Abstract
In Vibrio cholerae, the RNA binding protein and chaperone Hfq (VcHfq) facilitates the pairing of the quorum regulatory RNA (Qrr) small regulatory RNAs (sRNAs) to the 5′ untranslated regions of the mRNAs for a number of global regulators that modulate the expression of virulence genes. This Qrr-mediated sRNA circuit is an attractive antimicrobial target, but characterization at the molecular level is required for this to be realized. Here, we investigate the interactions between VcHfq and the Qrr sRNAs using a variety of biochemical and biophysical techniques. We show that the ring-shaped VcHfq hexamer binds the Qrrs with 1:1 stoichiometry through its proximal face, and the molecular envelope of the VcHfq-Qrr complex is experimentally determined from small angle scattering data to present the first structural glimpse of a Hfq-sRNA complex. This structure reveals that the VcHfq protein does not change shape on complex formation but the RNA does, suggesting that a chaperone role for VcHfq is a critical part of the VcHfq-Qrr interaction. Overall, these studies enhance our understanding of VcHfq-Qrr interactions.
INTRODUCTION
The bacterial protein Hfq regulates gene expression by facilitating the interaction of trans-encoded small regulatory RNAs (sRNAs) with mRNA targets to subsequently affect translation and/or mRNA turnover [reviewed in (1)]. sRNAs typically base-pair with the 5′ untranslated region (UTR) of an mRNA through short regions of complementarity. sRNA binding can bring about one of the three possible effects; firstly, and most commonly, they can prevent translation by directly blocking the ribosome binding site (RBS). Secondly, they can impede or disrupt the formation of inhibitory secondary structures in the 5′ UTR to expose the RBS and indirectly stimulate translation. Thirdly, they can modulate the susceptibility of the mRNA to cellular ribonucleases (RNases) by changing the RNA structure to create or remove RNase cleavage sites [reviewed in (2)]. The precise role that Hfq plays in these mechanisms remains unclear, but it includes binding of both the sRNA and the mRNA to increase their local concentrations and increase the probability of an interaction, and/or functioning as an RNA chaperone, remodeling the structure of one, or both, of the RNAs to facilitate productive pairing [reviewed in (1)]. Hfq could also affect the stability of the RNA(s) by altering their structures to promote or inhibit RNase activity (2) or by binding to canonical RNase cleavage sites and protecting the RNA(s) (3).
sRNAs are typically expressed as part of the global stress responses that allow bacteria to adapt their gene expression profiles to overcome environmental pressures such as starvation, osmotic stress, oxidative stress and low iron concentrations (2). For pathogenic bacteria, such as Vibrio cholerae, Hfq-dependent sRNA-mediated gene regulation tightly controls the expression of specific virulence factors (4,5). Consequently, virulence-associated sRNA-controlled circuits are attractive targets for the development of novel antibacterial strategies.
In V. cholerae, four sRNAs, the quorum regulatory RNAs 1–4 (Qrrs1–4), promote the expression of genes required for virulence and biofilm formation at low cell density (6). The Qrrs affect the translation of at least four mRNAs through Hfq-mediated base-pairing: hapR (6,7), vca0939 (8), aphA (9,10) and luxOU (11). Three of these targets are involved in the global regulation of V. cholerae pathogenicity: hapR encodes the HapR transcription factor that represses virulence genes (12) and is required for the production of hemagglutinin protease for release from the host (13), AphA is a transcription factor that enhances virulence-gene expression (14) and vca0939 encodes a GGDEF protein that synthesizes the second messenger cyclic di-guanosine monophosphate to stimulate biofilm formation (8). The four Qrrs were initially reported to be functionally redundant, with expression of any one Qrr resulting in a normal response to changing cell density (6,11). Furthermore, inactivation of one or more Qrr(s) leads to dosage compensation by the remaining Qrr(s) suggesting that their combined concentration is critical for their correct function (11). However, a recent study has shown that, unlike Qrrs2–4, Qrr1 is unable to stimulate translation of aphA, because it is shorter than the other Qrrs (99 nucleotides compared with 111 nucleotides for Qrr2 and 110 nucleotides for Qrr3 and Qrr4) and lacks part of the region required to base-pair with aphA mRNA (10). Qrr1 is also predicted to adopt a different secondary structure to Qrrs2–4 [(6) and Supplementary Figure S1). Therefore, it is possible that the Qrr sRNAs have preferential specificities for the different mRNA targets.
Given the importance of sRNA-mediated gene regulation in the pathogenicity of V. cholerae, the system is clearly an attractive target for new antibacterial agents. To design such a strategy and better understand how the components interact, it is necessary to visualize the interactions between Hfq, the sRNAs and the mRNAs at the molecular level. For V. cholerae Hfq (VcHfq) and the Qrrs, it is well-established that cognate base-pairing between the sRNAs and the 5′ UTRs of their mRNA targets is critical for regulation and that Hfq enhances this recognition (6–11). However, the protein-RNA interactions involved are poorly understood. We have recently shown that VcHfq binds Qrr1 tightly, with a dissociation constant (Kd) in the low-nanomolar range (15). This interaction is mediated primarily through the conserved N-terminal domain of the protein, with the C-terminal region potentially affecting the binding specificity (15). This interaction is examined here in more detail using a range of biochemical and biophysical techniques. VcHfq RNA-binding mutants were prepared and used to identify RNA-binding site(s) within the N-terminal domain of the protein, and a novel ab initio modelling process was developed to determine the structure of the VcHfq-Qrr1 complex from small angle scattering (SAS) data. Finally, VcHfq-Qrr interactions were investigated for Qrrs2–4 and compared with those in the VcHfq-Qrr1 complex, identifying their similarities and differences. Together, these data provide insights into the molecular recognition underlying the Qrr-mediated virulence of V. cholerae.
MATERIALS AND METHODS
Construction of Hfq expression plasmids
Construction of the pET28[VcHfq] plasmid for expression of wild-type V. cholerae Hfq has been described previously (15). pET28[VcHfqY25D], for expression of a distal face RNA-binding mutant, and pET28[VcHfqK56A], for expression of a proximal face RNA-binding mutant, were generated by standard site-directed mutagenesis protocols using pET28[VcHfq] as the template and the Y25D and K56A mutagenic primers, respectively (Supplementary Table S1). pET28[VcHfqY25DK56A], for expression of a distal and proximal face RNA-binding mutant, was similarly created using pET28[VcHfqY25D] as a template and the K56A mutagenic primers (Supplementary Table S1). Constructs were verified by DNA sequencing.
Protein expression and purification
BL21(DE3) was transformed with either pET28[VcHfq], pET28[VcHfqY25D], pET28[VcHfqK56A] or pET28[VcHfqY25DK56A] and was grown in Luria broth supplemented with 50 μg/ml kanamycin at 37°C, until the absorbance at 600 nm reached 0.6. Expression was induced with 1 mM isopropyl β-D-1-thiogalactopyranoside, and the cells were incubated for a further 3 hours at 37°C. Cells were harvested by centrifugation at 5000g for 20 minutes at 4°C, and the cell pellet was stored at −80°C.
The three Hfq mutant proteins were purified in a similar way to wild-type Hfq [described in (15)]. Cell pellets were thawed on ice and resuspended in 20-mM Tris pH 7.4, 500-mM NaCl, 10-mM MgCl2, 1-U/ml DNase I (Promega) and 0.5-mg/ml lysozyme (Sigma). Cells were disrupted by sonication, and the lysate clarified by centrifugation at 40 000g for 30 minutes at 4°C. After addition of 20-mM imidazole, the supernatants were loaded onto a HisTrap HP column (GE Healthcare) equilibrated in 20-mM Tris pH 7.4, 500-mM NaCl and 20-mM imidazole at 4°C using an ÄKTAxpress (GE Healthcare). Proteins were eluted with a linear gradient to 1-M imidazole, and VcHfq-containing fractions were pooled and buffer-exchanged into 20-mM Tris pH 8, 500-mM NaCl, 2.5-mM CaCl2 and 10% glycerol using a PD10 column (GE Healthcare). The proteins were incubated with 0.5-U thrombin (Sigma) at room temperature for 16 hours to cleave the N-terminal hexahistidine tag, buffer-exchanged into 20-mM Tris pH 8, 1-M NaCl, 1-M (NH4)2SO4 and 0.5-mM EDTA using a PD10 column and loaded onto a HiTrap Butyl-S FF column (GE Healthcare), equilibrated in the same buffer at 4°C using an ÄKTAxpress. Proteins were eluted with a linear gradient to 0-M (NH4)2SO4. VcHfqY25D (the distal face mutant) and VcHfqK56A (the proximal face mutant) were buffer-exchanged into 20-mM 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid (HEPES) pH 7, 100-mM NaCl and 0.5-mM EDTA using a PD10 column and loaded on a Mono S 4.6/100 PE column (GE Healthcare) equilibrated in the same buffer at 4°C using an ÄKTAxpress. VcHfqY25DK56A (the distal and proximal face double mutant) was buffer-exchanged into 20-mM 2-(N-morpholino)ethanesulfonic acid pH 6, 100-mM NaCl and 0.5-mM EDTA using a PD10 column, and loaded on a Mono S 4.6/100 PE column equilibrated in the same buffer at 4°C using an ÄKTAxpress. All three proteins were eluted in a linear gradient to 1-M NaCl, VcHfq-containing fractions were buffer-exchanged into 20-mM Tris pH 8, 500-mM NaCl, 0.5-mM EDTA and 10% glycerol using a PD10 column and concentrated to ∼10 mg/ml using a VivaSpin 2 centrifugal concentrator, with a molecular weight cut-off (MWCO) of 10 kDa. Concentrated proteins were loaded onto a Superdex 200 10/300 GL size exclusion column (GE Healthcare) equilibrated in 20-mM Tris pH 8, 500-mM NaCl, 0.5-mM EDTA and 10% glycerol using an ÄKTApurifier (GE Healthcare). VcHfq-containing fractions were pooled, concentrated to ∼5 mg/ml using a VivaSpin 2 centrifugal concentrator, with a MWCO of 10 kDa and stored at −80°C.
RNA synthesis
DNA templates encoding sRNAs Qrr1–4, preceded by the T7 promoter sequence, were generated by gene synthesis from overlapping primers (16) using the primers listed in Supplementary Table S1. Three guanine nucleotides were added to the 5′ end of each Qrr sequence to ensure efficient transcription by T7 RNA polymerase. RNAs were then synthesized by in vitro transcription using the MEGAScript T7 Kit (Applied Biosystems) for unlabelled RNA and the MAXIScript T7 Kit (Applied Biosystems) with [α-32P]UTP (PerkinElmer) for 32P-labelled RNA. RNA was purified using the MEGAClear Kit (Applied Biosystems) or MicroSpin G-25 columns (GE Healthcare) for unlabelled and 32P-labelled RNA, respectively.
Electrophoretic mobility shift assays (EMSAs)
RNA was heated to 80°C for 10 minutes and was allowed to cool to room temperature. Reactions (10-μl) were assembled containing 5-nM 32P-labelled or 30-nM unlabelled Qrr-RNA, the indicated concentration of VcHfq (hexamer), 20-mM Tris pH 8, 50-mM KCl, 50-mM NaCl, 0.5-mM EDTA and 10% glycerol. Reactions were incubated at room temperature for 30 minutes and analysed by 6% native PAGE using TBE buffer at room temperature. Gels with 32P-labelled RNA were visualized using a FLA-5000 Fluoro-phosphorimager (Fujifilm) and quantified using Multi Gauge software v2.2. Kds were determined from experiments repeated in triplicate, using a partition function for two non-identical independent sites as described by Lease and Woodson (17) in GraFit 5. Gels with unlabelled RNA were stained with SYBR Gold (Invitrogen) and visualized using a FLA-5000 Fluoro-phosphorimager.
Mass spectrometry (MS)
Non-denaturing MS (18) was performed using 5-μM solutions of wild-type VcHfq (hexamer) and 5-µM Qrr1, Qrr2, Qrr3 or Qrr4 sRNA in 250-mM aqueous ammonium acetate buffer pH 7. Samples were introduced using nano-electrospray ionization with a TriVersa Nanomate inlet system (Advion) and a Synapt T-wave Ion Mobility Mass Spectrometer (IM-MS; Waters) in positive ion mode. Experimental parameters were chosen to preserve intact non-covalent interactions during transfer of ions into the mass spectrometer (19). Key settings were: backing, 8 mbar; source, 12 × 10−3 mbar; sampling cone, 190 V; extraction cone, 5 V; trap and transfer collision energy, 60 and 12 V, respectively and bias, 30 V. For collision induced dissociation, trap collision energy was increased to 90 V. Data were smoothed using a window with ±50 channels using MassLynx software, 4.0.
Surface plasmon resonance (SPR)
SPR experiments were performed at 25°C using a Biacore T100 instrument (GE Healthcare). Approximately, 1000 resonance or response units (RUs) of purified VcHfq were immobilized onto a CM5 sensor chip (GE Healthcare) using amine coupling. The unit RU is defined as 1 RU = 1 pg mm−2 and is used to quantify sensor-surface coverage. Single cycle kinetic experiments were conducted to monitor Qrr binding with 4.7, 9.4, 18.8, 37.5 and 75-nM of each Qrr injected sequentially on the VcHfq surface in HBS-EP (HEPES buffered saline with EDTA and surfactant Tween 20) buffer (10-mM HEPES pH 7.4, 150-mM NaCl, 50-µM EDTA and 0.05% Tween 20). Kds were determined using a 1:1 binding model in T100 BiaEvaluation software v.2.0.2 (GE Healthcare).
SAS sample preparation
For both small angle X-ray scattering (SAXS) and small angle neutron scattering (SANS), VcHfq protein and the Qrr sRNAs were prepared in 20-mM Tris pH 8, 50-mM NaCl, 50-mM KCl and 0.5-mM EDTA. Complexes were prepared by mixing VcHfq protein and Qrr sRNA at equimolar concentrations. For SANS, VcHfq-Qrr1 complex was buffer exchanged into the equivalent buffer prepared in 0, 40, 73 or 100% D2O using a VivaSpin 2 centrifugal concentrator with a MWCO of 10 kDa.
SAXS data collection
SAXS data for the VcHfq-Qrr1 complex were collected using standard procedures on the bioSAXS beamline ID14-3 (20) at the European Synchrotron Radiation Facility (ESRF, Grenoble, France) at a wavelength of 0.931 Å and a camera length of 2.42 m. The detector was a Pilatus1M (Dectris), and the range of momentum transfer covered was 0.005 Å−1 < q < 0.5 Å−1. Data were collected for VcHfq-Qrr1 at a concentration of ∼2 mg/ml (∼20-µM, assuming 1:1 stoichiometry of VcHfq (hexamer):Qrr1 sRNA) in 10 successive 10-second frames with the sample continuously under flow. Data were normalized to the intensity of the incident beam, and the scattering of the buffer was subtracted in PRIMUS (21). Data were evaluated for aggregation (characterized by a sharp increase of the scattering curve at low q) and radiation damage (characterized by systematic deviations in consecutive scattering curves). Only data free from aggregation and radiation damage were processed further.
SAXS data for the Qrr sRNAs and the VcHfq-Qrr2, VcHfq-Qrr3 and VcHfq-Qrr4 complexes were collected using standard procedures on the I22 beamline at Diamond Light Source (DLS, Didcot, UK), equipped with a photon counting detector (22), at a wavelength of 1 Å and a camera length of 3.25 m. The covered range of momentum transfer was 0.003 < q < 0.55 Å−1. Data were collected for three concentrations, ∼1, 0.5 and 0.25 mg/ml (∼30, 15 and 7.5 µM) for the Qrr sRNAs alone and ∼3, 1.5 and 0.75 mg/ml (∼30, 15 and 7.5 µM, assuming 1:1 stoichiometry of VcHfq (hexamer):Qrr sRNA) for the VcHfq-Qrr complexes, in 120 successive 1-second frames. The data were normalized to the intensity of the incident beam using an in-house program, and the scattering of the buffer was subtracted using PRIMUS (21). Data were evaluated for interparticle effects (characterized by a downturn in the scattering curve at low q with decreasing sample concentration), aggregation and radiation damage. Only data free from aggregation and radiation damage were processed further.
SANS data collection
SANS data were collected using standard procedures on the D22 beamline at the Institute Laue-Langevin (ILL, Grenoble, France) at a wavelength of 6 Å and camera lengths of 2 and 11.2 m. The covered range of momentum transfer was 0.01 < q < 0.3 Å−1. Data were collected with VcHfq-Qrr1 complexes at a concentration of ∼2 mg/ml (∼20 µM, assuming 1:1 stoichiometry of VcHfq (hexamer):Qrr1 sRNA). Data were normalized to the intensity of the incident beam, and the scattering of the buffer was subtracted using GRASansP v5.09 (Dewhurst, ILL, Grenoble, France).
SAS data reduction
As the majority of information from the scattering data can be obtained from data with q < 0.2 Å−1, all SAS data were truncated at q = 0.2 Å−1 to eliminate the noise at high q before processing. The radius of gyration (Rg) and the forward scattering intensity, I(0), were evaluated with the program PRIMUS (21). Molecular masses for the SAXS data of the Qrr sRNAs were calculated from the I(0)s, normalized to the I(0) for phenylalanine transfer RNA. The distance distribution functions P(r)s were generated with the program GNOM (23) so that the Rg value agreed with that determined from the Guinier region in PRIMUS (21) to give the maximum particle dimension (Dmax).
Ab initio modeling of the Qrr sRNAs
Ab initio modelling of the Qrr sRNAs was performed with DAMMIF (24). Ten independent DAMMIF runs were carried out for each RNA. These generated similar, but not identical, shapes in each case. An averaged filtered shape was generated using DAMAVER and DAMFILT (25). De novo three-dimensional sRNA structures were predicted using iFoldRNA (26,27) and were superimposed on the ab initio models using SUPCOMB (28).
Ab initio modelling of the VcHfq-Qrr complexes
Ab initio modelling of the VcHfq-Qrr1 and VcHfq-Qrr2 complexes was performed using the multiphase dummy-atom modelling program MONSA (29,30). Default settings were used, with the exception that the looseness penalty was changed to 200 for VcHfq-Qrr2 modelling.
The protein phase, from SAXS data of the protein alone, was modelled as described previously (15). The model was moved to the origin with MASSHA (31) and a spherical search volume of either 155 Å or 200 Å diameters was created using the auxiliary ATSAS program pdb2dam4 (available upon request from ATSAS developers) for the VcHfq-Qrr1 and VcHfq-Qrr2 complexes, respectively. The diameters were chosen on the basis of a value slightly larger than that of the Dmax values determined from the P(r) functions, so as to minimize any constraint on the RNA structure during the modelling process. The following edits were made to the pdb outputs to maintain the Hfq phase during MONSA runs: ‘Hspace’ was replaced with ‘CA’, ‘0 1 201’ was replaced with ‘0 1 202’ and ‘0 2 201’ was replaced with ‘1 2 201’. These files were used as the starting search volume in MONSA and effectively ‘fixed’ the protein phase of the complex.
Two datasets were used during the modelling process to determine the structure of the RNA in the VcHfq-Qrr1 complex: SAXS data and 0% D2O SANS data. Only SAXS data were used for modelling the VcHfq-Qrr2 complex. Theoretical volumes for the individual phases, based on the amino acid for the protein and the nucleotide sequence for the RNA, and Rg values for the individual phases and the complex as a whole, determined from Guinier analysis, were used as constraints during the modelling process. The program MULCh (ModULes for the analysis of Contrast variation data) (32) was used to calculate the theoretical values of the scattering length density and scattering length difference (contrast), based on the amino acid sequence for the protein, the nucleotide sequence for the RNA and buffer composition. No symmetry constraints were imposed. The fits to the experimental data for the VcHfq-Qrr1 complex had χ2 values of 1.39 and 0.34 for the SAXS and SANS data, respectively, and the fit to the experimental data for the VcHfq-Qrr2 complex had a χ value of 0.19.
Circular dichroism (CD)
CD data were collected on an applied photophysics π*-180 spectrometer, using a cuvette with a 0.4-mm path length. Three experiments were conducted after Qrr sRNAs were prepared at 0.5–2 µM in 10-mM Tris pH 8 and 100-mM NaCl, heated to 80°C for 10 minutes and allowed to cool back to room temperature. Firstly, data for the Qrr sRNAs alone were collected at wavelengths spanning 200–360 nm in 1-nm steps at 20°C. Secondly, spectra for each Qrr sRNA were collected at temperatures from 20°C to 92°C, with 4°C increments. Finally, the spectra were collected for each Qrr sRNA as they were titrated with VcHfq to a ratio of 1:2 Qrr:VcHfq. Data from four or six scans were averaged, baseline-subtracted, smoothed using the Savitsky–Golay routine and calibrated against camphor sulphonic acid. Where appropriate, the protein contribution was also subtracted. Spectra were converted into molar ellipticity units (deg−1 cm2 dmol−1).
RESULTS
VcHfq binds Qrr sRNAs with 1:1 stoichiometry
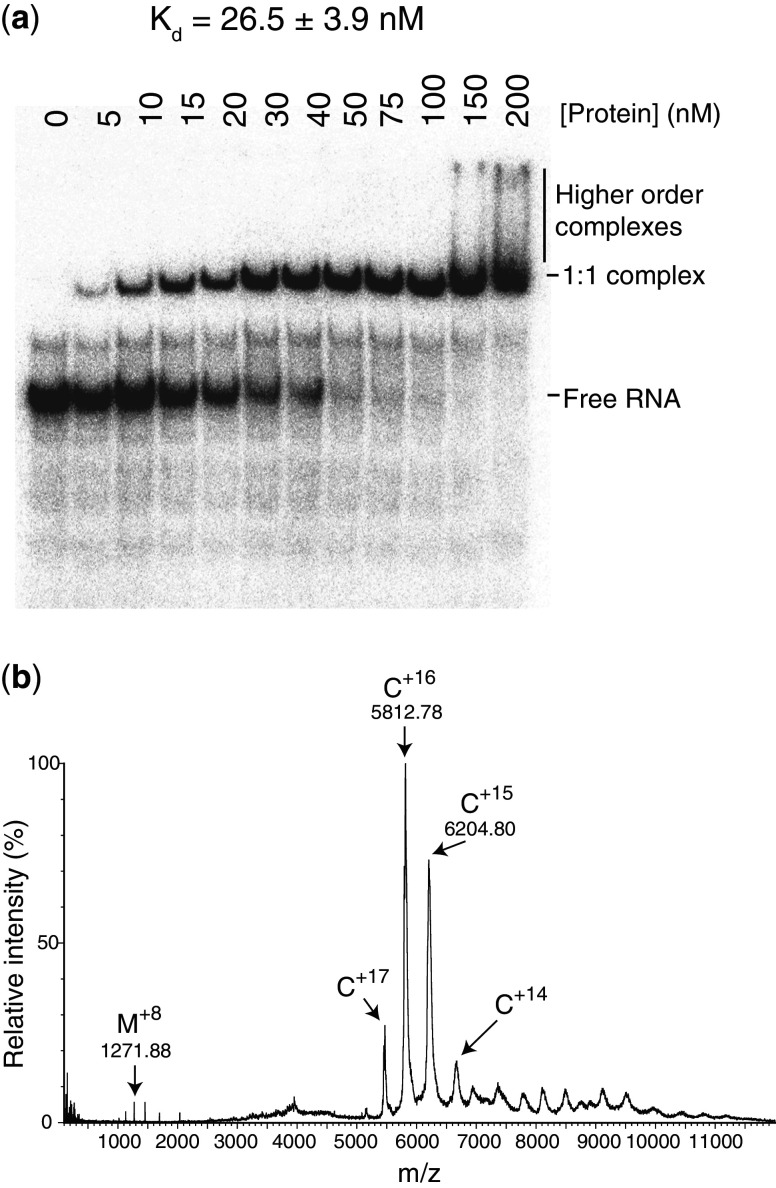
Escherichia coli Hfq is known to bind sRNAs with Kds, typically in the low-nanomolar range (33,34). In EMSAs, on addition of VcHfq to Qrr1, a distinct VcHfq-Qrr1 complex is formed and further addition of VcHfq leads to the formation of a higher order, less well-defined complex or complexes [(15) and Figure 1a]. Assuming that VcHfq is hexameric, the fraction of bound Qrr1 at each concentration of VcHfq was fit to a partition function for the protein bound to two independent sites on Qrr1 to give a Kd of ∼25 nM for the first complex formed [(15) and Figure 1a].
Figure 1.
VcHfq binds Qrr1 with 1:1 stoichiometry. (a) EMSA with VcHfq and 32P-labelled Qrr1 sRNA. Qrr1 was present at 5 nM in each lane and the concentration of hexameric VcHfq protein is indicated above the lane. The mobilities of free Qrr1 RNA, the 1:1 VcHfq:Qrr1 complex and higher order complexes are indicated on the right of the gel. The dissociation constant (Kd) for the 1:1 VcHfq:Qrr1 complex is reported above the gel and was calculated from three repeats. (b) Non-denaturing MS spectrum of VcHfq and Qrr1 sRNA. VcHfq and Qrr1 were mixed at equimolar concentrations. A peak series corresponding to the 14–17+ charges states of the 1:1 VcHfq (hexamer):Qrr1 complex is labelled with C (theoretical mass: 92 860.4 Da; experimental mass: 92 670 Da). A smaller series of peaks, centred on the 8+ charge state, corresponds to a small amount of monomeric VcHfq and is labelled with M (theoretical mass 10 163.6 Da; experimental mass: 10 166.9 Da).
To determine the likely stoichiometry of the VcHfq-Qrr1 complex(es), mixtures of VcHfq and Qrr1 sRNA were analysed using non-denaturing MS [Figure 1b; (18)], a technique that can be used to study non-covalent complexes. Experimental parameters were selected to preserve intact non-covalent interactions in the mass spectrometer (19). Protein-RNA complexes are expected to give rise to a series of peaks in the mass (m)/charge (z) spectrum that will be related by a common mass. This mass is the mass of the complex and allows determination of the stoichiometry of the complex, assuming that the complex constituents are known. Initially, VcHfq and Qrr1 were mixed at equimolar concentrations, assuming that VcHfq is present as a hexamer, and at a concentration above the Kd for the discrete complex observed in the EMSAs. A peak series corresponding to the 14–17+ charge states of a species with a mass of 92 670 Da were observed (Figure 1b). This species is consistent with a 1:1 VcHfq:Qrr1 complex, which has a theoretical mass of 92 860.4 Da. When VcHfq and Qrr1 were mixed with VcHfq in excess, again at concentrations above the Kd determined from the EMSAs, only a peak series corresponding to a 1:1 complex could be clearly detected. From these data we concluded that hexameric VcHfq only forms stable complexes with Qrr1 with 1:1 stoichiometry and propose that the discrete band observed in the EMSAs is a 1:1 VcHfq:Qrr1 complex.
Qrr1 is only one of four, functionally redundant, Qrr sRNAs present in V. cholerae (6), and the ability of VcHfq to form complexes with Qrr2, Qrr3 and Qrr4 was also examined by EMSAs (Supplementary Figure S2) and non-denaturing MS (Supplementary Figure S3). As can be seen from the left-most panels in Supplementary Figure S2, addition of VcHfq to each Qrr sRNA results in the formation of a discrete complex with poorly defined higher order complexes visible on continued addition of proteins. Non-denaturing MS of equimolar mixtures of VcHfq (hexameric) with Qrr2, Qrr3 or Qrr4, at concentrations above the Kd of the defined VcHfq:Qrr complexes observed in the EMSAs, produced peak series corresponding to complexes with 1:1 stoichiometry only (Supplementary Figure S3). Again this suggests that the stable species observed in the EMSAs is a 1:1 complex and that hexamericVcHfq binds to each Qrr with 1:1 stoichiometry.
VcHfq-Qrr complexes are of comparable stability
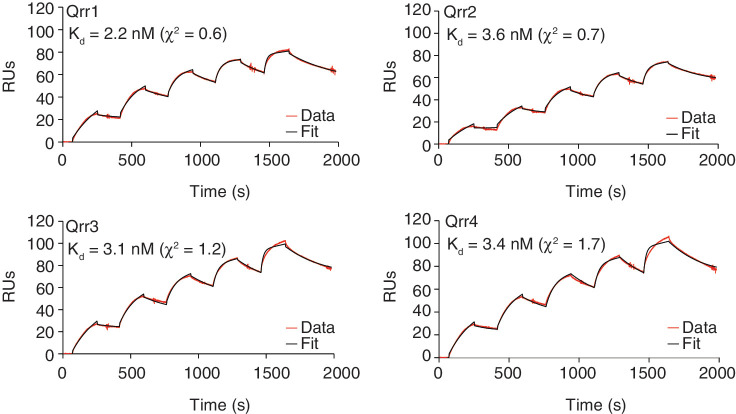
To investigate whether there might be subtle differences between the complexes formed between VcHfq and each of the four Qrrs, the binding affinity of each Qrr for VcHfq was measured by SPR. Single cycle kinetic experiments were performed in which VcHfq protein was immobilized to the sensor surface, and Qrr sRNA was injected at increasing concentrations in a single cycle without regenerating the surface between injections. The Qrr sRNA concentration range was selected such that, based on the EMSAs (Supplementary Figure S2), only 1:1 VcHfq (hexamer):Qrr complexes would be formed, allowing data to be fit to a 1:1 binding model. Sensorgrams of Qrr1, Qrr2, Qrr3 and Qrr4 binding to VcHfq are shown in Figure 2, together with the fits to the data and the calculated Kds. From these data, it is clear that all four Qrrs bind VcHfq tightly, with similar low-nanomolar Kds.
Figure 2.
VcHfq binds to all four Qrr sRNAs with low-nanomolar affinity. SPR sensorgrams for Qrr1, Qrr2, Qrr3 and Qrr4 binding to immobilized VcHfq. Experimental data is represented by a red line and the fit to a 1:1 binding model by a black line in each case. The calculated dissociation constant (Kd) is reported in the top left-hand corner of the sensorgram.
The relative stability of the 1:1 VcHfq (hexamer):Qrr complexes was also assessed by MS CID experiments. MS parameters were initially selected to observe intact 1:1 VcHfq (hexamer):Qrr complexes and then the trap collision energy was gradually increased to cause the complexes to dissociate. For each VcHfq-Qrr complex, the disappearance of the complex and the appearance of free VcHfq (monomer) were monitored. At a trap collision energy of 60 V, essentially all of the VcHfq were present as an intact 1:1 complex, with the Qrr sRNA for all four Qrrs (Figure 1b and Supplementary Figure S3). In contrast, at a trap collision energy of 90 V, a significant amount of free VcHfq (monomer) could be detected in each case (Supplementary Figure S4). Based on the relative heights of the VcHfq (monomer) and the VcHfq-Qrr complex peaks, slightly more VcHfq had dissociated for the Qrr1 and Qrr3 complexes than for the Qrr2 and Qrr4 complexes at 90 V. Small amounts of free VcHfq were also detected with the VcHfq-Qrr1 and VcHfq-Qrr3 complexes but not for the VcHfq-Qrr2 or VcHfq-Qrr4 complexes at the starting trap collision energy (Figure 1b and Supplementary Figure S3). This suggests that there may be subtle differences in the stability of each VcHfq-Qrr complex, dependent on the Qrr sRNA involved.
Qrr sRNAs bind to the proximal face of VcHfq
The conserved N-terminal domain of the Hfq protomer adopts the Sm fold of an N-terminal α-helix followed by five β-strands (35–40). Six of these monomers form a ring from which the disordered C-terminal regions extend outward to give an overall star-shaped structure (15,35–41). There are two principal RNA-binding sites on Hfq that have been well characterized through both structural (39) and mutagenesis studies (42): the proximal face and the distal face. The proximal face is the side with the exposed α-helices and C-terminal region and it preferentially binds single-stranded AU-rich RNA, which includes most sRNAs and their mRNA targets (37,40,42). Six consecutive nucleotides can bind to this face in a circular conformation around the central pore of the protein (37,38,40), and an additional uracil recognition site is located near to the N-terminus of the α-helix of each monomeric subunit (40). In contrast, the distal face is specific for poly(A) or A-R-N (where R is a purine and N is any nucleotide) triplets, both of which are often found in mRNAs (36,37,40,42). Again the binding topology is circular. However, the six binding pockets can each accommodate three nucleotides to bind up to a total of 18 nucleotides (36). The key residues involved in RNA binding for E. coli Hfq have been identified for each face and include Lys56 on the proximal face and Tyr25 on the distal face. Mutation of Lys56 to Ala and Tyr25 to Asp impairs proximal and distal face binding, respectively (42). As both of these amino acids are conserved in VcHfq, we tested whether mutating them would similarly impair the RNA-binding ability of the V. cholerae protein.
To investigate which face of VcHfq the Qrr sRNAs bind to, mutations were introduced on the distal face (Y25D), the proximal face (K56A) and both faces (Y25D K56A) of VcHfq. Their ability to bind to the Qrr sRNAs was then compared using EMSAs. Figure 3 shows gels from representative EMSAs for the three mutant VcHfq proteins binding to Qrr1. The distal face mutant binds Qrr1 with an affinity that is similar to, or perhaps slightly higher than, the wild-type protein (compare the top panel of Figure 3 with Figure 1a), suggesting that the distal face of VcHfq does not play a major role in Qrr1 binding. However, the proximal face mutant binds Qrr1 comparatively weakly (Kd 345 ± 130 nM) and the double mutant was essentially unable to detectably bind Qrr1 at the concentrations tested. This implies that VcHfq preferentially binds Qrr1 with its proximal face, although interactions are possible with the distal face. These results indicate that the RNA-binding modes employed by VcHfq, through the distal and proximal faces, are similar to those in E. coli Hfq.
Figure 3.
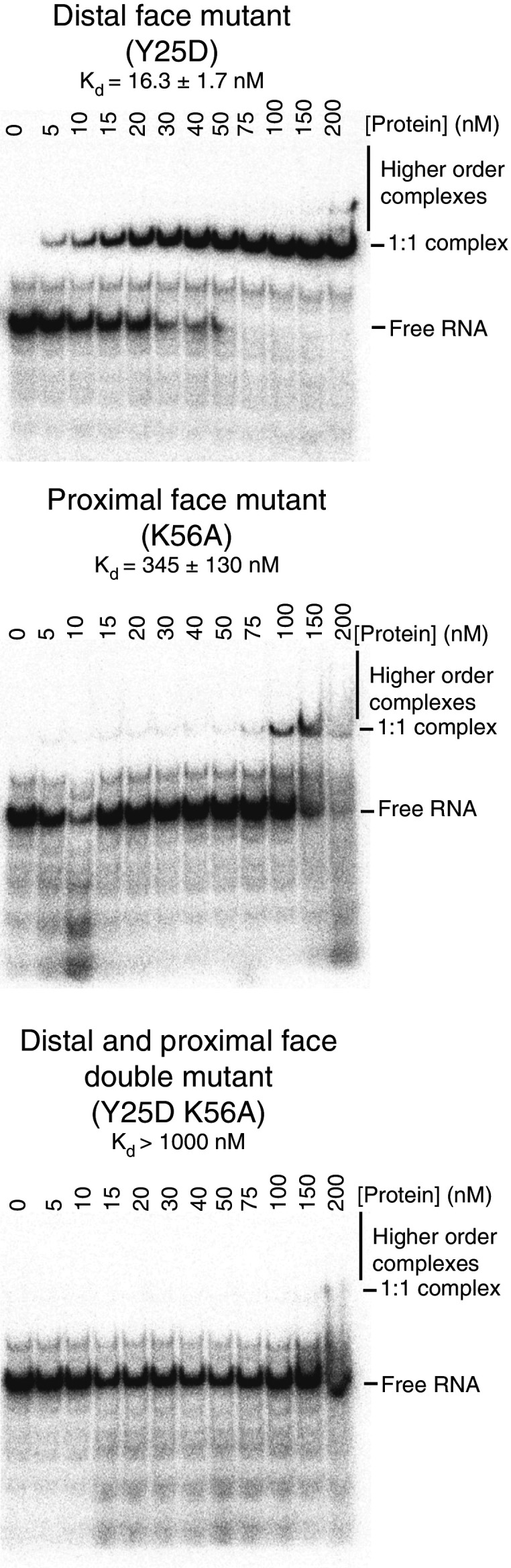
VcHfq preferentially binds Qrr1 with its proximal face. Representative EMSAs for a distal face mutant (Y25D), a proximal face mutant (K56A) and a distal and proximal face double mutant (Y25D K56A) of VcHfq binding to Qrr1 sRNA. Qrr1 was labelled with 32P and was present at 5 nM in each lane and the concentration of hexameric VcHfq protein is specified above the lane. The mobilities of free Qrr1 RNA, the 1:1 VcHfq:Qrr1 complex and higher order complexes are indicated on the right of the gels. The dissociation constant (Kd) for the 1:1 VcHfq:Qrr1 complex, calculated from three repeats, is reported above each gel.
Similar EMSAs were also carried out for Qrr2, Qrr3 and Qrr4, and representative gels are shown in Supplementary Figure S2. As for Qrr1, the distal face mutant of VcHfq is able to bind each Qrr with an affinity similar to wild-type, whereas the proximal face mutant binds more poorly. However, the effect of the proximal site mutation is not as pronounced for Qrrs2–4, as it is for Qrr1 suggesting that the preference for binding through the proximal face is not as strong for Qrrs2–4. Finally, the double mutant was unable to bind any of the Qrrs in the concentration range tested, confirming that RNA-binding interactions are mediated through the distal and/or proximal face of VcHfq.
Low-resolution solution structure of the VcHfq-Qrr1 complex
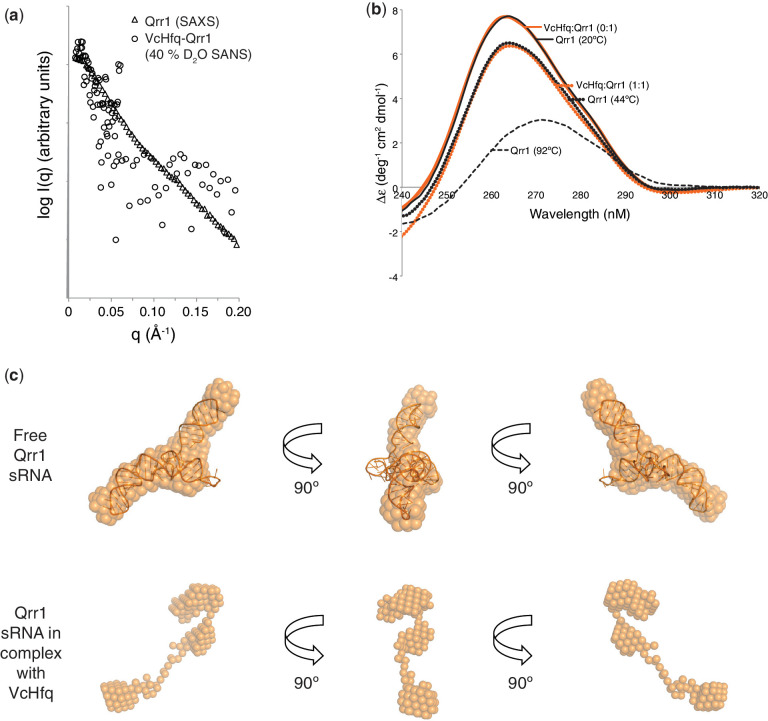
To investigate the organization of the VcHfq-Qrr1 complex, SAXS and SANS experiments were performed. VcHfq has been examined by SAXS previously and revealed that VcHfq hexamer adopts a six-pointed star-shaped conformation with six-fold symmetry (15). Here, Qrr1 sRNA, both free and in complex with VcHfq, has been examined by SAXS. The VcHfq-Qrr1 complex was prepared at a concentration where the stoichiometry would be expected to be 1:1 VcHfq (hexamer):Qrr1 based on the MS data (Figure 1b). The 1:1 VcHfq-Qrr1 complex was also examined in 0, 40, 73 and 100% D2O by SANS. SANS uniquely allows the spatial location of a single component of a protein–RNA complex, either the protein or the RNA, to be determined using contrast variation in a range of H2O/D2O mixtures to match out one of the components. In 0 and 100% D2O, scattering should be from the entire complex, whereas at 40 and 73% D2O scattering should be predominantly from the Qrr1 sRNA or the VcHfq protein component, respectively. When combined, these experiments, allowed the overall structure of the complex to be determined along with the conformations of the individual components in both their free forms and bound forms.
The parameters determined from the SAXS and SANS data are shown in Table 1. The Rg and Dmax values for the VcHfq-Qrr1 complex are 41 Å and 135–145 Å, respectively. These values are higher than for either the VcHfq protein component [Rg, 31.5 Å; Dmax, 103 Å (15)] or the Qrr1 sRNA component (Rg, 34.0 Å; Dmax, 130 Å) alone and would be consistent with VcHfq binding to a region of the RNA with unbound nucleotides extending away from the protein.
Table 1.
SAXS and SANS parameters
| D2O (%) | Rg (Å) | Dmax (Å) | |
|---|---|---|---|
| SAXS | |||
| VcHfqa | 0 | 31.5 ± 0.112 | 103 |
| Qrr1 | 0 | 34.0 ± 1.20 | 130 |
| Qrr2 | 0 | 41.1 ± 2.05 | 164 |
| Qrr3 | 0 | 46.3 ± 2.95 | 175 |
| Qrr4 | 0 | 41.1 ± 2.20 | 160 |
| VcHfq-Qrr1 | 0 | 41.5 ± 2.04 | 145 |
| VcHfq-Qrr2 | 0 | 52.3 ± 1.65 | 200 |
| VcHfq-Qrr3 | 0 | 52.7 ± 3.85 | 178 |
| VcHfq-Qrr4 | 0 | 50.4 ± 2.70 | 200 |
| SANS | |||
| VcHfq-Qrr1 | 0 | 41.0 ± 0.898 | 135 |
| VcHfq-Qrr1 | 100 | 40.9 ± 0.349 | 135 |
| VcHfq in situ | 73 | 31.7 ± 1.59 | 105 |
| Qrr1 in situ | 40 | 41.2 ± 4.95 | NDb |
aData from Vincent et al. (15) shown for comparison.
bNot determined.
MONSA (29,30) was selected for ab initio modelling of the VcHfq-Qrr1 complex. This program attempts to minimize the discrepancy between the fit of the model and the experimental data and describes the model as an assembly of beads within a spherical search volume. A caveat of this approach is that it is not possible to impose phase-specific symmetry during the modelling process, a significant disadvantage given the expected six-fold symmetry of the VcHfq protein.
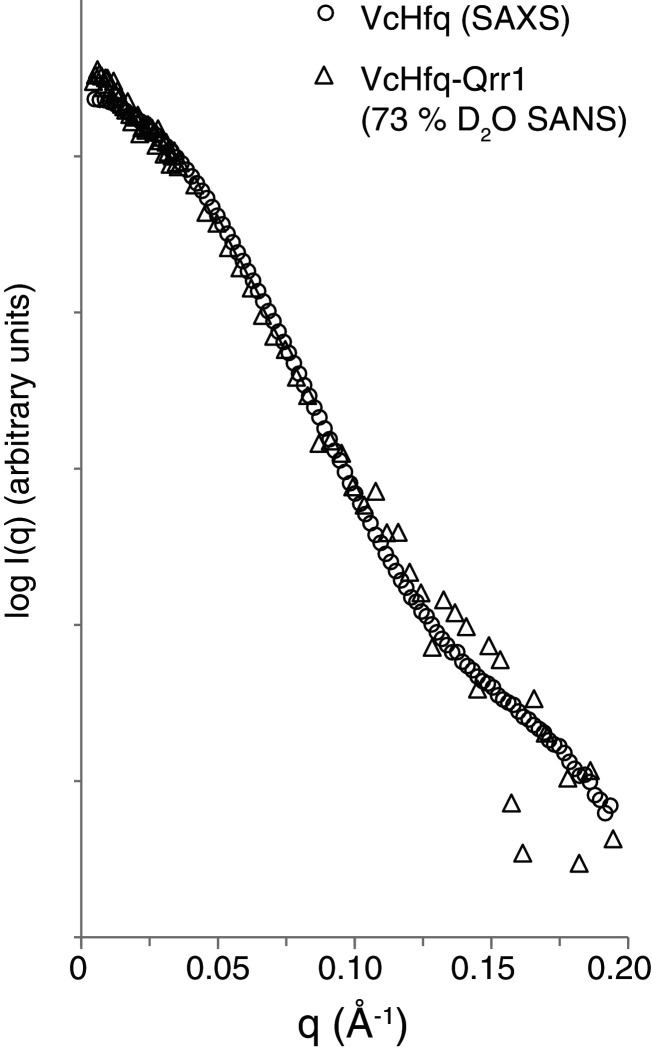
It was possible to overcome this difficulty because the protein does not seem to significantly change shape on complex formation. SAXS data of the free protein was compared with the SANS data of the VcHfq-Qrr1 complex in 73% D2O. This is the percentage of D2O at which the RNA phase of the complex is matched out and only scattering from the protein phase is observed. The parameters extracted from these data sets are similar (Table 1). Rgs were 31.5 Å and 31.7 Å and Dmaxs were 103 Å and 105 Å for the free VcHfq protein and VcHfq in complex with Qrr1 sRNA, respectively. Also, the profiles of the two scattering curves, shown in Figure 4, are almost identical. Together these data suggest that the protein adopts the same conformation in both its free form and in complex with Qrr1.
Figure 4.
The conformation of VcHfq does not change on complex formation. SAXS scattering curve of free VcHfq (circles) and SANS scattering curve of the VcHfq-Qrr1 complex in 73% D2O (triangles). SAS profiles have been normalized to the same intensity.
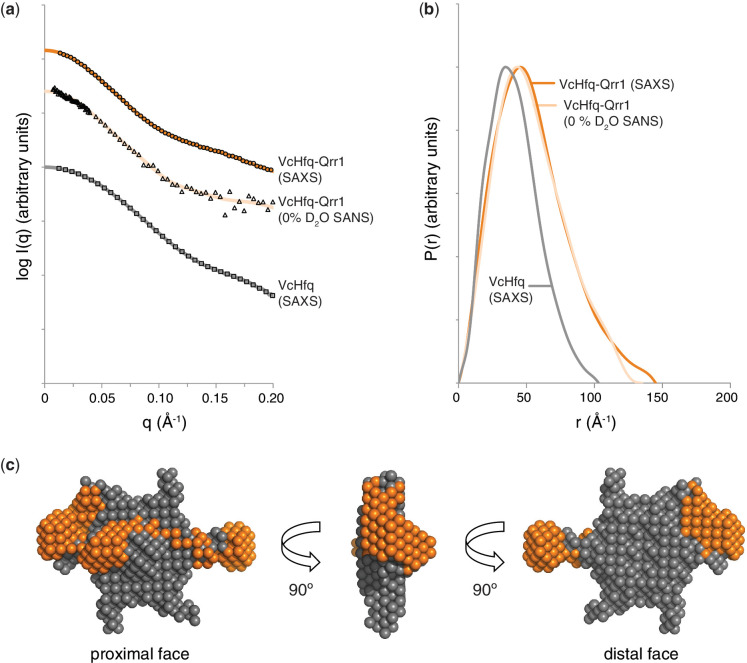
Given that the overall shape of the protein does not change on complex formation, it was decided that the model generated from the SAXS data of the free VcHfq protein (15) should be used as the input for MONSA to effectively ‘lock’ the symmetry and position of the protein in the model (see Materials and Methods for further details). The SAXS data of the VcHfq-Qrr1 complex and the SANS data of the complex in 0% D2O were then used for modelling. Scattering curves and distance distribution functions (P(r)s) for these data sets are shown in Figures 5a and b, respectively. The SANS data for the complex in 100% D2O was similar to that in 0% D2O and was excluded from the modelling based on redundancy. The SANS data for the VcHfq-Qrr1 complex in 40% D2O was also not included in the modelling process because of the poor signal-to-noise ratio in this dataset. The resultant ab initio model of the VcHfq-Qrr1 complex is presented in Figure 5c, and it clearly shows that VcHfq binds the Qrr1 sRNA across a single face. From the mutagenesis studies discussed previously, this is most likely the proximal face.
Figure 5.
Low-resolution model of the VcHfq-Qrr1 complex. (a) SAXS scattering curve for free VcHfq (squares), SAXS scattering curve for the VcHfq-Qrr1 complex (circles) and SANS scattering curve for the VcHfq-Qrr1 complex in 0% D2O (triangles). Back-transformed distance distribution functions, P(r), are shown as a solid grey line, a solid dark orange line and a solid light orange line for the SAXS data of free VcHfq, the SAXS data of the VcHfq-Qrr1 complex and the SANS data of the VcHfq-Qrr1 complex, respectively. (b) P(r) functions for the SAXS data of free VcHfq (grey), the SAXS data of the VcHfq-Qrr1 complex (dark orange) and the SANS data of the VcHfq-Qrr1 complex (light orange). (c) Ab initio model of the VcHfq-Qrr1 complex generated in MONSA (29,30) and visualized in PyMOL. VcHfq is shown as grey spheres and the Qrr1 sRNA as dark orange spheres.
VcHfq alters the structure of Qrr1 sRNA
Escherichia coli Hfq has been reported to be an RNA chaperone, changing the structure of RNAs on binding (43). To investigate whether VcHfq alters the structure of Qrr1 sRNA, a similar approach was taken to that used to prove that the protein adopts the same conformation when free and when in complex with Qrr1. SAXS data of the free Qrr1 sRNA was compared with the SANS data of the VcHfq-Qrr1 complex in 40% D2O, the percentage of D2O at which the protein phase of the complex is matched out and only scattering from the RNA phase is observed. Because of the poor signal-to-noise ratio in the SANS data, it is difficult to tell if the parameters extracted from the data (Table 1) or the profiles of the scattering curves for these datasets, shown in Figure 6a, really do differ.
Figure 6.
The conformation of the RNA changes on complex formation. (a) SAXS scattering curve of free Qrr1 (circles) and SANS scattering curve of the VcHfq-Qrr1 complex in 40% D2O (triangles). SAS profiles have been normalized to the same intensity. (b) CD spectra for VcHfq-Qrr1 complexes (orange lines) at 20°C at VcHfq:Qrr1 ratios of 0:1 (solid orange line) and 1:1 (dotted orange line) and for free Qrr1 sRNA (black lines) at 20°C (solid black line), 44°C (dotted black line) and 92°C (dashed black line). Qrr1 was present at 0.7 µM in each case. (c) The upper panels show a ribbon representation of Qrr1, predicted by iFoldRNA (26,27), superimposed with SUPCOMB (28) on the ab initio model of the sRNA (spheres) generated from the SAXS data of free Qrr1 with DAMMIF (24) and averaged with DAMAVER and DAMFILT (25). The lower panels show the ab initio model of the sRNA, as it appears in complex with VcHfq, generated from the SAXS and SANS data of the complex with MONSA (29,30). All models were visualized with PyMOL.
To further investigate potential VcHfq-induced conformational changes of Qrr1, the sRNA was examined by CD (Figure 6b). As VcHfq was titrated into Qrr1, the molar ellipticity at 265 nm decreased gradually up to a VcHfq:Qrr1 ratio of 1:1. To explore the extent of Qrr1 unfolding that this represented, heating experiments were conducted on the sRNA alone. As temperature was increased from 20°C to 92°C, a much larger change in ellipticity was observed relative to the addition of VcHfq. The position of the peak maxima also changed from 265 nm to 272 nm, consistent with the RNA transitioning toward a more single-stranded character. Therefore, the remodelling effect of VcHfq was equivalent to heating Qrr1 to ∼44°C, suggesting that VcHfq binding causes partial unwinding or restructuring of the RNA.
Ab initio models were generated for free Qrr1 sRNA and were in reasonable agreement with the three-dimensional structure predicted using iFoldRNA (26,27), as can be seen from the superposition of the predicted structure onto the average of the models in Figure 6c. This predicted structure could not be superimposed onto the ab initio model of Qrr1 in complex with VcHfq (Figures 5c and 6c) implying that the RNA has changed shape following VcHfq binding. Together, these data indicate that VcHfq does act as an RNA chaperone with respect to Qrr1 sRNA.
Comparison of the VcHfq-Qrr complexes
SAXS data were also collected for Qrr2, Qrr3 and Qrr4 in their free forms and in complex with VcHfq. The VcHfq-Qrr complexes were again prepared at a concentration where the stoichiometry would be expected to be 1:1 VcHfq (hexamer):Qrr based on the MS data (Supplementary Figure S3). The parameters that were extracted from these data are shown in Table 1. The Rg and Dmax values determined for Qrrs2–4 in their free form and in complex with VcHfq, are larger compared with Qrr1. This most likely reflects the differences in the lengths and structures of the RNAs, with Qrr1 containing 99 nucleotides, Qrr2 111 nucleotides and Qrrs3 and 4 110 nucleotides (Supplementary Figure S1). The scattering curves for the VcHfq-Qrr complexes, with that for VcHfq-Qrr1 shown again for comparison, are presented in Supplementary Figure S5a. The profiles of the scattering curves for the VcHfq-Qrr complexes for Qrr2–4 are similar, but they differ to that for VcHfq-Qrr1. Likewise, the P(r) functions in Supplementary Figure S5b are similar for Qrr2–4 in complex with VcHfq but different for VcHfq-Qrr1. This suggests that the VcHfq-Qrr complexes for Qrr2–4 will adopt a similar conformation but one that is different to that of VcHfq-Qrr1.
SANS data were not collected for Qrr2–4 in complex with VcHfq. However, modelling of the VcHfq-Qrr2 complex, as an example of the VcHfq-Qrr complexes for Qrr2–4, was performed with MONSA (29,30) by starting with the assumption that the VcHfq protein does not change shape on complex formation with any of the Qrrs. It was then possible to use the SAXS model of the free VcHfq as an input for MONSA, just as for VcHfq-Qrr1. The complex was then modelled using the SAXS data for the VcHfq-Qrr2 complex. The final ab initio model is shown in Supplementary Figure S5c. Similar to VcHfq-Qrr1, Qrr2 sRNA binds predominantly to one face of VcHfq and, from the mutagenesis studies, this is most likely the proximal face. However, more of the RNA seems to be exposed to the solvent in the VcHfq-Qrr2 complex, probably because of the longer length of the sRNA and VcHfq only being able to bind a finite number of nucleotides.
To investigate whether the structure of Qrr2 changes on VcHfq binding, CD and ab initio modelling were employed. The CD spectra shown in Supplementary Figure S6a show that the molar ellipticity of Qrr2 at 265 nm is lower in the presence of VcHfq, and that this decrease is equivalent to heating the sRNA to 44°C. This implies that VcHfq does partially unwind Qrr2 on binding. Similarly, ab initio modelling showed small changes in the conformation of Qrr2 in complex relative to the free form, such that the predicted structure of Qrr2 generated in iFoldRNA (26,27) could be superimposed on the model of the free Qrr2 with reasonable agreement, but not on the model of Qrr2 in complex with VcHfq (Supplementary Figure S6a).
Finally, the VcHfq-Qrr3 and VcHfq-Qrr4 complexes were also investigated by CD. The spectra are shown in Supplementary Figures S6b and S6c, respectively, and reveal that the molar ellipticity of these RNAs also decreases in the presence of VcHfq. For Qrr3, the VcHfq binding was equivalent to heating the RNA to 44°C, but heating to only 36°C could produce the same effect for Qrr4. This data suggest that VcHfq acts as an RNA chaperone with all four Qrr sRNAs, but the requirement for VcHfq may be slightly lower for Qrr4.
DISCUSSION
The RNA binding sites on Hfq have been well-defined, but the stoichiometry of Hfq-RNA complexes has been a controversial topic. The toroidal hexameric Hfq protein presents two binding faces (proximal and distal), each with six nucleotide-binding pockets (36,37,39,40). Consequently, each Hfq hexamer has the capability to bind multiple RNAs and, similarly, any given RNA can have multiple Hfq-binding sites. The situation is further complicated by the recent report that Hfq is not always hexameric (44). Panja and Woodson demonstrate that at monomer concentrations in the nM range, low-micromolar range and low-to-mid micromolar range E. coli Hfq is monomeric, hexameric or multimeric, respectively (44). Our non-denaturing MS analyses, performed at a VcHfq (monomer) concentration in the mid micromolar range, detected VcHfq-Qrr complexes with 1:1 VcHfq (hexamer):Qrr sRNA stoichiometry. Although VcHfq may have been expected to be multimeric under these conditions (44), it has been reported that multimer formation is disfavoured in the presence of RNA (17,45). The EMSA data are perhaps more difficult to interpret with respect to complex stoichiometry. These experiments were performed with a concentration of VcHfq (monomer) in the mid to high nanomolar range when the protein might be expected to be transitioning from a monomer-hexamer equilibrium to a hexamer (44). Nevertheless, we conclude that the discrete complexes observed in the gels do represent 1:1 VcHfq (hexamer):Qrr sRNA complexes, as hexameric VcHfq would be expected to be the predominant species present under our experimental conditions and a higher stoichiometry would be inconsistent with the MS results. Therefore, we suggest that hexameric VcHfq binds to each of the Qrr sRNAs with 1:1 stoichiometry. Previous studies have suggested that the C-terminal region of the protein is important for maintaining the specificity of binding and hence, the stoichiometry of Hfq:sRNA complexes (15).
Kds determined by SPR and the stability of each of the VcHfq-Qrr complexes assessed by collision induced MS were comparable. Only subtle differences between the different complexes could be detected, with the VcHfq-Qrr3 complex potentially being the least stable (Supplementary Figure S4). This suggests that the VcHfq-Qrr interaction is similar for each Qrr and is unlikely to account for the different mRNA target specificity reported for Qrr1 (10). The minor stability differences most likely reflect slight differences in the VcHfq binding site on each sRNA. Hfq has been shown to bind to the poly(U) tails formed by the Rho-independent transcriptional terminators (37) and VcHfq could potentially bind to the 3′ poly(U) tail of the Qrrs. Interestingly, the poly(U) tail is shortest for Qrr3 (Supplementary Figure S1) which could explain the lower stability for the VcHfq-Qrr3 complex.
Most sRNAs preferentially bind to the proximal face of Hfq (33,42), and this also seems to be the case for the Qrrs binding to VcHfq. The proximal face mutant has a far greater impact on sRNA binding than the distal face mutant for Qrr1. The findings that the distal face mutant can potentially bind Qrr1 more tightly than the wild-type protein and that both mutations on the distal and proximal face are required to prevent RNA binding indicates that Qrr1 can bind to both faces of VcHfq. It is possible that the 1:1 complex formed with the wild-type protein is actually a mixture of complexes with the sRNA bound to either the proximal or the distal face of VcHfq, but with Qrr1 bound predominantly to the proximal face. It has been reported that the K56A (proximal face) mutation, but not the Y25D (distal face) mutation, destabilizes the E. coli hexamer and this instability could impair the Hfq-RNA interaction (44). Although both the Y25D and K56A VcHfq mutants were hexameric based on size exclusion chromatography (data not shown), we cannot exclude the possibility that the differential effects of the distal and proximal face mutants arise partly from the proximal face mutation affecting the stability of the hexamer.
Similarly, binding to Qrrs2–4 was affected more by the proximal face mutant than the distal face mutant. However, the preference for forming the 1:1 complex with the sRNA binding to the proximal face is not so pronounced. Although there are no significant poly(A) stretches or ARN repeats in any of the four sRNAs to mediate distal-face binding, each Qrr does contain at least one ARN triplet. Based on secondary structure predictions [(6) and Supplementary Figure S1], these ARN triplets would be in a single-stranded region of the RNA and, therefore, accessible to VcHfq. Potentially, multiple ARN triplets could come together in the tertiary structure of the RNAs to strengthen the binding to the distal face, especially in the case of Qrrs2–4.
The low-resolution models of the VcHfq-Qrr1 and VcHfq-Qrr2 complexes are the first structures that show a full-length Hfq protein binding to a full-length sRNA. These structures support the mutagenesis studies that the Qrrs bind predominantly to one face of the VcHfq protein (presumably the proximal face). A number of crystal structures of Hfq bound to short oligoribonucleotides have revealed that Hfq can bind six consecutive nucleotides in a circular configuration around the central pore formed by the protein’s N-terminal core (36–38,40). Consistent with these structures, Figure 5c and Supplementary Figure S5c show that significant interactions occur between the VcHfq core and the sRNAs. However, there is little evidence that either Qrr1 or Qrr2 adopt the circular configuration observed for short oligoribonucleotides (36–38,40). This suggests that only a subset of the nucleotide-binding sites are actually occupied by the sRNA. Potentially, the tertiary structure of the longer sRNA prevents binding to all six nucleotide-binding sites simultaneously. The models of the VcHfq-Qrr1 and VcHfq-Qrr2 are most similar to the recent structure reported for E. coli Hfq bound to an eight-nucleotide fragment of DsrA (40). In this structure, seven nucleotides were bound on the proximal face, four nucleotides had the expected circular conformation, two nucleotides floated above the central pore and a single uracil was located near the N-terminus of the α-helix, distant from the central pore. Recent studies have suggested that sRNA-mRNA pairing is facilitated when the regions required for pairing are bound to the same face of Hfq (34,46). Secondary interactions between distant regions of the mRNA and the opposite face of VcHfq may help to stabilize the ternary complex. The models of the VcHfq-Qrr complexes could clearly accommodate an mRNA on the same face as the bound sRNA and/or on the opposite face.
In addition to the contacts between the VcHfq core and the Qrrs, the models also suggest that the C-terminal region of VcHfq could be involved in RNA binding. Investigations into the function of the disordered C-terminal region of Hfq proteins have suggested a role in RNA binding, with the C-terminus being required to maintain binding specificity and stoichiometry (15). It has been proposed that the C-terminus is able to make long-range interactions and possibly becomes more structured on RNA binding to stabilize the complex (41). However, the solution scattering studies described here revealed that the VcHfq protein does not significantly change conformation on complex formation with Qrr1. Therefore, any structural changes to the C-terminal region to facilitate RNA binding must be transient, such that the protein structure is essentially unchanged in the stable VcHfq-Qrr1 complex.
In addition to binding RNA, Hfq can function as a chaperone, altering the structure of the RNA (43,47). In contrast to the protein, the structure of each of the Qrr sRNAs does seem to change on Hfq-binding. Changes in Qrr structure, observed by CD and SAS, are consistent with a loss of double-stranded character in the RNA and it implies that Hfq has chaperone activity with the Qrrs. The temperature required to cause the same effect as VcHfq-binding is slightly lower for Qrr4 than for the other three Qrrs, but it is not clear why less structural rearrangement is possible or required for this sRNA. The requirement for Hfq-mediated unwinding could explain how Hfq enhances sRNA-mRNA pairing in this V. cholerae system.
Where exactly the VcHfq protein binds on the sRNAs, and the nature of the structural changes that it produces remain to be determined. Likewise, understanding the interactions between VcHfq and the mRNA targets of the Qrrs is important. Most likely, a ternary complex, consisting of VcHfq, a Qrr sRNA and an mRNA target, is formed for VcHfq to facilitate Qrr-mRNA pairing. What form this ternary complex will take is an intriguing question awaiting structural studies. Will the mRNA bind next to the sRNA on the proximal face or bind on the opposite, distal face? Will structural rearrangements of the mRNA be required?
Although many questions remain, significant progress has been made into understanding the interactions involved between VcHfq and the Qrr sRNAs. Furthermore, our novel approach for modelling SAS data of complexes in which at least one component does not change structure should allow similar studies to be completed for other Hfq-RNA complexes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–6.
FUNDING
Biotechnology and Biological Sciences Research Council [BB/F013140/1 to A.J.C.]; European Commission, Seventh Framework Programme [249154 to H.A.V.]; Institute of Biomedical and Biomolecular Sciences, University of Portsmouth, bursary (to C.A.H.). Funding for open access charge: BBSRC.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Petra Pernot (European Synchrotron Radiation Facility, Grenoble, France) and Marc Malfois (Diamond Light Source, Didcot, UK) for SAXS technical support, and Phil Callow (Institut Laue–Langevin, Grenoble, France) and Maxim Petoukhov and Haydyn Mertens (European Molecular Biology Laboratory, Hamburg, Germany) for help with SANS. We thank James McCullagh and Chris Schofield (Department of Chemistry, University of Oxford, UK) for access to the Synapt mass spectrometer. We thank Ben Luisi (University of Cambridge, UK) and T.J. Ragan (MRC National Institute for Medical Research, Mill Hill, UK) for helpful discussions and critical reading of the manuscript.
REFERENCES
- 1.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 2011;3:a003798. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moll I, Afonyushkin T, Vytvytska O, Kaberdin VR, Blasi U. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA. 2003;9:1308–1314. doi: 10.1261/rna.5850703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chao Y, Vogel J. The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 2010;13:24–33. doi: 10.1016/j.mib.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Papenfort K, Vogel J. Regulatory RNA in bacterial pathogens. Cell Host Microbe. 2010;8:116–127. doi: 10.1016/j.chom.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 7.Bardill JP, Zhao X, Hammer BK. The Vibrio cholerae quorum sensing response is mediated by Hfq-dependent sRNA/mRNA base-pairing interactions. Mol. Microbiol. 2011;80:1381–1394. doi: 10.1111/j.1365-2958.2011.07655.x. [DOI] [PubMed] [Google Scholar]
- 8.Hammer BK, Bassler BL. Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae. Proc. Natl. Acad. Sci. USA. 2007;104:11145–11149. doi: 10.1073/pnas.0703860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rutherford ST, van Kessel JC, Shao Y, Bassler BL. AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev. 2011;25:397–408. doi: 10.1101/gad.2015011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shao Y, Bassler BL. Quorum-sensing non-coding small RNAs use unique pairing regions to differentially control mRNA targets. Mol. Microbiol. 2012;83:599–611. doi: 10.1111/j.1365-2958.2011.07959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Svenningsen SL, Tu KC, Bassler BL. Gene dosage compensation calibrates four regulatory RNAs to control Vibrio cholerae quorum sensing. EMBO J. 2009;28:429–439. doi: 10.1038/emboj.2008.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu J, Miller MB, Vance RE, Dziejman M, Bassler BL, Mekalanos JJ. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA. 2002;99:3129–3134. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jobling MG, Holmes RK. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol. Microbiol. 1997;26:1023–1034. doi: 10.1046/j.1365-2958.1997.6402011.x. [DOI] [PubMed] [Google Scholar]
- 14.Skorupski K, Taylor RK. A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol. Microbiol. 1999;31:763–771. doi: 10.1046/j.1365-2958.1999.01215.x. [DOI] [PubMed] [Google Scholar]
- 15.Vincent HA, Henderson CA, Ragan TJ, Garza-Garcia A, Cary PD, Gowers DM, Malfois M, Driscoll PC, Sobott F, Callaghan AJ. Characterization of Vibrio cholerae Hfq provides novel insights into the role of the Hfq C-terminal region. J. Mol. Biol. 2012;420:56–69. doi: 10.1016/j.jmb.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao X, Yo P, Keith A, Ragan TJ, Harris TK. Thermodynamically balanced inside-out (TBIO) PCR-based gene synthesis: a novel method of primer design for high-fidelity assembly of longer gene sequences. Nucleic Acids Res. 2003;31:e143. doi: 10.1093/nar/gng143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lease RA, Woodson SA. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J. Mol. Biol. 2004;344:1211–1223. doi: 10.1016/j.jmb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 18.Sanglier S, Atmanene C, Chevreux G, Dorsselaer AV. Nondenaturing mass spectrometry to study noncovalent protein/protein and protein/ligand complexes: technical aspects and application to the determination of binding stoichiometries. Methods Mol. Biol. 2008;484:217–243. doi: 10.1007/978-1-59745-398-1_15. [DOI] [PubMed] [Google Scholar]
- 19.Sobott F, McCammon MG, Hernández H, Robinson CV. The flight of macromolecular complexes in a mass spectrometer. Philos. Transact. A Math. Phys. Eng. Sci. 2005;363:379–-391. doi: 10.1098/rsta.2004.1498. [DOI] [PubMed] [Google Scholar]
- 20.Pernot P, Theveneau P, Giraud T, Fernandes RN, Nurizzo D, Spruce D, Surr J, McSweeney S, Round A, Felisaz F. New beamline dedicated to solution scattering from biological macromolecules at the ESRF. J. Phys. Conf. Ser. 2010;247:012009. [Google Scholar]
- 21.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 22.Berry A, Helsby WI, Parker BT, Hall CJ, Buksh PA, Hill A, Clague N, Hillon M, Corbett G, Clifford P, et al. The Rapid2 X-ray detection system. Nucl. Instrum. Methods Phys. Res. Sect. A. 2003;513:260–263. [Google Scholar]
- 23.Svergun D. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- 24.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma S, Ding F, Dokholyan NV. iFoldRNA: three-dimensional RNA structure prediction and folding. Bioinformatics. 2008;24:1951–1952. doi: 10.1093/bioinformatics/btn328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding F, Sharma S, Chalasani P, Demidov VV, Broude NE, Dokholyan NV. Ab initio RNA folding by discrete molecular dynamics: from structure prediction to folding mechanisms. RNA. 2008;14:1164–1173. doi: 10.1261/rna.894608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 2001;34:33–41. [Google Scholar]
- 29.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Svergun DI, Nierhaus KH. A map of protein-rRNA distribution in the 70 S Escherichia coli ribosome. J. Biol. Chem. 2000;275:14432–14439. doi: 10.1074/jbc.275.19.14432. [DOI] [PubMed] [Google Scholar]
- 31.Konarev PV, Petoukhov MV, Svergun DI. MASSHA—a graphic system for rigid-body modelling of macromolecular complexes against solution scattering data. J. Appl. Crystallogr. 2001;34:527–532. [Google Scholar]
- 32.Whitten AE, Cai S, Trewhella J. MULCh: modules for the analysis of small-angle neutron contrast variation data from biomolecular assemblies. J. Appl. Crystallogr. 2008;41:222–226. [Google Scholar]
- 33.Olejniczak M. Despite similar binding to the Hfq protein regulatory RNAs widely differ in their competition performance. Biochemistry. 2011;50:4427–4440. doi: 10.1021/bi102043f. [DOI] [PubMed] [Google Scholar]
- 34.Fender A, Elf J, Hampel K, Zimmermann B, Wagner EG. RNAs actively cycle on the Sm-like protein Hfq. Genes Dev. 2010;24:2621–2626. doi: 10.1101/gad.591310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beich-Frandsen M, Večerek B, Sjöblom B, Bläsi U, Djinović-Carugo K. Structural analysis of full-length Hfq from Escherichia coli. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011;F67:536–540. doi: 10.1107/S174430911100786X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl. Acad. Sci. USA. 2009;106:19292–19297. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sauer E, Weichenrieder O. Structural basis for RNA 3′-end recognition by Hfq. Proc. Natl. Acad. Sci. USA. 2011;108:13065–13070. doi: 10.1073/pnas.1103420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sauter C, Basquin J, Suck D. Sm-like proteins in Eubacteria: the crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Res. 2003;31:4091–4098. doi: 10.1093/nar/gkg480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schumacher MA, Pearson RF, Møller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang W, Wang L, Zou Y, Zhang J, Gong Q, Wu J, Shi Y. Cooperation of Escherichia coli Hfq hexamers in DsrA binding. Genes Dev. 2011;25:2106–2117. doi: 10.1101/gad.16746011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beich-Frandsen M, Večerek B, Konarev PV, Sjöblom B, Kloiber K, Hämmerle H, Rajkowitsch L, Miles AJ, Kontaxis G, Wallace BA, et al. Structural insights into the dynamics and function of the C-terminus of the E coli RNA chaperone Hfq. Nucleic Acids Res. 2011;39:4900–4915. doi: 10.1093/nar/gkq1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat. Struct. Mol. Biol. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moll I, Leitsch D, Steinhauser T, Bläsi U. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep. 2003;4:284–289. doi: 10.1038/sj.embor.embor772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Panja S, Woodson SA. Hexamer to monomer equilibrium of E. coli Hfq in solution and its impact on RNA annealing. J. Mol. Biol. 2012;417:406–412. doi: 10.1016/j.jmb.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Updegrove TB, Correia JJ, Chen Y, Terry C, Wartell RM. The stoichiometry of the Escherichia coli Hfq protein bound to RNA. RNA. 2011;17:489–500. doi: 10.1261/rna.2452111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hwang W, Arluison V, Hohng S. Dynamic competition of DsrA and rpoS fragments for the proximal binding site of Hfq as a means for efficient annealing. Nucleic Acids Res. 2011;39:5131–5139. doi: 10.1093/nar/gkr075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geissmann TA, Touati D. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.