Abstract
Since the initial discovery of the catalytic capability of short DNA fragments, this peculiar enzyme-like property (termed DNAzyme) has continued to garner much interest in the scientific community because of the virtually unlimited applications in developing new molecular devices. Alongside the exponential rise in the number of DNAzyme applications in the last past years, the search for convenient ways to improve its overall efficiency has only started to emerge. Credence has been lent to this strategy by the recent demonstration that the quadruplex-based DNAzyme proficiency can be enhanced by ATP supplements. Herein, we have made a further leap along this path, trying first of all to decipher the actual DNAzyme catalytic cycle (to gain insights into the steps ATP may influence), and subsequently investigating in detail the influence of all the parameters that govern the catalytic efficiency. We have extended this study to other nucleotides and quadruplexes, thus demonstrating the versatility and broad applicability of such an approach. The defined exquisitely efficient DNAzyme protocols were exploited to highlight the enticing advantages of this method via a 96-well plate experiment that enables the detection of nanomolar DNA concentrations in real-time with the naked-eye (see movie as Supplementary Data).
INTRODUCTION
Alongside its fundamental role as repository of genetic information (1), DNA (2–4) has become in recent years a pivotal element for nanotechnological developments (5–7). Of particular importance are the enzyme-like properties that short oligonucleotides may display: this unusual activity was initially discovered in 1994 by Breaker and Joyce (8), when they demonstrated that short DNA fragments were effectual in promoting catalytic cleavage of RNA sequences they are associated with. This peculiar capability of DNA, initially called ‘catalytic DNA’ or ‘DNA enzyme’ activity, was subsequently termed deoxyribozyme or DNAzyme activity. A couple of years later, the scope of the DNAzyme process was extended far beyond the oligonucleotide cleavage activity by Li & Sen (9). upon the demonstration that DNA sequences (notably the 24-nucleotide PS5.M, vide infra) help catalyzing the metalation (copper, zinc) of some porphyrins (chiefly mesoporphyrin IX) (10). In the course of their studies, the authors made this interesting observation that this catalytic activity was subject to competitive inhibition by an already metalated porphyrin like hemin, thereby making these short DNA fragments interesting hemin aptamers. They further studied this DNA/hemin interaction, subsequently demonstrating that the hemin-binding properties—and so, the catalytic proficiency—of the studied DNA relied on their ability to adopt an intramolecular G-quadruplex structure (11); indeed, single-stranded G-rich sequences, like PS5.M (5’GTG3TCAT2GTG3TG3TGTG23’), have been shown to fold into a higher-order structure composed of stacked G-quartets and nucleated by physiologically relevant cations (like K+) (12–14). The curiosity made the authors investigating the possibility of the resulting quadruplex/hemin complex to display hemoprotein-like properties: they evaluated the ability of these complexes to facilitate the peroxidase-like H2O2-promoted oxidation of a chromogenic substrate (2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid), ABTS), and the observed catalytic activity provided the proof-of-concept that such system is capable of performing enzyme-like reactions (15). This pioneering work thus ushered in the modern era of what is nowadays called the ‘DNA enzymology’ field (5–7).
Over the last decade, the quadruplex-mediated DNAzyme process found dozens of applications, mainly as fluorescent, colorimetric or electrochemical sensors (16–18). Of particular interest among these applications was a method recently developed by Freeman et al. (19) to optically assess the activity of the telomerase (20–23): a closer look to this assay led us to delve into and decipher the quadruplex-mediated DNAzyme activity of multimeric quadruplexes (i.e. higher-order structures made of successive quadruplex units that result from the folding of long G-rich telomeric fragments) (24). In the course of our study, we found a way to improve the DNAzyme activities of both monomeric and multimeric quadruplexes by the use of a synthetic G-quartet (known as TASQ, for Template-Assembled Synthetic G-Quartet) (25,26), namely DOTASQ [for DOTA-templated Synthetic G-Quartet (27)]: the concomitant use of DNA and DOTASQ enables the hemin to be sandwiched at both extremities of quadruplex architectures, thereby being in a pocket whose hydrophobicity is optimal for catalyzing efficiently the peroxidatic process. The boosting effect of DOTASQ was efficient but modest (up to 6-fold) and enabled to lower the limit for reliable DNA detection at ∼500 nM scale.
Given the outstanding versatility of the DNAzyme applications (5–7,16–18), we believe that enhancing the peroxidatic activity of a quadruplex/hemin assembly may be highly useful. The above-mentioned DOTASQ approach is in this way interesting but suffers, despite its overall success, from two drawbacks: first, DOTASQ is itself a pre-catalyst for DNAzyme process (28), and consequently it is responsible for an elevated background reaction that makes the lowering of the detection limit hardly feasible; second, even if the chemical access of DOTASQ is straightforward (27), it is not commercially available, and consequently it will not be readily used in every common laboratories. Because we are convinced that it is worth finding efficient DNAzyme-enhancing agents, we decided to invest efforts to circumvent these drawbacks: results presented herein, which highlight the very positive role of ATP—and more generally of nucleotides—for boosting DNAzyme processes, are inspired by a recent report from D.-M. Kong and co-workers (29); ATP indeed fulfils the requirements of an ideal boosting agent since it is commercially available (from classical suppliers), cheap (∼10€/g at the highest purity grade) and practically convenient (notably in terms of both chemical stability and water-solubility). However, the precise role played by ATP remains to be elucidated: on the basis of ABTS oxidation studies, Kong and co-workers have highlighted its radical cation stabilizing properties (ABTS·+ being the final product of the catalysis) along with a rather ill-defined role of catalysis enhancer, also delineating its own trade-offs (notably its substrate- and quadruplex-dependency).
To further investigate and decipher the precise impact of ATP—and beyond this, of nucleotide supplements—and also to delineate the limits of its (their) use as DNAzyme-boosting agents, we propose herein the following stepwise progression: (i) we will firstly try to unravel the confusion that swirls around the DNAzyme catalytic cycle, to gain more accurate insights into how ATP can enhance its proficiency; (ii) we will secondly presents the results of comprehensive efforts to experimentally assess the proposed hypotheses about the ATP putative roles, notably via a study extended to the other nucleotides; (iii) and finally, we will draw conclusions about the role of nucleotide supplements in the DNAzyme catalytic cycle and, on this basis, try to optimize their use to make the DNA detection the most efficient possible.
MATERIALS AND METHODS
UV-Vis experiments were carried out in a 96-well plate with a ThermoScientific Multiskan GO microplate spectrophotometer. Oligonucleotides (DNA and RNA) were purchased from Eurogentec (Belgium) in OligoGold purity grade at ∼200 nmol scale (purified by RP-HPLC), except for 46AG and 70AG (at ∼1000 nmol, purified by IEX-HPLC and PAGE, respectively). Hemin, ABTS, 3,3′,5,5′-tetramethylbenzidine (TMB), adenosine diphosphate (ADP), nucleoside triphosphate (NTP) (N = A, T, C, G) were purchased from Sigma-Aldrich, and adenosine 5'-(β,γ-imido)triphosphate (ADP-N-P) from Jena Bioscience; all the chemicals were used without further purification.
Preparation of oligonucleotides
DNA and RNA structures were prepared in a Caco.K buffer, composed of 10 mM lithium cacodylate buffer (pH 7.2) plus 10 mM KCl/90 mM LiCl by mixing the stock solutions (500 µM in strands (except for TG4T at 2 mM in strands) in deionized water (18.2 MΩ.cm)) with the ad hoc volume of lithium cacodylate buffer solution (100 mM, pH 7.2), a KCl/LiCl solution (100 mM/900 mM) and water. The final concentrations expected were 25 µM, and diluted aliquots (2.5 µM or 0.25 µM) were obtained by addition of Caco.K buffer. The actual concentrations, expressed in motif concentration, were evaluated via UV-Vis spectra analysis at 260 nm and 90°C, using the molar extinction coefficient value provided by the manufacturer. The higher-order structures of the aliquots were obtained by heating the solutions at 90°C for 5 min, cooling in ice for 6 h to favour the intramolecular folding, and then were stored at least overnight at 4°C (except for the intermolecular TG4T obtained by heating the solution at 90°C for 5 min, cooling at 65°C for 120 min, 50°C for 90 min, 35°C for 60 min, 20°C for 60 min and finally stored at 4°C).
DNAzyme experiments
All the experiments were carried out at 25°C in a 200 µL volume with Caco.KTD buffer, composed of 10 mM lithium cacodylate buffer (pH 7.2) plus 10 mM KCl/90 mM LiCl, 0.05% Triton X-100, and 0.1% (dimethylsulfoxide (DMSO)). Stock solutions were hemin (100 µM in DMSO), ABTS (100 mM in water), TMB (5 mM in DMSO), H2O2 (60 mM in water), nucleotides (NTP, ADP or ADP-N-P, 20 mM Caco.KTD) and DNA aliquots (25, 2.5 or 0.25 µM). All the experiments were compared with a control experiment (‘background’) composed of the same volume of hemin, H2O2, ABTS (or TMB) and ATP (or not, depending of the experiment), completed with Caco.KTD up to 200 µL. The DNAzyme experiments were carried out with 100 nM DNA (25 µM stock solution), 1 µM hemin, 5 mM ABTS or 0.25 mM TMB, 6 mM H2O2, with or without 10 mM NTP, ADP or ADP-N-P, except for experiments carried out with variable amounts of 22AG: 0.2 nM to 200 nM of DNA aliquot (25, 2.5 and 0.25 µM stock solutions). Long-time experiments (24 hrs) were carried out with 5 nM (2.5 µM stock solution, with 5 mM ABTS) or 100 nM DNA (25 µM stock solution, with 0.25 mM TMB), 1 µM hemin, 0.6 mM or 6 mM H2O2 (for ABTS or TMB conditions, respectively) and 10 mM ATP or CTP.
Data treatment
The characteristic UV-Vis signals for oxidized ABTS (Abs@420 nm) and oxidized TMB (Abs@652 nm and Abs@450 nm for the intermediate and the final product, respectively) were plotted as a function of time with OriginPro.8 software (OriginLab Corp. Northampton, MA, USA); raw data of experiments were used as is, or subtracted from the corresponding control experiment (‘background’) and zeroed at their initial point.
RESULTS
The DNAzyme catalytic cycle: what is known? What is assumed?
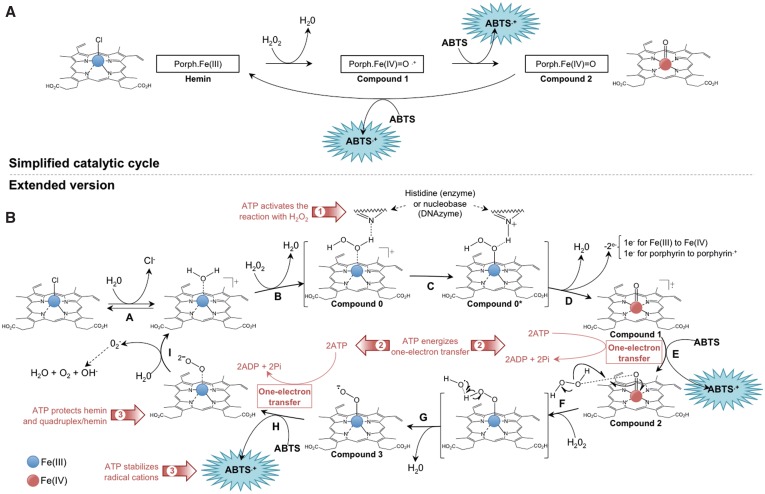
The accurate catalytic cycle of the DNAzyme process remains to be fully understood; it is usually—and conveniently—described as hinging on three main events (Figure 1A) (10,30): hemin (or porph.Fe(iii) complex) readily reacts with hydrogen peroxide (H2O2) to yield the highly reactive radical cation porph.Fe(iv)=O·+ (named Compound 1) that withdraws one electron from the substrate (herein, ABTS) to lead to the more stable ferryl-oxo porph.Fe(iv)=O intermediate (termed Compound 2). Removal of another electron from a second ABTS molecule enables the collapse of the ferryl intermediate (compound 2) back to the ferric resting state of the iron.
Figure 1.
Simplified (A) and detailed (B) mechanism of the DNAzyme catalytic cycle.
This mechanism, behind the advantage of being readily understandable, is rather ill-defined and above all not accurate enough to provide information as for the possible role(s) of ATP. On the basis of literature data (31,32), an extended version of a presumable DNAzyme catalytic cycle can be built (Figure 1B); this cycle is composed of 9 steps (from A to I) that can be summarized as follows: in the first step, the apical ligand of the iron (Cl− or H2O, equilibrium A) is displaced by binding of H2O2 (step B), whose cleavage (step C) depends upon the participation of an histidine residue (in the native enzyme) or of a nucleobase (for DNAzyme, whose origin (the surrounding loop?) remains to be firmly established) as proton scavenger. The loss of a water molecule leads to compound 1 (step D), concomitantly with the removal of two electrons, one from the iron atom, which becomes a ferryl intermediate, and the other one from the porphyrin ring that acquires a radical cation character. At this stage, a first electron is withdrawn (step E) from the substrate (ABTS) and this one-electron transfer leads both to ABTS·+ and compound 2; of note, in this process, the electron is accepted by the porphyrin that consequently loses its radical character. Upon interaction with a second molecule of H2O2, compound 2 is converted (step F) into a ferric-hydroperoxy (porph.Fe(iii)-O2H) intermediate that readily decomposes (step G) into a ferric-superoxy (porph.Fe(iii)-O2·−) species (compound 3), which may also be encountered under its isoelectronic ferrous-oxy (porph.Fe(ii)-O2) form. The delivery of a second electron (withdrawn from another molecule of ABTS) converts this intermediate to a ferric-peroxy (porph.Fe(iii)-O22–) species (step H) that releases its peroxy anion (which subsequently decomposes into oxygen, water and hydroxy anion) to restore the native iron/porphyrin complex (step I).
This mechanism is particularly focused on the iron/porphyrin complex since it is built on the basis on the deep investigations that have been carried out with hemoproteins (like peroxidase and catalase) (31,32); however, questions remain about the actual role of the quadruplex in this mechanism. It is currently suggested, but not unequivocally demonstrated, that one of the guanines constitutive of the G-quartet that interacts with the hemin flips out from the tetrad plane and supplies the fifth coordination required for the hemin to be catalytically competent (10,33). This is somewhat reminiscent of what has been observed during quadruplex-platination studies: Bombard and co-workers have indeed demonstrated that the platination of guanines involved in the external G-quartet of a quadruplex is possible after the guanine rotates out of the tetrad plane, thereby freeing its reactive N7 atom from the Hoogsteen H-bond network (34,35).
Even if already complex, this mechanism is however still simplified with regard to what might be the actual catalytic cycle, which still requires further experimental investigations to be firmly established. In light of this data, we can thus wonder about the effect(s) of ATP? There are likely to be multiple putative ATP roles involved, among which three reasonable ones can be postulated (red arrows in Figure 1B): (i) ATP acts via its nucleoside part and plays the role of the distal histidine of the native enzyme (step C), i.e. it interacts with and transiently accepts the leaving proton from H2O2 bound to porph.Fe(iii); (ii) the triphosphate tail of ATP is hydrolyzed thereby energizing the one-electron transfer steps (E and H), reminiscently of what is observed with nitrogenase (36) or reductase enzymes (37). This role actually corresponds to the classical ‘biofuel’ use of ATP; (iii) finally, ATP may also act as a stabilizing agent, both for radical cations produced by the catalysis and, as further detailed hereafter, for protecting the catalytic entity (hemin, quadruplex/hemin) from H2O2 injuries.
Of course, all these putative ATP roles are not mutually exclusive; herein, we will try to decipher the contribution of each of these roles, demonstrating that they have strong cross-talk between them and they collectively contribute to the positive effect of ATP (and NTP in general) on the DNAzyme process.
Preparatory works: defining the ATP-boosted DNAzyme standard protocols
Before tackling the many challenging aspects of our quest, we firstly need to define standard peroxidatic conditions supplemented with ATP, in an effort to make the comparison of the subsequent results the most systematic possible.
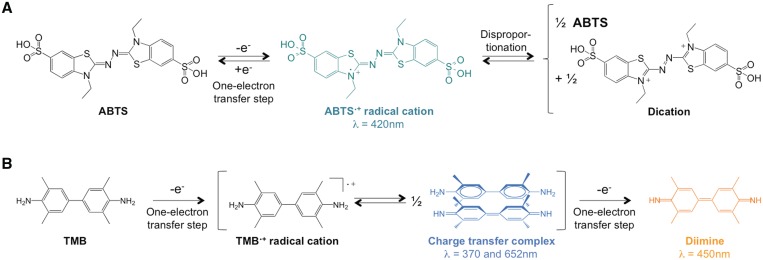
Optimizing the ATP-boosted conditions for 22AG/hemin-catalyzed oxidations: first step, the choice of the chromogenic substrates. Classically, two chromogenic substrates are employed to conveniently monitor the peroxidatic activity of the quadruplex/hemin systems, namely ABTS (38) and TMB (Figure 2) (39). The oxidation of ABTS (vide supra) leads to ABTS·+ that is a coloured product (deep green), relatively unstable (disproportionation, Figure 2A), characterized by a typical absorbance signal at 420 nm (38); the use of ABTS is generally preferred for experiments that are instrumentally monitored. The oxidation of TMB (Figure 2B) is a two-step procedure with a blue charge-transfer intermediate (with typical absorbance signals at 370 and 652 nm) and a yellow final product (which elicits an absorbance maximum at 450 nm) (39); the use of TMB is generally preferred for experiments that are monitored with the naked eye. We decided to perform quadruplex-mediated peroxidations with both probes, and to systematically vary each reactant (viz. ATP, H2O2, ABTS/TMB and hemin concentrations) in order to establish an ATP-boosted DNAzyme standard protocol (as a function of the probe). These evaluations were performed in our ‘classical’ conditions (24,28), i.e. in Caco.KTD buffer (10 mM lithium cacodylate buffer (pH 7.2) plus 10 mM KCl/90 mM LiCl, 0.05% Triton X-100 and 0.1% DMSO) with 22AG (a quadruplex-forming 22-nucleotide sequence d[AG3(T2AG3)3] that mimics the 3’-overhang of the human telomeres).
Figure 2.
Detailed oxidation pathway of ABTS (A) and TMB (B).
Optimizing the ATP-boosted conditions for 22AG/hemin-catalyzed oxidations: second step, the definition of the standard protocol for both ABTS and TMB oxidation, i.e. the study of the influence of the concentration of ATP, H2O2, hemin and chromogenic probes on the efficiency of the oxidation protocol. The catalytic capability of a system initially composed of 22AG (0.5 mM), hemin (1.0 µM) and H2O2 (0.6 mM) to oxidize ABTS (2.0 mM) is monitored via the appearance of the ABTS·+ UV-Vis signal at 420 nm (24,28). As further detailed in the Supplementary Data, several rows of experiments were conducted to optimize the ABTS oxidation protocol: experiments were performed with variable amounts of ATP (from 0 to 10 mM, see Supplementary Figure 1A), H2O2 (from 0.6 to 6.0 mM, see Supplementary Figure 1B), hemin (from 0.25 to 5.0 µM, see Supplementary Figure 1C) and ABTS (from 0.5 to 15 mM, see Supplementary Figure 1D), and it was concluded, in light of these results, that the standard DNAzyme protocol for 22AG-mediated ABTS oxidation is composed of 10 mM ATP, 6 mM H2O2, 1 µM hemin and 5 mM ABTS, in Caco.KTD buffer. Similar investigations were performed for optimizing the TMB oxidation protocol, with a catalytic system initially composed of hemin (1 µM), H2O2 (6 mM), 22AG (10 nM) and TMB (0.1 mM). The catalytic capability of the resulting system is monitored via the appearance of the UV-Vis signal of the diimine final product at 450 nm (39–41). According to a similar stepwise progression, experiments were performed with variable amounts of ATP (from 0 to 15 mM, see Supplementary Figure 2A), hemin (from 0.25 to 10.0 µM, see Supplementary Figure 2B), TMB (from 0.1 to 1.0 mM, see Supplementary Figure 2C) and H2O2 (from 1.2 to 6.0 mM, see Supplementary Figure 2D), and it was concluded, in light of these results, that the standard DNAzyme protocol for 22AG-mediated TMB oxidation is composed of 10 mM ATP, 6 mM H2O2, 1 µM hemin and 0.25 mM TMB, in Caco.KTD buffer.
Insights into the possible role(s) of ATP
As above-mentioned, ATP is suspected to enhance the proficiency of the DNAzyme catalytic cycle through three possible—and not mutually exclusive—roles (i.e. H2O2-activator, electron transfer-energizer and damage-controller), whose relevance is hereafter individually and sequentially assessed.
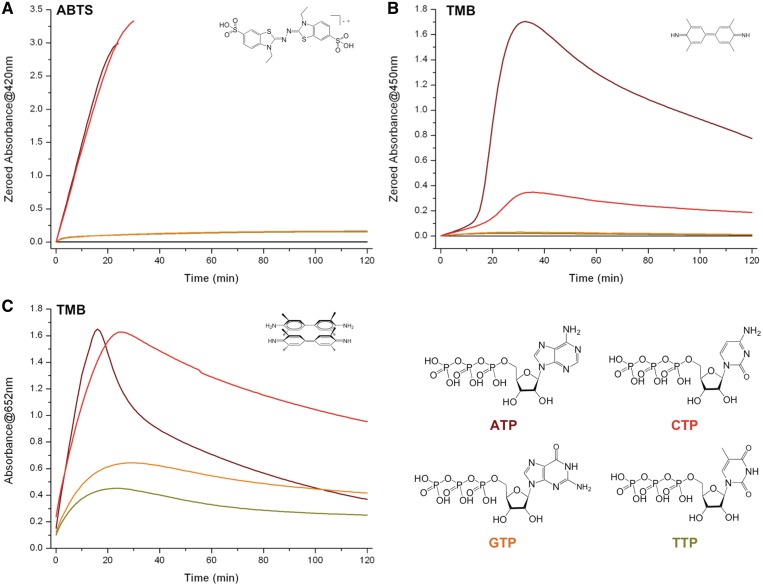
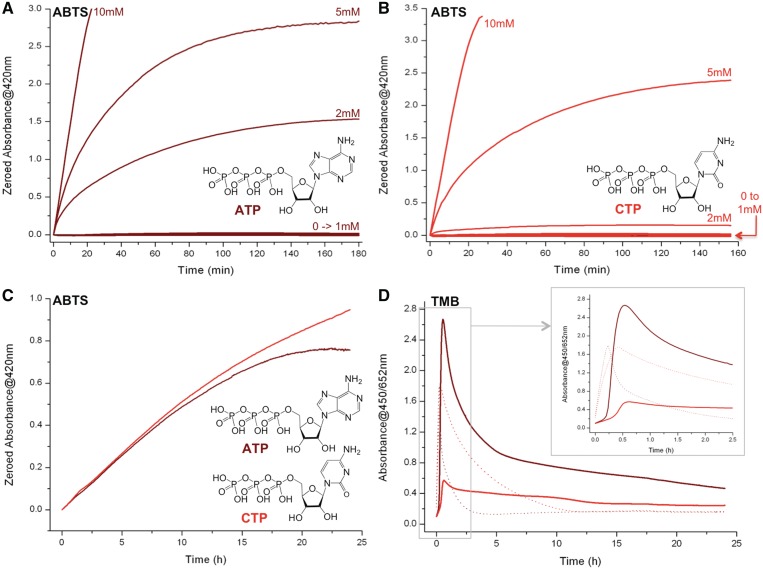
First putative ATP role: it mimics the distal histidine residue of the peroxidase enzyme, activating H2O2 for facilitating its transfer to the hemin iron atom (i.e. ATP is a H2O2-activator). In peroxidases, a distal histidine residue seems to play a crucial role in the activation of H2O2 bound to the iron atom (Step C, Figure 1B) (31,32); a pKa analysis approach of this transfer makes it rather paradoxical given that the protonation equilibria of the histidine side chain (an imidazole ring, i.e. the equilibrium His-Im.H+/His-Im) and H2O2 (i.e. the equilibrium H2O2/HO2−) have pKa values of 2.5 and 11.6, respectively, thus rendering the proton transfer very unlikely. However, the pKa of H2O2 bound to porph.Fe(iii) dramatically drops to 3.2–4, thereby making this transfer ‘less unfavorable’ (31). Herein, we postulate that externally added NTP may play an histidine-like role; a pKa analysis of the protonation pattern of the four NTP (ATP, CTP, GTP and TTP) reveals that only ATP and CTP display pKa values compatible with this transfer: the equilibria ATP.H+/ATP and CTP.H+/CTP are characterized by pKa values of 3.6 and 4.1, respectively (i.e. in the range or slightly superior to that of porph.Fe(iii)-H2O2/porph.Fe(iii)-HO2−), while that of the two others NTP makes them unable to assist this transfer (TTP/TTP−: pKa = 9.8 and GTP.H+/GTP/GTP−: pKa = 2.2 and 9.5). To demonstrate this, we performed 22AG-mediated ABTS/TMB oxidation reactions, boosted by the addition of 10 mM of ATP, CTP, GTP and TTP. As depicted in Figure 3, ATP and CTP were found equally active to oxidize ABTS (brown and red lines respectively, Figure 3A) while both GTP and TTP appeared poorly catalytically competent (orange and green lines respectively). Of note, the CTP results are surprising since in the initial report from Kong et al. (29), only ATP was found competent; as it will be further detailed hereafter, this discrepancy originates in our experimental conditions (notably in terms of CTP concentration: 10 versus 2 mM for the present and initial report, respectively), since it appears than the boosting ability of NTP is strongly dependent on its concentration (what we may name the critical nucleotide concentration, vide infra). The situation is more confused with TMB: ATP displayed obviously far better oxidative capabilities than all other NTP if only the formation of the final compound is monitored (Figure 3B); however, results dealing with the production of the intermediate (monitored at 652 nm, Figure 3C) are more in agreement with that of ABTS (i.e. ATP ≈ CTP ≫ GTP ≥ TTP). A possible explanation for such a discrepancy stands in the cationic nature of the synthesized entity: as stated above, the product of ABTS oxidation is ABTS·+ and that of the first step of TMB oxidation is TMB·+, which is in rapid equilibrium with the charge-transfer complex, while that of the second step is the neutral diimine (Figure 2). CTP appears thus as efficient as ATP to enhance the production of radical cations (Figure 3A and 3C) but, as seen in Figure 3C, the intermediate consumption is far more rapid in ATP-rich (brown line) than in CTP-rich conditions (red line). These results would indicate that CTP might display better radical cation stabilizing properties than ATP: however, given that saturated UV-Vis signals are readily reached for experiments carried out with ABTS, this hypothesis need to be further verified under different conditions (lower DNA loadings, longer reaction time, etc.); these results will be presented hereafter (see ‘Third putative ATP role’ below). Altogether these results thus tend to demonstrate that the ‘pKa hypothesis’ may be valid, i.e. that NTP supplements may play a histidine-like H2O2-activator role. To further bolster this, it was of interest to evaluate whether the direct addition of l-histidine supplement enhances the proficiency of the catalysis, reminiscently of what is observed with NTP. As depicted in Supplementary Figure 3, l-histidine indeed promotes the oxidation of ABTS (the higher the histidine concentration (10 mM), the better the catalysis), thereby supporting our hypothesis. However, the boosting capability of histidine is very low as compared with that of ATP (Supplementary Figure 3A and 3B for l-histidine and ATP respectively) and, most importantly, the ABTS·+ product is very rapidly degraded in this condition (as demonstrated by the fast decreasing of its UV-Vis signal, unlike ATP): this observation, alongside the residual boosting activity of both GTP and TTP (orange and green lines, Figure 3), implies that other mechanisms may be implicated: as discussed below, this might be related to the ability of NTP to power electron-transfer processes (see ‘Second putative ATP role’ below) and/or to protect the various reaction partners (notably the oxidation products, see ‘Third putative ATP role’ below).
Figure 3.
Compared DNAzyme activity of experiments carried out with 22AG (100 nM), hemin (1 µM), H2O2 (6 mM), ABTS (A, 5 mM) or TMB (B/C, 0.25 mM), unboosted (black lines, A/B) or boosted by the addition of 10 mM ATP (brown line), CTP (red line), GTP (orange line) and TTP (green line). The oxidation of ABTS is monitored via the appearance of the final product typical UV-Vis signal (420 nm, A), the oxidation of TMB via that of both the final product (450 nm, B) and the charge-transfer intermediate (652 nm, C).
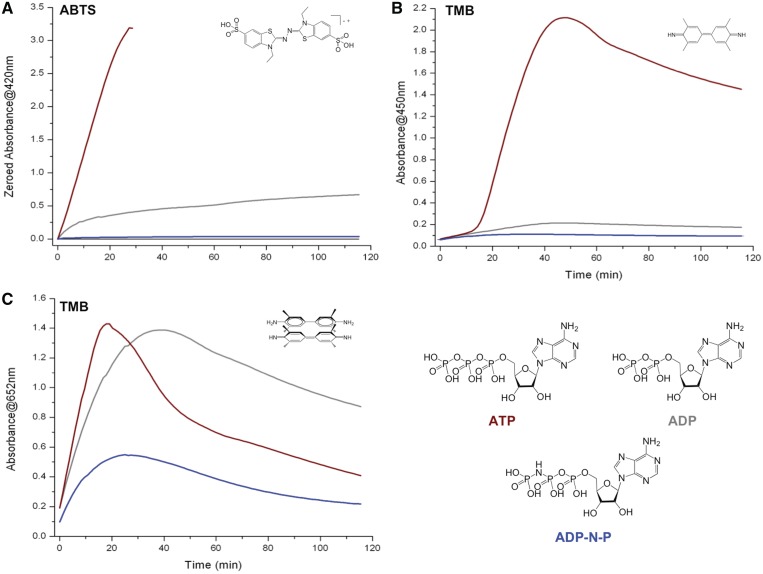
Second putative ATP role: its hydrolysis energizes the two one-electron transfer steps of the proposed DNAzyme catalytic cycle, thus fuelling the two steps in which the substrate is oxidized (i.e. ATP is a electron-transfer energizer). In the mechanism described in Figure 1B, steps E and H (one-electron transfer steps) are of notable importance: indeed, it has been recently shown that ATP hydrolysis may be intimately linked to electron transfer (ET) reaction (36,37). Again, the mechanisms that couple ATP hydrolysis to ET, which represents a quite unique energy transduction process, is not known in detail; Kurnikov et al. elegantly summarized it writing that ‘one of the mysteries (…) is how the energy of the phosphate bond hydrolysis is coupled to the electron-transfer (ET) reaction’ (36). This ATP-driven ET provides a novel method to produce organic radicals in enzyme-like catalysis, on the basis of the concomitant hydrolysis of two ATP for the transfer of a single electron. It is thus not unreasonable to wonder whether such ATP-assisted ET takes place herein; a first element of response is given by the series of experiments displayed in Figure 3: indeed, it is known that the NTP energy is related to the exergonic rupture of a phosphodiester bond, according to the following equilibrium NTP + H2O -> NDP + Pi (with Pi for inorganic phosphate), implying an identical energy release (Gibbs free energy ΔG°’ = −7.3 kcal/mol) for all NTP (42). Although ATP is the ubiquitous energy currency of the cell (43–46), other NTP are used in more specialized biogenesis pathways (GTP for protein synthesis (47), CTP for lipid synthesis, (48) etc.). What makes ATP so unique to power cell reactions is mainly the result of an evolutionary pressure that makes ATP bioavailability greater than all other NTP (49). Herein, the fact that all NTP enhance the catalytic oxidation of both ABTS and TMB (coloured lines versus black line, Figure 3) indicates that they energize to some extent the catalysis; however, the moderate activity of both GTP and TTP (orange and green lines, Figure 3) implies that the energy release linked to NTP hydrolysis plays a limited role to this enhancement. To examine this in depth, we decided to evaluate the influence of ADP-N-P (for adenosine 5′-(β,γ-imido)triphosphate, also known as AMP-PNP), which is a molecule structurally very close to ATP in which the terminal P-O-P bond is replaced by a more robust P-NH-P bond: ADP-N-P is thus considered—and widely used—as a non-hydrolysable ATP analogue (50). As depicted in Figure 4, the boosting capability of ATP (brown lines) is greatly diminished if ADP-N-P is used instead (blue lines), thereby unambiguously highlighting that the energetic contribution is an important feature of the ATP-boosting properties. However, the very low boosting capability of ADP-N-P (notably for ABTS oxidation, Figure 4A) is somewhat unexpected: this thus implies that the various parameters responsible for the boosting properties of ATP (H2O2-activation, electron-transfer powering) are complicatedly interlocked. This is further confirmed by the evaluation of the boosting ability of ADP: ADP is found less active than ATP for boosting the oxidation of ABTS (reminiscently of what has been reported by Kong et al. (29), grey line, Figure 4A), and far less active for the oxidation of TMB (especially for the production of the final product (Figure 4B), while that of the intermediate reaches elevated level (Figure 4C), rather like CTP, vide supra). These results are in agreement with that of ADP-N-P: indeed, ATP, ADP and ADP-N-P display similar pKa values; the discrepancies of results obtained with these three nucleotides thus enlightens that the H2O2-activation role is limitedly influential in the overall boosting capabilities of nucleotides. However, ADP is also a high-energy molecule that produces energy upon the exergonic hydrolysis ADP + H2O -> AMP + Pi (ΔG°’ = −7.3 kcal/mol) (42); thus, ADP is expected to energize the catalysis similarly to ATP. Given that the hydrolysable ADP elicits less boosting capabilities than ATP but more than non-hydrolysable ADP-N-P, the obtained results enlighten that the electron transfer-powering role influences the overall boosting capabilities of nucleotides, but only to a certain extent. Collectively, these results again underline that the various parameters responsible for the boosting properties of nucleotides are strongly tangled; the present study is thus useful in that it helps identifying these parameters, but additional investigations are needed to unravel the subtle relationships between them.
Figure 4.
Compared DNAzyme activity of experiments carried out with 22AG (100 nM), hemin (1 µM), H2O2 (6 mM), ABTS (A, 5 mM) or TMB (B/C, 0.25 mM) in the presence of 10 mM ATP (brown lines), ADP (grey lines) or ADP-N-P (blue lines). The oxidation of ABTS is monitored via the appearance of the final product typical UV-Vis signal (420 nm, A), the oxidation of TMB via that of both the final product (450 nm, B) and the charge-transfer intermediate (652 nm, C).
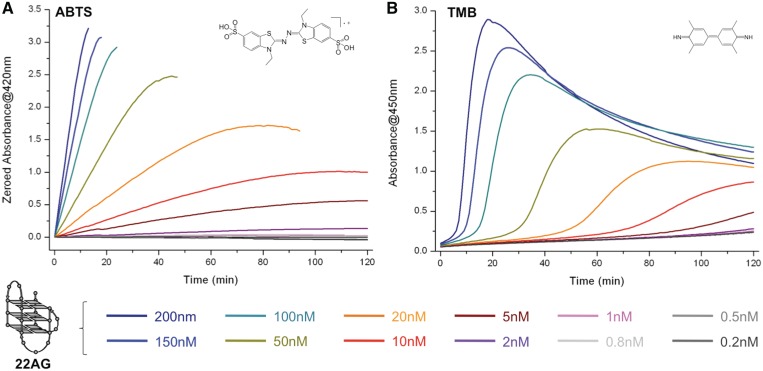
Third putative ATP role: it protects the various reaction partners, i.e. the hemin, the quadruplex/hemin adduct and the radical cations produced by the oxidation (ABTS·+, TMB·+), thereby influencing not only the catalysis but also the reaction products (i.e. ATP is a damage-controller). In their initial report, D.-M. Kong et al. indicated that ATP was the only NTP that elicits boosting ability; as indicated above, and somewhat contradictorily, we found that both ATP and CTP display boosting competencies. It was thus of interest, before going any further, to identify the origins of this discrepancy. To this end, we studied the boosting capability of both ATP and CTP as a function of their concentration (from 0.1 to 10 mM); as depicted in Figure 5A (for ATP) and 5B (for CTP), we interestingly found that this capability is not linearly related to the NTP concentration but appears triggered from a threshold concentration (that we thus may term ‘critical nucleotide concentration’): a satisfying activity is obtained from 2 mM of ATP and 5 mM of CTP, and this explains why CTP activity was not detected in the initial report (since used only up to 2 mM). The molecular basis of this critical nucleotide concentration is at present unknown (solubility? supramolecular self-assembly?); we have thus invested additional efforts to try to decipher this: to this end, and inspired by the reported works from both Kong (29,51) and Shangguan (30), we decided to gain insights into the protecting abilities of both ATP and CTP, i.e. the capability to protect firstly hemin and quadruplex/hemin adducts against H2O2-degradation, and secondly the radical cations produced by the catalysis (i.e. ABTS·+ and TMB·+) against disproportionation. For this purpose, a first series of UV-Vis titrations were performed with hemin alone, hemin in presence of H2O2 and 22AG/hemin adduct in presence of H2O2. Several interesting conclusions can be drawn from the results displayed in the Supplementary Data: (i) while hemin is mostly found disaggregated in Caco.KTD (λmax = 392 nm, Supplementary Figure 4A), both ATP and CTP favour its aggregated state (λmax = 354 nm, Supplementary Figure 4B and 4C); (ii) upon addition of H2O2, hemin is rapidly damaged (strong hypochromic shift, Supplementary Figure 4D) while its degradation is greatly slowed by both NTP (Supplementary Figure 4E and 4F), reminiscently of what has been reported by Kong and Shangguan (29,30,51); (iii) finally, the presence of 22AG does not modify the degradation kinetics of hemin alone (Supplementary Figure 4G), while it transiently disaggregates hemin in presence of ATP/CTP (before addition of H2O2, Supplementary Figure 4H and 3I) and does not modify the degradation kinetic of hemin (after addition of H2O2) afterward. To explain these results, our hypothesis is that hemin exists in solution both as monomeric (catalytically competent but H2O2-sensitive) and aggregated states (catalytically incompetent but H2O2-insensitive): without ATP/CTP, hemin is mainly found as monomer in Caco.KTD solution, which is rapidly damaged by the excess of H2O2, thus explaining the rapid catalysis breakdown observed in absence of NTP; on the contrary, the presence of ATP/CTP favours the hemin aggregated state, and the catalytically competent monomeric hemin is slowly released in presence of DNA to enter the catalysis, thereby cleverly counteracting the hemin inactivation process. A second series of experiments was carried out to investigate the cation-stabilizing properties of ATP, also evoked by Kong and Shangguan; as mentioned above, we believe that both ATP and CTP share this property. To further investigate this, ATP/CTP-boosted DNAzyme experiments were performed, under non-saturating conditions (i.e. lower DNA (5 versus 100 nM) and H2O2 (0.6 versus 6 mM) concentrations, notably for experiments carried out with ABTS) and longer reaction times (24 versus 2 h, to explore the robustness of the above property under harsh conditions). As depicted in Figure 5C and D, NTP-boosted experiments can be conveniently performed over 24 hrs, particularly with ABTS (Figure 5C): indeed, the DNAzyme process catalyzed with DNA concentration as low as 5 nM keeps on working over 20 hrs in presence of ATP, and even more (>24 h) with CTP; this indicates that there is no ABTS·+ degradations in these conditions, and consequently that NTP display outstanding radical cation-stabilizing properties. As above, the situation is more confused with TMB, since NTP seem to impact the two steps of the TMB oxidation differently: while the intermediate is produced to similar extents (dotted lines, Figure 5D and inset), its consumption is far slower in presence of CTP (∼10 h) than with ATP (∼2.5 h). Again, this may indicate that CTP displays better TMB·+-stabilization property than ATP; this certainly contributes but however not completely account for the differences of production of the final product (solid lines, Figure 5D), thereby highlighting that NTP influence the two steps differently, in a manner that still remains to be fully understood. Altogether, these results unambiguously demonstrate that NTP (chiefly ATP and CTP) indeed display enticing damage-controlling properties that collectively enhance the overall efficiency of the catalysis, stabilizing not only the catalytic system but also the reaction products.
Figure 5.
ATP- versus CTP-boosting capabilities: Compared DNAzyme activity of experiments carried out with 22AG (200 nM), hemin (1 µM), ABTS (5 mM), H2O2 (6 mM) and from 0.1 to 10 mM ATP (A) or CTP (B). Compared DNAzyme activity of experiments carried out with 22AG (5 nM (C) or 100 nM (D)), hemin (1 µM), ABTS (A, 5 mM) or TMB (B, 0.25 mM), H2O2 (0.6 and 6 mM with ABTS and TMB, respectively) and 10 mM ATP (brown lines) or CTP (red lines). The oxidation of ABTS (A–C) is monitored via the appearance of the final product typical UV-Vis signal (420 nm), the oxidation of TMB (D) via that of both the intermediate (652 nm, dotted lines) and the final product (450 nm, plain lines).
Implications in terms of DNA detection
Both ATP and CTP have thus been identified as valuable nucleotide supplements to boost the quadruplex-mediated DNAzyme catalysis; however, the greater availability of ATP (it can be bought on larger scale, at higher purity grade, and is ∼50-fold less expensive than CTP) makes it the NTP of choice for performing further investigations.
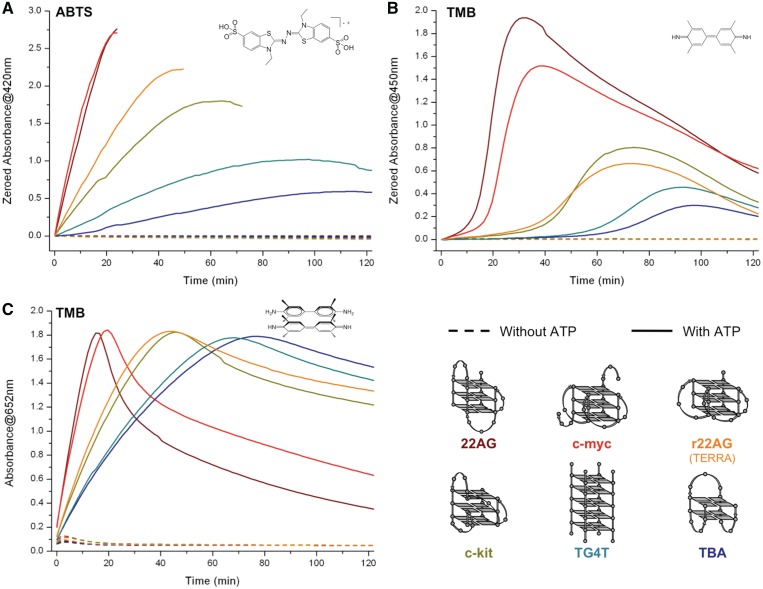
Extending the use of ATP as boosting agent for DNAzyme experiments catalyzed by hemin interacting with other quadruplex-DNAs: investigation of the quadruplex-dependency of the ATP-boosting capability. The present study enlightens that NTP mainly act as external factors, i.e. in a fashion that should be independent of the quadruplex architecture they are used with. To further demonstrate this, we extended the study of the ATP-boosting capability to other quadruplex architectures of clearly distinct nature (14,52–54): two other biologically relevant sequences that form intramolecular quadruplexes within the c-myc (d[GAG3TG4AG3TG4A2G]) and c-kit (d[CG3CG3CGCGAG3AG4]) promoter regions (55); a synthetic quadruplex-forming oligonucleotide that displays thrombin-binding aptamer properties (TBA, d[G2T2G2TGTG2T2G2]) (56); an intermolecular (tetramolecular) quadruplex TG4T ((d[TG4T])4) (57); and the RNA equivalent of 22AG (hereafter named r22AG, r[AG3(U2AG3)3] (58), also known as TERRA (for telomeric RNA repeat)) (59,60). As depicted in Figure 6, ATP has a strong positive impact in all instances (dashed line (for experiments without ATP) versus plain lines (for experiments with ATP)), for both chromogenic probes, thereby supporting the hypothesis according to which it mainly interacts as an external factor. The ATP effect is herein stronger for 22AG, c-myc, c-kit and r22AG than for TG4T and TBA: this observation could enable us to conclude that ATP helps to a greater extent the catalysis mediated by biologically relevant structures rather than by synthetic ones; however, to be firmly established, this conclusion has to be supported by results gained from investigations performed among a broader panel of quadruplex architectures. However, these results collectively support the hypothesis according to which NTP enhance the rate of DNAzyme catalysis via an external intervention, in a manner that is quadruplex-independent.
Figure 6.
Compared DNAzyme activity of experiments carried out with DNA (100 nM, i.e. 22AG (brown line), c-myc (red line), c-kit (dark yellow line), TG4T (light green line), TBA (blue line) and r22AG (orange line)), hemin (1 µM), H2O2 (6 mM), ABTS (A, 5 mM) or TMB (B/C, 0.25 mM), without (dashed lines) or with 10 mM ATP (plain lines). The oxidation of ABTS is monitored via the appearance of the final product typical UV-Vis signal (420 nm, A), the oxidation of TMB via that of both the final product (450 nm, B) and the charge-transfer intermediate (652 nm, C).
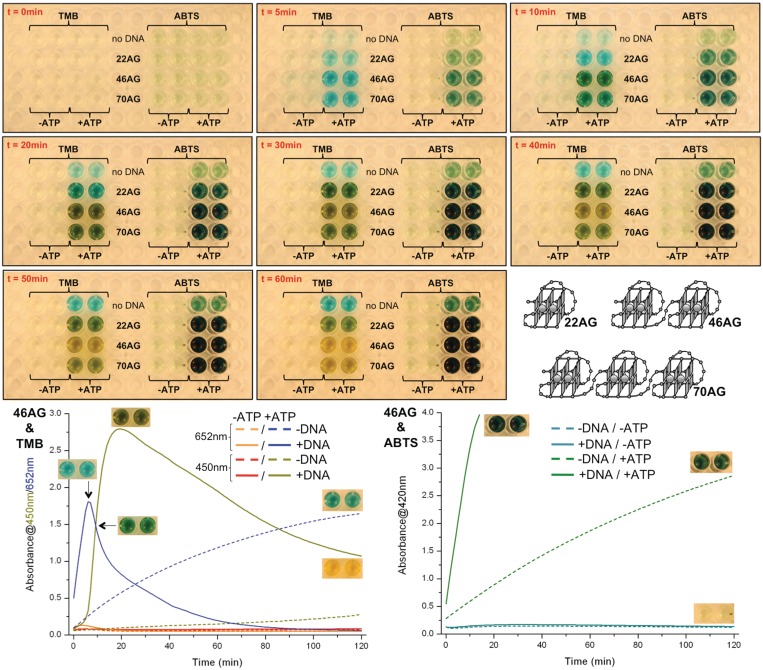
Extending the use of ATP as boosting agent for DNAzyme experiments catalyzed by hemin interacting with multimers of telomeric quadruplex-DNAs: investigation of the ATP influence on the detection of the lengthy telomeric stretches, for a possible application to improve the DNAzyme-based telomerase activity detection assay. To further benefit from the established ATP-based high-performance catalytic conditions, we decided to evaluate the peroxidatic capabilites of multimeric quadruplexes (namely 22AG d[AG3(T2AG3)3], 46AG d[AG3(T2AG3)7] and 70AG d[AG3(T2AG3)11]) (24), monitoring reactions both with a microplate reader and the naked eye (Figure 7). These human telomere mimicking fragments of various lengths (that thus fold into multimeric quadruplexes) were initially selected since they play a pivotal role in an in vitro assay that aims at assessing the telomerase activity via DNAzyme processes (24). These experiments were conducted with both ABTS and TMB, with or without ATP. Results are displayed in Figure 7 (the upper panel is composed of several pictures taken during the first hour of reaction—of note, a ∼3-min movie corresponding to the complete 2-hr reaction is available as Supplementary Data); several conclusions can be drawn from this series of experiments:
First and foremost, the obtained results firmly emphasize the outstanding positive impact of ATP since the wells corresponding to the unboosted experiments (labeled ‘-ATP’ for both ABTS and TMB) do not elicit any colour, even after 1 hr of reaction, implying an undetectable level of catalysis in absence of ATP in the conditions of the assay. In sharp contrast, the wells corresponding to the boosted experiments (labeled ‘+ATP’) all display colours from which a lot of information can be withdrawn (see the points below).
Second, these results clearly demonstrate the superiority of TMB over ABTS for experiments monitored with the naked eye since TMB offers a wide colour palette that corresponds to the various stages of the catalysis (detailed in the left lower panel, Figure 7): blue for the charge transfer intermediate (which appears within 5 min for the three DNA); green for a mixture of the blue intermediate and the yellow final product (particularly visible with 46AG at 10 min); and yellow for the final diimine (obtained with 46AG and 70AG after 60 min of reaction). In contrast, ABTS wells elicit only various tint levels for green colour. Moreover, the difference between DNA-catalyzed reactions and background controls (labeled ‘no DNA’) is also easier with TMB than with ABTS since, once more, it relies (e.g. after 60 min of reaction) on a difference of colours with TMB (blue versus yellow) and only of green intensity with ABTS (light versus dark green). Finally, the TMB-related colours enable also to gain deeper insights into the catalytic capabilities of the various studied DNA: the yellow colour of the final product is more rapidly obtained with 46AG than with 70AG, while 22AG wells stay green even after 60 min of reaction. Thus, TMB enables an easier ranking of catalytic proficiencies (herein 46AG > 70AG >> 22AG, in agreement with our previously reported results) (24) than ABTS.
Third, it is however important not to underestimate the interest of ABTS: indeed,—as it will be further detailed hereafter—ABTS provides generally a more rapid and intense response, with a higher signal-to-noise ratio. The lack of colour difference between the three DNA after 20 min of reaction originates in the fact that saturated levels of ABTS·+ are already reached, even at 100 nM DNA scale (as seen in the right lower panel, Figure 7). ABTS is also more practically convenient than TMB (i.e. its oxidation leads to a unique product; its use is far easier to optimize than TMB) and, last but not least, it provides a long-lasting response (>20 hrs), as unambiguously demonstrated by the experiments whose results are displayed in Figure 5.
Figure 7.
Compared DNAzyme activity of experiments carried out with multimeric quadruplexes (100 nM, i.e. 22AG, 46AG and 70AG), hemin (1 µM), H2O2 (6 mM), ABTS (5 mM) or TMB (0.25 mM), without (‘−ATP’) or with 10 mM ATP (‘+ATP’). Upper panel: selected pictures taken during the experiment (i.e. after 0, 5, 10, 20, 30, 40, 50 and 60 min of reaction). Lower panel: DNAzyme activity of experiments carried out without (dashed lines) or with 46AG (100 nM, plain lines), hemin (1 µM), H2O2 (6 mM), TMB (left, 0.25 mM) or ABTS (right, 5 mM), without or with 10 mM ATP. The oxidation of TMB is monitored via the appearance of both the final product (450 nm, dark yellow (with ATP) and red (without ATP) lines) and the charge-transfer intermediate (652 nm, blue (with ATP) and orange (without ATP) lines), the oxidation of ABTS via that of the final product typical UV-Vis signal (420 nm, light (without ATP) and dark green (with ATP) lines).
Benefitting from the optimized ATP-boosted 22AG/hemin-catalyzed DNAzyme protocol: lowering the limits for DNA detection to nanomolar ranges. We finally decided to benefit from these exquisitely efficient ATP-boosted conditions to determine the lower limits (in terms of concentration) at which DNA can be reliably detected. These investigations were performed with the moderately active 22AG, within a range of nanomolar (200 nM) to picomolar (200 pM) concentrations. Results displayed in Figure 8 clearly emphasize the high performances of the ATP-based DNAzyme catalyses, with a reliable detection in the low nanomolar range with both probes. A closer look to this series of results indicates the overall better performances of ABTS over TMB, in terms of both efficiency (ABTS enables a reliable detection of only 2 nM DNA (purple lines) while the threshold reached by TMB is at 5 nM (brown lines)) and—above all—rapidity of detection (for a given DNA concentration (e.g. 20 nM, orange lines), a reliable response is obtained within 5 min with ABTS while it requires ∼50 min with TMB).
Figure 8.
Compared DNAzyme activity of experiments carried out with various amount of 22AG (from 200 nM to 0.2 nM), hemin (1 µM), H2O2 (6 mM), ABTS (A, 5 mM) or TMB (B, 0.25 mM) and 10 mM ATP (plain lines). The oxidation of ABTS is monitored via the appearance of the typical final product UV-Vis signal (420 nm, A), the oxidation of TMB via that of the final diimine product (450 nm, B).
Altogether, these results support the initial postulate (vide supra) according to which ABTS and TMB should be respectively preferred for experiments that are instrumentally monitored (for the former) and monitored with the naked eye (for the later), both of them being anyway high-performance quadruplex-mediated DNAzyme chromogenic substrates.
DISCUSSION
Since its discovery in 1994 (8), the catalytic capability of DNA has continued to garner much interest in the scientific community because of the virtually unlimited applications in developing new molecular devices (5–7,10,16–18). However, the search for innovative applications has somewhat taken up too much scientist attention and has kept the limelight away from that of ways to improve its overall efficiency. Credence has been lent to this strategy by the recent works of D.-M. Kong and co-workers in which they demonstrate that the quadruplex-based DNAzyme proficiency can be enhanced by ATP supplements (29). Herein, we have made a further leap along this path, investigating in detail the influence of all the parameters that govern the catalytic efficiency (i.e. the concentrations of hemin, H2O2, ATP and substrate), extending the study to other nucleotides (ATP, CTP, GTP, TTP and ADP-N-P), various quadruplex forming oligonucleotides (22AG, c-myc, c-kit, TBA, TG4T, r22AG, 46AG and 70AG) and chromogenic probes (ABTS and TMB). The obtained results not only serve to acquaint us with the possible NTP roles as boosting agents (i.e. H2O2-activator, electron-transfer energizer and radical damage-controller), but also enable us to gain insights into what might be the actual DNAzyme catalytic cycle (which still remains a question open to debate). They also cast a new light on the importance of the choice of the substrates: both ABTS and TMB are high-performance probes, having however their own trade-offs (ABTS oxidation is faster and more sensitive than that of TMB, but TMB provides a more insightful colour response). The real-time 96-well plate experiment displayed in Figure 7 (and the corresponding movie as Supplementary Data) itself epitomizes the power of NTP (chiefly ATP) as boosting agents: even with a DNA concentration in the low nanomolar scale (100 nM), the wells corresponding to NTP-boosted reactions (‘+ATP’) all elicit a strong colour response, whatever the nature of the quadruplex catalyst, while the ones corresponding to the unboosted catalyses (‘−ATP’) stay colourless.
The development of DNAzyme conditions with increased effectiveness might provide a novel avenue to innovative quadruplex-based nanodevices; the strong emphasis on the positive impact of NTP reported herein clearly highlights the enticing advantages of this approach: since the DNAzyme catalysis proficiency is more than 100-fold enhanced by NTP supplements whatever the nature of both the quadruplex and the probes, there is little doubt that the present strategy may be applied to virtually all desired applications. Thus, we believe that the present study is an important step for the DNAzyme research field since it offers a scientific basis from which new experiments would not be attempted anymore without NTP supplements.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–4 and Supplementary Movie.
FUNDING
The Centre National de la Recherche Scientifique (CNRS), Université de Bourgogne (uB), and Conseil Régional de Bourgogne (CRB) via 3MIM project; the Agence Nationale de la Recherche (ANR) via ANR-10-JCJC-0709; and the CRB via PARI-SSTIC 6 (92-01). Funding for open access charge: The CNRS, uB, and CRB via 3MIM for funding.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Richard Decreau for fruitful discussions about the iron porphyrin-based oxidation mechanism and Francois Cuenot for his invaluable help with funding.
REFERENCES
- 1.McCarty M. A retrospective look: how we identified the pnemnococcal transforming substance as DNA. J. Exp. Med. 1994;179:385–394. doi: 10.1084/jem.179.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watson JD, Crick FHC. Molecular structure of nucleic acids. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 3.Wilkins MHF, Stokes AR, Wilson HR. Molecular structure of deoxypentose nucleic acids. Nature. 1953;171:738–740. doi: 10.1038/171738a0. [DOI] [PubMed] [Google Scholar]
- 4.Franklin RE, Gosling RG. Molecular configuration in sodium thymonucleate. Nature. 1953;171:740–741. doi: 10.1038/171740a0. [DOI] [PubMed] [Google Scholar]
- 5.Seeman NC. An overview of structural DNA nanotechnology. Mol. Biotechnol. 2007;37:246–257. doi: 10.1007/s12033-007-0059-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silverman SK. DNA as a versatile chemical component for catalysis, encoding, and stereocontrol. Angew. Chem. Int. Ed. 2010;49:7180–7201. doi: 10.1002/anie.200906345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krishnan Y, Simmel FC. Nucleic acid based molecular devices. Angew. Chem. Int. Ed. 2011;50:3124–3156. doi: 10.1002/anie.200907223. [DOI] [PubMed] [Google Scholar]
- 8.Breaker RR, Joyce GF. A DNA enzyme that cleaves RNA. Chem. Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Sen D. A catalytic DNA for porphyrin metallation. Nat. Struct. Biol. 1996;3:743–747. doi: 10.1038/nsb0996-743. [DOI] [PubMed] [Google Scholar]
- 10.Sen D, Poon LCH. RNA and DNA complexes with hemin [Fe(iii) heme] are efficient peroxidases and peroxygenases: how do they do it and what does it mean? Crit. Rev. Biochem. Mol. Biol. 2011;46:478–492. doi: 10.3109/10409238.2011.618220. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, Sen D. The modus operandi of a DNA enzyme: enhancement of substrate basicity. Chem. Biol. 1998;5:1–12. doi: 10.1016/s1074-5521(98)90082-5. [DOI] [PubMed] [Google Scholar]
- 12.Sen D, Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 13.Sen D, Geyer CR. DNA enzymes. Curr. Opin. Chem. Biol. 1998;2:680–687. doi: 10.1016/s1367-5931(98)80103-8. [DOI] [PubMed] [Google Scholar]
- 14.Collie GW, Parkinson GN. The application of DNA and RNA G-quadruplexes to therapeutic medicines. Chem. Soc. Rev. 2011;40:5867–5892. doi: 10.1039/c1cs15067g. [DOI] [PubMed] [Google Scholar]
- 15.Travascio P, Li Y, Sen D. DNA-enhanced peroxidase activity of a DNA aptamer-hemin complex. Chem. Biol. 1998;5:505–517. doi: 10.1016/s1074-5521(98)90006-0. [DOI] [PubMed] [Google Scholar]
- 16.Willner I, Shlyahocsky B, Zayats M, Willner B. DNAzymes for sensing, nanobiotechnology and logic gate applications. Chem. Soc. Rev. 2008;37:1077–1280. doi: 10.1039/b718428j. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Cao Z, Lu Y. Functional nucleic acid sensors. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kosman J, Juskowiak B. Peroxidase-mimicking DNAzymes for biosensing applications: a review. Anal. Chim. Acta. 2011;707:7–17. doi: 10.1016/j.aca.2011.08.050. [DOI] [PubMed] [Google Scholar]
- 19.Freeman R, Sharon E, Teller C, Henning A, Tzfati Y, Willner I. DNAzyme-like activity of hemin–telomeric G-quadruplexes for the optical analysis of telomerase and its inhibitors. Chembiochem. 2010;11:2362–2367. doi: 10.1002/cbic.201000512. [DOI] [PubMed] [Google Scholar]
- 20.Blackburn EH, Greider CW, Szostak JW. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nature Med. 2006;12:1133–1138. doi: 10.1038/nm1006-1133. [DOI] [PubMed] [Google Scholar]
- 21.Szostak JW. DNA ends: just the beginning. Angew. Chem. Int. Ed. 2010;49:7386–7404. doi: 10.1002/anie.201000635. [DOI] [PubMed] [Google Scholar]
- 22.Blackburn EH. Telomeres and telomerase: the means to the end. Angew. Chem. Int. Ed. 2010;49:7405–7421. doi: 10.1002/anie.201002387. [DOI] [PubMed] [Google Scholar]
- 23.Greider CW. Telomerase discovery: the excitement of putting together pieces of the puzzle. Angew. Chem. Int. Ed. 2010;49:7422–7439. doi: 10.1002/anie.201002408. [DOI] [PubMed] [Google Scholar]
- 24.Stefan L, Denat F, Monchaud D. Deciphering the DNAzyme activity of multimeric quadruplexes: insights into their actual role in the telomerase activity evaluation assay. J. Am. Chem. Soc. 2011;133:20405–20415. doi: 10.1021/ja208145d. [DOI] [PubMed] [Google Scholar]
- 25.Nikan M, Sherman JC. Template-Assembled Synthetic G-Quartets (TASQs) Angew. Chem. Int. Ed. 2008;47:4900–4902. doi: 10.1002/anie.200704199. [DOI] [PubMed] [Google Scholar]
- 26.Murat P, Bonnet R, Van der Heyden A, Spinelli N, Labbé P, Monchaud D, Teulade-Fichou M-P, Dumy P, Defrancq E. Template-Assembled Synthetic G-Quadruplex (TASQ): a useful system for investigating the interactions of ligands with constrained quadruplex topologies. Chem. Eur. J. 2010;16:6106–6114. doi: 10.1002/chem.200903456. [DOI] [PubMed] [Google Scholar]
- 27.Stefan L, Guédin A, Amrane S, Smith N, Denat F, Mergny J-L, Monchaud D. DOTASQ as a prototype of nature-inspired G-quadruplex ligand. Chem. Commun. 2011;47:4992–4994. doi: 10.1039/c0cc04960c. [DOI] [PubMed] [Google Scholar]
- 28.Stefan L, Xu H-J, Gros CP, Denat F, Monchaud D. Harnessing nature’s insights: synthetic small molecules with peroxidase-mimicking DNAzyme properties. Chem. Eur. J. 2011;17:10857–10862. doi: 10.1002/chem.201101337. [DOI] [PubMed] [Google Scholar]
- 29.Kong D-M, Xu J, Shen H-X. Positive effects of ATP on G-quadruplex-hemin DNAzyme-mediated reactions. Anal. Chem. 2010;82:6148–6153. doi: 10.1021/ac100940v. [DOI] [PubMed] [Google Scholar]
- 30.Yang X, Fang C, Mei H, Chang T, Cao Z, Shangguan D. Characterization of G-quadruplex/hemin peroxidase: substrate specificity and inactivation kinetics. Chem. Eur. J. 2011;17:14475–14484. doi: 10.1002/chem.201101941. [DOI] [PubMed] [Google Scholar]
- 31.Jones P, Dunford HB. The mechanism of Compound I formation revisited. J. Inorg. Biochem. 2005;99:2292–2298. doi: 10.1016/j.jinorgbio.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 32.Jantschko W, Furtmüller PG, Zederbauer M, Lanz M, Jakopitsch C, Obinger C. Direct conversion of ferrous myeloperoxidase to compound II by hydrogen peroxide: an anaerobic stopped-flow study. Biochem. Biophys. Res. Commun. 2003;312:292–298. doi: 10.1016/j.bbrc.2003.10.117. [DOI] [PubMed] [Google Scholar]
- 33.Travascio P, Witting PK, Mauk AG, Sen D. The peroxidase activity of a hemin-DNA oligonucleotide complex: free radical damage to specific guanine bases of the DNA. J. Am. Chem. Soc. 2001;123:1337–1348. doi: 10.1021/ja0023534. [DOI] [PubMed] [Google Scholar]
- 34.Ourliac-Garnier I, Elizondo-Riojas M-A, Redon S, Farrell NP, Bombard S. Cross-links of quadruplex structures from human telomeric DNA by dinuclear platinum complexes show the flexibility of both structures. Biochemistry. 2005;44:10620–10634. doi: 10.1021/bi050144w. [DOI] [PubMed] [Google Scholar]
- 35.Bertrand H, Bombard S, Monchaud D, Teulade-Fichou M-P. A platinum–quinacridine hybrid as a G-quadruplex ligand. J. Biol. Inorg. Chem. 2007;12:1003–1014. doi: 10.1007/s00775-007-0273-3. [DOI] [PubMed] [Google Scholar]
- 36.Kurnikov IV, Charnley AK, Beratan DN. From ATP to electron transfer: electrostatics and free-energy transduction in nitrogenase. J. Phys. Chem. B. 2001;105:5359–5367. [Google Scholar]
- 37.Buckel W, Hetzel M, Kim J. ATP-driven electron transfer in enzymatic radical reactions. Curr. Opin. Chem. Biol. 2004; 8:462–467. doi: 10.1016/j.cbpa.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 38.Childs RE, Bardsley WG. The steady-state kinetics of peroxidase with 2,2′-azino-di-(3-ethylbenzthiazoline-6-sulphonic acid) as chromogen. Biochem. J. 1975;145:93–103. doi: 10.1042/bj1450093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Josephy PD, Eling T, Mason RP. The horseradish peroxidase-catalyzed oxidation of 3,5,3′,5′-tetramethylbenzidine. J. Biol. Chem. 1982;257:3669–3675. [PubMed] [Google Scholar]
- 40.Marquez LA, Dunford HB. Mechanism of the oxidation of 3,5,3′,5′-tetramethylbenzidine by myeloperoxidase determined by transient- and steady-state kinetics. Biochemistry. 1997;36:9349–9355. doi: 10.1021/bi970595j. [DOI] [PubMed] [Google Scholar]
- 41.Li B, Du Y, Li T, Dong S. Investigation of 3,3′,5,5′-tetramethylbenzidine as colorimetric substrate for a peroxidatic DNAzyme. Anal. Chim. Acta. 2009;651:234–240. doi: 10.1016/j.aca.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 42.Stryer L. Biochemistry. 4th edn. New York: W. H. Freeman & Co.; 1995. [Google Scholar]
- 43.Lipmann F. Metabolic generation and utilization of phosphate bond energy. Adv. Enzymol. Rel. Sub. 1941;1:99–162. [Google Scholar]
- 44.Boyer PD. Energy, life and ATP. Angew. Chem. Int. Ed. 1998;37:2296–2307. doi: 10.1002/(SICI)1521-3773(19980918)37:17<2296::AID-ANIE2296>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 45.Walker JE. ATP synthesis by rotary catalysis. Angew. Chem. Int. Ed. 1998;37:2308–2319. doi: 10.1002/(SICI)1521-3773(19980918)37:17<2308::AID-ANIE2308>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 46.Skou JC. The identification of the sodium-potassium pump. Angew. Chem. Int. Ed. 1998;37:2320–2328. doi: 10.1002/(SICI)1521-3773(19980918)37:17<2320::AID-ANIE2320>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 47.Schimmel P. GTP hydrolysis in protein synthesis: two for Tu? Science. 1993;259:1264–1265. doi: 10.1126/science.8446896. [DOI] [PubMed] [Google Scholar]
- 48.Chang Y-F, Carman GM. CTP synthetase and its role in phospholipid synthesis in the yeast Saccharomyces cerevisiae. Prog. Lipid Res. 2008;47:333–339. doi: 10.1016/j.plipres.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson JE. Some thoughts on the evolutionary basis for the prominent role of ATP and ADP in cellular energy metabolism. J. Theor. Biol. 1984;111:615–623. doi: 10.1016/s0022-5193(84)80257-x. [DOI] [PubMed] [Google Scholar]
- 50.Yount RG, Babcock D, Ballantyne W, Ojala D. Adenylyl imidodiphosphate, an adenosine triphosphate analog containing a P-N-P linkage. Biochemistry. 1971;10:2484–2489. doi: 10.1021/bi00789a009. [DOI] [PubMed] [Google Scholar]
- 51.Jia S-M, Liu X-F, Kong D-M, Shen H-X. A simple, post-additional antioxidant capacity assay using adenosine triphosphate-stabilized 2,2′-azinobis(3-ethylbenzothiazoline)-6-sulfonic acid (ABTS) radical cation in a G-quadruplex DNAzyme catalyzed ABTS-H2O2 system. Biosens. Bioelectron. 2012;35:407–412. doi: 10.1016/j.bios.2012.03.029. [DOI] [PubMed] [Google Scholar]
- 52.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5’-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu Y. Chemistry in human telomere biology: structure, function and targeting of telomere DNA/RNA. Chem. Soc. Rev. 2011;40:2719–2740. doi: 10.1039/c0cs00134a. [DOI] [PubMed] [Google Scholar]
- 55.Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nature Rev. Drug Discov. 2011;10:175–261. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Macaya RM, Schultze P, Smith FW, Roe JA, Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. USA. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laughlan G, Murchie AI, Norman DG, Moore MH, Moody PC, Lilley DM, Luisi B. The high-resolution crystal structure of a parallel-stranded guanine tetraplex. Science. 1994;265:520–524. doi: 10.1126/science.8036494. [DOI] [PubMed] [Google Scholar]
- 58.Collie G, Parkinson GN, Neidle S, Rosu F, De Pauw E, Gabelica V. Electrospray mass spectrometry of telomeric RNA (TERRA) reveals the formation of stable multimeric G-quadruplex structures. J. Am. Chem. Soc. 2010;132:9328–9334. doi: 10.1021/ja100345z. [DOI] [PubMed] [Google Scholar]
- 59.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat-containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 60.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nature Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.