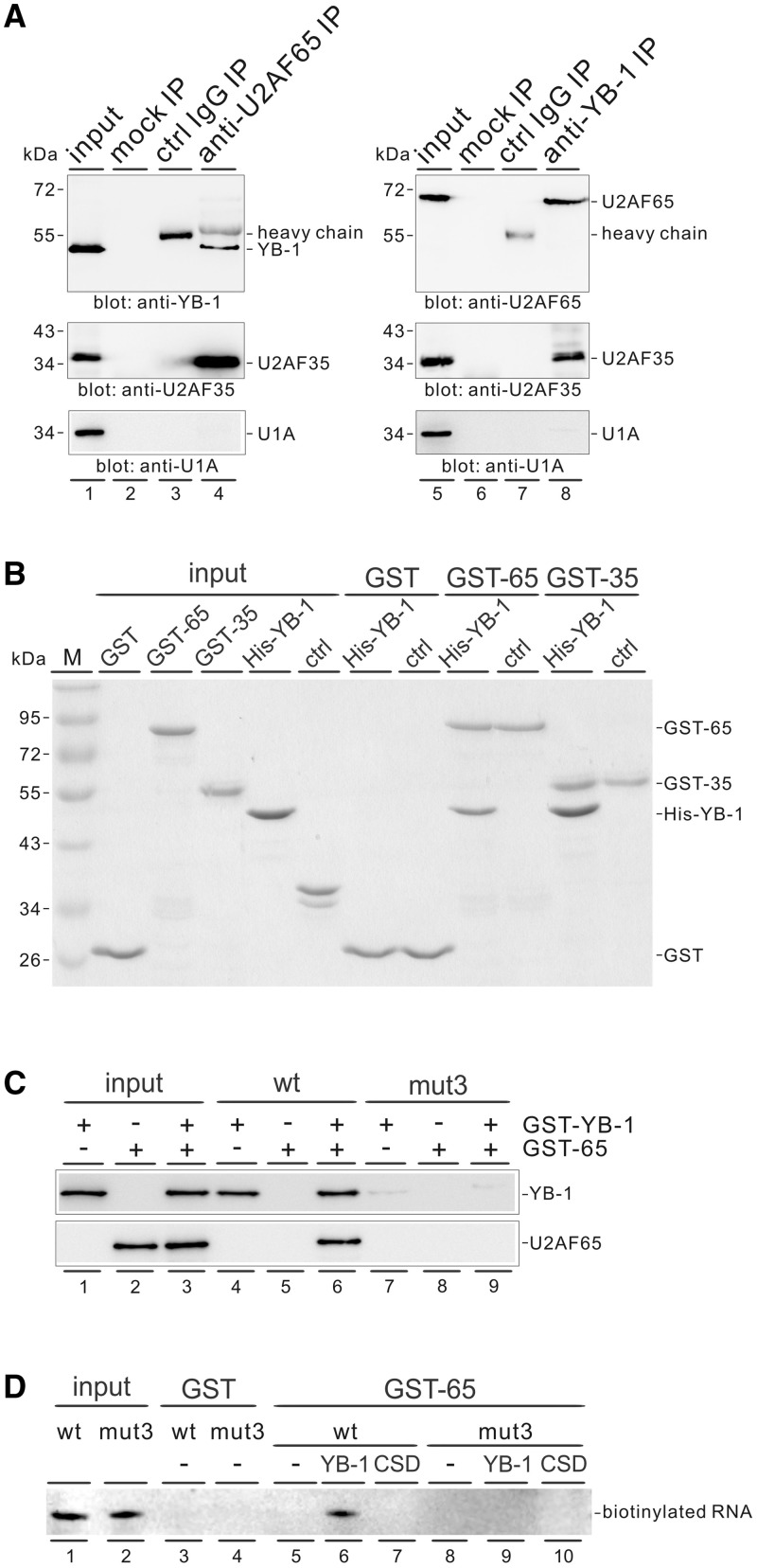
To explore the possibility that YB-1 may play a direct role in recruiting U2AF to a poor polypyrimidine tract, we analysed whether YB-1 associates with U2AF. We did immunoprecipitation experiments in HeLa cell nuclear extract using either anti-YB-1 or anti-U2AF65 antibody, followed by western blot analysis. U2AF65 antibody specifically precipitated YB-1 protein under stringent washing conditions containing 600 mM salt and vice versa (Figure 6A, upper panels). Because U2AF65 and U2AF35 form a heterodimer complex, it was not surprising that both U2AF65 and YB-1 antibodies immunoprecipitated U2AF35 as well (Figure 6A, middle panels). Western blotting detection of U1A was used as a negative control for immunoprecipitation (Figure 6A, lower panels). In addition, the association between YB-1 and U2AF proteins appears to be RNA-independent because HeLa cell nuclear extract was treated with RNase A before immunoprecipitation.
Figure 6.
YB-1 facilitates the recruitment of U2AF65 to a weak polypyrimidine tract. (A) Western blot analysis for the identification of YB-1 and U2AF in different immunoprecipitates. Protein G beads only (lanes 2 and 6) or bound with a control antibody (lanes 3 and 7), anti-U2AF65 antibody (lane 4) or anti-YB-1 antibody (lane 8) were incubated with nuclear extract pre-treated with RNase A and subsequently washed with buffer containing 600 mM KCl. Bound material was blotted with the indicated antibodies. Directly, 20% equivalents of the inputs were loaded (lanes 1 and 5). (B) In vitro GST pulldown assay. The GST, GST-U2AF65 and GST-U2AF-35 proteins immobilized on glutathione-Sepharose were incubated with recombinant His-YB-1 and a non-related His-tagged protein (ctrl). After washing with buffer containing 300 mM KCl, the bound proteins were separated by SDS-PAGE and visualized by Coomassie blue staining. (C) RNA affinity selection assay. The biotin-labeled CD44 wild-type- and mut3-derived RNAs immobilized on streptavidin beads were incubated with either GST-YB-1 or GST-U2AF65, or both proteins. The bound proteins were detected by anti-YB-1 or anti-U2AF65 antibody. (D) Detection of biotinylated RNAs associated with GST-U2AF65. The GST and GST-U2AF65 immobilized on glutathione-Sepharose were incubated with biotinylated CD44 wild-type- and mut3-derived RNAs in the absence or presence of His-YB-1 or His-CSD. The bound RNAs were detected by northern blotting using streptavidin-horseradish peroxidase conjugate and chemiluminescent system.
We further performed GST pulldown assays using recombinant proteins to confirm a direct interaction between YB-1 and U2AF proteins. Notably, recombinant GST-U2AF65 and GST-U2AF-35 protein specifically bound His-YB-1 in the presence of RNase A, but not an unrelated His-tagged protein (Figure 6B). As a control, GST protein did not pulldown YB-1 protein. We conclude that YB-1 interacts with both U2AF subunits directly, strongly suggesting that YB-1 facilitates U2AF recruitment through direct protein–protein interactions.
To provide direct evidence that YB-1 enhances the recruitment of U2AF65 to a weak polypyrimidine tract, we did RNA affinity selection assays using biotin-labeled RNAs. We observed that GST-YB-1 bound to the CD44 wild-type-derived RNA probes, which contain the upstream polypyrimidine tract of exon v5 and part of exon v5 (Figure 6C, lane 4), and GST-U2AF65 associated with the wild-type RNA probes only when GST-YB-1 was present (lanes 5 and 6). However, GST-YB-1 and GST-U2AF65 did not recognize the RNA probes when the intronic CAUC motifs were mutated (lanes 7–9). Vise versa, we detected the biotin-labeled RNAs associated with GST-U2AF65 protein. As expected, GST-U2AF65 was able to pulldown the wild-type RNAs only in the presence of full-length His-YB-1 (Figure 6D, lanes 5–7), whereas it did not pulldown the mutant RNAs, even when YB-1 was added (lanes 8-10). These results demonstrate that YB-1 facilitates the recruitment of U2AF65 to a weak 3′ splice site.
The C-terminal domain of YB-1 and the RS domains of U2AFs are responsible for YB-1–U2AF interaction
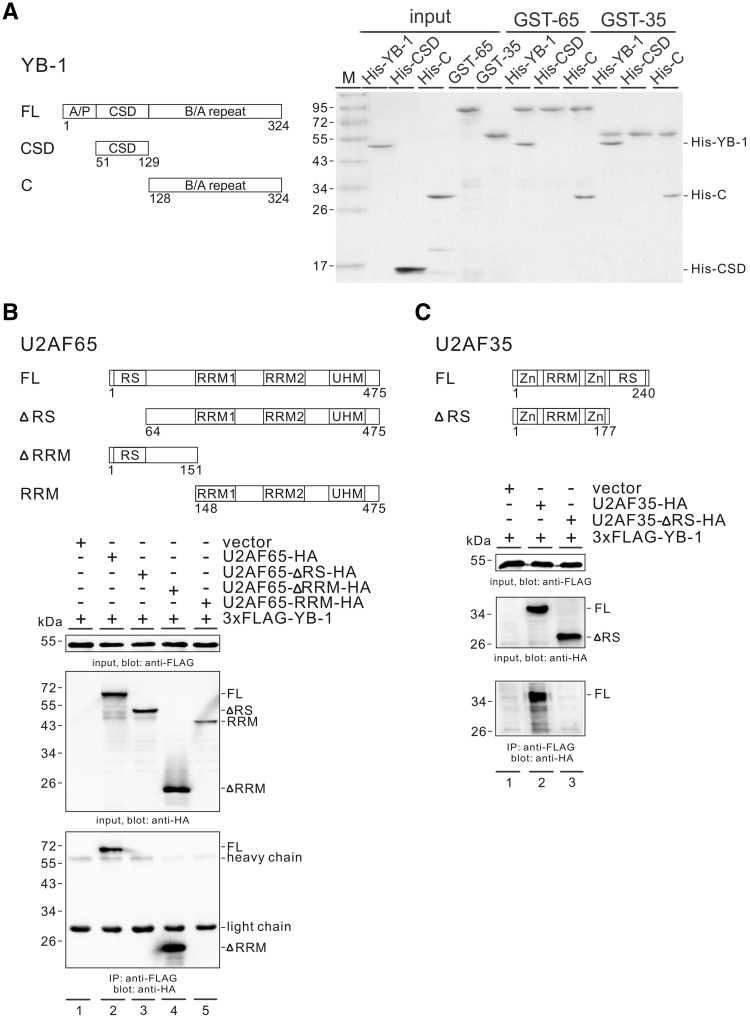
YB-1 protein is composed of three domains: the N-terminal alanine-proline rich (A/P) domain, the CSD and the C-terminal basic/acidic repeat (B/A repeat) domain (Figure 7A, left). To investigate which domain(s) of YB-1 is responsible for the interaction with U2AF, we used two His-tagged proteins. His-CSD contains the CSD (amino acids 51–129), whereas His-C carries the C-terminal B/A repeat domain (amino acids 128–324). We performed in vitro GST pulldown experiment as previously given. It showed that both U2AF subunits interact directly with the C-terminal domain of YB-1, but not the CSD domain (Figure 7A, right). This result also explains why GST-U2AF65 did not associate with the RNA probe when only the CSD domain of YB-1 was provided (Figure 6D, lane 7).
Figure 7.
Both the C-terminal domain of YB-1 and the RS domains of U2AF are important for the interaction between YB-1 and U2AF proteins. (A) In vitro GST pull down assay. Left panel: domain structure of full-length YB-1 and its mutants. Right panel: the GST-U2AF65 and GST-U2AF-35 proteins immobilized on glutathione-Sepharose were incubated with recombinant His-tagged full-length YB-1, its CSD and C-terminal domain proteins. After washing, the bound proteins were separated by SDS-PAGE and visualized by Coomassie blue staining. (B, C) Upper panels: domain structure of full-length U2AF and its mutants. RS: arginine/serine-rich domain; RRM: RNA recognition motif; UHM: U2AF homology motif; Zn: zinc finger domain. Lower panels: Co-IP of YB-1 and U2AF proteins. FLAG-tagged YB-1 was co-expressed with a vector (lane 1 in 7 A and 7B), HA-tagged U2AF65 (7 A, lane 2), HA-tagged U2AF65 mutants (7 A, lane 3–5), HA-tagged U2AF35 (7B, lane 2) or HA-tagged U2AF35-ΔRS (7B, lane 3) in HEK 293 cells. The expressed proteins in the cell lysates were detected by western blotting using anti-FLAG or anti-HA antibody. Cell lysates pre-treated with RNase A were immunoprecipitated with anti-FLAG antibody and immunoblotted with anti-HA antibody.
Both U2AF65 and U2AF35 contain two or one RRM-type RNA-binding domain, respectively, and a serine-arginine rich (RS) domain (Figure 7B and 7C, upper). To identify which domain(s) of U2AF proteins interacts with YB-1, we performed co-immunoprecipitation (co-IP) experiments. The FLAG-tagged YB-1 expression construct was co-transfected with HA-tagged U2AF65, U2AF35 or their deletion mutants into HEK 293 cells. RNase A was added to the lysis buffer during the co-IP assay, and the results indicated that FLAG-tagged YB-1 interacted with HA-tagged U2AF65 and U2AF35 in an RNA-independent manner (Figure 7B, lane 2, and 7C, lane 2). In addition, FLAG-tagged YB-1 interacted with U2AF65 mutant containing the RS, but lacking the RRM domain (ΔRRM; Figure 7B, lane 4). However, it did not interact with the mutant derivatives ΔRS nor RRM (Figure 7B, lanes 3 and 5). Similarly, FLAG-tagged YB-1 also did not interact with a U2AF35 mutant lacking the RS domain (ΔRS; Figure 7C, lane 3). Interestingly, YB-1 did not precipitate U2AF proteins lacking their RS domain via the endogenous other partner in the heterodimer, perhaps due to inefficient heterodimer formation between the HA-tagged U2AF proteins and the endogenous proteins. Taken together, these data reveal that both the C-terminal domain of YB-1 and at least one of the RS domains of the U2AF subunits are important for the interaction between YB-1 and U2AF subunits. Our results suggest a model for the activation of poor 3′ splice sites lacking U2AF65 binding sites, where YB-1 may recognize its binding motifs embedded in both exon and the upstream polypyrimidine tract via its CSD domain and recruit core spliceosomal factors via its C-terminal domain.
DISCUSSION
The RNA-binding specificity of an RNA-binding protein is important for understanding its roles in gene expression regulation. A number of earlier studies had reported that YB-1 functions in the regulation of splicing, mRNA stability and translation through binding to specific RNA sequences (for references, see Table 1). However, the RNA-binding specificity of YB-1 is not well defined. Using an SELEX approach, here we identify the CACC and CAUC motifs as high-affinity RNA-binding sites for YB-1. This was confirmed by gel shift assays. The RNA-binding specificity of YB-1 defined in this study explains older data in the literature, where most of previously characterized YB-1 binding sequences indeed contain CACC or CAUC motifs (underlined in Table 1). For example, it was reported that YB-1 UV crosslinks to the ACE sel RNA (CACCAGUCACCGC), but not to the ACE sel mut RNA (CUGGCGUCGUCGC) (15). Moreover, it was known that YB-1 negatively regulates its own translation through binding to a regulatory element in the 3′ untranslated region (UTR) of its mRNA (AGUCAUCCAACAA), which was identified by foot printing (38). The CAUC binding motif we identified in this study is also included in the AACAUC sequence, which had been defined as the consensus sequence recognized by the Xenopus YB-1 homologues FRGY1 and FRGY2 (41). Although the C A U/C C motif is defined as the high-affinity RNA-binding site for YB-1, the RNA-binding sequence of YB-1 can allow degeneracy and tolerance at certain positions like other splicing regulators. Our mutational analyses further suggest that the C1 in the consensus is not important for YB-1 recognition. This also explains that YB-1 can bind to a splicing enhancer element (ACAACCACAA) identified in CD44 exon v4 (15). We also showed that YB-1 did not bind to the CACC motif in CD44 exon v5 (Figure 4D), which indicated that the accessibility of YB-1 to RNA can be context-specific, depending on the local RNA secondary structure at its binding sites.
Table 1.
Known RNA sequences bound by YB-1
| Genes | Sequences | Regulation | References |
|---|---|---|---|
| GPX1 | UCUCGGGGGGGUUUUCAUCUAUGAGGGUG UUUCCUCUAAACCUACGAGGGAGGAACACCU GAUCUUACAGAAAAUACCACCUCGAGA | Translation | (33) |
| IL-2 | AUCACUCUCUUAAUCACUACU | mRNA stability | (34) |
| CD44 v4 | ACAACCACAA | Splicing | (15) |
| ACE sel | CACCAGUCACCGC | Splicing | (15) |
| Ferritin | UCUUGCUUCAACAGUGUUUGAACGGAAC | Translation | (35) |
| Renin | GCUGAGGCCCUCUGCCACCCAGGCAGGCCCUGCCU UCAGCCCUGGCCCAGAGCUGGAACACUCUCUGAGA UGCCCCUCUGCCUGGGCUUAUGCCCUCAGAUGG AGACAUUGGAUGUGGAGCUCCUGCUGGAUGCGUG CCCUGACCCCUGCACCAGCCCUUCCCUGCUUUGA GGACAAAGAGAAUAAAGACUUCAUGUUCAC | mRNA stability | (36) |
| YB-1 | GGGCUUAUCCCGCCUGUCCCGCCAUUCUCGCUAGU UCGAUCGGUAGCGGGAGCGGAGAGCGGACCCC AGAGAGCCCUGAGCAGCCCCACCGCCGCCGCCG GCCUAGUUACCAUCACACCCCGGGAGGAGCCGC AGCUGCCGCAGCCGGCCCCAGUCACCAUCACCG CAACCAUGAGCAGCGAGGCCGAGACCCAGCAGCC | Translation | (37) |
| YB-1 | AGUCAUCCAACAA | Translation | (38) |
| Dengue virus | UGACGCUGGGAAAGACCAGAGAUCCUGCUGCUC AGCAUCAUUCCAGGCACAGAACGCCAGAAAAUG GAAUGGUGCUGUUCAACAGGUUCU | Translation | (39) |
| TGF-β1 | GGGCACCCCCCCCGGCUCU | Translation | (40) |
| NF1 | ACAAC | Splicing | (14) |
The RNA-binding motifs identified in this study are underlined.
Using either in vitro or in vivo functional selection assays, several laboratories previously recovered sequences with enhancer activity from a pool of RNA sequences, resulting in the identification of two classes of exonic elements, purine-rich and the C/A-rich ESEs (42–44). YB-1 was identified as a splicing activator through recognizing a C/A-rich ESE in CD44 exon v4 (15). Here, we characterize the CAUC motif as a new type of ESE, consistent with previous studies indicating that some ESE activities are embedded in some general C/A-rich sequences. Importantly, we show that YB-1 regulates alternative splicing through binding to the core CAUC motifs in both exon and the upstream intron. Recently, the position-dependent effect of splicing regulators emerged as a key mechanism in splicing regulation (45–48). Most splicing regulators share some common positional principles. They often activate exon inclusion by binding to the downstream intron and silence exon inclusion by binding to the exon or the upstream intron region in the proximity to the alternative exon. The silencing effect could be explained by the competition between splicing regulators and the core spliceosomal factors. Particularly, it is known that PTB and Sxl can bind to polypyrimidine tracts and inhibit the use of 3′ splice sites by antagonizing the effect of U2AF65 (49). In contrast to most splicing factors, YB-1 appears to exert a positive influence on splice site selection through network interactions on its binding sites in both exon and the upstream polypyrimidine tract.
U2AF65 is an essential splicing factor that recognizes the polypyrimidine tract and promotes the annealing of U2 snRNA to the branch point. It has been shown that U2AF65 selectively binds sequences enriched in uridines that are frequently interrupted by cytidines (49). In the mammalian genome, the sequences at the 3′ splice site are highly degenerate. In particular, a large number of introns do not contain effective binding sites for U2AF65 in the polypyrimidine tracts (32). A fundamental question about how such poor 3′ splice sites are recognized by the splicing machinery has remained unclear. In this study, we provide evidence that YB-1 can facilitate the recruitment of U2AF65 to 3′ splice site through direct protein–protein interactions. We identified YB-1 as a new interacting partner for U2AF using three different approaches, including immunoprecipitation analyses in HeLa nuclear extract, in vitro GST pulldown assays using recombinant proteins, and in vivo co-immunoprecipitation experiments. We further characterized the C-terminal domain in YB-1 and the RS domains in U2AF as the interacting regions. Similar to SR proteins, YB-1 may bind to ESEs and interact with U2AF35, thereby recruiting U2AF65 to the poor polypyrimidine tracts (50). Based on our observation that YB-1 interacts with U2AF proteins via their RS domains, together with the previous finding that SRp30c is an interacting partner of YB-1 (51), we hypothesized that YB-1 might interact with other RS domain-containing splicing factors. We tested an SR-related protein, the U1 snRNP-specific protein U1-70K, and found that the interaction of YB-1 with U1-70K is indirect and RNA-dependent (Wei WJ and Mu SR, unpublished data). However, it is still possible that YB-1 may associate with U1-70K via other SR proteins and provide a bridge between the 5′ and 3′ splice sites.
Because it had been previously postulated that YB-1 may interact with the branch point binding protein, SF1 (15), we also tested this hypothesis by performing GST pulldown and co-immunoprecipitation (co-IP) assays in the presence of RNase A. As a result, we did not detect a direct interaction between YB-1 and SF1 (data not shown).
Our findings on the action of YB-1 binding sites within polypyrimidine tracts suggest that the interaction between U2AF65 and the polypyrimidine tract is dispensable for 3′ splice site recognition. YB-1 recognizes the weak polypyrimidine tract in a sequence-specific manner and mediates the recruitment of U2AF65 through protein–protein interactions. We have made several efforts to establish an in vitro splicing system for v5 inclusion, which, unfortunately was not efficient enough (data not shown). Considering a large number of introns in the mammalian genome contain a wide diversity of polypyrimidine tracts lacking U2AF65 binding sites, our study suggests that many RNA-binding proteins with distinct binding specificity may be involved in the recognition of weak 3′ splice sites.
In conclusion, we define the CACC and CAUC sequences as specific high-affinity RNA-binding motifs for YB-1. The core CAUC motif represents a novel splicing enhancer element that can function in both exon and intron. We provide evidence that YB-1 facilitates U2AF recruitment through direct interaction with U2AF, which establishes a first model for YB-1-mediated splicing regulation and provides new insights into the recognition of weak 3′ splice sites by the splicing machinery.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Material.
FUNDING
The National Basic Research Program of China [2011CBA01105, 2011CB811304]; National Natural Science Foundation of China [30970620]; “One Hundred Talented people” program of the Chinese Academy of Sciences; the Shanghai Municipal Council for Science and Technology [09PJ1411000 to J.H.]. Funding for open access charge: The National Natural Science Foundation of China.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Xiang-Dong Fu (University of California, San Diego) for critical reading and valuable comments on the manuscript and Reinhard Lührmann (Max Planck Institute for Biophysical Chemistry, Germany) for helpful discussion. We also thank Sandra E. Dunn (University of British Columbia, Canada), Kimitoshi Kohno (University of Occupational and Environmental Health, Japan) and Angela Krämer (University of Geneva, Switzerland) for the generous gifts of plasmid constructs, Gaoxiang Ge (Chinese Academy of Sciences, China) for providing the MDA-MB-231 cell line, Xiaojuan Yang and Ying Huang (Chinese Academy of Sciences, China) for providing purified YB-1 proteins.
REFERENCES
- 1.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 3.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 7.Matlin A, Clark JF, Smith CW. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 8.Kohno K, Izumi H, Uchiumi T, Ashizuka M, Kuwano M. The pleiotropic functions of the Y-box-binding protein, YB-1. Bioessays. 2003;25:691–698. doi: 10.1002/bies.10300. [DOI] [PubMed] [Google Scholar]
- 9.Bargou RC, Jürchott K, Wagener C, Bergmann S, Metzner S, Bommert K, Mapara MY, Winzer KJ, Dietel M, Dörken B, et al. Nuclear localization and increased levels of transcription factor YB-1 in primary human breast cancers are associated with intrinsic MDR1 gene expression. Nat. Med. 1997;3:447–450. doi: 10.1038/nm0497-447. [DOI] [PubMed] [Google Scholar]
- 10.Evdokimova V, Tognon C, Ng T, Sorensen PH. Reduced proliferation and enhanced migration: two sides of the same coin? Molecular mechanisms of metastatic progression by YB-1. Cell Cycle. 2009;8:2901–2906. doi: 10.4161/cc.8.18.9537. [DOI] [PubMed] [Google Scholar]
- 11.Fujii T, Yokoyama G, Takahashi H, Toh U, Kage M, Ono M, Shirouzu K, Kuwano M. Preclinical and clinical studies of novel breast cancer drugs targeting molecules involved in protein kinase C signaling, the putative metastasis-suppressor gene Cap43 and the Y-box binding protein-1. Curr. Med. Chem. 2008;15:528–537. doi: 10.2174/092986708783769759. [DOI] [PubMed] [Google Scholar]
- 12.Ray D, Kazan H, Chan ET, Peña Castillo L, Chaudhry S, Talukder S, Blencowe BJ, Morris Q, Hughes TR. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat. Biotechnol. 2009;27:667–670. doi: 10.1038/nbt.1550. [DOI] [PubMed] [Google Scholar]
- 13.Dong J, Akcakanat A, Stivers DN, Zhang J, Kim D, Meric-Bernstam F. RNA-binding specificity of Y-box protein 1. RNA Biol. 2009;6:59–64. doi: 10.4161/rna.6.1.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skoko N, Baralle M, Buratti E, Baralle FE. The pathological splicing mutation c.6792C > G in NF1 exon 37 causes a change of tenancy between antagonistic splicing factors. FEBS Lett. 2008;582:2231–2236. doi: 10.1016/j.febslet.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 15.Stickeler E, Fraser SD, Honig A, Chen AL, Berget SM, Cooper TA. The RNA binding protein YB-1 binds A/C-rich exon enhancers and stimulates splicing of the CD44 alternative exon v4. EMBO J. 2001;20:3821–3830. doi: 10.1093/emboj/20.14.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, Urlaub H, Lührmann R. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell. Biol. 2006;26:5528–5543. doi: 10.1128/MCB.00582-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartmuth K, Urlaub H, Vornlocher HP, Will CL, Gentzel M, Wilm M, Lührmann R. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16719–16724. doi: 10.1073/pnas.262483899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allemand E, Hastings ML, Murray MV, Myers MP, Krainer AR. Alternative splicing regulation by interaction of phosphatase PP2Cgamma with nucleic acid-binding protein YB-1. Nat. Struct. Mol. Biol. 2007;14:630–638. doi: 10.1038/nsmb1257. [DOI] [PubMed] [Google Scholar]
- 19.Dutertre M, Sanchez G, De Cian MC, Barbier J, Dardenne E, Gratadou L, Dujardin G, Le Jossic-Corcos C, Corcos L, Auboeuf D. Cotranscriptional exon skipping in the genotoxic stress response. Nat. Struct. Mol. Biol. 2010;17: 1358–1366. doi: 10.1038/nsmb.1912. [DOI] [PubMed] [Google Scholar]
- 20.Sutherland BW, Kucab J, Wu J, Lee C, Cheang MC, Yorida E, Turbin D, Dedhar S, Nelson C, Pollak M, et al. Akt phosphorylates the Y-box binding protein 1 at Ser102 located in the cold shock domain and affects the anchorage-independent growth of breast cancer cells. Oncogene. 2005;24:4281–4292. doi: 10.1038/sj.onc.1208590. [DOI] [PubMed] [Google Scholar]
- 21.Woerfel G, Bindereif A. In vitro selection of exonic splicing enhancer sequences: identification of novel CD44 enhancers. Nucleic Acids Res. 2001;29:3204–3211. doi: 10.1093/nar/29.15.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hui J, Reither G, Bindereif A. Novel functional role of CA repeats and hnRNP L in RNA stability. RNA. 2003;9:931–936. doi: 10.1261/rna.5660803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hui J, Hung LH, Heiner M, Schreiner S, Neumuller N, Reither G, Haas SA, Bindereif A. Intronic CA-repeat and CA-rich elements: a new class of regulators of mammalian alternative splicing. EMBO J. 2005;24:1988–1998. doi: 10.1038/sj.emboj.7600677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Xie WQ, Rothblum LI. Rapid, small-scale RNA isolation from tissue culture cells. BioTechniques. 1991;11:326–327. [PubMed] [Google Scholar]
- 26.Burtis KC, Baker BS. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell. 1989;56:997–1010. doi: 10.1016/0092-8674(89)90633-8. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka K, Watakabe A, Shimura Y. Polypurine sequences within a downstream exon function as a splicing enhancer. Mol. Cell Biol. 1994;14:1347–1354. doi: 10.1128/mcb.14.2.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat. Rev. Mol. Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 29.Ponta H, Wainwright D, Herrlich P. The CD44 protein family. Int. J. Biochem. Cell Biol. 1998;30:299–305. doi: 10.1016/s1357-2725(97)00152-0. [DOI] [PubMed] [Google Scholar]
- 30.Amir-Ahmady B, Boutz PL, Markovtsov V, Phillips ML, Black DL. Exon repression by polypyrimidine tract binding protein. RNA. 2005;11:699–716. doi: 10.1261/rna.2250405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chou MY, Underwood JG, Nikolic J, Luu MH, Black DL. Multisite RNA binding and release of polypyrimidine tract binding protein during the regulation of c-src neural-specific splicing. Mol. Cell. 2000;5:949–957. doi: 10.1016/s1097-2765(00)80260-9. [DOI] [PubMed] [Google Scholar]
- 32.Murray JI, Voelker RB, Henscheid KL, Warf MB, Berglund JA. Identification of motifs that function in the splicing of non-canonical introns. Genome Res. 2008;9:R97. doi: 10.1186/gb-2008-9-6-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen Q, Wu R, Leonard JL, Newburger PE. Identification and molecular cloning of a human selenocysteine insertion sequence-binding protein. A bifunctional role for DNA-binding protein B. J. Biol. Chem. 1998;273:5443–5446. doi: 10.1074/jbc.273.10.5443. [DOI] [PubMed] [Google Scholar]
- 34.Chen CY, Gherzi R, Andersen JS, Gaietta G, Jürchott K, Royer HD, Mann M, Karin M. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 2000;14:1236–1248. [PMC free article] [PubMed] [Google Scholar]
- 35.Ashizuka M, Fukuda T, Nakamura T, Shirasuna K, Iwai K, Izumi H, Kohno K, Kuwano M, Uchiumi T. Novel translational control through an iron-responsive element by interaction of multifunctional protein YB-1 and IRP2. Mol. Cell. Biol. 2002;22:6375–6383. doi: 10.1128/MCB.22.18.6375-6383.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skalweit A, Doller A, Huth A, Kähne T, Persson PB, Thiele BJ. Posttranscriptional control of renin synthesis: identification of proteins interacting with renin mRNA 3′-untranslated region. Circ. Res. 2003;92:419–427. doi: 10.1161/01.RES.0000059300.67152.4E. [DOI] [PubMed] [Google Scholar]
- 37.Fukuda T, Ashizuka M, Nakamura T, Shibahara K, Maeda K, Izumi H, Kohno K, Kuwano M, Uchiumi T. Characterization of the 5'-untranslated region of YB-1 mRNA and autoregulation of translation by YB-1 protein. Nucleic Acids Res. 2004;32:611–622. doi: 10.1093/nar/gkh223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skabkina OV, Lyabin DN, Skabkin MA, Ovchinnikov LP. YB-1 autoregulates translation of its own mRNA at or prior to the step of 40S ribosomal subunit joining. Mol. Cell. Biol. 2005;25:3317–3323. doi: 10.1128/MCB.25.8.3317-3323.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paranjape SM, Harris E. Y box-binding protein-1 binds to the dengue virus 3'-untranslated region and mediates antiviral effects. J. Biol. Chem. 2007;282:30497–30508. doi: 10.1074/jbc.M705755200. [DOI] [PubMed] [Google Scholar]
- 40.Fraser DJ, Phillips AO, Zhang X, van Roeyen CR, Muehlenberg P, En-Nia A, Mertens PR. Y-box protein-1 controls transforming growth factor-beta1 translation in proximal tubular cells. Kidney Int. 2008;73:724–732. doi: 10.1038/sj.ki.5002719. [DOI] [PubMed] [Google Scholar]
- 41.Bouvet P, Matsumoto K, Wolffe AP. Sequence-specific RNA recognition by the Xenopus Y-box proteins. An essential role for the cold shock domain. J. Biol. Chem. 1995;270:28297–28303. doi: 10.1074/jbc.270.47.28297. [DOI] [PubMed] [Google Scholar]
- 42.Coulter LR, Landree MA, Cooper TA. Identification of a new class of exonic splicing enhancers by in vivo selection. Mol. Cell. Biol. 1997;17:2143–2150. doi: 10.1128/mcb.17.4.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu HX, Zhang M, Krainer AR. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schaal TD, Maniatis T. Selection and characterization of pre-mRNA splicing enhancers: identification of novel SR protein-specific enhancer sequences. Mol. Cell. Biol. 19:1705–1719. doi: 10.1128/mcb.19.3.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 46.Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol. Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Llorian M, Schwartz S, Clark TA, Hollander D, Tan LY, Spellman R, Gordon A, Schweitzer AC, de la Grange P, Ast G, et al. Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nat. Struct. Mol. Biol. 2010;17:1114–1123. doi: 10.1038/nsmb.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeo GW, Coufal NG, Liang YT, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh R, Valcárcel J, Green MR. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 50.Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 51.Raffetseder U, Frye B, Rauen T, Jürchott K, Royer HD, Jansen PL, Mertens PR. Splicing factor SRp30c interaction with Y-box protein-1 confers nuclear YB-1 shuttling and alternative splice site selection. J. Biol. Chem. 2003;278:18241–18248. doi: 10.1074/jbc.M212518200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.